Open Access Article
Open Access ArticleDifferentiating urethane and urea bond activation in polyurethane foam acidolysis
Madeleine
Davis†
c,
Kelsey
Richardson†
 a,
Zach
Westman
a,
Alan L.
Stottlemyer
d,
Christopher
Letko
d,
Nasim
Hooshyar
e,
Vojtech
Vlcek
*bc,
Phillip
Christopher
a,
Zach
Westman
a,
Alan L.
Stottlemyer
d,
Christopher
Letko
d,
Nasim
Hooshyar
e,
Vojtech
Vlcek
*bc,
Phillip
Christopher
 *a and
Mahdi M.
Abu-Omar
*a and
Mahdi M.
Abu-Omar
 *ac
*ac
aDepartment of Chemical Engineering, University of California Santa Barbara, Santa Barbara, CA 93106-5080, USA. E-mail: pchristopher@ucsb.edu; mabuomar@ucsb.edu
bMaterials Department, University of California Santa Barbara, Santa Barbara, CA 93106, USA. E-mail: vlcek@ucsb.edu
cDepartment of Chemistry and Biochemistry, University of California Santa Barbara, Santa Barbara, CA 93106-9510, USA
dThe Dow Chemical Company, Midland, Michigan 48642, USA
eThe Dow Chemical Company, Herbert H Dowweg 5, Hoek 4542 NH, The Netherlands
First published on 10th December 2025
Abstract
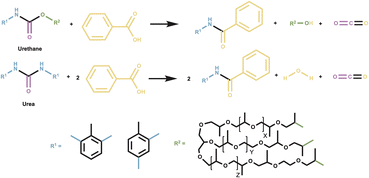
Polyurethane (PU), the sixth most produced plastic globally, is widely used as flexible foam synthesized from polyol, isocyanate, and water to form a thermoset polymer containing urethane and urea bonds. Chemical recycling of PU foams (PUFs), such as acidolysis by carboxylic acids, offers a sustainable route to recover polyol, the predominant mass component. Although PUFs contain both urea and urethane linkages, previous studies have not distinguished their relative acidolysis rates. Here, we use benzoic acid and its electronic analogues to distinguish urea and urethane acidolysis rates by tracking depolymerization via gas evolution, GPC, and NMR. Results reveal biphasic kinetic behavior characterized by rapid urethane bond cleavage and slower urea acidolysis. We find that urethane rates are insensitive to electronic modifications to benzoic acid, while urea bond depolymerization rates correlate with acid electronic structure, as shown by a Hammett reaction constant of (ρ = 3.00 ± 0.01). Density functional theory calculations reveal that the sensitivity of urea rates to acid electronic structure arises from bond elongation and charge delocalization in the transition state that is minimized in urethane acidolysis. These findings on urea and urethane reactivity in PU acidolysis inform more efficient chemical recycling strategies and guide the design of recyclable PU foam materials.
Introduction
Polyurethane (PU) is the sixth most used polymer globally, with a market size of 49.5 billion USD in 2023 and an annual production of approximately 26 million metric tons in 2022.1,2 In the synthesis of PU foams (PUFs), the most common application of PU, polyols react with isocyanate groups to form urethane bonds, while added water promotes the formation of urea bonds, resulting in a polymer containing both urethane and urea linkages. The relative amount of urea and urethane linkages is tailored for desired polymer properties; however, there are constraints to the ratio of urethane/urea linkages to ensure that PUF's desirable properties are maintained. Despite its widespread use, PU's end-of-life management remains a significant challenge. Currently, most post-consumer PUF is either landfilled or incinerated, with only 30% downcycled into lower-quality materials.3The limitations of mechanical recycling and environmental pressures to reduce landfilling highlight the need for efficient chemical recycling processes for PU waste.4–6 Chemical recycling not only reduces the environmental impact of PU disposal but also enables the recovery of high-value polyols—key components that account for the majority of a foam's mass—for use in the synthesis of new PU materials. Among known chemical recycling methods, including acidolysis, hydrolysis, aminolysis, glycolysis and other methods, acidolysis has shown particular promise for efficiently recovering high-purity polyols.7–12
Previous studies have explored PU acidolysis with organic dicarboxylic acids (DCAs) and methods for polyol recovery to replace virgin polyol in new PUF products.13–15 However, the rate of acidolysis, particularly the interplay between urethane and urea bond depolymerization and the influence of acid electronic structure, remains poorly understood. Recent work, including our own, suggests that the structural properties of DCAs, such as the distance between acid groups, can influence PU acidolysis rates.16 Yet, investigation into the rate of urethane versus urea bond cleavage and their dependence on acid electronics is lacking. Further, no previous studies have differentiated the rates of urea and urethane bond acidolysis, which is surprising given their chemically distinct nature.
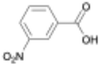
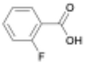
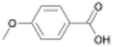
Here, we report the rate of PU acidolysis as a function of reaction time with a set of monocarboxylic acids (MCAs): benzoic acid (BA) and its electronic analogues (3-nitrobenzoic acid (3-NBA), 2-fluorobenzoic acid (2-FBA), 3-fluorobenzoic acid (3-FBA), 4-methylbenzoic acid (4-MBA), and 4-methoxybenzoic acid (4-MOBA)). Using quantitative CO2 measurements (∼1-second resolution), gel permeation chromatography (GPC), and nuclear magnetic resonance (NMR) spectroscopy, we identify biphasic kinetic behavior comprising two distinct regimes: (1) a rapid urethane bond cleavage step and (2) a slower urea bond acidolysis step. Notably, while urethane bond depolymerization rate is largely unaffected by acid electronics, the rate of urea bond scission is highly dependent on the electronic properties of BA derivatives. This is reflected in a Hammett plot with a positive reaction constant (ρ = 3) for urea bond acidolysis. Through computational analysis of transition states, we uncover the chemical origin of this difference, revealing distinct electronic sensitivities that govern the acidolysis of urea versus urethane bonds. We rationalize these results in the context of previous measurements of PUF acidolysis rates via DCAs and provide a mechanistic framework that accounts for the biphasic kinetic behavior in the case of BA and its derivatives, and the single-phase kinetic behavior seen with DCAs. These findings provide novel insights into the differential reactivity of urethane and urea bonds in PUFs, advancing our understanding of acidolysis mechanisms and informing the development of more effective chemical recycling strategies. Additionally, by enabling the deconvolution of urethane and urea bond decomposition rates, this information could be used to select reaction conditions that facilitate cleavage of predominantly urethane bonds, enabling rapid and efficient isolation of the high-value polyol product.
Results and discussion
Model system
The model PUF (M-PUF) used in this study was synthesized by The Dow Chemical Company using toluene diisocyanate (TDI) and VORANOL™ 8136 Polyol, a heteropolymer triol typical of flexible foam formulations. The resulting M-PUF is a representative foam for what is used in commercial applications. The reaction stoichiometry for M-PUF acidolysis with BA is shown in Scheme 1: urethane bond cleavage yields an amide, polyol, and CO2, while urea bond scission produces an amide, CO2, and an amine intermediate that further reacts with BA to form an additional amide and H2O.Based on the foam formulation, the theoretical CO2 release during acidolysis is 34 mL CO2 per g M-PUF from urea bonds and 16 mL CO2 per g M-PUF from urethane bonds (Tables S1–S3). We have previously reported on the use of CO2 evolution to measure the rate of polyurethane depolymerization with DCAs.16 GPC and 1H NMR spectroscopy were used to analyze the polyol (from urethane acidolysis) and amide products (from both urethane and urea acidolysis), respectively.
As shown in Table 1, we used BA and five of its derivatives (3-NBA, 2-FBA, 3-FBA, 4-MOBA, and 4-MBA) to study the influence of acid electronics on M-PUF acidolysis rates and compared the results to those from pimelic acid (PiA), a representative DCA that we previously studied.16 Reactions were conducted at 195![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C, below the M-PUF thermal degradation temperature (>210
°C, below the M-PUF thermal degradation temperature (>210![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C) but above the melting points of all acids.17 A 6
°C) but above the melting points of all acids.17 A 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mass ratio of acid
1 mass ratio of acid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M-PUF (23
M-PUF (23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio, signifying moles of acid to total moles of urethane and urea bonds) was used to ensure efficient wetting of the foam by the acids, minimize mass transport limitations, and enable measurement of intrinsic acidolysis reaction rates.16 The calculations for these ratios can be found in Tables S2–S4.
1 molar ratio, signifying moles of acid to total moles of urethane and urea bonds) was used to ensure efficient wetting of the foam by the acids, minimize mass transport limitations, and enable measurement of intrinsic acidolysis reaction rates.16 The calculations for these ratios can be found in Tables S2–S4.
Biphasic kinetic regimes of acidolysis with MCAs
Previous mechanistic and kinetic studies of PUF acidolysis have focused on DCAs, leaving MCAs largely unexplored. This is a significant knowledge gap, as MCAs may follow distinct mechanisms or reveal reactivity trends not observed with DCAs. For example, CO2 evolution from M-PUF acidolysis with BA and PiA exhibit distinct kinetic behavior (Fig. 1(a)). We note that both urea and urethane bond acidolysis stoichiometrically produce CO2. PiA shows a single kinetic regime, consistent with prior observations for other DCAs, implying that both urea and urethane acidolysis rates can be described by a single-phase kinetic expression.16 In contrast, BA exhibits biphasic kinetic behavior: an initial regime of faster CO2 evolution followed by a slower regime, which has not been reported in previous studies.To investigate the origin of the biphasic kinetic behavior for PUF acidolysis by BA, we monitored the reaction products using GPC and NMR. Since polyol is released exclusively through urethane bond cleavage, GPC provides a direct measure of urethane bond scission. Fig. 1(b) shows GPC chromatograms of polyol released from BA acidolysis as a function of time. The main peak at 3100 g mol−1 corresponds to the desired monomeric polyol product (VORANOL™ 8136 Polyol), while the larger molecular weight shoulder indicates polyol oligomers. We define the completion of urethane bond acidolysis as the time at which the oligomeric tail fully disappears and the chromatogram matches that of authentic virgin polyol, VORANOL™ 8136 (Fig. S11). Based on this criterion, urethane bond acidolysis with BA is complete in 120 minutes. However, at this time (marked by the black dot in Fig. 1(a)), only ∼65% of the expected CO2 has been produced. This indicates that although urethane decomposition is complete, full depolymerization of the M-PUF is not yet achieved at 120 minutes (Fig. 1(a)). This deviates from previous reports for acidolysis by DCAs, including PiA, which show full CO2 evolution at the same time as complete polyol release (Fig. S12).
We complemented the GPC analysis with 1H NMR analyses of the acidolysis products. Throughout acidolysis, as polyurethane bonds are chemically cleaved, products of a variety of molecular weights and reactive end groups are produced. Due to this heterogeneous intermediate product mixture, the 1H NMR spectra of reaction samples during acidolysis contain many overlapping signals. As a result, 1H NMR is most accurately applied to determine when all acidolysis is complete, since the reaction mixture at that point is homogeneous and of a known composition. NMR spectra were taken of reaction aliquots starting when the reaction mixture was fully solubilized and ending when gas evolution ceased. Small molecule models of urethane and urea bonds (Fig. S2), in addition to previous literature reports, were used to guide peak assignments. Amide signals (δ = 9.5–10.5 ppm) and oligomeric urea signals (δ = 8.3–9.3 ppm) help inform the extent of urethane and urea bond acidolysis (Fig. 2).13,14,18–20 Following completion of urethane bond acidolysis (as indicated by full polyol release in the GPC at 120 minutes), the NMR spectra continue to evolve as a function of time; amide product signals increase, and oligomeric urea signals persist until 300 minutes. Beyond this time, the 1H NMR spectra remain unchanged with only the amide signals expected from the products in this region. Fig. 2 assigns these peaks to amide products from the 2,4- and 2,6-TDI used in the M-PUF formulation. Peak integrals can be found in Fig. S3. This NMR analysis further supports the gas evolution data (Fig. 1(a)), which indicates that full acidolysis of both urethane and urea bonds is complete at 300 minutes.
To test the robustness of this proposed kinetic phenomenon, we ran additional experiments with added catalysts and different foam formulations. In the PUF synthesis process, catalysts such as Dabco T-9 and Dabco 33 LV are used and not removed. Even when adding double the amount used during PUF synthesis, the acidolysis kinetics, including the biphasic behavior, do not change (Fig. S14). This indicates that residual additives do not impact the biphasic kinetic behavior, likely because they are too dilute. Moreover, we carried out these reactions with two additional PUF formulations of varying amounts of urethane and urea bonds as well as an end-of-life (EOL) PUF. Once again, we observe biphasic gas evolution kinetic behavior, signifying that our observations and conclusions are robust to a wide range of commercially relevant foam formulations/mixtures (Fig. S15 and S16).
These results provide strong evidence that during M-PUF acidolysis by BA, urethane bond decomposition predominantly occurs during the initial, faster kinetic regime while urea bond decomposition predominantly occurs in the slower kinetic regime. GPC analysis confirms that polyol release is complete at 120 minutes, and the amount of CO2 evolved at this time corresponds to full stoichiometric release from urethane bonds (with a small additional amount from the onset of urea acidolysis). From 120 to 300 minutes, CO2 continues to evolve. Since urethane decomposition is complete, this gas evolution must be due to urea bond acidolysis. Finally, 1H NMR further confirms that acidolysis of both urethane and urea bonds is complete at this time. In summary, urethane cleavage occurs during the initial gas evolution regime while urea decomposition primarily occurs during the slower kinetic regime. This is not proposing that all urethane acidolysis uniquely occurs first, while all urea acidolysis occurs subsequently, but instead that the rate constant for urethane acidolysis is much faster than that for urea acidolysis. These analysis methods and conclusions are outlined in Table S5.
To determine whether this biphasic behavior is specific to BA or broadly applicable to MCAs, we compared the time required for urethane and urea bond decomposition across all MCAs studied, using the time of complete polyol and CO2 release, respectively (Fig. 3(a)). Due to experimental limitations with the slowest acids (4-MBA and 4-MOBA), we extrapolated the time of complete CO2 release from the initial slow regime behavior. More details on the fitting can be found in the SI. In all cases, except for 3-NBA which will be discussed in more detail below, we observe polyol release consistently preceding full CO2 evolution. Interestingly, the time required for complete decomposition of urea bonds varied significantly, from 1 h for 3-NBA to 9 h for 4-MOBA (Fig. S5–S9). Conversely, the time for urethane acidolysis only slightly varies with different acids tested and does not strongly trend with the acid electronics as discussed further in sections below. The CO2 plots for these acids also displayed biphasic behavior, as seen in Fig. 3(b), with 3-FBA as a representative electron withdrawing substituted acid and 4-MOBA as an example of electron donating substituted acid (see Fig. S13 for full gas evolution plots). To our knowledge, this is the first study to resolve the distinct kinetic regimes of urethane and urea bond decomposition in PUF depolymerization.
To extend this analysis, we compared the initial rates of CO2 release during M-PUF acidolysis, corresponding to the predominantly urethane regime, with the fastest MCAs (3-NBA, 2-FBA, and 3-FBA) to PiA, a representative DCA (Fig. 3(c)). Rates of urethane bond depolymerization were comparable across all MCAs and PiA, implying that urethane bond acidolysis is largely unaffected by electronic effects. In contrast, urea bond decomposition, represented by the second regime, was consistently slower for all MCAs relative to PiA.
To understand the kinetic implications of this behavior, we can compare the results of acidolysis with MCAs to those previously obtained by our group with DCAs. We hypothesize that in all PUF acidolysis reactions, not just those reported here for MCAs, urethane groups must be decomposed first to reveal urea linkages due to the structure of the material. Due to the polar nature of the urea linkages, these domains form strongly hydrogen bonded segments inside of softer, and more accessible, urethane domains.21,22 This phase-separation, in addition to the inherent reactivity differences of MCAs with urethane and urea bonds, is likely contributing to this observed behavior.
In MCA-mediated reactions, urea bond decomposition is slower than urethane bond decomposition, occurring in the second regime. Urethane bonds are cleaved primarily in the faster regime, exposing less-reactive urea bonds, leading to the observed biphasic kinetics. We can contrast DCA-mediated acidolysis. We can say definitively that urea bond decomposition is no longer rate limiting, since mono-phasic instead of biphasic kinetics is observed. Instead, we hypothesize that urethane bond acidolysis is rate-limiting. Once the urea bonds are exposed, they decompose with faster intrinsic rates than urethane bonds. As a result, CO2 from both urea and urethane bonds is released concurrently with polyol, resulting in a single kinetic regime. This mechanistic distinction emphasizes the importance of studying acidolysis with MCAs to develop a fuller understanding of bond-specific (urea vs. urethane) reactivity and kinetic behavior in PUF depolymerization. This also implies that urea bond acidolysis must be particularly sensitive to the nature of the acid, as this step must be much slower for BA than previously studied DCAs for this mechanistic picture to be true.
Conformational and electronic effects on urethane and urea acidolysis
As shown in Fig. 3(a), the time required for complete urethane bond acidolysis is largely independent of acid electronic structure, while that for urea bond acidolysis varies significantly with electronic substituents. To further support this observation, we fit the CO2 evolution data to a biexponential model and used the rate constants to construct a Hammett correlation. This assumes that urethane and urea acidolysis are fit by first order kinetics with distinct rate constants, where the faster process is urethane acidolysis. Details of the fitting procedure are provided in the SI. Note that 2-FBA is excluded from the Hammett correlations (Fig. 4(b) and (d)) because it is difficult to quantify a Hammett constant that captures only electronic effects, not steric effects, for an ortho substituent.23As hypothesized, the rates of the initial urethane regime are larger than that of the urea regime for all acids (Fig. 4(a) and (c)). Additionally, as shown in Fig. 4(b) and (d), the urethane acidolysis rate is mostly independent of acid electronics, while the urea rate shows a strong dependence on acid electronics, as demonstrated by a Hammett plot with a strong positive slope (ρ = 3.00 ± 0.01).
Notably, 3-NBA, the most electron withdrawing acid tested, appeared to display a near single kinetic regime. However, quantitative fitting showed that a biexponential model is necessary to provide a better fit than a single exponential model (Fig. S19). We hypothesize that the electronically accelerated urea bond acidolysis rate shifts the rate-limiting step closer to urethane bond acidolysis in 3-NBA, similar to DCA acidolysis. This comparison underscores the pronounced electronic sensitivity of urea bond acidolysis, in contrast to the electronic insensitivity of urethane bond cleavage.
While here we show that urethane bond acidolysis rate is largely insensitive to acid electronic effects, we have observed previously that urethane bond decomposition is sensitive to the chain length of the DCA acidolysis reagent.16,24 For example, urethane bond acidolysis is accelerated with acids with fewer carbons between the two acid groups. At longer chain lengths in DCAs, such as in PiA, this cooperative effect diminishes, and the urethane rate approaches that of MCAs.
In contrast, urea bond decomposition is now observed to be sensitive to both conformational and electronic influences. Although BA and PiA have similar acid strengths (pKa = 4.20 and 4.48, respectively), urea bond decomposition is consistently slower with BA and all MCAs than with DCAs. This suggests that the presence of only one acid group limits both the cooperative conformational and the electronic stabilization influences in urea bond acidolysis. Thus, urea bond acidolysis rates seem to be sensitive to both acid electronic and conformational effects, whereas urethane bond acidolysis is only sensitive to conformational effects observed as a function of DCA carbon chain length. The exact nature of the proposed conformational effect will be the subject of future studies.
To understand the molecular origin of this differential sensitivity to electronic effects in urea and urethane bonds, we conducted a computational analysis of the urethane and urea acidolysis reaction pathways using DFT [M06-2X/cc-pVTZ] with Gaussian-16.25 Through this analysis, we found that the activation barriers for urethane acidolysis were largely consistent regardless of acid reagent while urea barriers varied greatly (Fig. S21(a) and S21(b)). These barriers were used to replicate the experimentally observed Hammett correlations (Fig. S21(c) and S21(d)). Additionally, both urethane and urea bond decomposition proceed through six-centered transition states (see SI for more details). For urea, DFT-optimized geometries of the transition-states show that more electron withdrawing acids yield a later, more delocalized TS, as evidenced by elongation of the C6–N5 bond by ∼0.05 Å and the O3–H4 bond by 0.1 Å (Fig. 5(a)). Consistent with these geometric trends, we propose that the acceleration in urea acidolysis rates with electron withdrawing groups can be attributed to these groups stabilizing charge in the transition state, leading to the observed geometric elongation. Conversely, the urethane acidolysis transition state structure does not change with electronic substitutions, consistent with experimental observations (Fig. 5(c)). We hypothesize that this is due to the inherently higher electronegativity of the urethane oxygen acceptor (atom 5, Fig. 5(d)), which stabilizes the transition state and reduces dependence on acid electronics. This provides a structural basis for the experimentally observed electronic insensitivity of urethane acidolysis and the enhanced reactivity of urea bonds, further supporting the mechanistic regimes identified in our kinetic studies.
Conclusions
This work provides new insights into the kinetics of PU depolymerization, revealing unique reactivities and stabilizing influences for urethane versus urea bond acidolysis. Through acidolysis with BA and other MCAs, we identified biphasic kinetics and successfully deconvoluted the regimes into rapid urethane bond decomposition followed by slower urea bond cleavage.Urethane bond acidolysis was found to be directed exclusively by conformational factors; acidolysis rates increase with proximal carboxyl groups, as seen with short-chain DCAs, and are independent of the acid electronic structure or pKa. In contrast, urea bond cleavage is influenced by both conformational and electronic influences. Electron-withdrawing acids accelerate the urea acidolysis rate, as demonstrated by a Hammett plot with ρ = 3.00 ± 0.01. However, even the most electron withdrawing MCAs are slower than DCAs, emphasizing the additional conformational stabilization provided by a second acid group. Transition state analyses support this distinction; electron withdrawing groups stabilize the urea transition state through charge delocalization and bond elongation, while the urethane transition state remains structurally invariant with respect to acid electronics.
We further propose that DCA- and MCA-mediated acidolysis have a different rate-limiting step. For DCAs, we only observe one kinetic regime that correlates with acid chain length, but not pKa, indicating that urethane bond acidolysis is rate-limiting and conceals any electronic variations in the urea rate. In MCA systems, urethane bond decomposition occurs rapidly, exposing the less-reactive, rate-limiting urea bonds, and giving rise to the observed biphasic kinetics.
This study provides the first molecular-level understanding of the distinct reactivity of urethane and urea bonds during PU acidolysis with carboxylic acids, linking these differences to conformational, electronic and transition state (computational) insights. These insights deepen our understanding of PUF acidolysis mechanisms, providing valuable guidance for optimizing chemical recycling strategies for polyurethane waste.
Author contributions
M. D. preformed the experiments and analyzed the data. K. R. completed the kinetic fits and computational calculations and analyzed the data. Z. W. supervised the experimental design and analyzed the data. All authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
All data are available in the main text, in the supplementary information (SI) or upon request to the corresponding authors. Supplementary information: materials description, experimental procedures, gas evolution apparatus and kinetics, NMR spectra, GPC chromatograms, kinetic profiles and fitting, and DFT computational analyses. See DOI: https://doi.org/10.1039/d5sc05599g.Acknowledgements
This work was supported by the Dow Chemical Company. P. C. and M. M. A.-O. both acknowledge the Mellichamp Academic Initiative in Sustainability at UCSB for support. Use was made of computational facilities purchased with funds from the National Science Foundation (CNS-1725797) and administered by the Center for Scientific Computing (CSC). This work also used Bridges-2 at Pittsburgh Supercomputing Center through allocation CHM230006 from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296. Some experiments were performed using UC Santa Barbara's MRL Shared Experimental Facilities, supported by the MRSEC Program from NSF (award no. DMR 1720256). VORANOL is a trademark of The Dow Chemical Company (“Dow”) or an affiliated company of Dow.References
- Polyurethane Foam Market by Type (Rigid Foam, Flexible Foam, Spray Foam), End-use Industry (Building & Construction, Bedding & Furniture, Automotive, Electronics, Footwear, Packaging) and Region - Global Forecast to 2028, 2021, https://www.marketsandmarkets.com/Market-Reports/polyurethane-foams-market-1251.html, accessed Sept 15, 2024.
- Market Volume of Polyurethane Worldwide from 2015 to 2022, with a Forecast for 2023 to 2030; AgileIntel Research (ChemIntel360), Statista Research Department, 2023, https://www.statista.com/statistics/720341/global-polyurethane-market-size-forecast/, accessed Sept 15, 2024 Search PubMed.
- R. V. Gadhave, S. Srivastava, P. A. Mahanwar and P. T. Gadekar, Recycling and Disposal Methods for Polyurethane Wastes: A Review, Open J. Polym. Chem., 2019, 09, 39–51 CrossRef CAS.
- A. Kemona and M. Piotrowska, Polyurethane Recycling and Disposal: Methods and Prospects, Polymers, 2020, 12, 1752 CrossRef CAS.
- W. Yang, Q. Dong, S. Liu, H. Xie, L. Liu and J. Li, Recycling and Disposal Methods for Polyurethane Foam Wastes, Proc. Environ. Sci., 2012, 16, 167–175 CrossRef CAS.
- G. Rossignolo, G. Malucelli and A. Lorenzetti, Recycling of polyurethanes: where we are and where we are going, Green Chem., 2024, 26, 1132–1152 RSC.
- N. Gama, B. Godinho, P. Madureira, G. Marques, A. Barros-Timmons and A. Ferreira, Polyurethane Recycling Through Acidolysis: Current Status and Prospects for the Future, J. Polym. Environ., 2024, 32, 4777–4793 CrossRef CAS.
- X. Wu, R. C. Turnell-Ritson, P. Han, J. C. Schmidt, L. Piveteau, N. Yan and P. J. Dyson, Carbamate-bond breaking on bulk oxides realizes highly efficient polyurethane depolymerization, Nat. Commun., 2025, 16, 4322 CrossRef CAS PubMed.
- Y. Branson, S. Soltl, C. Buchmann, R. Wei, L. Schaffert, C. P. S. Badenhorst, L. Reisky, G. Jager and U. T. Bornscheuer, Urethanases for the Enzymatic Hydrolysis of Low Molecular Weight Carbamates and the Recycling of Polyurethanes, Angew. Chem., Int. Ed., 2023, 62, e202216220 CrossRef CAS PubMed.
- R. Villa, R. Salas, M. Macia, F. Velasco, B. Altava, E. Garcia-Verdugo and P. Lozano, How to Easily Depolymerize Polyurethane Foam Wastes by Superbase Catalysts in Ionic Liquids Below 100 °C, Angew. Chem., Int. Ed., 2025, 64, e202418034 CrossRef CAS PubMed.
- H. He, H. Hu, K. Du, M. Lu, F. Yang and L. Cui, et al., Prospects of high-value recycling methods for polyurethane based on the selective cleavage of C–O/C–N bonds, Green Chem., 2025, 27, 8467–8491 RSC.
- P. Paiva, L. M. C. Teixeira, R. Wei, W. Liu, G. Weber and J. P. Morth, et al., Unveiling the enzymatic pathway of UMG-SP2 urethanase: insights into polyurethane degradation at the atomic level, Chem. Sci., 2025, 16, 2437–2452 RSC.
- M. Grdadolnik, A. Drincic, A. Oreski, O. C. Onder, P. Utrosa, D. Pahovnik and E. Zagar, Insight into Chemical Recycling of Flexible Polyurethane Foam by Acidolysis, ACS Sustain. Chem. Eng., 2022, 10, 1323–1332 CrossRef CAS.
- N. Gama, B. Godinho, G. Marques, R. Silva, A. Barros-Timmons and A. Ferreira, Recycling of polyurethane by acidolysis: The effect of reaction conditions on the properties of the recovered polyol, Polymer, 2021, 219, 123561 CrossRef CAS.
- R. M. O'Dea, M. Nandi, G. Kroll, J. R. Arnold, L. T. J. Korley and T. H. Epps, 3rd, Toward Circular Recycling of Polyurethanes: Depolymerization and Recovery of Isocyanates, JACS Au, 2024, 4, 1471–1479 CrossRef PubMed.
- Z. Westman, B. Liu, K. Richardson, M. Davis, D. Lim, A. L. Stottlemyer, C. S. Letko, N. Hooshyar, V. Vlcek, P. Christopher and M. M. Abu-Omar, Influence of Carboxylic Acid Structure on the Kinetics of Polyurethane Foam Acidolysis to Recycled Polyol, JACS Au, 2024, 4, 3194–3204 CrossRef CAS.
- B. Liu, Z. Westman, K. Richardson, D. Lim, A. L. Stottlemyer, T. Farmer, P. Gillis, N. Hooshyar, V. Vlcek, P. Christopher and M. M. Abu-Omar, Polyurethane Foam Chemical Recycling: Fast Acidolysis with Maleic Acid and Full Recovery of Polyol, ACS Sustain. Chem. Eng., 2024, 12, 4435–4443 CrossRef CAS PubMed.
- D. T. J. Morris, S. M. Wales, D. P. Tilly, E. H. E. Farrar, M. N. Grayson and J. W. Ward, et al., A molecular communication channel consisting of a single reversible chain of hydrogen bonds in a conformationally flexible oligomer, Chem, 2021, 7, 2460–2472 CAS.
- W. K. A. Al-Ithawi, R. Aluru and A. V. Baklykov, et al., Mechanosynthesis of Polyureas and Studies of Their Responses to Anions, Polymers, 2023, 15, 4160 CrossRef CAS PubMed.
- X. Lu, Y. Wang and X. Wu, Molecular interaction in polyurea by 1-D and 2-D N.M.R, Polymer, 1993, 34, 56–60 CrossRef CAS.
- K. Aou, S. Ge, D. M. Mowery, R. C. Zeigler and R. R. Gamboa, Two-domain morphology in viscoelastic polyurethane foams, Polymer, 2015, 56, 37–45 CrossRef CAS.
- D. V. Dounis and G. L. Wilkes, Structure-property relationships of flexible polyurethane foams, Polymer, 1997, 38, 2819–2828 CrossRef CAS.
- M. Charton, Nature of the ortho effect V. ortho-Substituent Constants, J. Am. Chem. Soc., 1969, 91, 6649–6654 CrossRef CAS.
- K. Richardson, M. M. Abu-Omar, P. Christopher and V. Vlcek, Assessing the Conformational Landscape of Dicarboxylic Acids Using Ab Initio Molecular Dynamics: The Role of Phase and Intermolecular Interactions, J. Phys. Chem. B, 2025, 129, 7195–7202 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, and D. J. Fox, Gaussian 16, Revision C.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
Footnote |
| † Denotes equal contribution. |
| This journal is © The Royal Society of Chemistry 2026 |