Open Access Article
Open Access ArticleEfficient synthesis, dual anti-tubercular and antioxidant activity of triazole-acetophenone derivatives: enhanced efficacy via esterification and quantum mechanical validation of CYP121 binding
Rahul A. Jagtapa,
Bapu S. Jagdalea,
Sunil L. Dhonnara,
Samin A. Shaikh b,
Rahul A. Morec,
Rajasekhar Reddy Alavala
b,
Rahul A. Morec,
Rajasekhar Reddy Alavala d,
Ghazala Muteeb
d,
Ghazala Muteeb e,
Dennys Fernandez-Conde
e,
Dennys Fernandez-Conde f,
Tushar Janardan Pawar
f,
Tushar Janardan Pawar fg,
Rohan J. Meshramh,
Jesica Escobar-Cabrera
fg,
Rohan J. Meshramh,
Jesica Escobar-Cabrera *i and
Santosh S. Chobe
*i and
Santosh S. Chobe *a
*a
aP.G. Department of Chemistry and Research Centre, M.G.V's Loknete Vyankatrao Hiray Arts, Science and Commerce College (Affiliated to Savitribai Phule Pune University), Panchavati, Nashik, Maharashtra 422009, India
bDepartment of Chemistry, Kr. V. N. Naik Shikshan Prasarak Sanstha's Arts, Commerce and Science College (Affiliated to Savitribai Phule Pune University), Canada Corner, Nashik, Maharashtra 422002, India
cDepartment of Microbiology, Dayanand Science College (Affiliated to Swami Ramanand Teerth Marathwada University), Latur, Maharashtra 413512, India
dShobhaben Pratapbhai Patel School of Pharmacy & Technology Management, SVKM's NMIMS, V.L. Mehta Road, Vile Parle (W), Mumbai-400056, India
eDepartment of Nursing, College of Applied Medical Sciences, King Faisal University, Al-Ahsa, Saudi Arabia
fUniversidad Politécnica de Querétaro, Carretera Estatal 420 S/N, El Rosario, Querétaro 76240, Mexico
gEscuela de Ingeniería Química, Universidad Anahuac Querétaro, Circuito Universidades I, Fracción 2 S/N, Zibatá, El Marqués, Querétaro 76246, Mexico
hBioinformatics Centre, Savitribai Phule Pune University, Pune, Maharashtra 411007, India
iFacultad de Química, Universidad Autónoma de Querétaro, Querétaro 76010, Mexico
First published on 15th January 2026
Abstract
A series of twelve novel triazole-based acetophenone derivatives (3a–3f and 4a–4f) was efficiently synthesized via a catalyst-free 1,3-dipolar cycloaddition reaction, integrating a known CYP121 inhibiting scaffold with dual anti-tubercular and antioxidant properties. The anti-tubercular screening demonstrated that the ethyl ester derivatives (series 4) possessed significantly superior activity compared to their carboxylic acid counterparts (series 3). Specifically, compounds 4b, 4d, and 4e exhibited the highest potential, with MIC values (2.60 ± 1.12 to 3.25 ± 1.12 µg mL−1) nearly comparable to rifampicin (1.62 ± 0.56 µg mL−1). All derivatives also displayed moderate to excellent DPPH and hydroxyl radical scavenging activity, alongside minimal cytotoxicity toward human red blood cells. Comprehensive molecular docking against the MTB CYP121 enzyme established that this enhanced efficacy was due to a shift from polar carboxylate contacts to favorable non-polar interactions, including halogen bonding and π-stacking, especially in the fluorine and trifluoromethyl-substituted esters. This differential binding pattern was further validated and characterized at the electronic level using quantum mechanics (QM) based IGMplot analysis. The findings underscore that optimal anti-tubercular efficacy requires a synergistic balance of binding affinity, substituent nature, and physicochemical properties like predicted aqueous solubility.
Introduction
The escalating crisis of infectious diseases, particularly those caused by drug-resistant pathogens, represents one of the most formidable challenges to global public health in the 21st century.1 Despite significant advances in medicine and sanitation, infectious diseases remain a leading cause of morbidity and mortality worldwide, with the rapid emergence of antimicrobial resistance (AMR) progressively undermining the efficacy of existing therapeutic regimens. This necessitates an urgent and persistent effort to discover and develop novel, effective, and affordable small-molecule chemotherapeutic agents with unique mechanisms of action, capable of combating resistance and reducing treatment burden.1,2Tuberculosis (TB), caused by the bacterium Mycobacterium tuberculosis (MTB), stands out as a pathogen of critical global concern. MTB infection accounts for millions of new cases and deaths annually, and the emergence and spread of multi-drug-resistant (MDR), extensively drug-resistant (XDR), and totally drug-resistant (TDR) strains are jeopardizing decades of progress in disease control.2a,b The current standard treatment involves a protracted, multi-drug regimen lasting six months or more, leading to poor patient adherence, significant toxicity, and the further selection of resistant mutants.2a Therefore, the scientific community is actively seeking new chemical entities that can shorten the duration of therapy, simplify treatment protocols, and possess novel mechanisms to bypass established resistance pathways.2,3
A key challenge in the host–pathogen interaction is the immune response and the resulting physiological stress. Prolonged MTB infection is known to severely compromise the host's antioxidant defense mechanisms, leading to a surge of oxidative stress due to the accumulation of reactive oxygen species (ROS) and nitrogen species (RNS) in the lung microenvironment.4 While some of these ROS are utilized by the immune system to control infection, chronic, unchecked oxidative stress can exacerbate tissue damage, promote inflammation, and hinder the host's ability to clear the infection. Consequently, the discovery of novel small-molecule inhibitors with dual anti-tubercular and antioxidant properties has emerged as a paramount interest in modern TB drug discovery.4a,5 Such dual-functional compounds could not only kill the bacterium directly but also provide therapeutic benefit to the host by buffering oxidative damage, potentially improving treatment outcomes and reducing pathology.5
Heterocyclic compounds are pivotal in drug discovery, and among them, five-membered nitrogen heterocycles, particularly 1,2,3-triazoles, have attracted immense attention. The 1,2,3-triazole ring is a core component in many biologically active molecules and marketed drugs.6 This privileged scaffold, readily synthesized via the robust copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC) or its uncatalyzed variants, exhibits several favorable pharmacological characteristics: high chemical stability, excellent metabolic stability, strong hydrogen-bonding capability, and the ability to engage in crucial π-stacking interactions with biological targets.7 Previous studies have confirmed the broad utility of 1,2,3-triazole derivatives across various therapeutic areas, including antimicrobial, anticancer, antiviral, and, highly relevant to this work, anti-tubercular and antioxidant activities.8 The versatility of the 1,2,3-triazole moiety allows it to act as a bioisostere for amide bonds and as a linker to orient pharmacophores effectively within a protein binding site, making it an ideal precursor for generating novel drug candidates (Fig. 1a).6b,8c
The growing threat of resistance demands the exploration of new, non-conventional drug targets. Recent structural and genetic exploration of MTB has identified Cytochrome P450 121 (CYP121) as a highly attractive and indispensable drug target.9 CYP121 is an essential, mono-functional enzyme required for MTB viability and pathogenicity, as it catalyzes the final step in the biosynthesis of the cyclodityrosine moieties incorporated into the bacterial cell wall. Crucially, CYP121 is unique to mycobacteria and related species, meaning an inhibitor is highly unlikely to cross-react with human P450 enzymes, dramatically reducing the potential for host toxicity and adverse drug interactions. The pocket formed around the active site, proximal to the heme prosthetic group, presents a druggable patch highly suitable for small-molecule intervention.10 Based on fragment-based screening and crystallographic validation, triazole-based compounds have been experimentally proven to bind to and inhibit the MTB CYP121 enzyme, validating this enzyme as the primary molecular target for the current study.10,11
Given the therapeutic potential of the 1,2,3-triazole scaffold and the proven druggability of the CYP121 enzyme, we sought to design, synthesize, and evaluate two related series of novel 1,4-disubstituted-1H-1,2,3-triazolyl ketones. The key structural feature explored was the functional group at the C-4 position of the triazole ring, comparing a carboxylic acid series with their corresponding ethyl ester series, alongside systematic variation of the substituent on the acetophenone phenyl ring. This structure–activity relationship (SAR) study was specifically designed to provide in-depth correlation between minute structural changes and resultant biological activity, focusing on: (1) the effect of the acidic versus neutral functional group (carboxylic acid vs. ethyl ester) on anti-tubercular efficacy and CYP121 binding; (2) the influence of different acetophenone R-groups on activity; and (3) the inherent antioxidant potential of the entire library (Fig. 1b).
In this study, we report the catalyst-free synthesis of two related series of triazole-based acetophenone derivatives and their comprehensive evaluation as anti-tubercular agents against M. tuberculosis H37Ra, including minimum inhibitory concentration (MIC) determination. We further explore the therapeutic potential through DPPH and OH radical scavenging assays and determine the preliminary cytotoxicity using a hemolytic assay. The discussion is supported by a robust in silico component, beginning with classical molecular docking to understand CYP121 binding. Crucially, this is complemented by advanced quantum mechanics (QM) based Independent Gradient Model (IGMplot) analysis to characterize the electronic and non-covalent nature of the stabilizing interactions within the binding pocket.12 Finally, a unique correlation is established between binding affinity, experimental activity, and calculated physicochemical parameters, such as predicted aqueous solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S), aiming to provide a clear and rational path for future lead optimization.13
S), aiming to provide a clear and rational path for future lead optimization.13
Results and discussion
Synthesis and characterization
The synthesis of the triazole-based acetophenone derivatives was accomplished via an efficient, two-step procedure designed to minimize the use of transition-metal catalysts, aligning with principles of Green Chemistry. The ultimate goal was to produce two related series, the carboxylic acid derivatives (3a–3f) and the ethyl ester derivatives (4a–4f), enabling a definitive structure–activity relationship (SAR) study on the C4 functional group (Scheme 1).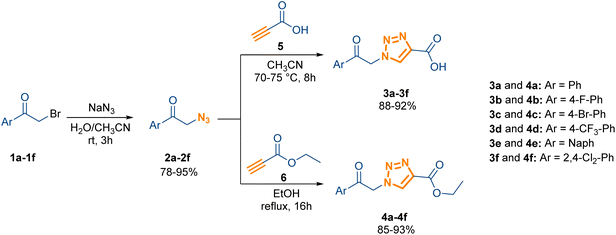 | ||
| Scheme 1 Synthetic route for the efficient, catalyst-free synthesis of 1-(2-oxo-2-arylethyl)-1H-1,2,3-triazole-4-carboxylic acid derivatives (3a–3f) and the corresponding ethyl esters (4a–4f). | ||
The first stage involved preparing the necessary α-azido ketone intermediates (2a–2f) from the readily available α-halo ketones (1a–1f) using sodium azide (NaN3). This nucleophilic substitution reaction was robust and performed under ambient conditions in an acetonitrile/water mixture, successfully accommodating various electron-donating and electron-withdrawing R-substituents (F, Br, CF3, etc.) to afford the intermediates in excellent yields (78–95%).
The pivotal second stage was the 1,3-dipolar cycloaddition to construct the 1,2,3-triazole ring. Instead of relying on traditional copper-catalyzed (CuAAC) methods, we successfully optimized a purely thermal, solvent-controlled approach that ensured strict 1,4-regioselectivity. Separate optimization was required for the two terminal functional groups:
The carboxylic acid series (3a–3f) was synthesized by reacting azides (2a–2f) with propiolic acid. Optimization favored acetonitrile as the sole solvent at 70–75 °C for 6–8 hours, yielding the products up to 92%.
The ethyl ester series (4a–4f), derived from azides (2a–2f) and ethyl propiolate, was optimized using ethanol as the solvent at a slightly higher temperature of 80 °C over 22–24 hours, resulting in similarly high yields, up to 93%. This successful catalyst-free route delivered the full library of twelve derivatives (3a–3f and 4a–4f) consistently and efficiently (Tables S1–S4, SI).
The structures of all final compounds were conclusively confirmed by a combination of spectroscopic and analytical techniques. The FT-IR spectra confirmed the presence of the characteristic acetophenone carbonyl (∼1700 cm−1) and, critically, the difference in the C4 functionality: the acid series (3a–3f) showed the broad O–H stretch (∼3300 cm−1) absent in the ester series (4a–4f).
The 1H-NMR and 13C-NMR analyses provided definitive proof of the desired 1,4-disubstitution regiochemistry and the successful attachment of the acetophenone moiety. A highly diagnostic singlet (∼6 ppm) in the 1H-NMR spectra was attributed to the methylene bridge (–CO–CH2–), confirming the N1 substitution on the triazole ring. The presence of two distinct carbonyl carbon signals (∼190 ppm for the ketone and ∼160 ppm for the acid/ester) in the 13C-NMR spectra further validated the structures.
Final confirmation of the molecular formulas was achieved via HRMS, with the observed mass-to-charge ratios (m/z) for all derivatives showing excellent correlation with the calculated exact masses.
Biological screening and cytotoxicity
| Compound | Substitution (Ar) | Zone of inhibition (mm) | Minimum inhibitory concentration (µg mL−1) | Binding free energy (kcal mol−1) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 mg | 0.1 mg | 0.01 mg | 1 mg | 0.1 mg | 0.01 mg | |||
| a MIC and zone of inhibition results are the average mean of three parallel experiments ± standard deviation (SD). Zone of inhibition criteria: ‘+’ = <5 mm, ‘++’ = >5 to <10 mm, ‘+++’ = >10 to <18 mm. TDH: co-crystallized triazole-diphenol ligand used for docking validation. | ||||||||
| 3a | Ph | +++ | +++ | +++ | 3.25 ± 1.12 | 3.25 ± 1.12 | 3.25 ± 1.12 | −6.27 |
| 3b | 4-F-Ph | +++ | +++ | +++ | 3.25 ± 1.12 | 3.25 ± 1.12 | 31.25 ± 0.00 | −6.2 |
| 3c | 4-Br-Ph | +++ | +++ | ++ | 7.81 ± 0.00 | 7.81 ± 0.00 | 7.81 ± 0.00 | −6.66 |
| 3d | 4-CF3-Ph | +++ | +++ | ++ | 3.25 ± 1.12 | 3.25 ± 1.12 | 7.81 ± 0.00 | −6.23 |
| 3e | Naph | +++ | ++ | + | 3.25 ± 1.12 | 3.25 ± 1.12 | 7.81 ± 0.00 | −6.34 |
| 3f | 2,4-Cl2-Ph | +++ | ++ | ++ | 3.25 ± 1.12 | 3.25 ± 1.12 | 7.81 ± 0.00 | −6.09 |
| 4a | Ph | ++ | ++ | ++ | 31.25 ± 0.00 | 31.25 ± 0.00 | 62.5 ± 0.00 | −5.94 |
| 4b | 4-F-Ph | +++ | +++ | +++ | 2.60 ± 1.12 | 2.60 ± 1.12 | 3.25 ± 1.12 | −7.23 |
| 4c | 4-Br-Ph | ++ | ++ | ++ | 31.25 ± 0.00 | 31.25 ± 0.00 | 62.5 ± 0.00 | −5.97 |
| 4d | 4-CF3-Ph | +++ | +++ | ++ | 2.60 ± 1.12 | 2.60 ± 1.12 | 7.81 ± 0.00 | −6.73 |
| 4e | Naph | +++ | +++ | ++ | 2.60 ± 1.12 | 2.60 ± 1.12 | 3.25 ± 1.12 | −6.58 |
| 4f | 2,4-Cl2-Ph | ++ | ++ | + | 3.25 ± 1.12 | 3.25 ± 1.12 | 7.81 ± 0.00 | −6.43 |
| Rifampicin | — | +++ | +++ | ++ | 1.62 ± 0.56 | 1.62 ± 0.56 | 3.25 ± 1.12 | — |
| TDH | — | — | — | — | — | — | — | −7.44 |
The analysis of the MIC data revealed a distinct and critical SAR revolving around the C-4 triazole functional group. The ethyl ester derivatives (series 4) demonstrated significantly superior anti-tubercular potency compared to their corresponding carboxylic acid derivatives (series 3). The most potent compounds were 4b (fluoro-substituted), 4d (trifluoromethyl-substituted), and 4e (naphthalene-substituted), all members of the ester series. These agents achieved excellent MIC values in the range of 2.60 ± 1.12 to 3.25 ± 1.12 µg mL−1. Importantly, this potency is highly competitive, nearly matching the standard first-line drug rifampicin (1.62 ± 0.56 µg mL−1).
In direct contrast, while the carboxylic acid derivatives (3a, 3b, 3d, 3e, and 3f) showed good activity, their MIC values were generally higher (3.25 ± 1.12 µg mL−1 at best). Moreover, certain ester compounds, such as 4a and 4c, showed poorer activity (MIC: 31.25 µg mL−1), highlighting that the substitution pattern on the acetophenone phenyl ring is a critical modulator of the ester group's overall efficacy. The general trend of enhanced activity upon esterification suggests that the ethyl ester moiety optimizes the compound's properties, either by improving membrane permeability (a key challenge for drug entry into MTB) or by engaging in more favorable non-polar interactions within the molecular target, CYP121, a hypothesis that is explored in detail in the subsequent in silico section.
While the H37Ra model is a standard surrogate for identifying structural leads due to its high enzymatic conservation with virulent strains, establishing the full translational potential of compounds 4b, 4d, and 4e will involve subsequent validation against M. tuberculosis H37Rv and multi-drug-resistant (MDR) clinical isolates. This preliminary screening effectively maps the SAR and identifies CYP121-targeting leads, providing a clear rationale for advanced testing in virulent and drug-resistant phenotypes.
The stability of the ethyl ester moiety under assay conditions is evidenced by the distinct MIC profiles of series 4 compared to the carboxylic acid series 3. The significantly higher potency of leads 4b and 4d (2.60 ± 1.12 µg mL−1) relative to their respective acids suggests that these molecules act in their intact ester forms. This is consistent with our computational findings, where the neutral ester promotes an optimal orientation for non-polar interactions within the CYP121 active site, a binding mode that is distinct from the polar contacts favored by the carboxylic acid derivatives.
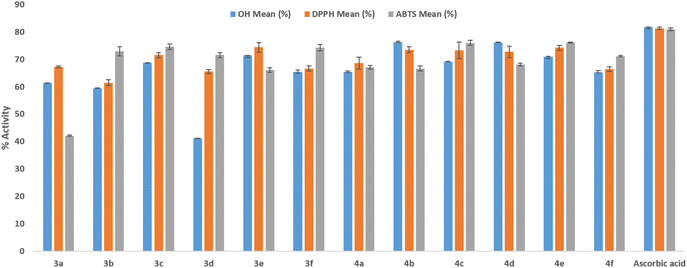
The ability to scavenge hydroxyl and DPPH radicals is particularly relevant in Tuberculosis, where the host immune response often triggers excessive oxidative stress, leading to pulmonary tissue necrosis. Specifically, OH scavenging activity ranged from 41.21% (3d) to 77.2% (4d), while DPPH scavenging ranged from 61.45% (3b) to 74.40% (3e). To further evaluate the antioxidant potential in biologically relevant aqueous conditions as suggested by recent literature, an ABTS assay was performed. The results confirm that the lead triazole-acetophenone derivatives retain potent scavenging activity in aqueous environments, with compounds 4b and 4d exhibiting significant ABTS scavenging percentages of 66.68 ± 0.93% and 68.11 ± 0.51%, respectively.
The presence of the triazole-ketone scaffold, coupled with systematically varied substitutions, provides a favorable electronic environment for scavenging free radicals. This multi-assay validation (DPPH, OH, and ABTS) reinforces the potential of these derivatives to mitigate oxidative stress across diverse physiological microenvironments. While these biochemical assays establish the intrinsic antioxidant capacity of the scaffold, future studies are intended to validate this benefit in cell-based infection models, focusing on the reduction of intracellular oxidative markers within MTB-infected macrophages.
In silico analysis: mechanism of CYP121 inhibition
The observed disparity in biological activity, particularly the superiority of the ester series, was investigated using a robust multi-scale computational approach. We focused on the essential M. tuberculosis enzyme, Cytochrome P450 121 (CYP121), known to be an excellent drug target due to its essentiality and uniqueness to mycobacteria.While CYP121 is an established essential target for triazole-based antimycobacterials, its role as the primary target in this study is supported by the strong correlation between experimental MIC values and the calculated electronic stabilization within the active site. The specificity of this interaction is distinguished from general antimicrobial effects by the low hemolytic potential observed (<1.5%), indicating that the compounds exert their activity through targeted enzymatic interference rather than non-specific membrane disruption.
The binding free energy (ΔGbind) derived from the docking analysis was correlated with the experimental MIC values (Table 1).
The docking results revealed a critical paradoxical trend: first, the carboxylic acid series (3a–3f) demonstrated moderate affinity (ΔGbind: −6.09 to −6.66 kcal mol−1). Despite the presence of a carboxylic acid group, which is capable of forming strong electrostatic interactions (salt bridges) with positively charged residues like Arg72 and Arg386 found near the CYP121 binding site, the acid series did not translate this structural potential into superior biological activity (e.g., 3c had the best affinity at −6.66 kcal mol−1 but weak MIC of 7.81 µg mL−1). This suggests that the orientation required for optimal electrostatic contact may be geometrically unfavorable. Second, the Ethyl Ester Series 4 displayed a varied, yet generally stronger, affinity profile (ΔGbind: −5.94 to −7.23 kcal mol−1). Crucially, the best affinity compounds, 4b, 4d, and 4e (ΔGbind: −7.23, −6.73, and −6.58 kcal mol−1 respectively), perfectly matched the top experimental anti-mycobacterial activity (MIC of 2.6 µg mL−1). This observation confirms that esterification is not detrimental to efficacy; rather, it is crucial for optimizing the binding mode in these derivatives. The loss of the highly polar carboxylic acid group is compensated for by the ethyl ester group, which promotes a more favorable lipophilic orientation, allowing the acetophenone R-substituents to engage in enhanced non-polar and halogen bonding interactions.
The R-substituents played a defining role in stabilizing the complexes, particularly through non-covalent halogen bonds. Compound 4d (trifluoromethyl), one of the most potent, was anchored via a network of halogen bonds involving its trifluoromethyl group and residues Gly232 and Val228. This type of interaction is crystallographically validated to be key in CYP121 inhibition. Similarly, the chlorinated phenyl ring in 4f also formed a stabilizing Halogen Bond with Val228, further demonstrating the importance of Halogen interactions in this binding pocket. Compound 4b (fluorophenyl) achieved its extremely favorable binding affinity (ΔGbind = −7.23 kcal mol−1) by utilizing a combination of conventional hydrogen bonds (Gln385) and effective π-stacking and alkyl interactions with Phe168 and Phe280. The observed SAR clearly indicates that esterification permits the compound to adopt an optimal orientation that strategically places the halogenated R-substituents into a hydrophobic sub-pocket, maximizing favorable non-polar forces over the polar contacts favored by the carboxylic acid.
Comparing the most active compound 4b and the standard ligand (TDH) to less active analogues (4a, 4c, 4d), two key structural and electronic insights were drawn concerning the interaction strength (δginter peak magnitude) and interaction positioning (peak separation). Compounds 4b and the TDH standard showed the highest δginter peak magnitude (reaching ∼0.06 on the Y-axis), signifying the strongest overall electronic stabilization. This directly correlates the superior bactericidal activity of 4b with its high electronic stability within the binding pocket. In contrast, compounds like 4a and 4f showed notably smaller peak magnitudes (∼0.015), indicating weaker non-covalent interactions. For Interaction Positioning, the lower peak separation observed in potent compounds like 4b and 4d indicates a closed, localized positioning of the interacting fragments, suggesting highly optimized, compact engagement within the active site. This QM analysis provides definitive evidence that the high experimental activity of the lead compounds is not merely a geometric fit but is supported by fundamentally strong and distinct non-covalent electronic stabilization forces that are critical for CYP121 inhibition.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S) calculated using ESOL, Ali, and Silicos-IT models (Table 2). A robust log
S) calculated using ESOL, Ali, and Silicos-IT models (Table 2). A robust log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S value, which is dependent on log
S value, which is dependent on log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (lipophilicity) and polar surface area (PSA), is crucial for MTB efficacy, as MTB possesses a unique, highly impermeable cell wall.
P (lipophilicity) and polar surface area (PSA), is crucial for MTB efficacy, as MTB possesses a unique, highly impermeable cell wall.
| Compound | ESOL log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
Ali log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
Silicos-IT log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
MIC (µg mL−1) | Binding affinity (kcal mol−1) |
|---|---|---|---|---|---|
a Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S values were calculated using three independent predictive models: ESOL log S values were calculated using three independent predictive models: ESOL log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, Ali log S, Ali log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, and Silicos-IT log S, and Silicos-IT log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S. MIC values are the average mean of three parallel experiments ± standard deviation (SD). Binding affinity (ΔGbind) is reported in kcal mol−1. S. MIC values are the average mean of three parallel experiments ± standard deviation (SD). Binding affinity (ΔGbind) is reported in kcal mol−1. |
|||||
| 4a | −2.58 | −2.92 | −3.39 | 31.25 | −5.94 |
| 4b | −2.74 | −3.03 | −3.67 | 2.6 | −7.23 |
| 4c | −3.48 | −3.64 | −4.21 | 31.25 | −5.97 |
| 4d | −3.42 | −3.85 | −4.25 | 2.6 | −6.73 |
| 4e | −3.73 | −4.22 | −5.05 | 2.6 | −6.58 |
| 4f | −3.76 | −4.23 | −4.6 | 3.25 | −6.43 |
The analysis of the ester series 4 yielded a key conclusion: optimal anti-tubercular efficacy is a synergistic function of high binding affinity and moderate solubility/lipophilicity, not an effect of a single factor. Compounds 4b (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = −2.74, ΔGbind = −7.23 kcal mol−1, MIC = 2.6 µg mL−1) and 4d (log
S = −2.74, ΔGbind = −7.23 kcal mol−1, MIC = 2.6 µg mL−1) and 4d (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = −3.42, ΔGbind = −6.73 kcal mol−1, MIC = 2.6 µg mL−1) demonstrated the perfect balance: sufficient lipophilicity (moderate log
S = −3.42, ΔGbind = −6.73 kcal mol−1, MIC = 2.6 µg mL−1) demonstrated the perfect balance: sufficient lipophilicity (moderate log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S values) for MTB cell wall penetration combined with very strong binding affinity for CYP121. In contrast, compound 4a was the most soluble (log
S values) for MTB cell wall penetration combined with very strong binding affinity for CYP121. In contrast, compound 4a was the most soluble (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = −2.58) but showed the weakest affinity (ΔGbind = −5.94 kcal mol−1), resulting in poor activity (MIC = 31.25 µg mL−1). Similarly, 4c showed reduced solubility (log
S = −2.58) but showed the weakest affinity (ΔGbind = −5.94 kcal mol−1), resulting in poor activity (MIC = 31.25 µg mL−1). Similarly, 4c showed reduced solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = −3.48) and weak affinity (ΔGbind = −5.97 kcal mol−1), leading to poor activity. These results confirm that for this scaffold, the enhanced activity of the lead compounds (4b, 4d, 4e) is a direct consequence of a precise physicochemical tuning achieved by the ethyl ester and the halogenated R-groups, which simultaneously promotes cell permeability and maximizes the electronic stabilization within the CYP121 active site.
S = −3.48) and weak affinity (ΔGbind = −5.97 kcal mol−1), leading to poor activity. These results confirm that for this scaffold, the enhanced activity of the lead compounds (4b, 4d, 4e) is a direct consequence of a precise physicochemical tuning achieved by the ethyl ester and the halogenated R-groups, which simultaneously promotes cell permeability and maximizes the electronic stabilization within the CYP121 active site.
To quantitatively validate these observations, a Pearson correlation analysis was conducted for the ester series. A strong correlation (r ≈ −0.84) was observed between the experimental pMIC and the calculated ΔGbind against CYP121, indicating that the target affinity is a primary driver of the observed activity. However, the ‘synergistic balance’ is evidenced by the fact that potency is maximized only when high affinity is paired with a specific lipophilicity range (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S −2.7 to −3.8), facilitating optimal cell wall penetration. This multivariate assessment confirms that neither binding affinity nor solubility alone can fully predict anti-tubercular efficacy for this triazole-acetophenone scaffold.
S −2.7 to −3.8), facilitating optimal cell wall penetration. This multivariate assessment confirms that neither binding affinity nor solubility alone can fully predict anti-tubercular efficacy for this triazole-acetophenone scaffold.
Experimental
General experimental procedures and materials
All chemical reagents, solvents, and starting materials were purchased commercially from Merck, BLD, Loba Chemical, and SD Fine Chemicals Reagent and were used without further purification.The progress of all reactions was monitored by thin-layer chromatography (TLC). Melting points (mp) were determined using a WRS-2A melting point apparatus and are reported uncorrected. Fourier-transform infrared (FT-IR) spectra were recorded using a KBr pellet. Nuclear Magnetic Resonance (NMR) spectra (1H at 500 MHz and 13C at 126 MHz) were recorded on a Bruker NMR spectrometer, using CDCl3 and DMSO-d6 as solvents, with chemical shifts (δ) reported in parts per million (ppm) and coupling constants (J) in Hertz (Hz). High-resolution mass spectrometry (HRMS) measurements were performed on a Thermo Fisher Micro TOF II using electrospray ionization (ESI) mode with CH3CN as the solvent.
Synthesis of triazole-based acetophenone derivatives
The synthesis was performed in two steps: a nucleophilic substitution reaction followed by a thermal 1,3-dipolar cycloaddition.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate = 1
ethyl acetate = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After completion, the MeCN was removed by vacuum distillation. The aqueous residue was extracted twice with 20 mL aliquots of ethyl acetate (EtOAc). The organic layer was combined, dried over anhydrous sodium sulfate (Na2SO4), filtered, and evaporated under reduced pressure to yield the α-azido ketone derivatives (2a–2f) as yellow to orange solids/liquids.
1). After completion, the MeCN was removed by vacuum distillation. The aqueous residue was extracted twice with 20 mL aliquots of ethyl acetate (EtOAc). The organic layer was combined, dried over anhydrous sodium sulfate (Na2SO4), filtered, and evaporated under reduced pressure to yield the α-azido ketone derivatives (2a–2f) as yellow to orange solids/liquids.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate = 1
ethyl acetate = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Upon completion, the solvent was removed by vacuum distillation. The resulting crude solid was filtered and dried under vacuum, affording the pure carboxylic acid derivatives (3a–3f).
1). Upon completion, the solvent was removed by vacuum distillation. The resulting crude solid was filtered and dried under vacuum, affording the pure carboxylic acid derivatives (3a–3f).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate = 1
ethyl acetate = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After reaction completion, the mixture was quenched with ice-cold water. The resulting solid precipitate was filtered and dried under vacuum, affording the pure ethyl ester derivatives (4a–4f).
1). After reaction completion, the mixture was quenched with ice-cold water. The resulting solid precipitate was filtered and dried under vacuum, affording the pure ethyl ester derivatives (4a–4f).Characterization data of synthesized compounds (2a–2f, 3a–3f, and 4a–4f)
All compounds were fully characterized by FT-IR, 1H-NMR, 13C-NMR, and HRMS.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 580; 1H-NMR (500 MHz, DMSO-d6): δ 13.13 (s, 1H), 8.62 (s, 1H), 8.17 (m, 2H), 7.49–7.45 (m, 2H), 6.26 (s, 2H); 13C-NMR (126 MHz, DMSO): δ 190.83, 167.12, 165.10, 162.19, 140.21, 131.88, 131.80, 131.25, 131.24, 131.08, 116.71, 116.53, 56.53; HRMS (m/z): [M + Na]+ calcd for C11H8FN3O3, 272.1949, found, 272.0445.
580; 1H-NMR (500 MHz, DMSO-d6): δ 13.13 (s, 1H), 8.62 (s, 1H), 8.17 (m, 2H), 7.49–7.45 (m, 2H), 6.26 (s, 2H); 13C-NMR (126 MHz, DMSO): δ 190.83, 167.12, 165.10, 162.19, 140.21, 131.88, 131.80, 131.25, 131.24, 131.08, 116.71, 116.53, 56.53; HRMS (m/z): [M + Na]+ calcd for C11H8FN3O3, 272.1949, found, 272.0445.Biological assays
where T is the absorbance of the test sample and C is the absorbance of the control sample.
where A0 is the control absorbance and A1 is the sample absorbance.
Computational methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) S). Molecular descriptors related to aqueous solubility (log
S). Molecular descriptors related to aqueous solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S) were calculated for the ester series (4a–4f) using the Swiss ADME web server and the Silicos-IT platform. The predictive models utilized for comparative analysis were ESOL log
S) were calculated for the ester series (4a–4f) using the Swiss ADME web server and the Silicos-IT platform. The predictive models utilized for comparative analysis were ESOL log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, Ali log
S, Ali log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, and Silicos-IT log
S, and Silicos-IT log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S. This data was used to establish the critical correlation between calculated solubility, calculated binding affinity, and experimental anti-tubercular activity (MIC).13
S. This data was used to establish the critical correlation between calculated solubility, calculated binding affinity, and experimental anti-tubercular activity (MIC).13Conclusions
In the present study, we successfully report the efficient, catalyst-free synthesis of two novel, structurally related series of 1,4-disubstituted-1H-1,2,3-triazolyl ketones: the carboxylic acid derivatives (3a–3f) and the ethyl ester derivatives (4a–4f). This work provides a comprehensive structure–activity relationship (SAR) study that clearly demonstrates the critical role of the C4 triazole functional group and the acetophenone R-substituents in determining anti-tubercular efficacy. Data from the biological screening against M. tuberculosis H37Ra revealed that the ester derivatives, particularly 4b, 4d, and 4e, were superior inhibitors, achieving MIC values highly comparable to Rifampicin. Furthermore, the entire library displayed favorable dual anti-tubercular and antioxidant potential, exhibiting moderate to excellent DPPH and hydroxyl radical scavenging activities, combined with minimal cytotoxicity toward human red blood cells. The observed structural paradox, where the neutral ethyl ester was more effective than the potentially polar carboxylic acid was fully rationalized through our advanced computational investigation. Molecular docking and interaction studies against CYP121 confirmed that the enhanced binding affinity of the lead compounds (4b with ΔGbind = −7.23 kcal mol−1) was due to a strategic shift in orientation that maximized non-polar forces, notably halogen bonding and π-stacking interactions. This superior electronic stabilization was rigorously validated by a subsequent Quantum Mechanics based IGMplot analysis. Ultimately, the correlation of binding affinity, predicted aqueous solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S), and experimental MIC data underscores that optimal efficacy is achieved through a precise synergistic balance of these three physicochemical and biological factors. Our lead compounds represent promising, dual-action drug candidates that merit further preclinical development for the treatment of drug-resistant tuberculosis.
S), and experimental MIC data underscores that optimal efficacy is achieved through a precise synergistic balance of these three physicochemical and biological factors. Our lead compounds represent promising, dual-action drug candidates that merit further preclinical development for the treatment of drug-resistant tuberculosis.
Author contributions
Conceptualization: S. S. C., R. J. M.; methodology: S. S. C., R. J. M., B. S. J., S. L. D., R. R. A., J. E.-C.; investigation: R. A. J., B. S. J., S. L. D., D. F.-C., S. A. S., R. A. M., G. M., M. S. A., T. J. P.; data curation: R. A. J., S. A. S., R. A. M.; formal analysis: R. J. M., R. A. J., S. A. S., R. R. A., M. S. A.; software: R. J. M., R. R. A., M. S. A., T. J. P.; visualization: R. A. J., M. S. A., T. J. P.; validation: S. S. C., R. A. J., B. S. J., S. L. D., S. A. S., R. A. M.; resources: S. S. C., R. J. M., R. R. A., J. E.-C.; supervision: S. S. C., J. E.-C.; project administration: S. S. C., J. E.-C.; funding acquisition: D. F.-C., G. M., J. E.-C.; writing – original draft: R. A. J., T. J. P.; writing – review & editing: S. S. C., R. J. M., R. A. M., M. S. A., T. J. P.Conflicts of interest
There are no conflicts to declare.Data availability
All relevant data supporting the findings of this article are contained within the manuscript and the supplementary information (SI). Supplementary information : detailed synthetic optimization tables, comprehensive spectroscopic characterization (FT-IR, NMR, HRMS), and complete datasets for the antioxidant and hemolytic assays. See DOI: https://doi.org/10.1039/d5ra09368f.Acknowledgements
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia [Grant No. KFU260125].Notes and references
- (a) S. S. Chobe, R. D. Kamble, S. D. Patil, A. P. Acharya, S. V. Hese, O. S. Yemul and B. S. Dawane, Green approach towards synthesis of substituted pyrazole-1,4-dihydro,9-oxa,1,2,6,8-tetrazacyclopentano[b]naphthalene-5-one derivatives as antimycobacterial agents, Med. Chem. Res., 2013, 22, 5197–5203, DOI:10.1007/s00044-013-0487-6; (b) M. A. Salam, M. Y. Al-Amin, M. T. Salam, J. S. Pawar, N. Akhter, A. A. Rabaan and M. A. A. Alqumber, Antimicrobial Resistance: A Growing Serious Threat for Global Public Health, Healthcare (Basel), 2023, 11, 1946, DOI:10.3390/healthcare11131946.
- (a) E. Kabir, M. Alam and M. I. Haque, Novel heterocycles as antitubercular drugs: A review, Diagn Microbiol Infect Dis, 2025, 113, 116978, DOI:10.1016/j.diagmicrobio.2025.116978; (b) D. B. Abdjul, F. Budiyanto, J. T. Wibowo, T. Murniasih, S. I. Rahmawati, D. W. Indriani, M. Y. Putra and A. Bayu, Unlocking potent anti-tuberculosis natural products through structure-activity relationship analysis, Nat Prod Bioprospect, 2025, 15, 44, DOI:10.1007/s13659-025-00529-4; (c) S. K. Bhagwat, S. S. Chobe, R. R. Alavala, A. Vora, R. A. More, V. D. Bobade, A. A. Patil, T. J. Pawar, F. Hernández-Rosas and S. V. Patil, Benzothiazole–thiazole hybrids as broad-spectrum antimicrobial agents: synthesis, SAR analysis, and molecular docking against bacterial and fungal targets, RSC Adv., 2025, 15, 31752–31762, 10.1039/d5ra04254b.
- K. Kisimba, K. Kasumbwe, F. Odun-Ayo and M. Faya, Flavonoids: Potential Novel Inhibitors of Mycobacterium tuberculosis, Infect Disord Drug Targets, 2025, 25, e18715265361578, DOI:10.2174/0118715265361578250504110100.
- (a) M. D. Shastri, S. D. Shukla, W. C. Chong, K. Dua, G. M. Peterson, R. P. Patel, P. M. Hansbro, R. Eri and R. F. O'Toole, Role of Oxidative Stress in the Pathology and Management of Human Tuberculosis, Oxid Med Cell Longev, 2018, 2018, 7695364, DOI:10.1155/2018/7695364; (b) F. Holguin, Oxidative stress in airway diseases, Ann Am Thorac Soc, 2013, 10, S150–S153, DOI:10.1513/annalsats.201305-116aw.
- S. K. Bhagwat, T. J. Pawar, S. A. Kulkarni, A. A. Patil, R. A. More, J. O. C. Jimenez-Halla, J. A. Alvarado-Salazar, J. L. Olivares-Romero, G. Muteeb, E. Delgado-Alvarado and S. V. Patil, Synthesis, characterization, biological activities, and computational studies of pyrazolyl-thiazole derivatives of thiophene, RSC Adv., 2024, 14, 39004–39016, 10.1039/d4ra06228k.
- (a) M. J. Vaishnani, S. Bijani, M. Rahamathulla, L. Baldaniya, V. Jain, K. Y. Thajudeen, M. M. Ahmed, S. A. Farhana and I. Pasha, Biological Importance and Synthesis of 1,2,3-Triazole Derivatives: A Review, Green Chem. Lett. Rev., 2024, 17, 2307989, DOI:10.1080/17518253.2024.2307989; (b) A. K. Kabi, S. Sravani, R. Gujjarappa, A. Garg, N. Vodnala, U. Tyagi, D. Kaldhi, V. Singh, S. Gupta and C. C. Malakar, An Overview on Biological Activities of 1,2,3-Triazole Derivatives, in Nanostructured Biomaterials: Basic Structures and Applications, 2022, pp 401–423, DOI:10.1007/978-981-16-8399-2_11; (c) M. S. Yadav, V. K. Pandey, M. K. Jaiswal, S. K. Singh, A. Sharma, M. Singh and V. K. Tiwari, Late-Stage Functionalization Strategies of 1,2,3-Triazoles: A Post-Click Approach in Organic Synthesis, J. Org. Chem., 2025, 90, 5731–5762, DOI:10.1021/acs.joc.5c00125.
- D. P. Vala, R. M. Vala and H. M. Patel, Versatile Synthetic Platform for 1,2,3-Triazole Chemistry, ACS Omega, 2022, 7, 36945–36987, DOI:10.1021/acsomega.2c04883.
- (a) Y. Nural, S. Ozdemir, M. S. Yalcin, B. Demir, H. Atabey, Z. Seferoglu and A. Ece, New Bis- and Tetrakis-1,2,3-Triazole Derivatives: Synthesis, DNA Cleavage, Molecular Docking, Antimicrobial, Antioxidant Activity and Acid Dissociation Constants, Bioorg. Med. Chem. Lett., 2022, 55, 128453, DOI:10.1016/j.bmcl.2021.128453; (b) Y. Belay, A. Muller, F. S. Mokoena, A. S. Adeyinka, L. R. Motadi and A. K. Oyebamiji, 1,2,3-Triazole and Chiral Schiff Base Hybrids as Potential Anticancer Agents: DFT, Molecular Docking and ADME Studies, Sci. Rep., 2024, 14, 6951, DOI:10.1038/s41598-024-57689-5; (c) S. A. Shaikh, S. R. Labhade, S. S. Chobe, M. V. Gaikwad, R. A. More, B. U. Patil and D. R. Boraste, Coumarin-Triazole-Thiazole Hybrids: A New Avenue in Antitubercular Agents, Bioorg. Chem., 2025, 161, 108567, DOI:10.1016/j.bioorg.2025.108567; (d) N. U. Hebbar, A. R. Patil, P. Gudimani, S. L. Shastri, L. A. Shastri, S. D. Joshi, S. K. Vootla, S. Khanapure and A. K. Shettar, Click Approach for Synthesis of 3,4-Dihydro-2(1H)-Quinolinone, Coumarin Moored 1,2,3-Triazoles as Inhibitor of Mycobacterium tuberculosis H37Rv, Their Antioxidant, Cytotoxicity and In Silico Studies, J. Mol. Struct., 2022, 1269, 133795, DOI:10.1016/j.molstruc.2022.133795.
- (a) C. Brengel, A. Thomann, A. Schifrin, G. Allegretta, A. A. M. Kamal, J. Haupenthal, I. Schnorr, S. H. Cho, S. G. Franzblau, M. Empting, J. Eberhard and R. W. Hartmann, Biophysical Screening of a Focused Library for the Discovery of CYP121 Inhibitors as Novel Antimycobacterials, ChemMedChem, 2017, 12, 1519–1528, DOI:10.1002/cmdc.201700363; (b) T. Padayachee, D. C. Lamb, D. R. Nelson and K. Syed, Structure–Function Analysis of the Essential Mycobacterium tuberculosis P450 Drug Target, CYP121A1, Int. J. Mol. Sci., 2024, 25, 4886, DOI:10.3390/ijms25094886.
- M. E. Kavanagh, A. G. Coyne, K. J. McLean, G. G. James, C. W. Levy, L. B. Marino, L. P. S. De Carvalho, D. S. H. Chan, S. A. Hudson, S. Surade, D. Leys, A. W. Munro and C. Abell, Fragment-Based Approaches to the Development of Mycobacterium tuberculosis CYP121 Inhibitors, J. Med. Chem., 2016, 59, 3272–3300, DOI:10.1021/acs.jmedchem.6b00007.
- S. A. Hudson, K. J. McLean, S. Surade, Y. Q. Yang, D. Leys, A. Ciulli, A. W. Munro and C. Abell, Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121, Angew. Chem., Int. Ed., 2012, 51, 9311–9315, DOI:10.1002/anie.201202544.
- (a) C. Lefebvre, G. Rubez, H. Khartabil, J. C. Boisson, J. Contreras-García and E. Hénon, Accurately Extracting the Signature of Intermolecular Interactions Present in the NCI Plot of the Reduced Density Gradient: Versus Electron Density, Phys. Chem. Chem. Phys., 2017, 19, 17928–17936, 10.1039/c7cp02110k; (b) C. Lefebvre, H. Khartabil, J. C. Boisson, J. Contreras-García, J. P. Piquemal and E. Hénon, The Independent Gradient Model: A New Approach for Probing Strong and Weak Interactions in Molecules from Wave Function Calculations, ChemPhysChem, 2018, 19, 724–735, DOI:10.1002/cphc.201701325.
- (a) J. S. Delaney, ESOL: Estimating Aqueous Solubility Directly from Molecular Structure, J. Chem. Inf. Comput. Sci., 2004, 44, 1000–1005, DOI:10.1021/ci034243x; (b) F. Ahmed and H. Xiong, Recent Developments in 1,2,3-Triazole-Based Chemosensors, Dyes Pigm., 2021, 185, 108905, DOI:10.1016/j.dyepig.2020.108905; (c) J. Ali, P. Camilleri, M. B. Brown, A. J. Hutt and S. B. Kirton, Revisiting the General Solubility Equation: In Silico Prediction of Aqueous Solubility Incorporating the Effect of Topographical Polar Surface Area, J. Chem. Inf. Model., 2012, 52, 420–428, DOI:10.1021/ci200387c.
- (a) G. G. Mandawad, R. D. Kamble, S. V. Hese, R. A. More, R. N. Gacche, K. M. Kodam and B. S. Dawane, An Efficient Synthesis of Isoxazoline Libraries of Thiophene Analogs and Its Antimycobacterial Investigation, Med. Chem. Res., 2014, 23, 4455, DOI:10.1007/s00044-014-1016-y; (b) R. N. Jones, C. H. Ballow and D. J. Biedenbach, Multi-Laboratory Assessment of the Linezolid Spectrum of Activity Using the Kirby–Bauer Disk Diffusion Method: Report of the Zyvox® Antimicrobial Potency Study (ZAPS) in the United States, Diagn. Microbiol. Infect. Dis., 2001, 40, 59–65, DOI:10.1016/S0732-8893(01)00235-8.
- R. A. Khalifa, M. S. Nasser, A. A. Gomaa, N. M. Osman and H. M. Salem, Resazurin Microtiter Assay Plate Method for Detection of Susceptibility of Multidrug-Resistant Mycobacterium tuberculosis to Second-Line Anti-Tuberculous Drugs, Egypt. J. Chest Dis. Tuberc., 2013, 62, 281–287, DOI:10.1016/j.ejcdt.2013.05.008.
- E. Gerasimova, E. Gazizullina, S. Kolbaczkaya and A. Ivanova, The Novel Potentiometric Approach to Antioxidant Capacity Assay Based on the Reaction with Stable Radical 2,2′-Diphenyl-1-Picrylhydrazyl, Antioxidants, 2022, 11, 1974, DOI:10.3390/antiox11101974.
- S. B. Rajput, R. B. Shinde, M. M. Routh and S. M. Karuppayil, Anti-Candida Properties of Asaronaldehyde of Acorus gramineus Rhizome and Three Structural Isomers, Chin. Med., 2013, 8, 18, DOI:10.1186/1749-8546-8-18.
- S. B. Rajput, R. B. Shinde, M. M. Routh and S. M. Karuppayil, Anti-Candida Properties of Asaronaldehyde of Acorus gramineus Rhizome and Three Structural Isomers, Chin. Med., 2013, 8, 18, DOI:10.1186/1749-8546-8-18.
- (a) R. J. Meshram, V. B. Baladhye, R. N. Gacche, B. K. Karale and R. B. Gaikar, Pharmacophore Mapping Approach for Drug Target Identification: A Chemical Synthesis and In Silico Study on Novel Thiadiazole Compounds, J. Clin. Diagn. Res., 2017, 11, KF01–KF08, DOI:10.7860/JCDR/2017/22761.9925; (b) K. K. Patil, R. J. Meshram, S. H. Barage and R. N. Gacche, Dietary Flavonoids Inhibit the Glycation of Lens Proteins: Implications in the Management of Diabetic Cataract, 3 Biotech, 2019, 9, 61, DOI:10.1007/s13205-019-1581-3.
- (a) S. Dallakyan and A. J. Olson, Small-Molecule Library Screening by Docking with PyRx, Methods Mol. Biol., 2015, 1263, 243–250, DOI:10.1007/978-1-4939-2269-7_19; (b) G. M. Morris, H. Ruth, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility, J. Comput. Chem., 2009, 30, 2785–2791, DOI:10.1002/jcc.21256.
- P. P. Mogle, R. J. Meshram, S. V. Hese, R. D. Kamble, S. S. Kamble, R. N. Gacche and B. S. Dawane, Synthesis and Molecular Docking Studies of a New Series of Bipyrazol-Yl-Thiazol-Ylidene-Hydrazinecarbothioamide Derivatives as Potential Antitubercular Agents, MedChemComm, 2016, 7, 1415–1421, 10.1039/c6md00085a.
- (a) A. Daina, O. Michielin and V. Zoete, SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules, Sci. Rep., 2017, 7, 42717, DOI:10.1038/srep42717; (b) S. K. Bhagwat, F. Hernández-Rosas, A. Vidal-Limon, J. O. C. Jimenez-Halla, B. K. Ghotekar, V. D. Bobade, E. Delgado-Alvarado, S. V. Patil and T. J. Pawar, Iodinated Salicylhydrazone Derivatives as Potent α-Glucosidase Inhibitors: Synthesis, Enzymatic Activity, Molecular Modeling, and ADMET Profiling, Chemistry, 2025, 7, 117, DOI:10.3390/chemistry7040117.
| This journal is © The Royal Society of Chemistry 2026 |