Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unconventional chalcogen-containing azolylidene metal complexes as potential anticancer therapeutics
Jan
Romano-deGea
 *a,
Irina L.
Sinenko
*a,
Irina L.
Sinenko
 a,
Peter M. F.
Pânzar
a,
Peter M. F.
Pânzar
 a,
Adriana
Neves Vieira
a,
Adriana
Neves Vieira
 a,
Lindsey E. K.
Frederiksen
a,
Lindsey E. K.
Frederiksen
 a,
Kseniya
Glinkina
a,
Kseniya
Glinkina
 a,
Farzaneh
Fadaei-Tirani
a,
Farzaneh
Fadaei-Tirani
 a,
Rosario
Scopelliti
a,
Rosario
Scopelliti
 a,
Fabien
Kuttler
a,
Fabien
Kuttler
 b,
Kelvin
Lau
b,
Kelvin
Lau
 c and
Paul J.
Dyson
c and
Paul J.
Dyson
 *a
*a
aInstitute of Chemical Sciences and Engineering, École Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland. E-mail: jan.romanodegea@epfl.ch; paul.dyson@epfl.ch
bBiomolecular Screening Facility, École Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland
cProtein Production and Structure Core Facility, École Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland
First published on 29th December 2025
Abstract
Organometallic compounds with N-heterocyclic carbene (NHC) ligands have been studied for their anticancer and antimicrobial properties, with imidazole and benzimidazole derivatives being the predominant scaffolds for potential NHC-containing drugs. In contrast, chalcogen-containing azolylidene ligands, (N,Y)HCs (Y = O, S, Se), remain largely unexplored in both medicinal inorganic chemistry and, more generally, in inorganic chemistry. Consequently, to study the effect of the incorporation of a chalcogen atom in the ligand, classical (N,N)HC complexes of platinum, gold and ruthenium were selected based on their previously reported biological activity and proposed mechanisms of action, and their (N,Y)HC (Y = O, S, Se) analogues were synthesised. The electronic and steric properties of the ligands and complexes were explored and their biological activity was evaluated. The introduction of a chalcogen atom within the heterocyclic scaffold of the ligands was found to modulate their interaction with biomolecules and regulate the cytotoxicity of the metal complexes towards ovarian cancer cells.
Introduction
N-heterocyclic carbene (NHC) metal complexes display remarkable stability and tuneability, which explains their widespread use in many areas of chemistry.1,2 In recent years, NHC complexes, in particular platinum, gold, and ruthenium compounds, have been evaluated as anticancer, antimicrobial, antiviral and antiparasitic agents.3–7Platinum-NHC complexes display cytotoxic effects comparable, or superior, to cisplatin against a variety of cancer cell lines.8 Traditionally, the mechanism of action (MoA) of platinum-based anticancer compounds has been related to their ability to bind the minor groove of DNA to then form 1,2-intrastrand crosslinks between nucleobases, blocking the translation and replication of DNA.4 In contrast, due to geometric constraints, trans-(NHC)PtX2(amine) complexes presumably form long-range DNA intra- and inter-strand adducts.9,10 These alternative crosslinks are less likely to be recognised as defects by repair proteins in cisplatin-resistant tumours.11 Therefore, such complexes are more likely to be active against cisplatin-resistant cell lines.12
In comparison, gold-NHC complexes are reported to inhibit proteins, such as thioredoxin reductase (TrxR), an enzyme overexpressed in some solid tumours.13,14 TrxR inhibition is associated with inhibition of mitochondrial respiration, potentially inducing apoptosis via mitochondria-mediated pathways.15,16 Furthermore, gold-NHC complexes tend to display high antiproliferative activity.4,13,17
Ruthenium-NHC complexes have been reported as inhibitors of cysteine- and selenocysteine-containing biomolecules, including TrxR and cathepsin B (CatB).18 The latter is a cysteine protease for which elevated expression levels are often associated with the progression of various tumours.19 Additionally, (p-cymene)(NHC)RuCl2 complexes act as antiproliferative agents, with IC50 values frequently in the low micromolar range.20,21
The modulation of the biological properties of metal NHC complexes is usually achieved through structural modifications introduced on the nitrogen atoms, also known as wingtips, or through the substituents on the heterocyclic backbone.4,5 Other carbene ligand classes such as triazoles and cyclic(alkylamino)carbenes (cAACs) have also been employed as scaffolds in medicinal inorganic chemistry.22,23 In contrast, metal complexes with chalcogen-containing azolylidene ligands, (N,Y)HCs (Y = O, S, Se), are rare,24 and studies of their biological properties are very scarce.25,26 In particular, only a single selenium-containing carbene metal complex has been previously reported.27 The effect on the biological activity of substituting the nitrogen atom in (N,N)HC ligands by a chalcogen atom remains, to the best of our knowledge, unexplored. (N,Y)HC (Y = O, S, Se) ligands present different steric and electronic properties compared to their classical nitrogen-containing counterparts. The chalcogen atoms are not alkylated, and hence they are more exposed than nitrogen atoms in (N,N)HCs, resulting in the characteristic “missing-wingtip” shape of these ligands.28 The metal atoms are also more exposed, albeit to a lesser extent. Besides modulating the electron donating abilities, the chalcogen atoms affect the aromaticity of the heterocycles.29 The chalcogen atoms in the azolylidene ligands have lone pairs that can act as acceptors in hydrogen bonds (HBs).29–31 Additionally, sulphur- and selenium-containing molecules can form intra- and intermolecular chalcogen bonds (ChBs).31 The presence of these interactions has ramifications in a wide range of fields and applications, including catalysis and biology, particularly in substrate and ligand–protein binding.32–36
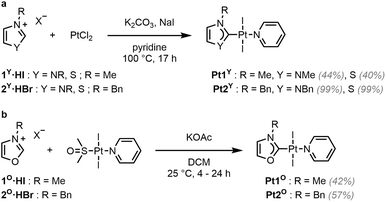
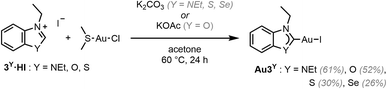
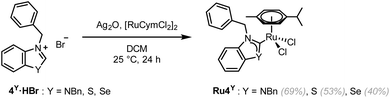
To explore the effect of the introduction of a chalcogen atom to the cytotoxicity and to evaluate structure–activity relationships (SAR) in unconventional chalcogen azolylidene metal complexes, four parent (N,N)HC metal complexes were selected based on reported examples in the literature demonstrating considerable cytotoxic effects and with a hypothesis on the mechanism of action (Fig. 1a): two trans-(NHC)PtI2(amine) complexes bearing non-fused 1,3-dimethylimidazolylidene or 1,3-dibenzylimidazolylidene ligands (Pt1NMe and Pt2NBn) capable of overcoming cisplatin-acquired resistance;12 a highly antiproliferative TrxR inhibitor gold(I) iodido complex bearing a benzoannulated 1,3-diethylbenzimidazolylidene ligand (Au3NEt);37 and a ruthenium cymene complex bearing a fused 1,3-dibenzylbenzimidazolylidene ligand (Ru4NBn) with TrxR and CatB inhibition properties.38 Their azolylidene analogues were successfully synthesised (Fig. 1b) and their biological behaviour was evaluated.
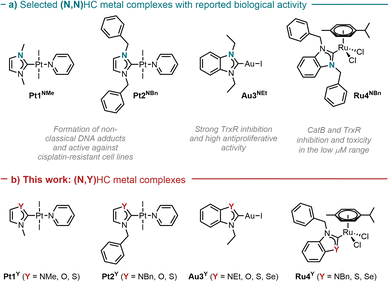 | ||
| Fig. 1 Selected parent (N,N)HC metal complexes (a) and their azolylidene (N,Y)HC (Y = NR, S, O, Se) analogues (b). | ||
Results and discussion
Synthesis and characterisation
The non-fused and benzoannulated azolium (N,Y)HC proligands, 1Y·HX–4Y·HX (Y = NR, O, S or Se; X = halide), were prepared in one step by heating the appropriate azole in acetonitrile (or neat) with the corresponding alkyl or benzyl halide to obtain the desired products in good to excellent yields. The azolium salts are hygroscopic and, in some cases, deliquescent. Therefore, special care should be taken with their isolation, purification and storage. The yields of the oxazolium salts are consistently lower than the others due to their tendency to ring open when exposed to heat, moisture or basic conditions.39 The corresponding (N,Y)HC metal complexes were synthesised using either silver transmetalation or a weak base as described previously.1,24,40,41 All compounds were characterised by 1H and 13C NMR spectroscopy and high-resolution mass spectrometry (HRMS).Platinum (N,Y)HC complexes
Complexes Pt1Y (Y = NMe, S) and Pt2Y (Y = NBn, S) were synthesised in one step using a slightly modified literature procedure (Scheme 1a).42 The respective proligands (1Y·HI and 2Y·HBr), PtCl2, K2CO3 (the base required to deprotonate the carbene precursors), and NaI (as the iodide source) were heated under reflux in pyridine (acting as both the solvent and ligand). When applying these conditions to the synthesis of Pt1O and Pt2O, only trans-[Pt(py)2I2] was isolated from the mixtures, and ring opening of the oxazolium salts was observed by 1H NMR spectroscopy.39 Instead, in order to obtain Pt1O and Pt2O, trans-[Pt(pyridine)(dmso)I2] was reacted in DCM with the corresponding oxazolium salt in the presence of potassium acetate (KOAc), a weaker base, to yield the desired products in acceptable yields (Scheme 1b).The desymmetrisation of the (N,Y)HC ligand by the introduction of the chalcogen atom is apparent in the 1H NMR spectra of Pt1Y and of Pt2Y (Y = NR, O, S). One signal is present for the protons in the heterocyclic backbone in Pt1NMe and Pt2NBn, whereas two distinct signals are observed in Pt1Y and Pt2Y (Y = O, S) (SI Fig. 1). No major differences were observed in the 1H NMR spectra for the peaks corresponding to the pyridine ligand. The 195Pt NMR chemical shift of Pt1Y and Pt2Y (Y = NR, O, S) ranges between −4100 and −4400 ppm, in keeping with previous reports on platinum(II) (N,N)HC complexes.43 The introduction of the chalcogen atom into the carbene ligands leads to a slight upfield shift of the 195Pt NMR signals of Pt1O and Pt2O compared to Pt1NMe and Pt2NBn, whereas a considerable downfield shift is observed for Pt1S and Pt2S. The upfield shift is indicative of a more electron-rich metal centre, due to a more electron-donating and less π-accepting ligand.44,45 Therefore, it could be expected that the carbene ligands in Pt1O and Pt2O form weaker bonds with the Pt(II) centre.
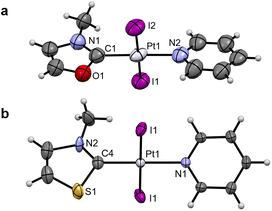
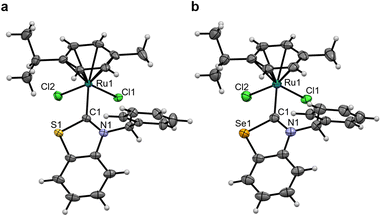
Crystals suitable for single-crystal X-ray diffraction (scXRD) were grown for Pt1NMe, Pt1O, Pt1S and Pt2NBn (see Fig. 2 and SI Fig. 4a and b). The structures confirmed the presence of the coordinated (N,Y)HC ligands and of the expected trans-configuration. In Pt1Y (Y = NMe, O, S), the Pt–CNYHC bond length decreases from Pt1O to Pt1NMe and then to Pt1S (Table 1), consistent with the 195Pt NMR chemical shift. Additionally, the (N,Y)HC ligands in Pt1O and Pt1S are slightly tilted with respect to the platinum atom (172.2° and 174.5°, respectively), deviating from the ideal “linear” structure observed in Pt1NMe and Pt2NBn (180°). Presumably, the elongated Y–CNYHC (Y = O, S) bond results in the distorted geometries. All other bonds and angles are within the expected ranges.
| Pt1NMe | Pt1O | Pt1S | |
|---|---|---|---|
| Pt–CNYHC | 1.961(5) | 2.079(18) | 1.948(7) |
| Pt–NPyr | 2.082(4) | 1.960(18) | 2.071(5) |
| Pt–IAvg | 2.5935(3) | 2.5953(21) | 2.5932(3) |
| NR–CNYHC | 1.347(5) | 1.37(3) | 1.251(11) |
| Y–CNYHC | 1.347(5) | 1.31(3) | 1.801(5) |
| CNYHC–Pt–NPyr | 180.0 | 178.07(7) | 180.0 |
| NR–CNYHC–Y | 105.1(5) | 107(2) | 106.8(7) |
| NYHC centroid–CNYHC–Pt | 180.0 | 172.2 | 174.5 |
| NYHC–Pyr | 35.7 | 11.9 | 22.2 |
Gold (N,Y)HC complexes
Au3NEt, Au3S and Au3Se were prepared from the reaction between the corresponding proligand, Au(SMe2)Cl, and K2CO3 in acetone at 60 °C (Scheme 2).41 A modified approach, using KOAc in place of K2CO3, was employed in the preparation of the oxazolylidene complex Au3O, to avoid the ring opening of the (N,O)HC ligand.The benzothiazole peaks split in the 1H NMR spectra of Au3O due to the desymmetrisation of the ligand, and become fully resolved in Au3S and Au3Se (SI Fig. 2). Additionally, the ethyl “wingtips” peaks shift downfield, indicating a more deshielded environment due to the introduction of the chalcogen atom or close proximity to the metal due to the lower steric hindrance. As observed in the platinum complexes, the Au–CNYHC bond length in the single-crystal structures decreases from Au3O to Au3NEt and then to Au3S (Fig. 3, Table 2, and SI Fig. 4c). Similarly, the (N,Y)HC ligands in Au3O and Au3S are slightly tilted with respect to the gold atom compared to the ideal structure in Au3NEt (173.0° and 174.5°, respectively, vs. 180°). All other bonds and angles are within the expected ranges.
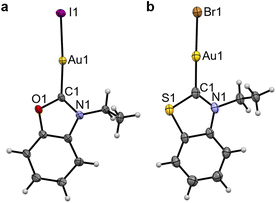 | ||
| Fig. 3 Single-crystal XRD structures of Au3O (a) and Au3S (X = Br) (b). Thermal ellipsoids are drawn with a 50% probability. The structure of Au3NEt can be found in SI Fig. 4. | ||
| Au3NEt | Au3O | Au3S (X = Br) | |
|---|---|---|---|
| Au–CNYHC | 2.008(10) | 2.05(4) | 1.981(3) |
| Au–X | 2.5449(8) | 2.556(3) | 2.4023(3) |
| NR–CNYHC | 1.330(8) | 1.33(5) | 1.331(4) |
| Y–CNYHC | 1.330(8) | 1.35(5) | 1.718(3) |
| CNYHC–Au–X | 180.0 | 174.7(11) | 176.45(8) |
| NR–CNYHC–Y | 107.8(8) | 110(3) | 110.4(2) |
| NYHC centroid–CNYHC–Au | 180.0 | 173.0 | 174.5 |
Ruthenium (N,Y)HC complexes
Ru4NBn was synthesised in two steps using a transmetalation route from a silver-(N,N)HC intermediate.46 As the isolation of the silver-(N,S)HC and silver-(N,Se)HC intermediate was unsuccessful, a previously reported one-pot route was employed to obtain Ru4S and Ru4Se (Scheme 3).47 Unfortunately, the synthesis of Ru4O was unsuccessful both via the one-step or two-step transmetalation routes, as well as by using the weak base (KOAc) route.41 A similar approach to the synthesis of Pt1O or Pt2O was also attempted, using KOAc and a ruthenium complex bearing a labile ligand, (η6-p-cymene)RuCl2(DMSO). However, the (N,O)HC ruthenium complex was not isolated.The rotation of the Ru–CNYHC bond is restricted in Ru4NBn, with the benzylic N–CH2–Ph 1H NMR peaks appearing as two coupling doublets (at 5.84 and 6.56 ppm). In contrast, these peaks converge into singlets in Ru4S and Ru4Se (at 6.36 and 6.37 ppm, respectively), suggesting that the carbene is able to freely rotate (SI Fig. 3). Single-crystals of Ru4Y (Y = NBn, S, Se) suitable for scXRD were grown (Fig. 4, Table 3 and SI Fig. 4d). To the best of our knowledge, the structure of Ru4Se is the first example of a selenazolylidene complex to be reported. Only one other selenium-containing carbene complex has been previously reported, i.e. a chromium(0) (arylseleno)(diethylamino)carbene Fischer complex.27Ru4Se features a central Ru(II) ion in the typical distorted pseudo-octahedral geometry of piano-stool complexes also exhibited by Ru4NBn and Ru4S. The ruthenium atom is coordinated by two chlorido ligands, an η6-bound cymene ligand and 4Se, a selenium-containing (N,Se)HC ligand. The CNYHC–Se distance is 1.875(3) Å and the N–CNYHC–Y angle is 109.5(2)°. All other bonds and angles are within range of the values previously reported in Ru(II)-arene NHC complexes. The crystal packing is stabilised by π–π stacking of adjacent benzoselenazolylidene rings. Close Cl⋯H contacts may contribute to the packing stability, and a close contact between the selenium atom and the oxygen atom in THF is also observed (dSe–O = 3.157 Å and θC–Se–O = 162.6°), constituting evidence of a ChB.48 The Ru–CNYHC bond length decreases from 2.097(7) Å in Ru4NBn to 2.038(2) and 2.032(2) Å in Ru4S and Ru4Se, respectively, indicating that the interaction between the metal and the (N,Y)HC ligands might be stronger.
| Ru4NBn | Ru4S | Ru4Se | |
|---|---|---|---|
| Ru–CNYHC | 2.097(7) | 2.038(2) | 2.032(2) |
| Ru–Arene | 1.699 | 1.702 | 1.705 |
| Ru–Clavg | 2.422(2) | 2.412(9) | 2.413(1) |
| NR–CNYHC | 1.359(8) | 1.346(3) | 1.338(3) |
| Y–CNYHC | 1.390(9) | 1.725(2) | 1.875(3) |
| CNYHC–Ru–Arene | 126.3 | 129.1 | 129.4 |
| Cl1–Ru–Cl2 | 84.32(6) | 86.48(2) | 86.89(2) |
| NR–CNYHC–Y | 105.6(5) | 109.2(2) | 109.5(2) |
| N–CH2–Phenyl | 86.6 ± 0.5 | 79.1 | 79.5 |
| NYHC centroid–CNYHC–Ru | 178.5 | 176.2 | 175.0 |
Electronic and steric analysis of the ligands and complexes
The electronic and steric properties of 1Y–4Y (Y = NR, O, S, Se) were evaluated.49 The net electronic influence was assessed using Tolman electronic parameter (TEP) values, further separated into the σ-donating ability and π-accepting contributions by analysing the one-bond coupling constant, 1JCH, of the carbene carbon atom in the 1H NMR of the azolium salts and the 77Se NMR chemical shift, δSe, of the selenium adducts.50 Additionally, DFT calculations were performed to support the experimental findings.51 Experimental TEP (TEPexp) were determined from the CO stretching frequencies of Rh[(N,Y)HC](CO)2Cl, Rh1Y–Rh4Y (Fig. 5a), with the donor ability of the carbene atom following the trend, N ≫ O > S > Se. The σ-donor strength was extracted from the one-bond coupling constant, 1JCH, of the carbene atom of 1Y·HX–4Y·HX (Y = NR, O, S or Se; X = halide) in d6-DMSO.52,53 The extent of σ-donation of the ligands is slightly lower in (N,O)HCs than in classical (N,N)HCs, whereas (N,S)HCs and (N,Se)HCs are slightly stronger σ-donors (SI Fig. 5a). The π-backbonding properties were assessed using a method based on the 77Se NMR chemical shift, δSe, of the selenium adducts Se1Y–Se4Y (Y = NR, O, S, Se).54 Based on the magnitude of δSe, the π-accepting properties of the carbene ligands increases according to the following sequence N < O < S < Se (SI Fig. 5b). Calculated TEP values (TEPcomp), obtained from the analysis of the molecular electrostatic potential surface as reported previously, are in agreement with the trend observed from the experimental data (SI Fig. 6a).55 The HOMO and LUMO energy of 1Y–4Y (Y = NR, O, S, Se) correlate well with the experimentally measured σ-donating and π-accepting character of the ligands (SI Fig. 6b).56 The electron occupancy in the M–CNYHC bond decreases in the order (N,O)HC > (N,N)HC > (N,Se)HC > (N,S)HC (SI Table 8), in agreement with the 195Pt NMR chemical shifts and crystallographically determined bond lengths.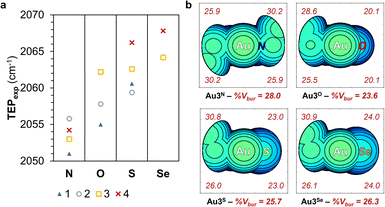 | ||
| Fig. 5 TEPexp of carbene ligands 1Y–4Y (Y = NR, O, S, Se) (a) and steric map of Au3Y (Y = NEt, O, S, Se) showing total and per quadrant buried volume (%Vbur) (b). | ||
The study of the electrostatic surface of the optimised 4Y (Y = O, S, Se) structures reveals the presence of a highly polarised p-orbital shaped lone pair on the chalcogen atom and perpendicular to the ring (SI Fig. 7a). These lone pairs, which become more diffuse when descending Group 16, could engage in HB interactions. Furthermore, sigma-holes, which are directly related to the ChB ability of the molecules, are observed in 4S and 4Se (SI Fig. 7b and SI Table 6).31 The potential to form ChB is evidenced by the Se–O close contact present in the crystal structure of Ru4Se, which adheres to the crystallographic definition of a ChB.57,58
Complementary to the electronic characterisation, the steric effects of the ligands were evaluated. The percentage of buried volume (Vbur) around the metal centre was estimated from the experimentally determined or optimised structures of Pt1Y, Pt2Y, Au3Y, and Ru4Y (Y = NR, O, S, Se) (Fig. 5b and SI Fig. 10).59 The total %Vbur decreases upon substitution of the alkylated nitrogen for a chalcogen, and then increases with the increasing atomic size of the Group 16 element, following the trend N ≈ Se < S < O. Furthermore, the %Vbur in the chalcogen-containing quadrants display less buried volume, highlighting that the coordination sphere surrounding the metal is less sterically crowded in the (N,Y)HCs (Y = O, S, Se) complexes.28,60
The steric and electronic analysis indicates that the unconventional azolylidene carbenes ligands, 1Y–4Y (Y = NR, O, S, Se), are stronger π-acceptor ligands than classical (N,N)HCs with similar σ-donation ability (with the exception of the oxazolylidenes). Furthermore, they present a less congested binding sphere and the incorporation of chalcogen atoms potentially enables HB and ChB interactions.
In vitro studies
The cytotoxicity of Pt1Y, Pt2Y, Au3Y, and Ru4Y (Y = NR, O, S, Se) was evaluated against the A2780 ovarian cancer cell line, A2780cis cells with acquired resistance to cisplatin, and non-cancerous human embryonic kidney HEK293T cells (Table 4), using the PrestoBlue assay.61 FDA-approved drugs cisplatin and auranofin, and the experimental drug RAPTA-C were included as controls. Note that the cytotoxicity of proligands 1Y·HX–4Y·HX (Y = NR, O, S, Se; X = halide) is lower than that of the related complexes against the screened cell lines (SI Table 9). Complexes Pt1Y and Pt2Y (Y = NR, O, S) exhibit cytotoxicity values in the range of 0.6 to 30 µM against the ovarian cancer cell lines. Complexes based on the benzylazolylidene scaffold, Pt2Y (Y = NBn, O, S), are more cytotoxic than the methylazolylidene analogues, Pt1Y (Y = NMe, O, S), probably as a consequence of their higher lipophilicity or the presence of a benzyl moiety capable of intercalating DNA bases.65,66 In both these cell lines, the most cytotoxic complexes are Pt1NMe and Pt2NBn (≈3 µM and ≈1 µM, respectively). The toxicity of the complexes diminished when replacing the imidazole ring for an oxazole (Pt1O and Pt2O, ≈10 µM and ≈5 µM), and further decreased when exchanging the oxygen atom for a sulphur atom (Pt1S and Pt2S, ≈15 µM and ≈8 µM). Although less cytotoxic than cisplatin to the A2780 cell line, Pt1Y and Pt2Y (Y = NR, O, S) are able to overcome acquired cisplatin-resistance in the A2780cis cell line (resistance index, RI = 1.0–2.4 vs. 24.1 for cisplatin), indicating that they probably operate via a different MoA to cisplatin. In particular, Pt2NBn presents comparable efficacy and selectivity for cancer cells to cisplatin, while being much more effective in the cisplatin-resistant cell line (1.2 ± 0.1 µM vs. 8.4 ± 4.6 µM).| Compound | IC50 (µM) after 72 h | RIa | logPOW | ||
|---|---|---|---|---|---|
| A2780 | A2780cis | HEK293T | |||
| a Resistance index (RI) = (IC50 A2780cis/IC50 A2780). | |||||
| Pt1NMe | 2.9 ± 0.2 | 2.8 ± 0.7 | 4 ± 1 | 1.0 | 1.3 ± 0.6 |
| Pt1O | 8 ± 2 | 16 ± 4 | 18 ± 6 | 2.0 | 1.0 ± 0.5 |
| Pt1S | 13 ± 8 | 30 ± 29 | 47 ± 23 | 2.4 | 1.3 ± 1.1 |
| Pt2NBn | 0.6 ± 0.2 | 1.2 ± 0.1 | 1.4 ± 0.1 | 1.9 | 2.1 ± 0.8 |
| Pt2O | 4 ± 1 | 5 ± 2 | 6 ± 2 | 1.5 | 1.6 ± 0.2 |
| Pt2S | 6 ± 1 | 12 ± 3 | 9 ± 2 | 2.2 | 1.8 ± 1.0 |
| Cisplatin | 0.4 ± 0.1 | 8 ± 5 | 1.5 ± 0.2 | 24.1 | −2.19 (ref. 62) |
| Au3NEt | 0.6 ± 0.2 | 2.7 ± 0.1 | 2.1 ± 0.6 | 4.9 | 1.6 ± 1.1 |
| Au3O | 0.2 ± 0.2 | 5 ± 2 | 1.0 ± 0.1 | 31.5 | 1.2 ± 0.4 |
| Au3S | 0.2 ± 0.1 | 6.4 ± 0.4 | 1.9 ± 0.3 | 29.2 | 1.5 ± 0.1 |
| Au3Se | 0.1 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.1 | 3.4 | 1.4 ± 0.3 |
| Auranofin | 0.1 ± 0.1 | 1.9 ± 0.4 | 0.4 ± 0.1 | 20.6 | 1.6 (ref. 63) |
| Ru4NBn | 5.9 ± 1.0 | 12.8 ± 0.9 | 13 ± 2 | 2.2 | 2.7 ± 0.3 |
| Ru4S | 10 ± 1 | 26 ± 5 | 20 ± 3 | 2.7 | 2.2 ± 0.2 |
| Ru4Se | 3.8 ± 0.8 | 9 ± 3 | 5 ± 1 | 2.2 | 2.4 ± 0.7 |
| RAPTA-C | >100 µM | >100 µM | >100 µM | −1.8 (ref. 64) | |
The gold complexes, Au3Y (Y = NEt, O, S, Se) have IC50 values in all three cell lines ranging from 0.1 to 6.4 µM, with the complexes having different behaviour depending on the cell line. In the ovarian cancer A2780 cell line, Au3Se (0.1 ± 0.1 µM), Au3S (0.2 ± 0.1 µM) and Au3O (0.2 ± 0.2 µM) exhibit cytotoxicity comparable to auranofin (0.1 ± 0.1 µM), whereas Au3NEt (0.6 ± 0.2 µM) is less cytotoxic. In contrast, in the cisplatin-resistant cell line A2780cis, Au3Se (0.2 ± 0.1 µM) has the lowest IC50 value, followed by Au3NEt, Au3O and then Au3S. With the exception of Au3NEt and Au3Se (RI = 4.9 and 3.4), the complexes did not overcome acquired cisplatin resistance (RI = 20–31), indicating that the MoA of Au3Y (Y = O, S) likely involves interactions with DNA (that would be more efficiently repaired in the cisplatin resistant cells and would result in lower cytotoxicity).
Compared to RAPTA-C, which is not cytotoxic in vitro (IC50 > 100 µM), but effective in vivo,64,67Ru4Y (Y = NBn, S, Se) are considerably more cytotoxic, with IC50 values in a similar range to Pt1Y (Y = NMe, O, S), i.e. 0.6 to 30 µM in the three cell lines. Ru4Se is the most cytotoxic compound of the series in all three cell lines, which was also observed in Au3Se. Additionally, all the complexes are active against cisplatin-resistant cells (RI = 2.2–2.7).
The lipophilicity of the complexes Pt1Y, Pt2Y, Au3Y, and Ru4Y (Y = NR, O, S, Se) decreases with the introduction of the chalcogen atom (Table 4 and SI Table 6), with the (N,O)HCs complexes exhibiting the highest hydrophilic character, likely due to the formation of HBs with water. Although the cytotoxicity of some NHC complexes has been previously linked with their lipophilicity,68 this does not appear to be the case for Pt1Y, Pt2Y, Au3Y, and Ru4Y (Y = NR, O, S, Se). SAR analysis (based on the experimental and computational data from this study) was performed to identify major chemical, physical, structural and electronic properties modulating the cytotoxicity of Pt1Y, Pt2Y, Au3Y and Ru4Y (Y = NR, O, S, Se). Notably, typical factors such as lipophilicity or aromaticity do not appear to affect the cytotoxicity of the complexes to a great extent (SI Fig. S13). Instead, cytotoxicity appears to be correlated with the charge at the metal, the energy of the σ-hole and lone pair energy, or the Y–CNYHC bond length, and inversely correlated with percentage of Vbur in the NR quadrant or the energy of the π-donor orbital. These properties are directly linked to the nature of the azolylidene ligand. Despite the potential interest in Pt1Se and Pt2Se given the high cytotoxicity exhibited by Au3Se and Ru4Se, we did not synthesise them due to stability and accessibility challenges of unsubstituted selenazoles.69
The cytotoxicity of the platinum complexes, Pt1NMe and Pt2NBn, is similar to other previously reported trans-(NHC)PtX2(amine) complexes (between 0.9 and 3.1 µM).12 Additionally, similar to the platinum compounds in this study, some of the related complexes reported also overcome acquired cisplatin-resistance. The cytotoxicity of Ru4NBn in A2780 cells is also comparable to those reported in the literature (2.1 ± 0.9 µM and 2.4 ± 1.0 µM against MCF-7 breast adenocarcinoma and HT-29 colon carcinoma cells, respectively).38 In contrast, a 10-fold increase in the toxicity is observed in Au3NEt compared to the same complex bearing a chlorido ligand instead of an iodido ligand (0.6 ± 0.2 µM vs. 6.4 ± 2.0 µM).37Au3S demonstrated comparable activity to a previously-reported peptide-derivatised (N,S)HC gold complex in lung carcinoma (A549) (IC50 = 0.4 ± 0.01 µM).26 We were not able to find any reports of the cytotoxicity of (N,O)HC complexes or of (N,Se)HC complexes, which had remained unexplored until now.
Mechanistic studies
As previously mentioned in the introduction, the parent (N,N)HC complexes, Pt1NMe, Pt2NBn, Au3NEt, and Ru4NBn, were selected as they had demonstrated anticancer activity and a MoA had been proposed. Interactions with DNA were proposed for Pt1NMe and Pt2NBn,12 whereas Au3NEt and Ru4NBn were reported to interact with thioredoxin reductase (TrxR) and cathepsin B (CatB).37,38The inhibitory effect of Pt1Y and Pt2Y (Y = NR, O, S) on DNA synthesis during cell proliferation was quantified in cellulo using an EdU incorporation assay and fluorescence cell microscopy (Fig. 6 and SI Fig. 12).70 Gemcitabine, a clinically-approved DNA synthesis inhibitor,71 was used as a positive control (100% inhibition), and the cells treated with an equivalent amount of DMSO served as a negative control (“untreated”, 0% inhibition). Cisplatin and transplatin were included as references. DNA synthesis was blocked to various degrees in the cells treated with the different platinum complexes. As expected based on their structure and cytotoxicity, cisplatin was a better DNA synthesis inhibitor than transplatin (62% vs. 23%), and Pt1Y (Y = NMe, O, S) displayed a lower degree of inhibition than Pt2Y (Y = NBn, O, S), presumably as a consequence of the ability of the benzyl wingtip to intercalate DNA. In particular, Pt2NBn, the compound with the highest cytotoxicity, inhibited DNA synthesis more effectively than cisplatin under the tested conditions and reached comparable inhibition to the positive control (gemcitabine, 100%). Overall, the extent of inhibition of DNA synthesis correlates well with the cytotoxicity of the complexes, suggesting that the inhibition of DNA synthesis, likely by the formation of non-classical DNA adducts, is a key MoA of Pt1Y and Pt2Y (Y = NR, O, S).
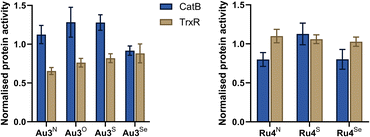
The inhibitory activity of Au3Y (Y = NEt, O, S, Se) and Ru4Y (Y = NBn, S, Se) against CatB and TrxR was studied in A2780 cells using commercially available assays (Fig. 7). Au3Y (Y = NEt, O, S, Se) inhibit the enzymatic activity of TrxR but are inactive against CatB. However, the degree of inhibition does not correlate with the cytotoxicity of the complexes, which further highlights that Au3Y (Y = O, S, Se) may have other molecular targets. It is worth noting that other gold complexes, such as auranofin, have promiscuous multi-target activity.72,73 In contrast, Ru4Y (Y = NBn, S, Se) are inactive for TrxR, whereas Ru4Y (Y = NBn, Se) inhibit CatB activity, correlating well with their cytotoxicity and indicating that cathepsin B is a likely biological target of Ru4Y (Y = NBn, Se), although other targets cannot be excluded.74
Structural studies
Crystals of adducts between Ru4Y (Y = S, Se) and hen egg white lysozyme (HEWL), a model protein, were obtained by crystal soaking and analysed by single-crystal X-ray diffraction (Fig. 8). Both crystals presented anomalous electron density around Asp101, where the ruthenium atoms and some of the Ru4Y (Y = S, Se) ligands were modelled.The ruthenium centre is coordinated in a bidentate fashion to the carboxylic acid of Asp101. The average Ru–OAsp101 bond length is 2.227 and 2.152 Å for Ru4S and Ru4Se, respectively, indicating that Ru4Se might bind the protein more strongly. It is conceivable that binding of Ru4S to CatB is also weaker than that of Ru4Se, which might explain the weaker CatB inhibition exhibited by Ru4S (Fig. 7), also supported by the electronic description of 4S as a weaker π-acceptor. In both structures, the (N,Y)HC ligand remains coordinated to the metal centre, but the arene and chlorido ligands have been substituted by solvent molecules and by the oxygen atoms in Asp101. The Ru–CNYHC distance in the protein crystals adducts is 1.987 and 1.990 Å for Ru4S and Ru4Se, respectively, slightly shorter than in the intact Ru4S and Ru4Se structures (2.038(2) and 2.032(2) Å, respectively). In both crystallised adducts, the metal drug fragments have an occupancy of 1.0, and no further additional isomers were observed. A comparison of the ruthenated HEWL structures to the native protein crystals (PDB:4NHI) revealed no major structural perturbations (RMSD of 0.920 and 0.894 Å).
Presumably, the binding in Ru4Y (Y = S, Se) is driven by the electrostatic interactions between the resulting cationic complex from the substitution of the chlorido ligands in Ru4Y (Y = S, Se) and the negatively charged catalytic site of HEWL. It should be noted that the different soaking times (5–10 s vs. 3 days) could affect the binding sites. Other ruthenium complexes have been shown to interact with Asp101 in HEWL, however, only as naked ruthenium ions.75,76 Crystals of adducts between HEWL and dichloro(1,3-dimethylbenzimidazol-2-ylidene)(η6-p-cymene)ruthenium(II), a complex related to Ru4NBn, revealed His15 and Lys33 as the preferred binding sites.75 Hence, replacement of the nitrogen atom for a chalcogen atom in Ru4Y (Y = NBn, S, Se) modulates the preferential binding site of the ruthenium complexes, likely a consequence of the more electron-poor ruthenium centres. Previously, other (N,N)HC ruthenium complexes have been reported to bind to both nitrogen- and oxygen-containing residues in several proteins,77–80 as well as to interact with carboxylic acid-containing amino acid residues.81
In order to elucidate the molecular interaction between the complexes and their studied biological targets, molecular docking was employed. However, the prediction of the binding site of metal complexes is non-trivial due to the reactivity of metal complexes with respect to nucleophiles (for example, water, amino acids, or nucleobases) and the scarcity of force fields able to describe metal atoms. Several solutions have been reported or adapted to address this challenge.82–85 We developed and validated an approach to the docking of metal-containing compounds based on Autodock86 (see SI for the further details and discussion). The developed protocol yields reasonable redocking results in terms of the prediction of the metal binding site, and of the position, orientation, and conformation of the ligands.
Blind molecular docking (the whole biomolecule structure is used without any bias towards specific binding sites) was performed using the approach to evaluate potential binding sites and affinities of Pt1Y and Pt2Y (Y = NR, O, S) to DNA (from a simulated trans-bound DNA interstrand adduct),87 of Au3Y (Y = NEt, O, S, Se) to TrxR (PDB:2J3N),88 and of Ru4Y (Y = NBn, S, Se) to CatB (PDB:3AI8).89
Pt1Y and Pt2Y (Y = NR, O, S) formed N7-guanine interstrand DNA adducts (SI Fig. 17, 18 and SI Table 12). The binding energies for Pt2Y (Y = NBn, O, S) were lower than for Pt1Y (Y = NMe, O, S), consistent with the higher cytotoxicity and higher degree of inhibition of DNA synthesis (Fig. 6) exhibited by Pt2Y (NBn, O, S), arising from the additional π–π interactions between the benzyl group and the nucleobases (SI Fig. 18). Both chalcogen atoms in Pt2O and Pt2S are positioned towards a hydrogen bond donor area around the amine group of an adjacent adenine, which likely establishes a HB and stabilises the conformation.
Despite predicting binding sites for Au3Y (Y = NEt, O, S, Se) not typically linked to TrxR inhibition (Sec498),14 a HB was observed Au3O between the oxygen atom in the oxazolylidene ligand and the amine of Lys67 (3.01 Å, SI Fig. 18). This was not observed in Au3S or Au3Se, consistent with the expected strength of the HB (SI Table 4). In contrast, a ChB was present between the carbonyl oxygen in Thr58 and the chalcogen atom in Au3S or Au3Se (3.60 Å and 3.39 Å, SI Fig. 18). These results highlight that HB and ChB interactions are likely to occur in (N,O)HC, and in (N,S)HC and (N,Se)HC complexes, respectively.
Docking studies of Ru4Y (Y = NBn, S, Se) to CatB indicated that the complexes are likely to form adducts with residues Cys29 and His199 simultaneously, which are part of the catalytic pocket and active site of the protein.90 Other ruthenium complexes have previously been reported to interact with Cys29.18 For Ru4Se, two different poses binding Cys29:His199 were observed (SI Fig. 19), which could explain the higher cytotoxicity of Ru4Se (Fig. 7).
Conclusions
Unconventional chalcogen-containing azolylidene (N,Y)HC (Y = NR, O, S, Se) metal complexes based on (benz)oxazole, (benzo)thiazole and benzoselenazole were successfully synthesised, including the first reported transition metal selenazolylidene complexes. The electronic and steric properties of these unconventional, chalcogen-based carbene ligands were investigated. The ligands were found to be overall weaker donors with enhanced π-acceptor ability, which could prove useful in the synthesis of low-valent or main group complexes and potentially favouring catalytic reactions benefiting from electron-deficient metal centres. The cytotoxicity of the (N,Y)HC complexes was evaluated against ovarian cancer cells. Distinct trends in cytotoxicity emerged, depending on the metal centre. In the platinum complexes, substitution of the alkylated nitrogen in the azolylidene ligand for a chalcogen atoms decreased cytotoxicity following the trend (N,N)HC > (N,O)HC > (N,S)HC. For gold and ruthenium, the (N,Se)HC complexes, being the only reported examples of this class, were more cytotoxic than, in order, the respective (N,O)HC, (N,N)HC, and (N,S)HC analogues, suggesting a new avenue for putative metallocarbene anticancer drug candidates. In many cases, the complexes were able to overcome cisplatin-related resistance. Mechanistic and structural studies were performed in vitro, in cellulo, in crystallo, and in silico, revealing that incorporation of a chalcogen atom into the heterocyclic scaffold can modulate biological targets, activity and biomolecular interactions of the complexes. These findings highlight the potential of unconventional chalcogen azolylidene ligands as tuneable scaffolds to fine-tune the biological activity of metal complexes.Author contributions
J. R.-dG. conceived the idea and led the project. J. R.-dG., P. M. F. P, A. N. V. and L. E. K. F. conducted the synthesis and characterisation of the proligands and metal complexes. I. L. S. and K. G. carried out the biological experiments. F. F.-T. and R. S. collected and processed the single-crystal X-ray data. F. K. performed the cell fluorescence microscopy measurements and K. G. solved the protein crystal structures. All authors contributed to the discussion of the results and revision of the manuscript. P. J. D. directed and supervised the project.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article has been included in the supplementary information (SI). See DOI: https://doi.org/10.1039/d5sc05555e.CCDC 2130715, 2270445–2270450, 2271662, 2395164 and 2395165 contain the supplementary crystallographic data for this paper.91a–j
Protein crystallographic data for the adducts of HEWL with Ru4S and Ru4Se has been deposited at the PDB under accession numbers 9HTI and 9HTJ, respectively.92a,b
Additional data generated in this study is openly available at https://doi.org/10.5281/zenodo.16159145.
Acknowledgements
This research has been funded by the Swiss National Science Foundation and the École Polytechnique Fédérale de Lausanne. The Mass Spectrometry and Elemental Analysis service, and the Nuclear Magnetic Resonance platform of EPFL are acknowledged for their support. We thank Dr S. A. Tellite and Dr R. Laplaza for the fruitful DFT discussions, and Prof. C. A. Davey for advice on structural biology. The authors acknowledge Dr F. Pojer and Dr A. N. Larabi of the Protein Production and Structure Core Facility and the staff at the Biomolecular Screening Facility of EPFL. We are grateful for the encouragement of Dr R. J. Somerville, Dr D. Savary, and Dr R.-C. Turnell-Ritson, and the support of Dr A. Egurbide-Sifre and Dr H. Solé-Àvila.Notes and references
- M. N. Hopkinson, C. Richter, M. Schedler and F. Glorius, Nature, 2014, 510, 485–496 Search PubMed.
- P. Bellotti, M. Koy, M. N. Hopkinson and F. Glorius, Nat. Rev. Chem., 2021, 5, 711–725 CrossRef CAS PubMed.
- K. M. Hindi, M. J. Panzner, C. A. Tessier, C. L. Cannon and W. J. Youngs, Chem. Rev., 2009, 109, 3859–3884 Search PubMed.
- L. Oehninger, R. Rubbiani and I. Ott, Dalton Trans., 2013, 42, 3269–3284 RSC.
- I. Ott, in Advances in Inorganic Chemistry, Elsevier, 2020, vol. 75, pp. 121–148 Search PubMed.
- S. A Patil, A. P Hoagland, S. A Patil and A. Bugarin, Future Med. Chem., 2020, 12, 2239–2275 CrossRef PubMed.
- Q. Zhao, B. Han, C. Peng, N. Zhang, W. Huang, G. He and J. Li, Med. Res. Rev., 2024, 44, 2194–2235 Search PubMed.
- S. Zhao, Z. Yang, G. Jiang, S. Huang, M. Bian, Y. Lu and W. Liu, Coord. Chem. Rev., 2021, 449, 214217 CrossRef CAS.
- P. Heringova, J. Woods, F. S. Mackay, J. Kasparkova, P. J. Sadler and V. Brabec, J. Med. Chem., 2006, 49, 7792–7798 CrossRef CAS.
- T. Kishimoto, Y. Yoshikawa, K. Yoshikawa and S. Komeda, Int. J. Mol. Sci., 2019, 21, 34 CrossRef PubMed.
- J. Kasparkova, Nucleic Acids Res., 2004, 32, 5546–5552 CrossRef CAS PubMed.
- M. Skander, P. Retailleau, B. Bourrié, L. Schio, P. Mailliet and A. Marinetti, J. Med. Chem., 2010, 53, 2146–2154 CrossRef CAS PubMed.
- M. Mora, M. C. Gimeno and R. Visbal, Chem. Soc. Rev., 2019, 48, 447–462 RSC.
- A. Pratesi, C. Gabbiani, E. Michelucci, M. Ginanneschi, A. M. Papini, R. Rubbiani, I. Ott and L. Messori, J. Inorg. Biochem., 2014, 136, 161–169 Search PubMed.
- P. J. Barnard and S. J. Berners-Price, Coord. Chem. Rev., 2007, 251, 1889–1902 CrossRef CAS.
- H. Ghareeb and N. Metanis, Chem. - Eur. J., 2020, 26, 10175–10184 CrossRef CAS.
- B. Dominelli, J. D. G. Correia and F. E. Kühn, J. Organomet. Chem., 2018, 866, 153–164 CrossRef CAS.
- A. Casini, C. Gabbiani, F. Sorrentino, M. P. Rigobello, A. Bindoli, T. J. Geldbach, A. Marrone, N. Re, C. G. Hartinger, P. J. Dyson and L. Messori, J. Med. Chem., 2008, 51, 6773–6781 CrossRef CAS.
- N. Aggarwal and B. F. Sloane, Proteomics:Clin. Appl., 2014, 8, 427–437 CAS.
- A. Catalano, A. Mariconda, M. S. Sinicropi, J. Ceramella, D. Iacopetta, C. Saturnino and P. Longo, Antibiotics, 2023, 12, 365 Search PubMed.
- K. J. Kilpin, S. Crot, T. Riedel, J. A. Kitchen and P. J. Dyson, Dalton Trans., 2014, 43, 1443–1448 RSC.
- C. Vanucci-Bacqué, M. Wolff, B. Delavaux-Nicot, A. M. Abdallah, S. Mallet-Ladeira, C.-L. Serpentini, F. Bedos-Belval, K. W. Fong, X. Y. Ng, M. L. Low, E. Benoist and S. Fery-Forgues, Dalton Trans., 2024, 53, 11276–11294 RSC.
- B. Bertrand, A. S. Romanov, M. Brooks, J. Davis, C. Schmidt, I. Ott, M. O'Connell and M. Bochmann, Dalton Trans., 2017, 46, 15875–15887 RSC.
- H. V. Huynh, The Organometallic Chemistry of N-heterocyclic Carbenes, Wiley, 1st edn, 2017 Search PubMed.
- Y. Gothe, I. Romero-Canelón, T. Marzo, P. J. Sadler, L. Messori and N. Metzler-Nolte, Eur. J. Inorg. Chem., 2018, 2018, 2461–2470 CrossRef CAS.
- A. Gutiérrez, M. C. Gimeno, I. Marzo and N. Metzler-Nolte, Eur. J. Inorg. Chem., 2014, 2014, 2512–2519 CrossRef.
- E. O. Fischer, D. Himmelreich, R. Cai, H. Fischer, U. Schubert and B. Zimmer-Gasser, Chem. Ber., 1981, 114, 3209–3219 CrossRef CAS.
- J. Zhang, T. Li, X. Li, A. Lv, X. Li, Z. Wang, R. Wang, Y. Ma, R. Fang, R. Szostak and M. Szostak, Commun. Chem., 2022, 5, 60 CrossRef CAS.
- K. E. Horner and P. B. Karadakov, J. Org. Chem., 2015, 80, 7150–7157 CrossRef CAS PubMed.
- Z. Kelemen, O. Hollóczki, J. Oláh and L. Nyulászi, RSC Adv., 2013, 3, 7970 Search PubMed.
- M. A. A. Ibrahim and E. M. Z. Telb, ACS Omega, 2020, 5, 21631–21640 CrossRef CAS PubMed.
- W. Wang, H. Zhu, S. Liu, Z. Zhao, L. Zhang, J. Hao and Y. Wang, J. Am. Chem. Soc., 2019, 141, 9175–9179 Search PubMed.
- Z. Alamiddine, S. Thany, J. Graton and J. Le Questel, ChemPhysChem, 2018, 19, 3069–3083 CrossRef CAS PubMed.
- L. Vogel, P. Wonner and S. M. Huber, Angew. Chem., Int. Ed., 2019, 58, 1880–1891 CrossRef CAS.
- A. Chand, D. K. Sahoo, A. Rana, S. Jena and H. S. Biswal, Acc. Chem. Res., 2020, 53, 1580–1592 CrossRef CAS.
- K. Konidaris, A. Daolio, A. Pizzi, P. Scilabra, G. Terraneo, S. Quici, J. S. Murray, P. Politzer and G. Resnati, Cryst. Growth Des., 2022, 22, 4987–4995 CrossRef CAS.
- R. Rubbiani, I. Kitanovic, H. Alborzinia, S. Can, A. Kitanovic, L. A. Onambele, M. Stefanopoulou, Y. Geldmacher, W. S. Sheldrick, G. Wolber, A. Prokop, S. Wölfl and I. Ott, J. Med. Chem., 2010, 53, 8608–8618 CrossRef CAS PubMed.
- L. Oehninger, M. Stefanopoulou, H. Alborzinia, J. Schur, S. Ludewig, K. Namikawa, A. Muñoz-Castro, R. W. Köster, K. Baumann, S. Wölfl, W. S. Sheldrick and I. Ott, Dalton Trans., 2013, 42, 1657–1666 RSC.
- J. M. Duelos and P. Haake, Biochemistry, 1974, 13, 5358–5362 CrossRef.
- T. Scattolin and S. P. Nolan, Trends Chem., 2020, 2, 721–736 CrossRef CAS.
- N. V. Tzouras, F. Nahra, L. Falivene, L. Cavallo, M. Saab, K. Van Hecke, A. Collado, C. J. Collett, A. D. Smith, C. S. J. Cazin and S. P. Nolan, Chem.–Eur. J., 2020, 26, 4515–4519 CrossRef CAS PubMed.
- E. Chardon, G. Dahm, G. Guichard and S. Bellemin-Laponnaz, Organometallics, 2012, 31, 7618–7621 CrossRef CAS.
- M. Bouché, B. Vincent, T. Achard and S. Bellemin-Laponnaz, Molecules, 2020, 25, 3148 CrossRef PubMed.
- T. M. Gilbert and T. Ziegler, J. Phys. Chem. A, 1999, 103, 7535–7543 CrossRef CAS.
- L. Falivene and L. Cavallo, Coord. Chem. Rev., 2017, 344, 101–114 CrossRef CAS.
- N. Y. S. Lam, D. Truong, H. Burmeister, M. V. Babak, H. U. Holtkamp, S. Movassaghi, D. M. Ayine-Tora, A. Zafar, M. Kubanik, L. Oehninger, T. Söhnel, J. Reynisson, S. M. F. Jamieson, C. Gaiddon, I. Ott and C. G. Hartinger, Inorg. Chem., 2018, 57, 14427–14434 CrossRef CAS PubMed.
- Z. İ. Oruç, L. Gök, H. Türkmen, O. Şahin, O. Büyükgüngör and B. Çetinkaya, J. Organomet. Chem., 2016, 807, 36–44 CrossRef.
- P. Scilabra, G. Terraneo and G. Resnati, Acc. Chem. Res., 2019, 52, 1313–1324 CrossRef CAS PubMed.
- H. V. Huynh, Chem. Rev., 2018, 118, 9457–9492 CrossRef CAS PubMed.
- R. Dorta, E. D. Stevens, N. M. Scott, C. Costabile, L. Cavallo, C. D. Hoff and S. P. Nolan, J. Am. Chem. Soc., 2005, 127, 2485–2495 Search PubMed.
- D. J. Nelson and S. P. Nolan, Chem. Soc. Rev., 2013, 42, 6723 Search PubMed.
- G. Meng, L. Kakalis, S. P. Nolan and M. Szostak, Tetrahedron Lett., 2019, 60, 378–381 CrossRef CAS.
- K. Verlinden, H. Buhl, W. Frank and C. Ganter, Eur. J. Inorg. Chem., 2015, 2015, 2416–2425 CrossRef CAS.
- A. Liske, K. Verlinden, H. Buhl, K. Schaper and C. Ganter, Organometallics, 2013, 32, 5269–5272 CrossRef CAS.
- J. Mathew and C. H. Suresh, Inorg. Chem., 2010, 49, 4665–4669 CrossRef CAS PubMed.
- H. Jacobsen, A. Correa, A. Poater, C. Costabile and L. Cavallo, Coord. Chem. Rev., 2009, 253, 687–703 Search PubMed.
- C. B. Aakeroy, D. L. Bryce, G. R. Desiraju, A. Frontera, A. C. Legon, F. Nicotra, K. Rissanen, S. Scheiner, G. Terraneo, P. Metrangolo and G. Resnati, Pure Appl. Chem., 2019, 91, 1889–1892 CrossRef CAS.
- A. S. Lundemba, D. D. Bibelayi, P. V. Tsalu, P. A. Wood, J. Cole, J. S. Kayembe and Z. G. Yav, Cryst. Struct. Theory Appl., 2021, 10, 57–69 CAS.
- L. Falivene, R. Credendino, A. Poater, A. Petta, L. Serra, R. Oliva, V. Scarano and L. Cavallo, Organometallics, 2016, 35, 2286–2293 CrossRef CAS.
- A. C. Hillier, W. J. Sommer, B. S. Yong, J. L. Petersen, L. Cavallo and S. P. Nolan, Organometallics, 2003, 22, 4322–4326 CrossRef CAS.
- M. Xu, D. J. McCanna and J. G. Sivak, J. Pharmacol. Toxicol. Methods, 2015, 71, 1–7 Search PubMed.
- A. K. Nath, X. Shi, D. L. Harrison, J. E. Morningstar, S. Mahon, A. Chan, P. Sips, J. Lee, C. A. MacRae, G. R. Boss, M. Brenner, R. E. Gerszten and R. T. Peterson, Cell Chem. Biol., 2017, 24, 565–575.e4 Search PubMed.
- T. Marzo, D. Cirri, C. Gabbiani, T. Gamberi, F. Magherini, A. Pratesi, A. Guerri, T. Biver, F. Binacchi, M. Stefanini, A. Arcangeli and L. Messori, ACS Med. Chem. Lett., 2017, 8, 997–1001 CrossRef CAS PubMed.
- C. Scolaro, A. Bergamo, L. Brescacin, R. Delfino, M. Cocchietto, G. Laurenczy, T. J. Geldbach, G. Sava and P. J. Dyson, J. Med. Chem., 2005, 48, 4161–4171 CrossRef CAS PubMed.
- I. Buß, D. Garmann, M. S. Galanski, G. Weber, G. V. Kalayda, B. K. Keppler and U. Jaehde, J. Inorg. Biochem., 2011, 105, 709–717 CrossRef PubMed.
- T. Mahata, A. Kanungo, S. Ganguly, E. K. Modugula, S. Choudhury, S. K. Pal, G. Basu and S. Dutta, Angew. Chem., Int. Ed., 2016, 55, 7733–7736 CrossRef CAS.
- P. Nowak-Sliwinska, J. R. Van Beijnum, A. Casini, A. A. Nazarov, G. Wagnières, H. Van Den Bergh, P. J. Dyson and A. W. Griffioen, J. Med. Chem., 2011, 54, 3895–3902 CrossRef CAS PubMed.
- G. Lv, L. Guo, L. Qiu, H. Yang, T. Wang, H. Liu and J. Lin, Dalton Trans., 2015, 44, 7324–7331 RSC.
- P. Langer, Synlett, 2022, 33, 728–736 CrossRef CAS.
- N. Talarek, J. Petit, E. Gueydon and E. Schwob, in DNA Replication, ed. S. Vengrova and J. Dalgaard, Springer New York, New York, NY, 2015, vol. 1300, pp. 105–112 Search PubMed.
- S. Yang, D. Luo, N. Li, C. Li, S. Tang and Z. Huang, Biochemistry, 2020, 59, 4344–4352 Search PubMed.
- C. S. Allardyce and P. J. Dyson, Dalton Trans., 2016, 45, 3201–3209 RSC.
- S. Rodríguez-Enríquez, D. X. Robledo-Cadena, S. C. Pacheco-Velázquez, J. L. Vargas-Navarro, J. A. Padilla-Flores, T. Kaambre and R. Moreno-Sánchez, PLoS One, 2024, 19, e0309331 CrossRef.
- R. F. S. Lee, A. Chernobrovkin, D. Rutishauser, C. S. Allardyce, D. Hacker, K. Johnsson, R. A. Zubarev and P. J. Dyson, Sci. Rep., 2017, 7, 1590 CrossRef PubMed.
- M. P. Sullivan, M. K. Nieuwoudt, G. A. Bowmaker, N. Y. S. Lam, D. Truong, D. C. Goldstone and C. G. Hartinger, Chem. Commun., 2018, 54, 6120–6123 Search PubMed.
- L. Messori and A. Merlino, Dalton Trans., 2014, 43, 6128 Search PubMed.
- M. P. Sullivan, M. Cziferszky, I. Tolbatov, D. Truong, D. Mercadante, N. Re, R. Gust, D. C. Goldstone and C. G. Hartinger, Angew. Chem., Int. Ed., 2021, 60, 19928–19932 Search PubMed.
- A. Annunziata, M. E. Cucciolito, M. Di Ronza, G. Ferraro, M. Hadiji, A. Merlino, D. Ortiz, R. Scopelliti, F. Fadaei Tirani, P. J. Dyson and F. Ruffo, Organometallics, 2023, 42, 952–964 Search PubMed.
- B. Y. T. Lee, M. P. Sullivan, E. Yano, K. K. H. Tong, M. Hanif, T. Kawakubo-Yasukochi, S. M. F. Jamieson, T. Soehnel, D. C. Goldstone and C. G. Hartinger, Inorg. Chem., 2021, 60, 14636–14644 Search PubMed.
- M. P. Sullivan, M. Groessl, S. M. Meier, R. L. Kingston, D. C. Goldstone and C. G. Hartinger, Chem. Commun., 2017, 53, 4246–4249 RSC.
- Z. Adhireksan, G. Palermo, T. Riedel, Z. Ma, R. Muhammad, U. Rothlisberger, P. J. Dyson and C. A. Davey, Nat. Commun., 2017, 8, 14860 Search PubMed.
- S. L. Dürr, A. Levy and U. Rothlisberger, Nat. Commun., 2023, 14, 2713 Search PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 Search PubMed.
- E. Ortega-Carrasco, A. Lledós and J.-D. Maréchal, J. Comput. Chem., 2014, 35, 192–198 Search PubMed.
- M. L. A. Hakkennes, F. Buda and S. Bonnet, J. Chem. Inf. Model., 2023, 63, 7816–7825 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS.
- H.-C. Tai, R. Brodbeck, J. Kasparkova, N. J. Farrer, V. Brabec, P. J. Sadler and R. J. Deeth, Inorg. Chem., 2012, 51, 6830–6841 CrossRef CAS.
- K. Fritz-Wolf, S. Urig and K. Becker, J. Mol. Biol., 2007, 370, 116–127 CrossRef CAS.
- B. Mirković, M. Renko, S. Turk, I. Sosič, Z. Jevnikar, N. Obermajer, D. Turk, S. Gobec and J. Kos, ChemMedChem, 2011, 6, 1351–1356 CrossRef PubMed.
- J. Schmitz, E. Gilberg, R. Löser, J. Bajorath, U. Bartz and M. Gütschow, Bioorg. Med. Chem., 2019, 27, 1–15 CrossRef CAS.
- (a) CCDC 2130715: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc29j5rs; (b) CCDC 2270445: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2g6l5g; (c) CCDC 2270446: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2g6l6h; (d) CCDC 2270447: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2g6l7j; (e) CCDC 2270448: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2g6l8k; (f) CCDC 2270449: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2g6l9l; (g) CCDC 2270450: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2g6lbm; (h) CCDC 2271662: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2g7vf0; (i) CCDC 2395164: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2ldccr; (j) CCDC 2395165: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2ldcds.
- (a) J. Romano-deGea, K. Lau and P. J. Dyson, Adduct formed during the incubation of dichloro(3-benzylbenzoselenazol-2-ylidene)(eta6-p-cymene)ruthenium(II) with HEWL, 9HTI, 2026, DOI:10.2210/pdb9HTI/pdb; (b) J. Romano-deGea, K. Lau and P. J. Dyson, Adduct formed during the incubation of dichloro(3-benzylbenzothiazol-2-ylidene)(eta6-p-cymene)ruthenium(II) with HEWL, 9HTJ, 2026, DOI:10.2210/pdb9HTJ/pdb.
| This journal is © The Royal Society of Chemistry 2026 |

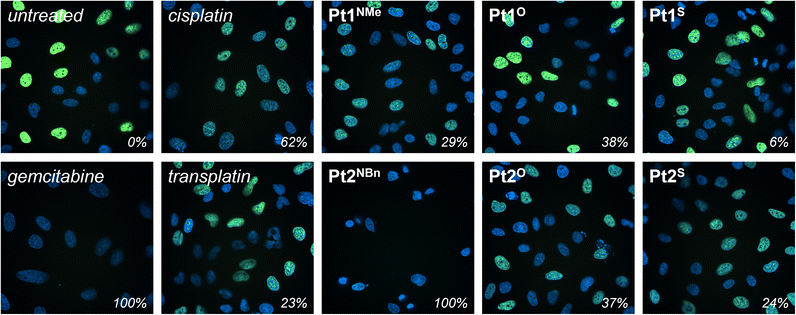
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (blue), and nuclei undergoing DNA synthesis with Edu-AlexaFluor-488 (green). Gemcitabine and DMSO were used as a positive (100%) and negative (0%) control, respectively.
342 (blue), and nuclei undergoing DNA synthesis with Edu-AlexaFluor-488 (green). Gemcitabine and DMSO were used as a positive (100%) and negative (0%) control, respectively.