Open Access Article
Open Access ArticleSingle-cell metabolic accumulation analysis by microfluidic hydrogel microspheres combined with mass spectrometry
Gaowa
Xing†
 ab,
Hongren
Yao†
b,
Zengnan
Wu
*b,
Yuxuan
Li
b,
Ying
Hou
ab,
Hongren
Yao†
b,
Zengnan
Wu
*b,
Yuxuan
Li
b,
Ying
Hou
 b and
Jin-Ming
Lin
b and
Jin-Ming
Lin
 *b
*b
aFujian Provincial Key Laboratory of Ecological Impacts and Treatment Technologies for Emerging Contaminants, Key Laboratory of Ecological Environment and Information Atlas (Putian University) Fujian Provincial University, College of Environmental and Biological Engineering, Putian University, Putian 351100, China
bDepartment of Chemistry, Beijing Key Laboratory of Microanalytical Methods and Instrumentation, Key Laboratory of Bioorganic Phosphorus Chemistry & Chemical Biology (Ministry of Education), Tsinghua University, Beijing 100084, P. R. China. E-mail: jmlin@mail.tsinghua.edu.cn
First published on 28th November 2025
Abstract
Metabolic analysis of tumor cells is crucial for understanding tumor metabolic reprogramming, and single-cell metabolic accumulation studies provide precise insights into tumor biology. In this work, we established a microfluidic assay for single-cell metabolic accumulation analysis by combining droplet-generated hydrogel microspheres for single-cell encapsulation with a microchamber culture chip for microsphere isolation and 3D cultivation. This system enables efficient accumulation of secreted metabolites from individual cells for downstream analysis. Coupled with mass spectrometry, this system revealed metabolic accumulation heterogeneity among individual cells of the same tumor type. Moreover, metabolic profiling of different tumor types, including A549, HepG2, and HCT116, demonstrated distinct metabolic patterns, enabling cell classification based on accumulation differences. The method supports high-throughput, multiplexed, and dynamic metabolic analysis in a 3D single-cell context, offering a scalable strategy for decoding tumor heterogeneity with potential application in cell-state characterization and advancing oncology research.
Introduction
Cancer is a complex genetic disorder characterized by the uncontrolled growth of abnormal cells, primarily driven by intricate genomic alterations involving mutations in oncogenes and tumor suppressor genes.1 These genetic mutations instigate metabolic reprogramming in cancer cells, leading to distinct metabolic pathways compared to normal cells.2 A well-known phenomenon in this context is the “Warburg effect”,3 wherein tumor cells, even in the presence of sufficient oxygen, prominently convert a substantial amount of glucose into lactate. Metabolic reprogramming is a pivotal process that governs the survival, metastasis, and resistance activities of cancer cells.4–6 Understanding these intricate processes not only contributes to a comprehensive comprehension of cancer cells but also provides a crucial avenue for therapeutic interventions.Investigating the metabolism of tumor cells provides a direct avenue for understanding tumor reprogramming. However, the predominant approach in studying human tumor metabolism involves collective measurements obtained from bulk tumors. Nevertheless, heterogeneity arises within the same tumor mass due to diverse external microenvironments at different locations and hierarchical levels. As a result, each tumor cell may exhibit unique metabolic states.7,8 Thus, it is crucial to examine tumor cell metabolism at the single-cell level.
Methods for studying single-cell metabolism typically involve the initial isolation of individual cells. Researchers have developed various techniques for single-cell isolation, including laser capture microdissection,9,10 flow cytometry,11,12 microfluidic chips,13–15 and others.16,17 Among these, microfluidics-based single-cell analysis for various applications, including single-cell proteomics,18 transcriptomics,19 metabolomics,20 lipidomics,21 multiomics22 and post-translational modifications.23 And droplet microfluidic methods, due to their integration of the high operability and the throughput advantages of flow cytometry, have found widespread applications in single-cell genomics,24,25 transcriptomics,26,27 proteomics,28,29 and other fields. They also hold significant potential for single-cell metabolomics research.30 In particular, the recent development of microfluidic hydrogel microspheres has emerged as a valuable tool for constructing three-dimensional (3D) culture systems. Compared with traditional 2D culture systems, hydrogel microspheres with 3D porous structure can maintain cell morphology and activity for a long time and can conduct bidirectional interactions with the extracellular microenvironment. This advancement allows for better simulation of the 3D extracellular matrix environment surrounding cells.31–34
Approaches for single-cell metabolism analysis encompass Raman spectroscopy,35 fluorescence spectroscopy,36 mass spectrometry,37,38 and other detection methods. However, these techniques commonly provide information on the metabolic state of cells at particular time points rather than offering insights into metabolic accumulation.39 The substances measured often constitute intracellular components rather than metabolites released by the cells. The cumulative measurement of metabolites from single cells over time could facilitate the quantification of single-cell metabolism, providing more comprehensive information than that obtained from analysis at specific time points.
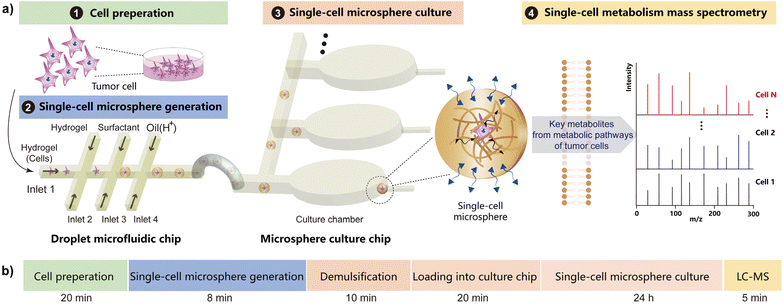
Here, we present a microfluidic method for analyzing single-cell metabolic accumulation by integrating a droplet microfluidic device with a hydrogel microsphere manipulation chip (Fig. 1a). Single-cell encapsulated hydrogel microspheres were generated using droplet microfluidics and subsequently isolated on a dedicated single-microsphere culture chip. This system enables downstream mass spectrometry analysis of metabolites, allowing in-depth investigation of metabolic accumulation at the single-cell level. The duration of each operational step for single-cell analysis is illustrated in Fig. 1b. This microfluidic hydrogel assisted mass spectrometry method was applied to analyze the tumor cell metabolism in 3D culture, revealing heterogeneity in single-cell metabolic accumulation. Expanding upon previous studies,40–42 we applied our method to various tumor cell types, including A549, HepG2 and HCT116 (form National Infrastructure of Cell Line Resource, Beijing, China), and classified the cells based on distinct metabolic profiles. Our device supports multiplexed analysis of metabolites in individual cells, offering new insights into the dynamic metabolic states of single cells. This method holds significant promise for tumor analysis and stem cell research, highlighting the importance and versatility of our approach.
Results and discussion
Single-cell hydrogel microspheres generation by droplet microfluidics
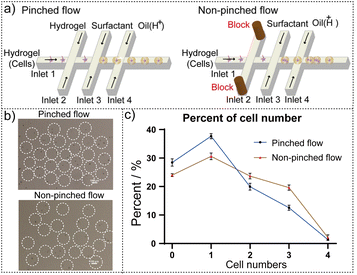
It is crucial to examine tumor cell metabolism at the single-cell level because each tumor cell may exhibit unique metabolic states. The microfluidic hydrogel microspheres can maintain cell morphology and activity for a long time, realizing 3D cell culture.31,32 In addition, the 3D porous network structure of the hydrogel microspheres allows for the free diffusion of water molecules, ions, oxygen, and small molecule nutrients, without hindering the transport of cellular metabolites into the culture media.33,34 Therefore, we use microfluidic hydrogel microspheres to encapsulate single cells and investigate the metabolism of single cells. The hydrogel microspheres containing individual cells were generated using a droplet microfluidic chip (Fig. S1a and b), which was fabricated according to our previous work.43,44 To optimize encapsulation rates and achieve uniform cell distribution, we employed a pinched flow setup in a co-flow arrangement (left structure in Fig. 2a). Here, a cell-containing hydrogel (Inlet 1, 5, 10, 20 and 40 µL h−1) was introduced, flanked by sheath flows of cell-free hydrogel solution (Inlet 2, 55, 50, 40 and 20 µL h−1), creating a laminar flow that aligned single cells into a single column. At these relatively low flow rates, the shear stress exerted on cells is negligible.45 The hydrogel in Inlet 1 and Inlet 2 was the mixture of sodium alginate and EDTA-Ca solution, acting as the dispersed phase for droplet generation. The sodium alginate, with the advantage of acid-triggered crosslinking properties, encapsulates cells into sodium alginate microcapsules that allow for free diffusion of metabolites and nutrients, simulating the extracellular matrix.46,47 The continuous phase, consisting of a fluorinated oil with surfactant (Inlet 3) and acetic acid (Inlet 4), enabled droplet detachment. The acetic acid can release calcium ions to complete alginate cross-linking. The hydrogel microspheres, whose size can be adjusted over a wide range, are suitable for cell cultivation and facilitate the manipulation of cells for metabolic analysis using mass spectrometry methods,48,49 and their size can be adjusted over a wide range. The size of the hydrogel microspheres can be adjusted by modulating the continuous-to-dispersed phase flow rate ratio. By increasing the flow rate of the continuous phase from 200 to 400 µL h−1 while maintaining a constant dispersed phase flow rate (from Fig. S1c–g). The smallest size (approximately 140 µm) was achieved at a flow rate of 350 µL h−1. Beyond this flow rate, the uniformity of hydrogel microspheres began to decline (Fig. S1h). Therefore, we set the optimal continuous phase flow rate at 350 µL h−1 to ensure consistent microsphere size and quality.The pinched flow method of droplet generating, compared with direct cutting (right structure in Fig. 2a), minimized cell number dispersion and enhanced the rate of single-cell encapsulation in the microfluidic hydrogel microspheres. As shown in Fig. 2b, there are more single-cell microspheres generated by the pinched flow method when the flow rate ratio between the inner and outer phases was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 at the same cell concentration (1 × 106). Lower cell concentrations made it easier to obtain single-cell microspheres (Fig. S2). The percentages of single-cell microspheres were calculated under a microscope at a cell concentration of 1 × 106 according to the two droplet generation methods. As shown in Fig. 2c, the pinched flow was better for single-cell encapsulation with higher single-cell encapsulation efficiency (approaching 40%). Furthermore, this structure allowed for precise control over cell encapsulation and the production of hydrogel microspheres containing different numbers of cells.
2 at the same cell concentration (1 × 106). Lower cell concentrations made it easier to obtain single-cell microspheres (Fig. S2). The percentages of single-cell microspheres were calculated under a microscope at a cell concentration of 1 × 106 according to the two droplet generation methods. As shown in Fig. 2c, the pinched flow was better for single-cell encapsulation with higher single-cell encapsulation efficiency (approaching 40%). Furthermore, this structure allowed for precise control over cell encapsulation and the production of hydrogel microspheres containing different numbers of cells.
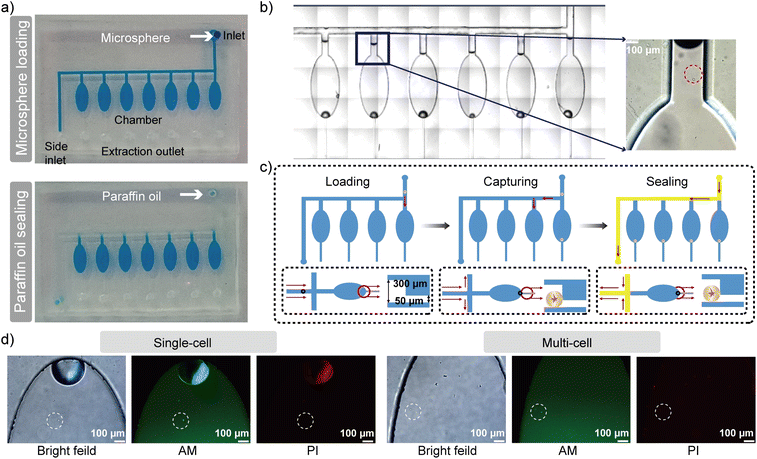
Single-cell microsphere cultivation in the culture chip
The single-microsphere culture chip was designed to isolate and culture single-cell microspheres (Fig. S3). Here, we designed 7 chambers on the chip for representative cell analysis (Fig. 3a). Furthermore, the number of chambers can be expanded on a single chip or connecting multiple chips in series to increase the throughput (Fig. S4). When the culture medium containing the single-cell microspheres was injected into the culture chip channel, microspheres (approximately 140 µm in diameter) flowed smoothly through side channels (Fig. 3b) into the chambers. Notably, due to their size, these microspheres could not pass through the 50 µm high extraction channels, leading to their retention at the end of the chambers (Fig. 3c). This retention also sealed the channel to prevent the medium containing the single-cell microspheres from flowing into the current and subsequent chambers, achieving isolation of individual microspheres. Reducing microsphere concentration, decreasing the injection flow rate, and gently shaking the culture chip can prevent multiple microspheres from entering a single chamber. After all chambers were loaded, paraffin oil was introduced into the inlet channels to partition them. The entire process can be observed under the microscope (Fig. S5).The loaded culture chip was then placed in a culture dish with PBS, creating a saturated vapor environment to prevent the evaporation of the aqueous phase inside the chambers, and incubated in a CO2 incubator. After 24 h incubation, the cell viability was evaluated after live/dead staining of cells. The staining reagents (Calcein AM/PI) were injected into the culture chip and then permeated in the chamber within 20 min (Fig. S6). The cell viability was confirmed for both single-cell and multiple cells without cell division (Fig. 3d), demonstrating robust biological compatibility of our approach. In addition, the single-cell viability rate was statistically calculated to be over 95% (Fig. S7). By combining droplet microfluidics with microscopes, the single cells were accurately encapsulated and cultured in the culture chip, suitable for further studies on single-cell metabolic activity.
Mass spectrometry detection of single-cell metabolism
The proposed single-microsphere culture chip enables the accumulation of metabolites from individual cells in a controlled 1.5 µL chamber volume over 24 h, allowing these metabolites to concentrate for mass spectrometry analysis, which is more suitable compared with oil–water droplets for cell analysis with high cell activity. After 24 h of cultivation, the culture medium from each chamber was extracted and adjusted to 10 µL for subsequent mass spectrometry testing. All tests were conducted under the same conditions, ensuring that any potential metabolite adsorption or degradation would be consistent across samples. For metabolic profiling, all metabolites of cells can be determined by our method through establishing separate mass spectrometry detection methods for each metabolite. As a proof of concept, we selected 10 key metabolites from common tumor cell metabolic pathways, including glucose from glycolysis, glutamate and glutamine from amino acid metabolism, arginine, ornithine, and citrulline from guanidino metabolism, arachidonic acid from arachidonic acid metabolism, and stearic acid and palmitic acid from fatty acid metabolism.50,51 These key metabolites were selected, based on previous reports, from representative metabolic pathways of common tumor cells to validate our single-cell analysis method.50,51 Mass spectrometry methods for each metabolite were validated with pre-experimental checks confirming the feasibility of the assays (Fig. S8, S9 and Table S1). To this end, hydrogel microsphere concentrations in the Petri dish were controlled to match those within the chip chambers (one microsphere per 1.5 µL), and culture medium samples were collected at 0 h and 24 h (Fig. S10). Also, the arachidonic acid metabolism form single A549 cells cultured on the chip was analyzed (Fig. S11). Furthermore, the glutamic acid metabolism of microspheres encapsulating one or two A549 cells cultured on the chip was analyzed (Fig. S12). The 24 h variations in metabolite levels revealed detectable heterogeneity in single-cell metabolic activity.Subsequently, metabolic accumulation analysis was conducted on individual A549 cells using our method, which revealed significant metabolic differences within the same cell line (Fig. 4). As shown in Fig. 4a, certain cells (cell 1 and cell 11) exhibited higher glutamate metabolism, while others showed increased fatty acid metabolism (cell 7 and cell 10). Additionally, some cells displayed substantial citrulline cycling (cell 15 and cell 22), while others demonstrated higher overall metabolic activity (cell 4) or generally lower activity (cell 14). These findings highlight the metabolic heterogeneity among single cells. Analysis of single-cell metabolic distribution revealed distinct patterns (Fig. 4b). Some metabolites, such as arginine, ornithine, and citrulline, showed single-peaked distributions, indicating that most cells shared similar metabolic levels. Others, like arachidonic acid and lactate, displayed bimodal distributions, indicating heterogeneity across the cell population. These results illustrate the diversity in metabolic characteristics among individual cells, highlighting the significance of single-cell analysis in decoding the intricacies of cellular biology. This metabolic diversity forms a solid scientific foundation for forthcoming research on cellular states, functions, and potential applications in personalized medicine. Notably, the system enables repeated metabolite collections. Following an initial extraction, the chambers can be refilled with culture media or therapeutic compounds for subsequent treatment, thereby facilitating analysis of the real-time metabolic effects of treatments (Fig. S13).
Metabolic accumulation differences across different cell types
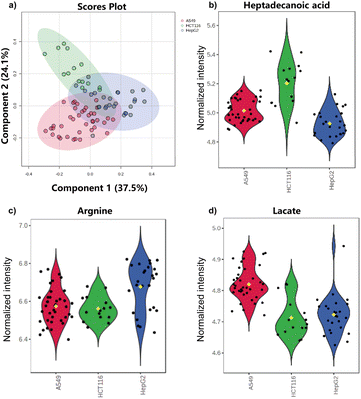
Single-cell variability in parameters like cell size, migration capacity, protein expression, and metabolic function is commonly used for cell sorting,52 including the isolating of circulating tumor cells.53 While metabolic distinctions between normal and tumor cells have been well investigated,54 research on metabolic differences among different tumor cell types often focuses on functional proteins and specific metabolites. Using our microfluidic hydrogel microspheres combined with mass spectrometry method, we analyzed single-cell metabolic accumulation across several cancer cell types to probe the cellular heterogeneity in the following experiment.The single-microsphere culture chip enabled single-cell metabolic accumulation studies across human lung cancer A549 cells, human colon cancer HCT116 cells, and human hepatocellular carcinoma HepG2 cells. Ten compounds were analyzed after logarithmic transformation and normalization, followed by Partial Least Squares-Discriminant Analysis (PLS-DA). The results showed that component I accounted for 37.5% and component II for 24.1%, suggesting distinct metabolic accumulation among the three tumor cell types (Fig. 5a). However, partial overlap of plots indicated that metabolic accumulation alone may not fully discriminate the cell types. The heatmap (Fig. S14) indicated that the metabolic accumulation alone partially discriminated the cell types. Importantly, further analysis of individual metabolites revealed distinct distributional features. For example, heptadecanoic acid accumulation in HepG2 cells displayed a unimodal profile, while A549 cells showed a bimodal distribution (Fig. 5b). Arginine accumulation showed cell type-dependent variation, with HepG2 cells exhibiting a wider distribution range (6.3–7.0) compared with the narrower range observed in HCT116 cells (6.4–6.8) (Fig. 5c). Lactate accumulation also differed in distributional shape, with HepG2 cells exhibiting a broader, multimodal distribution (Fig. 5d). Overall, while metabolic differences among various tumor cell types were observed, they were less pronounced than those between normal and tumor cells. These results emphasize that classifying tumor cells by common metabolite accumulation alone may require additional data for a more comprehensive analysis.
Conclusions
In conclusion, we developed an integrated system that combines droplet microfluidics with a cell culture chip to enable the generation, isolation, and subsequent collection of metabolic products, facilitating detailed metabolic accumulation analysis at the single-cell level. By using the proposed method, we reveal metabolic heterogeneity, providing insights into cellular metabolic processes and regulatory mechanisms. Through the analysis of multiple tumor cell types, our system not only uncovered metabolic diversity among individual tumor cells but also established a versatile experimental framework for studying metabolic distinctions across different tumor types. With the increasing more types of cellular metabolites analysis, this system is poised for advanced 3D single-cell metabolomic analysis across diverse cell types and may be extended to other single-cell omics applications, including those related to therapeutic research. Furthermore, this system can be used for probing the interfering factors of cellular heterogeneity. When combined with microscopic imaging, it establishes the correlation between heterogeneity and key factors such as cell cycle or size. In future iterations, a cell screening module can be integrated into the droplet generation chip to improve the efficiency of single-cell encapsulation. Analytical throughput can also be expanded by increasing the number of culture chambers on a single chip or linking multiple chips in series. Moreover, incorporating microfluidic structures for real-time metabolite collection and continuous flow culture will enable dynamic monitoring of single-cell metabolic accumulation, providing a robust foundation for studying cellular dynamics and developing personalized medical strategies.Author contributions
J.-M. L. obtained the funds, provided conceptualization and supervised the project. G. X. obtained the funds, designed, fabricated and characterized the microchip. Y. H. performed all cell experiments. G. X. and H. Y. wrote the original manuscript. Z. W. edited the manuscript and supervised the project. H. Y. performed the HPLC-MS/MS experiments and analyzed the data. All authors reviewed the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: materials and methods; the droplet microfluidic device and the statistics of microsphere sizes; cell numbers in hydrogel microspheres at different cell densities; mask design of the single-microsphere culture chip; schematic diagram of increasing throughput; photomicrograph of loading microsphere into the culture chip; diffusion of sodium fluorescein over time in the culture chips; viability of single cells cultured on the culture chip (n = 100); mass spectrometry condition optimization process; chromatograms of single-cell metabolites samples; pre-experimental procedure of cellular metabolites; metabolic accumulation of arachidonic acid from A549 cultured on the chip (*P < 0.05); metabolic accumulation of glutamic acid from A549 cultured on the chip (**P<0.01, *P<0.05); schematic diagram of repeating metabolite collections; heatmap of metabolic accumulation differences among HCT116 cells (n = 41), A549 cells (n = 47) and HepG2 cells (n = 35); MRM conditions of single-cell metabolite. See DOI: https://doi.org/10.1039/d5sc05261k.Acknowledgements
This work was supported by the National Key R&D Program of China (No. 2022YFC3400700), the National Natural Science Foundation of China (No. 22034005), the Young and Middle-aged Teacher Education Research Project of Fujian Province (No. JZ240051), Research Projects of Putian University (No. 2024172), and the Startup Fund for Advanced Talents of Putian University (No. 2024046).Notes and references
- S. Nong, X. Han, Y. Xiang, Y. Qian, Y. Wei, T. Zhang, K. Tian, K. Shen, J. Yang and X. Ma, MedComm, 2023, 4, e218 CrossRef CAS.
- X. Xu, Q. Peng, X. Jiang, S. Tan, Y. Yang, W. Yang, Y. Han, Y. Chen, L. Oyang and J. Lin, Exp. Mol. Med., 2023, 55, 1357–1370 CrossRef CAS PubMed.
- Y. Wang and G. J. Patti, Trends Cell Biol., 2023, 33, 1014–1024 CrossRef CAS PubMed.
- M. Parlani, C. Jorgez and P. Friedl, Trends Cell Biol., 2023, 33, 388–402 CrossRef PubMed.
- M. H.-R. Bao and C. C.-L. Wong, Cells, 2021, 10, 1715 CrossRef CAS PubMed.
- X. Chen, S. Chen and D. Yu, Metabolites, 2020, 10, 289 CrossRef CAS PubMed.
- C. A. Lyssiotis and A. C. Kimmelman, Trends Cell Biol., 2017, 27, 863–875 Search PubMed.
- E. Reznik, A. Luna, B. A. Aksoy, E. M. Liu, K. La, I. Ostrovnaya, C. J. Creighton, A. A. Hakim and C. A. Sander, Cell Syst., 2018, 6, 301–313 CrossRef CAS PubMed.
- G. Jia, J. Preussner, X. Chen, S. Guenther, X. Yuan, M. Yekelchyk, C. Kuenne, M. Looso, Y. Zhou and S. Teichmann, Nat. Commun., 2018, 9, 4877 CrossRef PubMed.
- P. Ellis, L. Moore, M. A. Sanders, T. M. Butler, S. F. Brunner, H. Lee-Six, R. Osborne, B. Farr, T. H. H. Coorens and A. R. J. Lawson, Nat. Protoc., 2021, 16, 841–871 CrossRef CAS PubMed.
- B. B. Tournier, S. Tsartsalis, K. Ceyzériat, Z. Medina, B. H. Fraser, M.-C. Grégoire, E. Kövari and P. Millet, J. Cereb. Blood Flow Metab., 2020, 40, 1242–1255 CrossRef CAS.
- J. P. Robinson, R. Ostafe, S. N. Iyengar, B. Rajwa and R. Fischer, Cells, 2023, 12, 1875 CrossRef CAS.
- X. Zeng, X. Guo, S. Jiang, X. Yang, Z. Zhong, S. Liu, Z. Zhu, L. Song and C. Yang, Anal. Chem., 2023, 95, 13313–13321 CrossRef CAS.
- L. Yang, J. George and J. Wang, Proteomics, 2020, 20, 1900226 CrossRef CAS.
- A. M. Xu, Q. Liu, K. L. Takata, S. Jeoung, Y. Su, I. Antoshechkin, S. Chen, M. Thomsonc and J. R. Heath, Lab Chip, 2018, 18, 3251–3262 RSC.
- M. Stevens, P. Harder and L. W. M. M. Terstappen, Lab Chip, 2024, 24, 460–466 RSC.
- D. Zheng, J. Zhang, W. Jiang, Y. Xu, H. Meng, C. L. Poh and C.-H. Chen, Lab Chip, 2024, 24, 137–147 RSC.
- D. Feng, H. Li, T. Xu, F. Zheng, C. Hu, X. Shi and G. Xu, Anal. Chim. Acta, 2022, 1221, 340116 CrossRef CAS.
- S. T. Gebreyesus, A. A. Siyal, R. B. Kitata, E. S.-W. Chen, B. Enkhbayar, T. Angata, K.-I. Lin, Y.-J. Chen and H.-L. Tu, Nat. Commun., 2022, 13, 37 CrossRef CAS PubMed.
- L. Zhang, T. Xu, J. Zhang, S. C. C. Wong, M. Ritchie, H. W. Hou and Y. Wang, Anal. Chem., 2021, 93, 10462–10468 Search PubMed.
- A. Kontiza, J. V. Gerichten, K. D. G. Saunders, M. Spick, A. D. Whetton, C. F. Newman and M. J. Bailey, Anal. Chem., 2024, 96, 17594–17601 CrossRef CAS PubMed.
- J. Lamanna, E. Y. Scott, H. S. Edwards, M. D. Chamberlain, M. D. M. Dryden, J. Peng, B. Mair, A. Lee, C. Chan, A. A. Sklavounos, A. Heffernan, F. Abbas, C. Lam, M. E. Olson, J. Moffat and A. R. Wheeler, Nat. Commun., 2020, 11, 5632 CrossRef CAS.
- Y. Zhu, P. D. Piehowski, R. Zhao, J. Chen, Y. Shen, R. J. Moore, A. K. Shukla, V. A. Petyuk, M. Campbell-Thompson, C. E. Mathews, R. D. Smith, W.-J. Qian and R. T. Kelly, Nat. Commun., 2018, 9, 882 CrossRef PubMed.
- H. K. Komori, S. A. LaMere, A. Torkamani, G. T. Hart, S. Kotsopoulos, J. Warner, M. L. Samuels, J. Olson, S. R. Head, P. Ordoukhanian, P. L. Lee, D. R. Link and D. R. Salomon, Genome Res., 2011, 21, 1738–1745 CrossRef CAS PubMed.
- A. Rotem, O. Ram, N. Shoresh, R. A. Sperling, A. Goren, D. A. Weitz and B. E. Bernstein, Nat. Biotechnol., 2015, 33, 1165–1172 CrossRef CAS.
- E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev and S. A. McCarroll, Cell, 2015, 161, 1202–1214 Search PubMed.
- X. Zhang, T. Li, F. Liu, Y. Chen, J. Yao, Z. Li, Y. Huang and J. Wang, Mol. Cell, 2019, 73, 130 CrossRef PubMed.
- E. P. Mimitou, A. Cheng, A. Montalbano, S. Hao, M. Stoeckius, M. Legut, T. Roush, A. Herrera, E. Papalexi, Z. Ouyang, R. Satija, N. E. Sanjana, S. B. Koralov and P. Smibert, Nat. Methods, 2019, 16, 409–412 CrossRef CAS PubMed.
- M. Stoeckius, C. Hafemeister, W. Stephenson, B. Houck-Loomis, P. K. Chattopadhyay, H. Swerdlow, R. Satija and P. Smibert, Nat. Methods, 2017, 14, 865–868 CrossRef CAS.
- D. S. Wishart, Y. D. Feunang, A. Marcu, A. C. Guo, K. Liang, R. Vázquez-Fresno, T. Sajed, D. Johnson, C. Li, N. Karu, Z. Sayeeda, E. Lo, N. Assempour, M. Berjanskii, S. Singhal, D. Arndt, Y. Liang, H. Badran, J. Grant, A. Serra-Cayuela, Y. Liu, R. Mandal, V. Neveu, A. Pon, C. Knox, M. Wilson, C. Manach and A. Scalbert, Nucleic Acids Res., 2018, 46, D608–D617 Search PubMed.
- X. Liu, S. Yan, H. Wu, M. Chen, H. Dai, Z. Wang, M. Chai, Q. Hu, D. Li, L. Chen, R. Diao, S. Chen, L. Wang and X. Shi, Adv. Funct. Mater., 2024, 34, 2315907 Search PubMed.
- Y. Peng, X. Chen, Q. Zhang, S. Liu, W. Wu, K. Li, H. Lin, X. Qing, Y. Xiao, B. Wang, D. Quan, S. Feng, Z. Rao, Y. Bai and Z. Shao, Adv. Sci., 2024, 11, 2304761 Search PubMed.
- Z. Wu, Y. Zheng, L. Lin, Y. Lin, T. Xie, W. Liao, S. Chen, Y. Zhang and J.-M. Lin, Matter, 2024, 7, 3645–3657 CrossRef CAS.
- Z. Wu, Y. Zheng, J.-M. Lin, Y. Li, Y. Lin, X. Wang and L. Lin, Chem. Eng. J., 2024, 481, 148403 CrossRef CAS.
- D. Sun, F. Cao, Y. Tian, A. Li, W. Xu, Q. Chen, W. Shi and S. Xu, Anal. Chem., 2019, 91, 15484–15490 CrossRef CAS PubMed.
- F. D. Ben, M. Turetta, G. Celetti, A. Piruska, M. Bulfoni, D. Cesselli, W. T. S. Huck and G. Scoles, Angew. Chem., 2016, 128, 8723–8726 CrossRef.
- Q. Liu, W. Ge, T. Wang, J. Lan, S. Martínez-Jarquín, C. Wolfrum, M. Stoffel and R. Zenobi, Angew. Chem., 2021, 133, 24739–24747 CrossRef.
- Q. Ruan, J. Yang, F. Zou, X. Chen, Q. Zhang, K. Zhao, X. Lin, X. Zeng, X. Yu, L. Wu, S. Lin, Z. Zhu and C. Yang, Anal. Chem., 2022, 94, 1108–1117 CrossRef CAS PubMed.
- M. Pelletier, L. K. Billingham, M. Ramaswamy and R. M. Siegel, Methods Enzymol., 2014, 542, 125–149 Search PubMed.
- C. Jin, Z. Cao, H.-L. Zhu and Z. Li, Biosens. Bioelectron., 2024, 261, 116484 CrossRef CAS PubMed.
- D. Sun, J. Lu, D. Chen, Y. Jiang, Z. Wang, W. Qin, Y. Yu, Z. Chen and Y. Zhang, Sens. Actuators, B, 2018, 268, 359–367 CrossRef CAS.
- L. Kan, D. Cui, Y. Chai, L. Ma, X. Li and M. Zhao, Int. J. Biol. Macromol., 2020, 165, 1755–1764 CrossRef CAS PubMed.
- G. Xing, Y. Shang, X. Wang, Z. Wu, Q. Zhang, J. Ai, Q. Pu and L. Lin, Chin. Chem. Lett., 2024, 35, 109491 CrossRef CAS.
- G. Xing, Y. Shang, X. Wang, H. Lin, S. Chen, Q. Pu and L. Lin, Biosens. Bioelectron., 2023, 220, 114885 CrossRef CAS.
- Y. Zhao, Q. Li and X. Hu, Anal. Methods, 2018, 10, 3489–3497 RSC.
- B. Li, L. Zhang, Y. Yin, A. Chen, B. R. Seo, J. Lou, D. J. Mooney and D. A. Weitz, Matter, 2024, 7, 3447–3468 CrossRef CAS.
- Q. Wei, J. Zhou, Y. An, M. Li, J. Zhang and S. Yang, Int. J. Biol. Macromol., 2023, 232, 123450 CrossRef CAS.
- S. Chen, Y. Li, Y. Zhang, H. Yao, T. Xu, Y. Zhang, J.-M. Lin and X.-L. Meng, J. Pharm. Anal., 2024, 14, 100967 Search PubMed.
- H. Yao, Y. Li, Y. Zheng and J.-M. Lin, Anal. Chem., 2023, 95, 10999–11006 CrossRef CAS.
- L. Wu, Y. Jin, X. Zhao, K. Tang, Y. Zhao, L. Tong, X. Yu, K. Xiong, C. Luo, J. Zhu, F. Wang, Z. Zeng and D. Pan, Cell Metab., 2023, 35, 1580–1596 CrossRef CAS PubMed.
- Z. N. Ling, Y. F. Jiang, J. N. Ru, J. H. Lu, B. Ding and J. Wu, Targets Ther., 2023, 8, 345 Search PubMed.
- M. Nian, B. Chen, M. He and B. Hu, Anal. Chem., 2024, 96, 766–774 CrossRef CAS.
- Y. Xu, B. Chen, M. He, Z. Cui and B. Hu, Anal. Chem., 2023, 95, 14061–14067 Search PubMed.
- S. Ansaryan, Y. C. Liu, X. Li, A. M. Economou, C. S. Eberhardt, C. Jandus and H. Altug, Nat. Biomed. Eng., 2023, 7, 943–958 CrossRef CAS.
Footnote |
| † These authors contribute to the manuscript equally. |
| This journal is © The Royal Society of Chemistry 2026 |