Open Access Article
Open Access ArticleMedicinal chemistry meets nanotechnology: machine learning assisted colorimetric sensing platform for oxalic acid based on drug mediated copper oxide nanoparticles
Muhammad Ishtiaq Jan *a,
Abda Khatoona,
Wei Sunb,
Naeem Khana,
Sajid Alic,
Ibrahim A. Shaaband,
Mansoor Khana,
Amir Badshah
*a,
Abda Khatoona,
Wei Sunb,
Naeem Khana,
Sajid Alic,
Ibrahim A. Shaaband,
Mansoor Khana,
Amir Badshah a,
Imran Rabbanie and
Umar Nishan
a,
Imran Rabbanie and
Umar Nishan *a
*a
aDepartment of Chemistry, Kohat University of Science and Technology, Kohat 26000, Khyber Pakhtunkhwa, Pakistan. E-mail: umarnishan85@gmail.com; mishtiaqjan@kust.edu.pk
bHainan International Joint Research Center of Marine Advanced Photoelectric Functional Materials, College of Chemistry and Chemical Engineering, Hainan Normal University, Haikou 571158, P. R. China
cDepartment of Chemistry, Bacha Khan University, Charsadda 24420, Khyber Pakhtunkhwa, Pakistan
dCentral Labs, King Khalid University, P.O. Box 960, AlQura’a, Abha, 61413, Saudi Arabia
eDepartment of Pharmacy, Kohat University of Science and Technology, Kohat 26000, Khyber Pakhtunkhwa, Pakistan
First published on 15th January 2026
Abstract
Increased levels of oxalic acid are associated with an increased risk of kidney stone formation, which can lead to renal failure. In addition, its high concentration in the blood can lead to cardiovascular diseases. Therefore, it is vital to detect and quantify oxalic acid economically and rapidly. Copper oxide nanoparticles (CuONPs) are gaining importance as colorimetric nanosensors due to their intrinsic color change, cost-effectiveness, and easy synthesis. Paracetamol-mediated CuO NPs were synthesized through a new approach and characterized through various spectroscopic and morphological techniques. UV-visible spectroscopy confirmed the synthesis of CuO NPs through surface plasmon resonance at 225 nm. The peak at 850 cm−1 corresponds to the stretching vibration of CuO NPs. The XRD and SEM characterization techniques confirmed the particle size of 27.51 nm with a spherical morphology. A machine learning-assisted strategy was developed with four prediction models: Random Forest, Linear Regression, XGBoost, and Decision Tree Regression. The intrinsic colorimetric features of CuO NPs were observed through the naked eye and quantified through spectroscopy with the addition of oxalate. The developed platform selectively detected oxalate levels in concentrations ranging from 1 to 120 µM, with a limit of detection (LOD) of 0.23 µM and a limit of quantification (LOQ) of 0.78 µM. The developed biosensor successfully quantified oxalate, crucial for diagnosing hyperoxaluria and preventing calcium oxalate stone formation in the kidneys. The machine learning complementary tools further bolster the accuracy of colorimetric concentration prediction.
1. Introduction
The most significant sources of oxalic acid are plants, and it is critical to various biological systems.1 However, due to its acidic nature, the potential of oxalic acid to form metallic complexes is very high, and this condition provides substantial support for the formation of kidney stones.2 Generally, small quantities of oxalic acid are produced within the body through the metabolism of some amino acids, vitamins, and minerals, which are usually excreted through the urine at a rate of 10 to 50 mg/day3. Hyperoxaluria affects many more serious conditions other than the mere development of kidney stones; it leads to the dysregulation of minerals and also cardiovascular complications, even affecting neurological disorders.4 The process initiated by hyperoxaluria promotes renal cell apoptosis directly during the formation of stones.5 Hence, hyperoxaluria is a grave health risk around the world and requires effective treatment methods after proper diagnosis.6 The medicinal industry greatly relies on oxalic acid as an excipient during the formulation of drugs. The food industry also applies oxalic acid as a preservative and in the purification of food material.7 However, due to its ability to scavenge free radicals, oxalic acid can be effectively incorporated into formulations for skin care lesions, thereby preventing oxidative damage to cells.8The sensing of oxalic acid has previously been reported through electrochemical,9 spectroscopic chromatographic10,11 and enzymatic methods.12 These mentioned techniques, despite their merits, suffer from factors like high cost, difficult operation and maintenance, and sample pretreatment. On the other hand, the colorimetric sensing approach can offer a more viable alternative in terms of much lower cost, easy operation, and portability and does not require personnel with special skills.13,14 The rise of nanotechnology has resulted in a renewed interest in the field of colorimetric nanosensors.
Preceding nanomaterials, various sensing platforms were reported based on natural enzymes such as horseradish peroxidase and glucose oxidase, etc. However, natural enzymes suffer from various shortcomings, such as lower stability, difficult handling, high cost, and low shelf life.15 Various nanomaterials, such as manganese dioxide (MnO2) nanosheets,16 silver (Ag) NPs have been explored for the determination of oxalic acid.17 The strong surface plasmon resonance, distance-dependent optical properties, and the ability to generate visually detectable signals make nanomaterials excellent candidates for colorimetric sensing applications. Specifically, the copper ion complex acts as a sensor, which indicates the presence of oxalate through a distinct color change upon selective binding.18 This innovative system uses a pyrrole-containing copper complex and chromeazurol S to visually detect oxalate, changing color from blue to yellow upon addition of oxalate. However, the use of chromeazurol S dye is associated with skin and eye irritation.19 The literature study revealed that oxalic acid was quantified with an electrochemical Ag-NPs and nitrogen-doped graphene oxide (N-GO) nanocomposite. The exceptional performance of the sensor, including its current response, excellent selectivity, and high stability, is attributed to the beneficial synergistic interaction between the Ag-NPs and N-GO.9 An electrochemical sensor for oxalic acid detection was developed using palladium nanoparticle-loaded carbon nanofiber (Pd/CNF) composites. The well-dispersed Pd nanoparticles on the carbon nanofibers gave the sensor high electrocatalytic performance and rapid voltametric responses with a complex procedure.20 However, high-cost metal nanoparticle-based systems that use expensive elements like Au, Pt, or Pd are not desirable because they elevate the cost of testing. In addition, different nanoparticles have been developed for the colorimetric sensing of oxalic acid due to several advantages, including low cost, rapidity, and being easy availability.17,21–24
The safety record of paracetamol in clinical settings makes it a promising candidate for biomedical applications of nanoparticle synthesis.25 Its rich electron density can help in the reduction of the metal salt in the synthesis of the nanoparticles. The paracetamol drug stabilizes nanoparticles, mitigating nanoparticle agglomeration and enhancing colloidal stability in diverse media, including biological fluids. Additionally, its antioxidant activity synergizes with the intrinsic properties of nanoparticles such as CuO NPs, potentially providing a dual therapeutic advantage in addressing oxidative stress-related pathologies.
The novelty of this work lies in the synthesis and use of paracetamol-mediated CuO NPs for the colorimetric sensing of oxalic acid. Comprehensive characterization of the synthesized nanoparticles and their functionalization was performed. The synthesized platform was successfully used for the detection and quantification of oxalic acid with excellent precision, sensitivity, and selectivity. It was also applied for the detection and quantification of oxalic acid in urine samples. The method further advances innovation by coupling these tailored nanoparticles with machine-learning algorithms to optimize colorimetric response patterns, enabling precise quantification, improved limit of detection, robust prediction accuracy, and accurate prediction of oxalic acid concentrations with improved sensitivity and precision. The data-driven computational layer transforms the sensor from a qualitative color-change system into a robust quantitative analytical tool.
2. Materials and methods
2.1 Reagents
CuO NPs were synthesized using paracetamol, copper(II) sulfate pentahydrate (CuSO4·5H2O), dimethyl sulfoxide (DMSO), acetic acid, and sodium hydroxide (NaOH). The oxalic acid was used as an analyte. The reagents were of analytical grade and used as received from Sigma-Aldrich, USA. All the solutions were prepared using deionized water.2.2 Synthesis of CuO NPs
CuO NPs were synthesized using 500 mg of paracetamol dissolved in DMSO. The resulting solution was magnetically stirred for 10 minutes at room temperature and filtered. The filtrate was then added dropwise to a 50 mM copper(II) sulfate solution and magnetically stirred for 4 hours. The resulting solution was centrifuged at 1100 rpm for 20 minutes to isolate the CuO NPs as a pellet. The obtained CuO NPs were subsequently dried at 50 °C and stored at room temperature for further analysis.262.3 Instrumentation
The characteristic peak for the synthesis of CuO NPs was investigated by UV-vis spectroscopy. The functional groups of synthesized CuO NPs were identified by Fourier transform infrared spectroscopy (FTIR) using an MX-300 spectrometer, with characteristic stretching vibrations at characteristic peaks.27 The scanning electron microscopy (SEM) (JSM-5910) was used for studying the surface morphology of synthesized CuO NPs. High-resolution images of the nanoparticles were taken to examine the shape and morphology.28 The X-ray diffraction (XRD) (Bruker Smart Apex) was used to investigate the crystalline nature of the developed CuO NPs. The particle size of NPs was calculated using the Debye–Scherrer equation.29 The X-ray photoelectron spectroscopy (XPS) (EscaLab 250Xi Thermo, USA, with Al Kα X-ray radiation) analysis was used for the oxidation state and elemental composition of CuO NPs by measuring the binding energy of the released electrons.302.4 Functionalization of CuO NPs with acetic acid
The stability and prevention of agglomeration of CuO NPs were performed through functionalization with acetic acid. For this purpose, 6 mg of the NPs were taken in 1 mL of acetic acid for 30 minutes with continuous stirring till a homogeneous solution was obtained. The bluish dispersion of functionalized CuO NPs was stored for further study.312.5 Colorimetric sensing of oxalic acid
The colorimetric sensing of oxalate was investigated at ambient temperature and pH 7. The assay was performed with 100 µL of 10 µM oxalate solution mixed with 250 µL of CuO NPs in an Eppendorf tube. The blue color of the capped CuO NPs was rapidly transitioned to a colorless solution with the addition of oxalate. The assay response was readily observed by the naked eye, with a visual change, and subsequently confirmed using a UV-vis spectrophotometer. The real urine samples were collected from Kohat Development Hospital (KDA), Kohat, Khyber Pakhtunkhwa, Pakistan. The research work was approved by the ethical committee of Kohat University of Science and Technology, Kohat, Pakistan, under Reference No: Ref-No/KUST/Ethical Committee/528.2.6 Machine learning analysis of RGB-concentration relationship
Colorimetric images of oxalic acid solutions were resized uniformly, and their red (R), blue (B), and green (G) values were determined by using the Python Imaging Library.32 RGB values were then correlated with the known concentrations of oxalic acid solutions. To perform machine learning, four prediction models were selected: Random Forest, Linear Regression, XGBoost, and Decision Tree Regression.33 These models were trained on a dataset that consisted of 01 blank and 16 samples of different concentrations (from 1 to 120 µM) of oxalic acid solutions along with their respective RGB values. The train-test technique was used to evaluate the performance of models. For this purpose, the data was divided into 80% training and 20% testing subsets. Performance of the models was evaluated using evaluation metrics such as Mean Absolute Error (MAE), Root Mean Square Error (RMSE), and the coefficient of determination (R2).34 The machine learning approach indicates its potential to combine with colorimetric analysis as a quick and portable supporting tool for concentration estimation of oxalic acid.3. Results and discussion
3.1 Potential of the calorimetric method for sensing oxalic acid
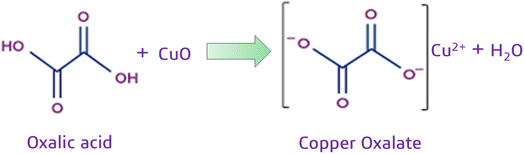
The reaction of capped CuO NPs with acetic acid involves the coordination of oxalic acid with the Cu2+ ions on the surface of the CuO NPs, effectively converting the CuO into copper oxalate, CuC2O4. This leads to a change in the optical properties of the nanoparticle, which is demonstrated by a color change in the NP solution and hence forms a basis for a colorimetric assay. This is a reliable method for quantifying oxalic acid concentration in various samples, as illustrated in Scheme 1.3.2 Characterization of CuO NPs
The characterizations of the synthesized CuO NPs were performed for optical, structural, size, and morphological properties. Moreover, the identification of surface functional groups is a necessity for a comprehensive understanding of their nature.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, a characteristic feature of carbonyl groups and, precisely, carboxyl groups belonging to the drug. The peak at 1120 cm−1 corresponds to C–O stretching. It suggests the presence of esters, ethers, or alcohols. The peak at 980 cm−1 indicates C–O–C stretching. It shows the ether linkages within the drug structure. The peak at 884 cm-1 corresponds to the C–C stretching, which indicates the organic backbone of the drug. Lastly, the peak at 850 cm−1 has confirmed the presence of Cu–O stretching vibration and hence validates the synthesis of the CuO NPs27,38 as shown in Fig. 2.
O stretching, a characteristic feature of carbonyl groups and, precisely, carboxyl groups belonging to the drug. The peak at 1120 cm−1 corresponds to C–O stretching. It suggests the presence of esters, ethers, or alcohols. The peak at 980 cm−1 indicates C–O–C stretching. It shows the ether linkages within the drug structure. The peak at 884 cm-1 corresponds to the C–C stretching, which indicates the organic backbone of the drug. Lastly, the peak at 850 cm−1 has confirmed the presence of Cu–O stretching vibration and hence validates the synthesis of the CuO NPs27,38 as shown in Fig. 2.
3.3 Colorimetric sensing of the proposed reaction
The addition of 100 µL oxalic acid with a concentration of 10 µM was added to 250 µL acetic acid-capped CuO NPs with 50 mM concentration, resulting in a distinct colorimetric transition from blue to light blue, confirming the reaction. The response between capped CuO NPs and oxalic acid was further optimized with respect to various parameters to obtain a transparent solution. The visual change was observed with a decrease in the peak intensity at 225 nm through spectrophotometric analysis. The consistent and reproducible relationship between the increasing oxalic acid concentration and the observed reduction in peak intensity at 225 nm provides strong validation for the use of acetic acid-capped CuO NPs as a colorimetric sensor for detecting oxalic acid, as shown in Fig. 6. | ||
| Fig. 6 UV-visible spectrum of CuO NPs. A: CuO NPs without the addition of oxalate solution have high absorbance. B: CuO NPs with the addition of oxalate have low absorbance. | ||
3.4 The optimization of various parameters for the colorimetric sensing of oxalic acid
Several factors affecting the performance of CuO NPs were optimized. This was done using a standard approach, where one variable was changed while the others remained constant. However, all the different factors influencing the performance of the proposed sensor were successfully optimized.3.5 The analytical characteristics of the fabricated CuO NPs biosensor
The analytical performance of the fabricated sensor for oxalic acid detection was evaluated, and the corresponding spectroscopic data are presented in Fig. 8(A). The incremental addition of oxalic acid led to a discernible colorimetric transition from blue to colorless, accompanied by a concomitant decrease in the spectroscopic absorption peak intensity at 225 nm. The systematic investigation was conducted across a concentration range of oxalic acid spanning 1 to 120 µM, revealing a direct interaction between the analyte concentration and the optical response of the fabricated sensor. Moreover, the developed sensor accurately detected oxalic acid concentration within the LOD of 0.23 µM and LOQ of 0.78 µM, as shown in Fig. 8(B). The oxalic acid was also colorimetrically quantified with curcumin-mediated nanoparticles in the presence of Fe(III). The method was complex and based on the inhibitory effect of oxalate ion on the reaction with Fe(III) in acidic media within a linear range of 0.15 to 1.70 µM21. Moreover, oxalic acid is predominantly present in natural products, and different approaches were used to quantify its concentration and cope with its drastic effects.22 The oxalic acid was also determined using oxalate oxidase and sensors based on the injection of the recognition element technology, with a linear range of 0 and 5 mM.50 The amperometry biosensor with a complex approach was prepared using a multi-walled carbon nanotube-gold nanoparticle composite for oxalate determination. The electrocatalytic response revealed a linear range of oxalic acid concentrations from 1 to 800 µM, with a good detection limit of 1 µM. However, the utilization of gold nanoparticles increased the sensor's cost.51 The present proposed sensor was compared with previously reported literature, as shown in Table 1. The proposed developed sensor demonstrated robust quantitative capabilities for oxalic acid determination. Notably, the sensor exhibited rapid kinetic behavior, achieving a response time within 100 seconds. Therefore, the analytical merits obtained from the present study have several advantages over previously reported sensor systems for oxalic acid detection in the context of cost-effectiveness, easy approach synthesis, and rapid response with lower LOQ.| Sr # | Material used | Method | Linear range (µM) | LOD (µM) | References |
|---|---|---|---|---|---|
| 1 | CURNs | Colorimetry | 1.6–18.8 | 0.77 | 21 |
| 2 | AgNPs | Colorimetry | 10–40 | 3.3 | 22 |
| 3 | c-MWCNT/Au GNPs | Colorimetry | 1–800 | 1 | 51 |
| 4 | FeSSA | Colorimetry | 4.5–888 | 4 | 23 |
| 5 | PtNPGNs | Colorimetry | 100–50000 | 5 | 52 |
| 6 | Ag-Nps + N-GO | Colorimetry | 10–300 | 2 | 9 |
| 7 | CuO NPs@CH3COOH | Colorimetry | 1–120 | 0.23 | Present work |
3.6 Interference study
The interference study of the developed biosensor was performed with various metal ions, including calcium and magnesium. In addition, different biomolecules, including glucose, lysine, trypsin, and uric acid. The selectivity of the present sensor was also investigated in structurally similar compounds, including oxalic acid, pyruvic acid, maleic acid, and glutamic acid, to assess potential interference. Moreover, an equal concentration (100 µM) of the other tested species did not elicit a similar response or interfere with detection under the experimental conditions optimized for oxalic acid. The developed sensor exhibited a significant colorimetric response only for oxalic acid and observed a notable decrease in absorbance at 225 nm. Therefore, the high sensitivity of CuO NPs toward oxalic acid makes them excellent colorimetric probes. Thus, the sensor is highly selective toward the detection of oxalic acid, which promises the applicability of the sensor for selective detection, as schematically shown in Fig. 9. | ||
| Fig. 9 The selectivity of CuO NPs for the sensing of oxalic acid in the presence of various potential interfering species. | ||
3.7 Real sample analysis of urine
The high percent recovery of oxalic acid from the real urine sample using the CuO NPs sensor confirms its accuracy and efficiency for oxalic acid detection. Within the complex biological matrix, the detection of oxalic acid indicates minimal interference and reliable quantification under optimal conditions. The CuO NPs colorimetric sensor was used to directly measure the oxalate ions in real samples, as shown in Fig. 10. The proposed sensor was validated by assessing the recovery of spiked oxalate solutions at concentrations of 0.7, 2, 3.5, 11.5, and 16 µM, given in Table 2. The results were calculated using the percentage recovery formula and are summarized as follows:| Samples | Spike level (µM) | Spike added (µM) | Found (µM) | Spike recoveries (%) | RSD (%) |
|---|---|---|---|---|---|
| 1 | 0.1 | 0.7 | 0.8 | 114.28 | 1.25 |
| 2 | 0.4 | 2 | 2.2 | 110 | 4.54 |
| 3 | 0.5 | 3.5 | 3.9 | 111.428 | 2.56 |
| 4 | 0.51 | 11.5 | 12.4 | 107.88 | 2.41 |
| 5 | 2.23 | 16 | 18.1 | 113.12 | 1.1 |
Recovery% = oxalate found/oxalate added × 100.18
3.8 Predictive modeling of concentration from RGB values
The performance of predictive machine learning models was evaluated by comparison of predicted values of oxalic acid concentrations with actual concentrations, as shown in Fig. 11(A–D). Decision tree (DT) and XGBoost (XGB) regressors have shown apparently the best performance due to slightly lower RMSE and MAE and higher R2 (Table 3 and Fig. 12). However, the overall predicted results of these two models revealed overfitting of models due to the limited dataset. In contrast, Random Forest (RF) and Linear Regression (LR) models performed well by producing more stable and chemically interpretable results. The RF model handled the non-linear relationship in the data without becoming too complex, while the simpler model, LR, revealed a steady trend between actual and predicted concentrations. Overall, both models have given consistent predictions of all concentrations of oxalic acid. Although XGB and DT mathematically overtook other models, in the context of colorimetric analysis, where reproducibility and interpretation are critical, RF and LR were chosen as reliable models regardless of little higher error values. The relationship between RGB values and oxalic acid concentrations demonstrated a clear trend, as shown in Fig. 13. These results indicate that simpler models like random forest and linear regression can perform well compared to complex models, especially when the data set is small and linearity is evident. The performance gap observed for XGB and DT may be attributed to insufficient data points for these algorithms to generalize effectively. These results specify that machine learning can be a complementary and supportive tool to colorimetric methods for concentration prediction. | ||
| Fig. 11 Actual values of oxalic acid dilutions against predicted values obtained by ML models (A) linear regression, (B) Random Forest, (C) XGBoost Regression, (D) Decision Tree. | ||
| Model | MAE | RMSE | R2 |
|---|---|---|---|
| Linear regression | 14.629 | 16.993 | 0.861 |
| XGBoost regression | 10.571 | 11.065 | 0.941 |
| Random forest | 11.490 | 14.836 | 0.894 |
| Decision tree regression | 9.750 | 10.404 | 0.948 |
4. Conclusions
The present research work demonstrates a successful, facile, and cost-effective drug-mediated synthesis of CuO NPs through a drug-mediated approach. All the spectroscopic and morphological analyses confirmed their synthesis. The surface functionalization, deagglomeration, and conductivity enhancement of the synthesized nanoparticles were successfully achieved through their capping with acetic acid. The acetic acid-capped CuO NPs revealed a colorimetric response to oxalic acid at a surface plasmon resonance of 220 nm, unequivocally establishing their potential as a colorimetric sensor. The readily observable color change from blue to transparent resulted from the interaction between CuO NPs and oxalic acid and was observed at 250 µL of capped nanoparticles, 100 µL of oxalic acid (10 µM), and pH 7, with a reaction time of 100 seconds at room temperature. The fabricated colorimetric CuO NPs sensor selectively detected oxalic acid with an LOQ of 0.78 µM. The research findings unveil a promising pathway for the development of rapid, on-site oxalic acid detection that can potentially be used for diverse applications. In addition, these findings confirmed the machine learning capability as a swift, portable enhancement to colorimetric methods for accurate oxalic acid concentration quantification.Author contributions
Muhammad Ishtiaq Jan: writing – original draft, methodology, investigation, formal analysis, conceptualization, supervision. Abda Khatoon: writing – review & editing, investigation, formal analysis. Wei Sun: writing – review & editing, data curation, formal analysis. Naeem Khan: writing – review & editing, formal analysis, conceptualization. Sajid Ali: writing – review & editing, methodology, formal analysis. Ibrahim A. Shaaban: writing – review & editing, validation, funding acquisition. Mansoor Khan: writing – review & editing, methodology, formal analysis. Amir Badshah: writing – review & editing, data curation, formal analysis. Imran Rabbani: writing – review & editing, formal analysis, methodology. Umar Nishan: writing – review & editing, methodology, formal analysis, conceptualization, supervision.Conflicts of interest
The authors declare no conflict of interest.Data availability
All relevant data are presented within the manuscript; however, additional information is available upon request to the corresponding author based on reasonable demand.Acknowledgements
The authors would like to extend their appreciation to University Higher Education Fund for funding this research work under Research Support Program for Central labs at King Khalid University through the project number CL/PRI/C/16.References
- M. U. Hasan, Z. Singh, H. M. S. Shah, J. Kaur, A. Woodward, E. Afrifa-Yamoah and A. U. Malik, Postharvest Biol. Technol., 2023, 206, 112574 Search PubMed.
- S. Milardović, Z. Grabarić and B. S. Grabarić, Food Technol. Biotechnol., 2000, 38, 203–210 Search PubMed.
- B. Beck, P. Cochat and Y. Frishberg, in Pediatric Nephrology, 8th edn, 2022, pp. 831–846 Search PubMed.
- N. Stepanova, V. Driianska, L. Korol, L. Snisar and L. Lebed, Korean J. Intern. Med., 2022, 37, 167–178 Search PubMed.
- L. Li, Y. Peng, M. Liu, Z. Wang, Q. Wang, S. Ming, X. Gao and Y. Sun, Discov. Med., 2019, 28, 75–85 Search PubMed.
- L. Frassetto and I. Kohlstadt, Am. Fam. Physician, 2011, 84, 1234–1242 Search PubMed.
- A. N. Amenaghawon, J. E. Ayere, U. O. Amune, I. C. Otuya, E. C. Abuga, C. L. Anyalewechi, O. V. Okoro, J. A. Okolie, P. K. Oyefolu, S. O. Eshiemogie, B. E. Osahon, M. Omede, S. A. Eshiemogie, S. Igemhokhai, M. O. Okedi, H. S. Kusuma, O. E. Muojama, A. Shavandi and H. Darmokoesoemo, Environ. Res., 2024, 251(Part 2), 118703 CrossRef CAS.
- T. Kayashima and T. Katayama, Biochim. Biophys. Acta, Gen. Subj., 2002, 1573, 1–3 CrossRef CAS.
- M. A. Zafar, Y. Liu and M. Jacob, SSRN Electron. J., 2022, 8, 100188 CAS.
- Q. Zhai, Spectrochim. Acta, Part A, 2008, 71(2), 332–335 CrossRef PubMed.
- D. T.-S. A, K. Kawamura and L. A. Barrie, Atmos. Environ., 2010, 44, 5316–5319 CrossRef.
- S. G. Yan Zhang, C. Wu and J. Zhang, J. Electrochem. Soc., 2017, 164, B29 CrossRef.
- U. Nishan, A. Ahmed, N. Muhammad and M. Shah, RSC Adv., 2024, 14, 7022–7030 RSC.
- A. Badshah, S. Noreen, M. Shah, M. Asad and R. Ullah, RSC Adv., 2024, 14, 19539–19549 RSC.
- F. Rigoldi, S. Donini, A. Redaelli, E. Parisini and A. Gautieri, APL Bioeng., 2018, 2(1), 1–18 CrossRef.
- P. Ying Gan, N. Hu, C. He, S. Zhou, J. Tu, T. Liang, Y. Pan, D. Kirsanov, A. Legin, H. Wan and P. Wang, Biosens. Bioelectron., 2019, 130, 254–261 CrossRef.
- P. F. Basharzad, K. Farhadi, M. Forough and R. Molaei, Sens. Lett., 2016, 14, 906–912 CrossRef.
- H. Tavallali, G. Deilamy-Rad and N. Mosallanejad, Food Technol. Biotechnol., 2018, 56, 329–336 Search PubMed.
- L. J. Tang and M. H. Liu, Bull. Korean Chem. Soc., 2010, 31, 3159–3162 CrossRef CAS.
- Y. Liu, J. Huang, D. Wang, H. Hou and T. You, Anal. Methods, 2010, 2, 855–859 RSC.
- N. Pourreza, N. Lotfizadeh and H. Golmohammadi, Spectrochim. Acta, Part A, 2018, 192, 251–256 CrossRef CAS PubMed.
- R. N. Juine and A. Das, ACS Sustain. Chem. Eng., 2020, 8, 11579–11587 CrossRef CAS.
- L. Wu, F. Li, H. Yu, L. Shen and M. Wang, Spectrochim. Acta, Part A, 2023, 284, 121784 CrossRef CAS PubMed.
- C. Cesar Souza Machado, J. F. da Silveira Petruci and S. G. Silva, Microchem. J., 2021, 168, 106466 CrossRef CAS.
- U. Nishan, I. Ullah, R. Gul, A. Badshah, N. Muhammad, N. Khan, M. Shah, M. Asad, S. Afridi, R. Ullah, E. A. Ali and S. C. Ojha, ACS Omega, 2023, 8, 44931–44941 CrossRef CAS.
- U. Nishan, R. Gul, N. Muhammad, M. Asad, A. Rahim, M. Shah, J. Iqbal, J. Uddin, A. ul. H. Ali Shah and S. Shujah, Microchem. J., 2020, 159, 105382 CrossRef CAS.
- J. Singh, G. Kaur and M. Rawat, Journal of Bioelectronics and Nanotechnology, 2016, 1(1), 9 Search PubMed.
- G. Ren, D. Hu, E. W. C. Cheng, M. A. Vargas-reus, P. Reip and R. P. Allaker, Int. J. Antimicrob., 2009, 33, 587–590 CrossRef CAS.
- N. Verma and N. Kumar, ACS Biomater. Sci. Eng., 2019, 5, 1170–1188 CrossRef CAS.
- C. K. Wu, M. Yin, S. O'Brien and J. T. Koberstein, Chem. Mater., 2006, 18, 6054–6058 CrossRef CAS.
- D. Deng, Y. Jin, Y. Cheng, T. Qi and F. Xiao, ACS Appl. Mater. Interfaces, 2013, 5, 3839–3846 CrossRef CAS.
- S. Raschka, J. Patterson and C. Nolet, Information, 2020, 11(4), 193 CrossRef.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and É. Duchesnay, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- F. Zehra, T. Fatima and M. Khanuja, Adv. Mater. Technol., 2025, 00519, 1–18 Search PubMed.
- D. Renuga, J. Jeyasundari, A. S. S. Athithan and Y. B. A. Jacob, Mater. Res. Express, 2020, 7, 1–7 Search PubMed.
- M. Bin Mobarak, M. S. Hossain, F. Chowdhury and S. Ahmed, Arab. J. Chem., 2022, 15(10), 104117 Search PubMed.
- U. Nishan, S. Rehman, R. Ullah, A. Bari, S. Afridi, M. Shah, J. Iqbal, H. U. Khan and N. Muhammad, Front. Mater., 2023, 10, 1–10 Search PubMed.
- A. S. Ethiraj and D. J. Kang, Nanoscale Res. Lett., 2012, 7, 70 CrossRef PubMed.
- O. A. Bulavchenko and Z. S. Vinokurov, Catalysts, 2023, 13(11), 1421 CrossRef CAS.
- G. Ravi and N. Patra, J. Mater. Sci. Mater. Eng., 2025, 20(34), 1–8 Search PubMed.
- W. Z. Shia, Y. S. Lianga, B. Lua, M. Chena, Y. Lia and Z. Yanga, Quim. Nova, 2019, 42, 638–641 Search PubMed.
- M. C. Biesinger, Surf. Interface Anal., 2017, 49, 1325–1334 CrossRef CAS.
- M. Piñon-Espitia, D. Lardizabal-Gutiérrez, M. L. Camacho-Ríos, G. Herrera-Pérez and M. T. Ochoa-Lara, Mater. Chem. Phys., 2021, 272, 124981 CrossRef.
- R. F. Roberts, J. Electron Spectrosc. Relat. Phenom., 1974, 4, 273–291 CrossRef CAS.
- N. Baig and T. A. Saleh, Glob. Challenges, 2019, 3, 1800115 CrossRef PubMed.
- X. Sun and Y. Luo, Mater. Lett., 2005, 59, 3847–3850 CrossRef CAS.
- T. Li, Y. Li, Y. Zhang, C. Dong, Z. Shen and A. Wu, Analyst, 2015, 140, 1076–1081 RSC.
- P. Aggarwal, J. S. Rana, M. Chitkara and A. Kumar, J. Clust. Sci., 2024, 35, 2093–2103 CrossRef CAS.
- W. L. Daniel, M. S. Han, J. S. Lee and C. A. Mirkin, J. Am. Chem. Soc., 2009, 131, 6362–6363 CrossRef CAS PubMed.
- F. Hong, N. O. Nilvebrant and L. J. Jönsson, Biosens. Bioelectron., 2003, 18, 1173–1181 CrossRef CAS.
- C. S. Pundir, N. Chauhan, Rajneesh, M. Verma and Ravi, Sens. Actuators, B, 2011, 155, 796–803 CrossRef CAS.
- X. Chen, Z. Cai, Z. Huang, M. Oyama, Y. Jiang and X. Chan, Nanoscale, 2013, 5, 5779–5783 RSC.
| This journal is © The Royal Society of Chemistry 2026 |