Open Access Article
Open Access ArticleDesign, synthesis, biological evaluation, and in silico studies of novel 1,3-thiazole derivatives
Nguyen Duy Khanha,
Nguyen Thi Hong Anhb,
Nguyen Van Sona,
Nguyen Thi Mai Tho a,
Nguyen Thi Nhat Thanga,
Tran Thuy Quyena and
Tran Nguyen Minh An
a,
Nguyen Thi Nhat Thanga,
Tran Thuy Quyena and
Tran Nguyen Minh An *a
*a
aFaculty of Chemical Emgineering (FCE), Industrial University of Ho Chi Minh City (IUH), 12 Nguyen Van Bao, Hanh Thong Ward, Ho Chi Minh City, Vietnam. E-mail: trannguyenminhan@iuh.edu.vn
bFaculty of Chemical Engineering, Ho Chi Minh City University of Industry and Trade, 140 Le Trong Tan Street, Tay Thanh Ward, Ho Chi Minh, 70000, Vietnam
First published on 9th January 2026
Abstract
A series of novel 1,3-thiazole heterocyclic derivatives has been synthesized and structurally characterized using melting point, FT-IR, 1D and 2D NMR, and HR-MS. The compounds were screened for antibacterial activities, three human cancer cell lines (A549, HepG2, and MCF-7), and anti-inflammatory and α-glucosidase inhibitory activities, utilising MIC, MTT, α-glucosidase inhibitory, and NO assays, respectively. Entry 4e exhibited the most potent antibacterial activity (MIC = 83.5 µM). Entry 4g exhibited significant inhibitions against three cancer cell lines: MCF-7, HepG2, and A549 (IC50 = 2.6–6.6 µM). Entry 4f exhibited significant anti-inflammatory activity (IC50 = 15.4 ± 1.0 µM), comparable to the dexamethasone drug (IC50 = 13.7 ± 1.2 µM). Notably, 4a and 4l exhibited potent α-glucosidase inhibitory activity (IC50 = 46 ± 2 µM and 41 ± 2 µM, respectively), surpassing acarbose (IC50 = 117 ± 8 µM). Molecular docking studies revealed energetically favourable interactions between the ligands and the active sites of the target enzymes, in good agreement with the predicted biological activities obtained from in silico analyses. A 100 ns molecular dynamics simulation of the 4l-4J5T complex further confirmed its structural stability after 60 ns, with key residues Glu428 and Asn452 maintaining hydrogen bond interactions for approximately 80% of the simulation time. In addition, ADMET predictions indicated that compound 4l possesses favourable pharmacokinetic properties. Collectively, these findings highlight a novel thiazole-based scaffold exhibiting multi-target biological activity and underscore its promising potential for further drug development.
1. Introduction
Thiazole is a sulfur- and nitrogen-containing five-membered heterocycle known for broad pharmacological significance. Thiazole derivatives have demonstrated antibacterial,1 anti-inflammatory,2 anticancer,3 and antidiabetic effects,4 owing to their strong ability to interact with target enzymes and receptors.5,6 Clinically approved thiazole-containing drugs, including the thiazolidinediones pioglitazone and rosiglitazone, highlight the therapeutic importance of this scaffold in type-II diabetes management through peroxisome proliferator-activated receptor gamma (PPARγ) activation and subsequent enhancement of insulin sensitivity.7–10Carbazole is a tricyclic aromatic system found in natural and synthetic bioactive molecules.11 Its rigid π-conjugated framework and favourable electronic properties enable interactions with diverse biological targets.12–15 Carbazole derivatives have been widely reported to show anticancer, anti-inflammatory, antioxidant, antimicrobial, neuroprotective, and antidiabetic properties, underscoring their pharmacological versatility.16 Carvedilol, a U.S. Food and Drug Administration (FDA)-approved carbazole-based β-blocker, further exemplifies the clinical relevance of this scaffold, also demonstrating beneficial effects on insulin sensitivity and glucose regulation.17,18 Recent investigations have shown that carbazole derivatives may modulate key diabetic targets, including α-glucosidase, dipeptidyl peptidase-4 (DPP-4), and PPARγ, supporting their potential in the treatment of type 2 diabetes.19–21 Given the established bioactivity of thiazole and carbazole systems, the hybridisation of both motifs represents a rational approach to generate multifunctional therapeutic scaffolds. Recent studies have reported carbazole–thiazole hybrids with promising activity profiles, reinforcing the attractiveness of this strategy. Several studies, such as those by Krawczyk et al. (2024), Marufa et al., Çapan et al., and Donarska et al. (2025), have reported hybrid derivatives with promising in vitro and in silico activities.22–25
Bacterial replication and cell-wall integrity depend on DNA gyrase, topoisomerase IV, and β-lactamase. Potent antibacterial agents often employ planar aromatic frameworks or metal-binding heterocycles to disrupt these catalytic systems.26 Tyrosine kinases, tubulin, and topoisomerases regulate cancer proliferation. Carbazole provides a rigid π-surface for DNA and protein interactions, whereas thiazole introduces polarity and facilitates the orientation of H-bonding. Their hybrid enables complementary hydrophobic and polar contacts, supporting its promise as an anticancer scaffold.27 Inducible nitric oxide synthase (iNOS) mediated nitric oxide production drives chronic inflammation. Ligands with π-rich skeletons and heteroatom donors can engage the heme centre and L-arginine pocket, enabling efficient iNOS modulation.28 α-Glucosidase regulates carbohydrate breakdown and postprandial glucose levels. Heterocycle-bearing ligands mimic substrate interactions. Thiazole substitution enhances polarity and binding orientation, highlighting relevance for glycaemic modulation.29 Based on this rationale, we designed, synthesised, and examined a series of previously unreported 1,3-thiazole derivatives. The compounds underwent comprehensive characterisation through Fourier transform infrared spectroscopy (FT-IR), nuclear magnetic resonance spectroscopy (NMR), and high-resolution mass spectrometry (HR-MS) analyses. Their biological potential was evaluated using in vitro assays, which included assessments of α-glucosidase inhibition, antibacterial activity, anti-inflammatory properties, and anticancer effects. Simultaneously, in silico studies involving molecular docking against chosen therapeutic targets, absorption, distribution, metabolism, excretion, and toxicity (ADMET) prediction, and molecular dynamics simulations were conducted to clarify binding behaviour and pharmacokinetic appropriateness. This combined experimental and computational method facilitates the identification of potential multi-target lead candidates.
2. Materials and methods
2.1. Materials
2.2. Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nbromoketone = 1
nbromoketone = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in ethanol at 60–70 °C for 6 h under reflux to form the target 1,3-thiazole derivatives (4a–4q), which were purified by filtration, washed with ethanol, and confirmed for purity by TLC prior to structural characterization.32
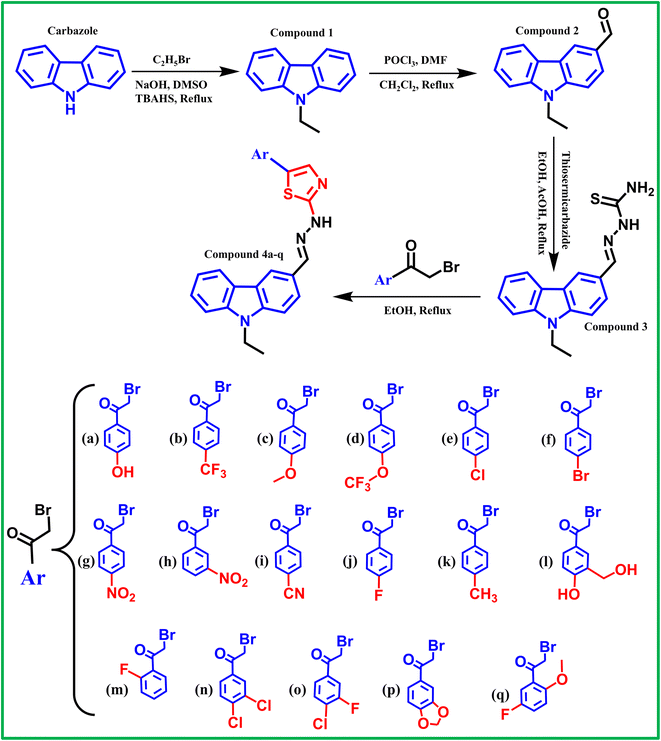
1) in ethanol at 60–70 °C for 6 h under reflux to form the target 1,3-thiazole derivatives (4a–4q), which were purified by filtration, washed with ethanol, and confirmed for purity by TLC prior to structural characterization.322.2.3.1 Compound 1 (9-ethyl-9H-carbazole). Colorless amorphous powder (10.9805 g), yield of 93.75%. 1H-NMR (500 MHz, DMSO-d6, 25 °C, δ ppm): 1.27 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.38–4.40 (m, 2H, –CH2–CH3); 7.20 (t, J = 6.0 Hz, 2H, aromatic–H (H-2; H-7)); 7.45 (t, J = 6.0 Hz, 2H, Ar–H (H-3, H-6)); 7.56 (d, J = 6.5 Hz, 2H, Ar–H (H-1, H-8)); 8.14 (d, J = 6.0 Hz, 2H, Ar–H (H-4, H-5)). 13C-NMR (125 MHz, DMSO-d6, 25 °C, δ ppm) 139.5; 125.6; 122.1; 120.2; 118.9; 108.9; 36.8 (–CH2–CH3); 13.5 (–CH2–CH3). Observation on the 1H-NMR spectrum showed the appearance of a triplet signal at δ = 1.286 ppm corresponding to three protons and a multiplet signal at δ = 4.38–4.40 ppm corresponding to two protons.32 In the 13C-NMR spectrum, two signals were observed at δ = 36.8 and 13.5 ppm (Fig. S1–S4). These NMR signals confirm the presence of a –CH2–CH3 group attached to the carbazole framework.
2.2.3.2 Compound 2 (9-ethyl-9H-carbazole-3-carbaldehyde). Colorless amorphous powder (6.0612 g), yield of 51.14%. 1H-NMR (500 MHz, DMSO-d6, 25 °C, δ ppm): 1.32 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.52 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.32 (t, J = 6.0 Hz, 1H, Ar–H); 7.55 (t, J = 5.8 Hz, 1H, Ar–H); 7.79 (d, J = 7.0 Hz, 1H, Ar–H); 8.01 (dd, J1 = 6.0 Hz, J2 = 1.5 Hz, 1H, Ar–H); 8.76 (d, J = 1 Hz, 1H, Ar–H); 10.07 (s, 1H, –CHO). 13C-NMR (125 MHz, DMSO-d6, 25 °C, δ ppm) 191.8 (–CHO); 143.3; 140.0; 128.7; 126.7; 126.6; 123.9; 122.3; 122.2; 120.8; 120.1; 109.8; 109.5; 37.3 (–CH2–CH3); 13.6 (–CH2–CH3).34 Observation of the 1H-NMR spectrum, in addition to the characteristic signals of the ethyl group at δ = 1.35 ppm (t, J = 6.0 Hz, 3H, –CH2–CH3) and δ = 4.52 ppm (q, J = 6.0 Hz, 2H, –CH2–CH3), reveals the appearance of a singlet corresponding to one proton of the –CHO group at δ = 10.07 ppm (Fig. S5).32 Similarly, the 13C-NMR spectrum exhibits a signal at δ = 191.8 ppm, characteristic of the –CHO carbon. These results confirm the successful introduction of the carbonyl group at the C3 position of 9-ethyl-9H-carbazole, resulting in the formation of compound (2), 9-ethyl-9H-carbazole-3-carbaldehyde.
2.2.3.3 Compound 3 (((9-ethyl-9H-carbazol-3-yl)methylene)hydrazine-1-carbothioamide). Light yellow powder (6.0133 g), yield of 92.34%. 1H-NMR (500 MHz, DMSO-d6, 25 °C, δ ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.46 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H, Ar–H); 7.48 (t, J = 6.0 Hz, 1H, Ar–H); 7.63 (d, J = 7.0 Hz, 3H, Ar–H, –NH2); 7.97 (dd, J1 = 6.0 Hz, J2 = 1.5 Hz, 1H, Ar–H); 8.14 (s, 1H); 8.21 (d, J = 6.5 Hz, 1H); 8.23 (s, 1H, –CH
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 8.58 (d, J = 1.0 Hz, 1H, Ar–H); 11.36 (s, 1H,
N–); 8.58 (d, J = 1.0 Hz, 1H, Ar–H); 11.36 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–).32
N–NH–).3213C-NMR (125 MHz, DMSO-d6, 25 °C, δ ppm) 177.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S); 143.6 (Ar–CH
S); 143.6 (Ar–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); 140.5; 140.0; 126.1; 125.1; 122.2; 120.6; 120.2; 119.2; 109.4; 37.1 (–CH2–CH3); 13.7 (–CH2–CH3).32 The formation of the thiosemicarbazone group (Fig. S6–S9) was confirmed by the disappearance of the –CHO proton signal (δ = 10.07 ppm) and the appearance of a new singlet signal at δ = 11.36 ppm corresponding to the
N); 140.5; 140.0; 126.1; 125.1; 122.2; 120.6; 120.2; 119.2; 109.4; 37.1 (–CH2–CH3); 13.7 (–CH2–CH3).32 The formation of the thiosemicarbazone group (Fig. S6–S9) was confirmed by the disappearance of the –CHO proton signal (δ = 10.07 ppm) and the appearance of a new singlet signal at δ = 11.36 ppm corresponding to the ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH– proton, as well as another singlet at δ = 8.23 ppm assigned to the –CH
N–NH– proton, as well as another singlet at δ = 8.23 ppm assigned to the –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– group in the 1H-NMR spectrum.32 Similarly, in the 13C-NMR spectrum, signals at δ = 177.5 (C
N– group in the 1H-NMR spectrum.32 Similarly, in the 13C-NMR spectrum, signals at δ = 177.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) and 143.6 (Ar–CH
S) and 143.6 (Ar–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) were observed. These results confirmed the successful formation of compound (3) (((9-ethyl-9H-carbazol-3-yl)methylene)hydrazine-1-carbothioamide) with no detectable impurities.
N–) were observed. These results confirmed the successful formation of compound (3) (((9-ethyl-9H-carbazol-3-yl)methylene)hydrazine-1-carbothioamide) with no detectable impurities.
The physicochemical properties of compounds 4a–q (Fig. S10–S81) are as follows:
2.2.3.4 4-(2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazol-4-yl)phenol (4a). Chemical formula: C24H20N4OS; colorless amorphous powder (0.1743 g); yield of 62.63%; melting point: 277–278 °C; Rf = 0.27 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3238 (–OH, –CH stretching vibration); 3076 (–CH3); 2988 (–CH2–); 2934, 2888 (CH aromatic); 1615 (C
3); IR (λmax, cm−1, KBr): 3238 (–OH, –CH stretching vibration); 3076 (–CH3); 2988 (–CH2–); 2934, 2888 (CH aromatic); 1615 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1231 (Ar–O–); 1091 (
N thiazole); 1231 (Ar–O–); 1091 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 830, 743 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.46 (q, J = 6.0 Hz, 3H, –CH2–CH3; R–OH); 6.84 (d, J = 7.5 Hz, 2H); 7.08 (s, 1H, H–C5-thiazole); 7.25 (t, J = 6.0 Hz, 1H), 7.49 (t, J = 6.3 Hz 1H), 7.63–7.69 (m, 4H), 7.89 (d, J = 7.5 Hz, 1H), 8.21 (d, J = 6.5 Hz, 1H); 8.34 (s, 1H); 8.41 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.2 (quanternary carbon, C2-thiazole); 157.2 (quanternary carbon, C–OH); 140.2 (–CH
N–NH–); 830, 743 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.46 (q, J = 6.0 Hz, 3H, –CH2–CH3; R–OH); 6.84 (d, J = 7.5 Hz, 2H); 7.08 (s, 1H, H–C5-thiazole); 7.25 (t, J = 6.0 Hz, 1H), 7.49 (t, J = 6.3 Hz 1H), 7.63–7.69 (m, 4H), 7.89 (d, J = 7.5 Hz, 1H), 8.21 (d, J = 6.5 Hz, 1H); 8.34 (s, 1H); 8.41 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.2 (quanternary carbon, C2-thiazole); 157.2 (quanternary carbon, C–OH); 140.2 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 139.9 (quanternary carbon, C9a-carbazole); 127.0, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4-phenyl, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.3; 115.3; 109.7; 109.4; 100.3 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H20N4OS: 413.1436, found: 413.1434.
N–); 139.9 (quanternary carbon, C9a-carbazole); 127.0, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4-phenyl, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.3; 115.3; 109.7; 109.4; 100.3 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H20N4OS: 413.1436, found: 413.1434.
2.2.3.5 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(4-(trifluoromethyl)phenyl)thiazole (4b). Chemical formula: C25H19F3N4S; light brown powder (0.1744 g), yield of 55.64%; melting point: 177–178 °C; Rf = 0.27 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3357 (–CH stretching vibration); 3072 (–CH3); 2934, 2891 (–CH2–); 1629 (C
3); IR (λmax, cm−1, KBr): 3357 (–CH stretching vibration); 3072 (–CH3); 2934, 2891 (–CH2–); 1629 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1481, 1568 (C
N thiazole); 1481, 1568 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1324, 1115 (–CF3); 1066 (
C aromatic); 1324, 1115 (–CF3); 1066 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 844, 748 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.49 (t, J = 6.5 Hz,1H); 7.56 (s, 1H, (H–C5-thiazole)); 7.64 (d, J = 6.5 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.77 (d, J = 7.0 Hz, 2H); 7.87 (d, J = 7.0 Hz, 1H); 8.08 (d, J = 7.0 Hz, 2H, –CH
N–NH–); 844, 748 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.49 (t, J = 6.5 Hz,1H); 7.56 (s, 1H, (H–C5-thiazole)); 7.64 (d, J = 6.5 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.77 (d, J = 7.0 Hz, 2H); 7.87 (d, J = 7.0 Hz, 1H); 8.08 (d, J = 7.0 Hz, 2H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–; H–C1-carbazole); 8.22 (d, J = 6.5 Hz, 1H); 8.24 (s, 1H); 8.38 (s, 1H); 12.10 (s, 1H,
N–; H–C1-carbazole); 8.22 (d, J = 6.5 Hz, 1H); 8.24 (s, 1H); 8.38 (s, 1H); 12.10 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.7 (quanternary carbon, C2-thiazole); 148.9 (quanternary carbon, C4-thiazole); 143.1 (–CH
N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.7 (quanternary carbon, C2-thiazole); 148.9 (quanternary carbon, C4-thiazole); 143.1 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9 (quanternary carbon, C1-phenyl); 138.4 (quanternary carbon, C4-phenyl); 127.6; 127.3; 126.1 (quanternary carbon, C4a, 8a-carbazole); 126.0; 125.5; 125.5; 125.3; 125.2 (quanternary carbon, C4b-carbazole); 123.5 (quanternary carbon, C3-carbazole); 123.4 (quanternary carbon, –CF3); 122.3; 122.1; 120.5; 119.6; 119.2; 109.6; 109.4; 106.0 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H19F3N4S: 465.1361, found: 465.1357.
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9 (quanternary carbon, C1-phenyl); 138.4 (quanternary carbon, C4-phenyl); 127.6; 127.3; 126.1 (quanternary carbon, C4a, 8a-carbazole); 126.0; 125.5; 125.5; 125.3; 125.2 (quanternary carbon, C4b-carbazole); 123.5 (quanternary carbon, C3-carbazole); 123.4 (quanternary carbon, –CF3); 122.3; 122.1; 120.5; 119.6; 119.2; 109.6; 109.4; 106.0 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H19F3N4S: 465.1361, found: 465.1357.
2.2.3.6 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(4-methoxyphenyl)thiazole (4c). Chemical formula: C25H22N4OS; light orange powder (0.2802 g); yield of 97.29%; melting point: 267–268 °C; Rf = 0.26 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 8
Vethyl acetate = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2); IR (λmax, cm−1, KBr): 3349 (–CH stretching vibration); 3013 (–CH3); 2961 (–CH2–); 2765 (–O–CH3); 1620 (C
2); IR (λmax, cm−1, KBr): 3349 (–CH stretching vibration); 3013 (–CH3); 2961 (–CH2–); 2765 (–O–CH3); 1620 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1508, 1471 (C
N thiazole); 1508, 1471 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1239, 1184, 1025 (Ar–O–CH3); 821, 750 (aromatic ring bearing substituents).1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 3.80 (s, 3H, –O–CH3); 4.47 (t, J = 6.0 Hz, 2H, –CH2–CH3); 6.98–7.00 (m, 2H), 7.15 (s, 1H, H–C5-thiazole); 7.25 (t, J = 6.3 Hz, 1H); 7.49 (t, J = 6.5, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.69 (d, J = 7.0 Hz, 1H); 7.78–7.81 (m, 2H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.39 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.3 (quanternary carbon, C2-thiazole); 158.9 (quanternary carbon, C4-phenyl); 143.5 (quanternary carbon, C4-thiazole); 140.3 (–CH
C aromatic); 1239, 1184, 1025 (Ar–O–CH3); 821, 750 (aromatic ring bearing substituents).1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 3.80 (s, 3H, –O–CH3); 4.47 (t, J = 6.0 Hz, 2H, –CH2–CH3); 6.98–7.00 (m, 2H), 7.15 (s, 1H, H–C5-thiazole); 7.25 (t, J = 6.3 Hz, 1H); 7.49 (t, J = 6.5, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.69 (d, J = 7.0 Hz, 1H); 7.78–7.81 (m, 2H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.39 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.3 (quanternary carbon, C2-thiazole); 158.9 (quanternary carbon, C4-phenyl); 143.5 (quanternary carbon, C4-thiazole); 140.3 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.0 (quanternary carbon, C9a-carbazole), 127.0; 126.2 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole; C1-phenyl); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.3; 114.0; 109.7; 109.4; 101.2 (C5-thiazole); 55.2 (–O–CH3); 37.2 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H22N4OS: 427.1592, found: 427.1595.
N–); 140.0 (quanternary carbon, C9a-carbazole), 127.0; 126.2 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole; C1-phenyl); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.3; 114.0; 109.7; 109.4; 101.2 (C5-thiazole); 55.2 (–O–CH3); 37.2 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H22N4OS: 427.1592, found: 427.1595.
2.2.3.7 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(4-(trifluoromethoxy)phenyl)thiazole (4d). Chemical formula: C25H19F3N4OS; white orange powder (0.2731 g); yield of 84.19%; melting point: 253–254 °C; Rf = 0.66 (VHexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) VEthyl acetate = 8
VEthyl acetate = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2); IR (λmax, cm−1, KBr): 3079 (–CH3); 2977, 2938 (–CH2–); 1626 (CH
2); IR (λmax, cm−1, KBr): 3079 (–CH3); 2977, 2938 (–CH2–); 1626 (CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1598, 1564, 1491 (C
N thiazole); 1598, 1564, 1491 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1263, 1229, 1162 (Ar–O–CF3); 1046, 1017 (
C aromatic); 1263, 1229, 1162 (Ar–O–CF3); 1046, 1017 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 848, 737 (aromatic ring bearing substituents).1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.0 Hz, 1H); 7.41 (s, 2H); 7.42 (s, 1H, H–C5-thiazole); 7.49 (t, J = 6.3 Hz, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.69 (d, J = 6.0 Hz, 1H); 7.87 (dd, J1 = 7.0 Hz, J2 = 1.0 Hz, 1H); 7.97–8.00 (m, 2H, –CH
N–NH–); 848, 737 (aromatic ring bearing substituents).1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.0 Hz, 1H); 7.41 (s, 2H); 7.42 (s, 1H, H–C5-thiazole); 7.49 (t, J = 6.3 Hz, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.69 (d, J = 6.0 Hz, 1H); 7.87 (dd, J1 = 7.0 Hz, J2 = 1.0 Hz, 1H); 7.97–8.00 (m, 2H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–; H–C1-carbazole); 8.22 (s, 1H); 8.23 (s,1H); 8.38 (d, J = 6.5 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 148.9 (quanternary carbon, C4-thiazole); 143.1 (–CH
N–; H–C1-carbazole); 8.22 (s, 1H); 8.23 (s,1H); 8.38 (d, J = 6.5 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 148.9 (quanternary carbon, C4-thiazole); 143.1 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 133.9 (quanternary carbon, R–CF3); 127.3, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole; C1-phenyl); 123.5 (quanternary carbon, C3-carbazole); 122.3; 122.1; 121.1; 120.5; 119.6; 119.2; 109.6; 109.4; 104.3 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H19N4OSF3: 481.1310, found: 481.1308.
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 133.9 (quanternary carbon, R–CF3); 127.3, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole; C1-phenyl); 123.5 (quanternary carbon, C3-carbazole); 122.3; 122.1; 121.1; 120.5; 119.6; 119.2; 109.6; 109.4; 104.3 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H19N4OSF3: 481.1310, found: 481.1308.
2.2.3.8 4-(4-Chlorophenyl)-2-(2-((9-ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazole (4e). Chemical formula: C24H19ClN4S; white orange powder (0.2783 g); yield of 85.47%; melting point: 259–260 °C; Rf = 0.56 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 8
Vethyl acetate = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2); IR (λmax, cm−1, KBr): 3294 (–CH stretching vibration); 3091, 3021 (–CH3–); 2971, 2932 (–CH2–); 1606 (C
2); IR (λmax, cm−1, KBr): 3294 (–CH stretching vibration); 3091, 3021 (–CH3–); 2971, 2932 (–CH2–); 1606 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1481 (C
N thiazole); 1481 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1044, 1013 (
C aromatic); 1044, 1013 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 820, 740 (aromatic ring bearing substituents); 616, 551 (Ar–Cl).1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.46 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.39 (s, 1H, H–C5-thiazole); 7.46–7.50 (m, 3H); 7.63 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.85–7.89 (m, 3H); 8.22 (d, J = 6.5 Hz, 1H); 8.24 (s, 1H); 8.38 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.5 (quanternary carbon, C2-thiazole); 149.0 (quanternary carbon, C4-thiazole); 143.2 (–CH
N–NH–); 820, 740 (aromatic ring bearing substituents); 616, 551 (Ar–Cl).1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.46 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.39 (s, 1H, H–C5-thiazole); 7.46–7.50 (m, 3H); 7.63 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.85–7.89 (m, 3H); 8.22 (d, J = 6.5 Hz, 1H); 8.24 (s, 1H); 8.38 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.5 (quanternary carbon, C2-thiazole); 149.0 (quanternary carbon, C4-thiazole); 143.2 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 133.5 (quanternary carbon, C4-phenyl); 131.9 (quanternary carbon, C1-phenyl); 128.6; 127.2; 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole); 123.6; (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.6; 119.2; 109.7; 109.4; 104.1 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SCl: 431.1091, found: 431.1091.
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 133.5 (quanternary carbon, C4-phenyl); 131.9 (quanternary carbon, C1-phenyl); 128.6; 127.2; 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole); 123.6; (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.6; 119.2; 109.7; 109.4; 104.1 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SCl: 431.1091, found: 431.1091.
2.2.3.9 4-(4-Bromophenyl)-2-(2-((9-ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazole (4f). Chemical formula: C24H19BrN4S; white orange powder (0.2911 g); yield of 90.89%; melting point: 259–260 °C; Rf = 0.54 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 8
Vethyl acetate = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2); IR(λmax, cm−1, KBr): 3298 (–CH stretching vibration); 3094 (–CH3); 2969 (–CH2–); 1622 (C
2); IR(λmax, cm−1, KBr): 3298 (–CH stretching vibration); 3094 (–CH3); 2969 (–CH2–); 1622 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1483 (C
N thiazole); 1483 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1233, 1192 (C–N); 1043, 1009 (
C aromatic); 1233, 1192 (C–N); 1043, 1009 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 819, 740 (aromatic ring bearing substituents); 617, 520 (Ar–Br).1H-NMR (500 MHz, DMSO-d6 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.46 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.0 Hz, 1H); 7.40 (s, 1H, H–C5-thiazole); 7.49 (t, J = 6.5 Hz, 1H); 7.60–7.65 (m, 3H); 7.68 (d, J = 7.0 Hz, 1H); 7.81–7.83 (m, 2H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.39 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 149.0 (quanternary carbon, C4-thiazole); 143.2 (–CH
N–NH–); 819, 740 (aromatic ring bearing substituents); 617, 520 (Ar–Br).1H-NMR (500 MHz, DMSO-d6 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.46 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.0 Hz, 1H); 7.40 (s, 1H, H–C5-thiazole); 7.49 (t, J = 6.5 Hz, 1H); 7.60–7.65 (m, 3H); 7.68 (d, J = 7.0 Hz, 1H); 7.81–7.83 (m, 2H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.39 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 149.0 (quanternary carbon, C4-thiazole); 143.2 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 140.0; 133.8; 131.5 (quanternary carbon, C1-phenyl); 127.6, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.4 (quanternary carbon, C3-carbazole); 123.6 (quanternary carbon, C4b-carbazole); 122.3 (quanternary carbon, C4-phenyl); 122.3; 120.6; 120.5; 119.6; 119.3; 109.7; 109.4; 104.2 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SBr: 475.0592, found: 475.0588.
N–); 140.2 (quanternary carbon, C9a-carbazole); 140.0; 133.8; 131.5 (quanternary carbon, C1-phenyl); 127.6, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.4 (quanternary carbon, C3-carbazole); 123.6 (quanternary carbon, C4b-carbazole); 122.3 (quanternary carbon, C4-phenyl); 122.3; 120.6; 120.5; 119.6; 119.3; 109.7; 109.4; 104.2 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SBr: 475.0592, found: 475.0588.
2.2.3.10 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(4-nitrophenyl)thiazole (4g). Chemical formula: C24H19N5O2S; orange powder (0.2441 g); yield of 81.92%; melting point: 264–265 °C; Rf = 0.72 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3313 (–CH stretching vibration); 3058 (–CH3–); 2976, 2928 (–CH2–); 1584 (C
3); IR (λmax, cm−1, KBr): 3313 (–CH stretching vibration); 3058 (–CH3–); 2976, 2928 (–CH2–); 1584 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1501, 1336, 1235 (–NO2); 1476 (C
N thiazole); 1501, 1336, 1235 (–NO2); 1476 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1050 (
C aromatic); 1050 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 854, 748 (aromatic ring bearing substituents).1H NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.3 Hz, 1H); 7.49 (t, J = 6.3 Hz, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.0 Hz, 1H); 7.71 (s, 1H); 7.87 (dd, J1 = 7.0 Hz, J2 = 1.0 Hz, 1H); 8.13 (d, J = 7.5 Hz, 2H); 8.22 (d, J = 6.5 Hz, 1H); 8.24 (s, 1H); 8.28 (d, J = 7.0 Hz, 2H); 8.38 (s, 1H); 12.15 (s, 1H,
N–NH–); 854, 748 (aromatic ring bearing substituents).1H NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.3 Hz, 1H); 7.49 (t, J = 6.3 Hz, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.0 Hz, 1H); 7.71 (s, 1H); 7.87 (dd, J1 = 7.0 Hz, J2 = 1.0 Hz, 1H); 8.13 (d, J = 7.5 Hz, 2H); 8.22 (d, J = 6.5 Hz, 1H); 8.24 (s, 1H); 8.28 (d, J = 7.0 Hz, 2H); 8.38 (s, 1H); 12.15 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.8 (quanternary carbon, C2-thiazole); 148.5 (quanternary carbon, C4-thiazole); 143.3 (–CH
N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.8 (quanternary carbon, C2-thiazole); 148.5 (quanternary carbon, C4-thiazole); 143.3 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.8 (quanternary carbon, C9a-carbazole); 140.2 (quanternary carbon, C4-phenyl); 139.9 (quanternary carbon, C1-phenyl); 126.3 (quanternary carbon, C4a, 8a-carbazole); 126.1 (quanternary carbon, C1-carbazole); 125.3; 124.1 (quanternary carbon, C3-carbazole); 123.6; 122.3; 122.1; 120.6; 119.7; 119.2; 109.7; 109.4; 108.1 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N5SO2: 442.1337, found: 442.1332.
N–); 140.8 (quanternary carbon, C9a-carbazole); 140.2 (quanternary carbon, C4-phenyl); 139.9 (quanternary carbon, C1-phenyl); 126.3 (quanternary carbon, C4a, 8a-carbazole); 126.1 (quanternary carbon, C1-carbazole); 125.3; 124.1 (quanternary carbon, C3-carbazole); 123.6; 122.3; 122.1; 120.6; 119.7; 119.2; 109.7; 109.4; 108.1 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N5SO2: 442.1337, found: 442.1332.
2.2.3.11 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(3-nitrophenyl)thiazole (4h). Chemical formula: C24H19N5O2S; light gray black powder (0.1945 g); yield of 65.29%; melting point: 268–269 °C; Rf = 0.34 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3239 (–CH stretching vibration); 3064 (–CH3–); 2983, 2880 (–CH2–); 1630 (C
3); IR (λmax, cm−1, KBr): 3239 (–CH stretching vibration); 3064 (–CH3–); 2983, 2880 (–CH2–); 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1531, 1346 (–NO2); 1596, 1486 (C
N thiazole); 1531, 1346 (–NO2); 1596, 1486 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1088 (
C aromatic); 1088 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 890, 737 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.48 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.0 Hz, 1H); 7.49 (t, J = 6.4 Hz, 1H); 7.64–7.66 (m, 2H); 7.69 (d, J = 7.0 Hz, 1H); 7.72 (t, J = 6.8 Hz, 1H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.5 Hz, 1H); 8.15–8.17 (m, 1H); 8.22–8.24 (m, 2H); 8.32 (d, J = 6.5 Hz, 1H); 8.39 (s, 1H); 8.70 (t, J = 1.8 Hz, 1H); 12.18 (s, 1H,
N–NH–); 890, 737 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.48 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.25 (t, J = 6.0 Hz, 1H); 7.49 (t, J = 6.4 Hz, 1H); 7.64–7.66 (m, 2H); 7.69 (d, J = 7.0 Hz, 1H); 7.72 (t, J = 6.8 Hz, 1H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.5 Hz, 1H); 8.15–8.17 (m, 1H); 8.22–8.24 (m, 2H); 8.32 (d, J = 6.5 Hz, 1H); 8.39 (s, 1H); 8.70 (t, J = 1.8 Hz, 1H); 12.18 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.8 (quanternary carbon, C2-thiazole); 148.3 (quanternary carbon, C4-thiazole); 148.0 (quanternary carbon, C3-phenyl); 143.3 (–CH
N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.8 (quanternary carbon, C2-thiazole); 148.3 (quanternary carbon, C4-thiazole); 148.0 (quanternary carbon, C3-phenyl); 143.3 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.3 (quanternary carbon, C9a-carbazole); 140.0; 136.2 (quanternary carbon, C1-phenyl); 131.6; 130.2, 126.2 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.9; 119.7; 119.3; 109.7; 109.4; 106.0 (C5-thiazole); 37.2 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N5SO2: 442.1337, found: 442.1341.
N–); 140.3 (quanternary carbon, C9a-carbazole); 140.0; 136.2 (quanternary carbon, C1-phenyl); 131.6; 130.2, 126.2 (quanternary carbon, C4a, 8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.9; 119.7; 119.3; 109.7; 109.4; 106.0 (C5-thiazole); 37.2 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N5SO2: 442.1337, found: 442.1341.
2.2.3.12 4-(2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazol-4-yl)benzonitrile (4i). Chemical formula: C25H19N5S; light orange powder (0.2746 g); yield of 96.54%; melting point: 273–274 °C; Rf = 0.50 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3242 (–CH stretching vibration); 3123, 3054 (–CH3); 2986 (–CH2–); 2888 (CH aromatic); 2224 (–C
3); IR (λmax, cm−1, KBr): 3242 (–CH stretching vibration); 3123, 3054 (–CH3); 2986 (–CH2–); 2888 (CH aromatic); 2224 (–C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N); 1616 (C
N); 1616 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1489 (C
N thiazole); 1489 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1385, 1337 (C–S); 1235, 1193 (C–N); 1087, 1033 (
C aromatic); 1385, 1337 (C–S); 1235, 1193 (C–N); 1087, 1033 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 839, 749 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 5.8 Hz, 3H, –CH2–CH3); 4.47 (q, J = 5.3 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.49 (t, J = 6.5 Hz, 1H); 7.63–7.65 (m, 2H); 7.68 (dd, J1 = 7.0 Hz, J2 = 1.5 Hz, 1H); 7.86–7.88 (m, 3H, Ar–H); 8.04–8.06 (m, 2H, Ar–H); 8.22 (d, J = 6.5 Hz, 1H, Ar–H); 8.25 (s, 1H); 8.38 (s, 1H); 12.11 (s, 1H,
N–NH–); 839, 749 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 5.8 Hz, 3H, –CH2–CH3); 4.47 (q, J = 5.3 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.49 (t, J = 6.5 Hz, 1H); 7.63–7.65 (m, 2H); 7.68 (dd, J1 = 7.0 Hz, J2 = 1.5 Hz, 1H); 7.86–7.88 (m, 3H, Ar–H); 8.04–8.06 (m, 2H, Ar–H); 8.22 (d, J = 6.5 Hz, 1H, Ar–H); 8.25 (s, 1H); 8.38 (s, 1H); 12.11 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.7 (quanternary carbon, C2-thiazole); 148.7 (quanternary carbon, C4-thiazole); 143.3 (–CH
N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.7 (quanternary carbon, C2-thiazole); 148.7 (quanternary carbon, C4-thiazole); 143.3 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.95; 138.8 (quanternary carbon, C4-phenyl); 132.6, 126.1 (quanternary carbon, C4a, 8b-carbazole), 125.3 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.3; 119.0 (quanternary carbon, –C
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.95; 138.8 (quanternary carbon, C4-phenyl); 132.6, 126.1 (quanternary carbon, C4a, 8b-carbazole), 125.3 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.3; 119.0 (quanternary carbon, –C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N); 109.7 (quanternary carbon, C1-phenyl); 109.5; 107.1 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H19N5S: 422.1439, found: 422.1445.
N); 109.7 (quanternary carbon, C1-phenyl); 109.5; 107.1 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H19N5S: 422.1439, found: 422.1445.
2.2.3.13 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(4-fluorophenyl)thiazole (4j). Chemical formula: C24H19FN4S; pale yellow powder (0.1908 g); yield of 68.22%; melting point: 257–258 °C; Rf = 0.75 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3234 (–CH stretching vibration); 3059 (–CH3); 2976, 2932 (–CH2–); 1615 (C
3); IR (λmax, cm−1, KBr): 3234 (–CH stretching vibration); 3059 (–CH3); 2976, 2932 (–CH2–); 1615 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1494 (C
N thiazole); 1494 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1234 (C–N aromatic, Ar–F); 1131 (Ar–F); 1086, 1044 (
C aromatic); 1234 (C–N aromatic, Ar–F); 1131 (Ar–F); 1086, 1044 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 837, 748 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.23–7.26 (m, 3H); 7.29 (s, 1H, H–C5-thiazole); 7.49 (t, J = 6.4 Hz, 1H); 7.64 (d, J = 6.5 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.86 (dd, J1 = 7.0 Hz, J2 = 1.5 Hz, 1H); 7.89–7.92 (m, 2H (–CH
N–NH–); 837, 748 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.23–7.26 (m, 3H); 7.29 (s, 1H, H–C5-thiazole); 7.49 (t, J = 6.4 Hz, 1H); 7.64 (d, J = 6.5 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.86 (dd, J1 = 7.0 Hz, J2 = 1.5 Hz, 1H); 7.89–7.92 (m, 2H (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– and H–C1-carbazole)); 8.21–8.23 (m, 2H); 8.37 (s, 1H); 12.05 (s, 1H,
N– and H–C1-carbazole)); 8.21–8.23 (m, 2H); 8.37 (s, 1H); 12.05 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.5 (quanternary carbon, C2-thiazole); 160.7 (quanternary carbon, –CF3); 143.0 (–CH
N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.5 (quanternary carbon, C2-thiazole); 160.7 (quanternary carbon, –CF3); 143.0 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 131.3; 127.5 (quanternary carbon, C1-phenyl); 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.4 (quanternary carbon, C4b-carbazole); 123.5 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.5; 119.6; 119.2; 115.4; 115.3; 109.6; 109.4; 103.0 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SF: 415.1392, found: 415.1397.
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 131.3; 127.5 (quanternary carbon, C1-phenyl); 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.4 (quanternary carbon, C4b-carbazole); 123.5 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.5; 119.6; 119.2; 115.4; 115.3; 109.6; 109.4; 103.0 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SF: 415.1392, found: 415.1397.
2.2.3.14 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(p-tolyl)thiazole (4k). Chemical formula: C25H22N4S; brown powder (0.2101 g); yield of 75.85%; melting point: 279–280 °C; Rf = 0.65 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3240 (–CH stretching vibration); 3045 (–CH3); 2976 (–CH2–); 2756, 1491 (Ar-CH3); 1617 (C
3); IR (λmax, cm−1, KBr): 3240 (–CH stretching vibration); 3045 (–CH3); 2976 (–CH2–); 2756, 1491 (Ar-CH3); 1617 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1491 (C
N thiazole); 1491 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1233 (C–N); 1087, 1034 (
C aromatic); 1233 (C–N); 1087, 1034 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 814, 743 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 2.33 (s, 3H, phenyl–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.22–7.26 (m, 4H, H–C5-thiazole; H–C6-carbazole; H–C3,C5-tolyl); 7.48–7.50 (m, 1H); 7.64 (d, J = 6.5 Hz,1H); 7.68 (d, J = 7.0 Hz, 1H); 7.75 (d, J = 7.0 Hz, 2H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 2H); 8.24 (s, 1H); 8.38 (d, J = 1.5 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.3 (quanternary carbon, C2-thiazole); 143.1 (–CH
N–NH–); 814, 743 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 2.33 (s, 3H, phenyl–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.22–7.26 (m, 4H, H–C5-thiazole; H–C6-carbazole; H–C3,C5-tolyl); 7.48–7.50 (m, 1H); 7.64 (d, J = 6.5 Hz,1H); 7.68 (d, J = 7.0 Hz, 1H); 7.75 (d, J = 7.0 Hz, 2H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 2H); 8.24 (s, 1H); 8.38 (d, J = 1.5 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.3 (quanternary carbon, C2-thiazole); 143.1 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 140.0; 136.8 (quanternary carbon, C4-phenyl); 129.1 (quanternary carbon, C1-phenyl); 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.5; 125.4 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.6; 119.2; 109.7; 109.4; 102.3 (C5-thiazole); 56.0; 37.1 (–CH2–CH3); 20.8 (–CH3); 18.5; 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H22N4S: 411.1643, found: 411.1651.
N–); 140.2 (quanternary carbon, C9a-carbazole); 140.0; 136.8 (quanternary carbon, C4-phenyl); 129.1 (quanternary carbon, C1-phenyl); 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.5; 125.4 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.6; 119.2; 109.7; 109.4; 102.3 (C5-thiazole); 56.0; 37.1 (–CH2–CH3); 20.8 (–CH3); 18.5; 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H22N4S: 411.1643, found: 411.1651.
2.2.3.15 4-(2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazol-4-yl)-2-(hydroxymethyl)phenol (4l). Chemical formula: C25H22N4O2S; stone brown powder (0.2432 g); yield of 81.45%; melting point: 280–281 °C; Rf = 0.22 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 5
Vethyl acetate = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5); IR (λmax, cm−1, KBr): 3417, 3309 (–OH, –CH stretching vibration); 3064 (-CH3); 2978, 2905 (–CH2–); 1610 (C
5); IR (λmax, cm−1, KBr): 3417, 3309 (–OH, –CH stretching vibration); 3064 (-CH3); 2978, 2905 (–CH2–); 1610 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1498 (C
N thiazole); 1498 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1388 (–CH2–OH); 1268, 1233, 1205 (Ar–OH, C–N); 1092, 1031 (
C aromatic); 1388 (–CH2–OH); 1268, 1233, 1205 (Ar–OH, C–N); 1092, 1031 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–; –CH2–OH); 876, 807, 738 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 4.53 (s, 2H, –CH2OH); 6.82 (d, J = 7.0 Hz, 1H, H–C6-phenyl); 7.02 (s, 1H, –CH2OH); 7.25 (t, J = 6.1 Hz, 1H); 7.50 (t, J = 7.0 Hz, 1H); 7.54 (dd, J1 = 7.0 Hz, J2 = 2.0 Hz, 1H); 7.64 (d, J = 6.5 Hz, 1H); 7.69 (d, J = 7.0 Hz, 1H); 7.83 (d, J = 1.5 Hz, 1H); 7.88 (d, J = 6.5 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.28 (s, 1H); 8.40 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.1 (quanternary carbon, C2-thiazole); 154.2 (quanternary carbon, C1-phenyl); 140.3 (–CH
N–NH–; –CH2–OH); 876, 807, 738 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 4.53 (s, 2H, –CH2OH); 6.82 (d, J = 7.0 Hz, 1H, H–C6-phenyl); 7.02 (s, 1H, –CH2OH); 7.25 (t, J = 6.1 Hz, 1H); 7.50 (t, J = 7.0 Hz, 1H); 7.54 (dd, J1 = 7.0 Hz, J2 = 2.0 Hz, 1H); 7.64 (d, J = 6.5 Hz, 1H); 7.69 (d, J = 7.0 Hz, 1H); 7.83 (d, J = 1.5 Hz, 1H); 7.88 (d, J = 6.5 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.28 (s, 1H); 8.40 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.1 (quanternary carbon, C2-thiazole); 154.2 (quanternary carbon, C1-phenyl); 140.3 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.0 (quanternary carbon, C9a-carbazole); 128.7 (quanternary carbon, C2-phenyl); 126.2 (quanternary carbon, C4a, 8a-carbazole); 125.1 (quanternary carbon, C4b-carbazole); 124.9 (quanternary carbon, C4-phenyl); 123.7 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.8; 119.3; 114.6; 109.7; 109.4; 100.3 (C5-thiazole); 58.2 (–CH2–OH); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H22N4SO2: 443.1541, found: 443.1538.
N–); 140.0 (quanternary carbon, C9a-carbazole); 128.7 (quanternary carbon, C2-phenyl); 126.2 (quanternary carbon, C4a, 8a-carbazole); 125.1 (quanternary carbon, C4b-carbazole); 124.9 (quanternary carbon, C4-phenyl); 123.7 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.8; 119.3; 114.6; 109.7; 109.4; 100.3 (C5-thiazole); 58.2 (–CH2–OH); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H22N4SO2: 443.1541, found: 443.1538.In addition, tandem mass spectrometry (MS/MS) analysis was performed for compound 4l. The MS/MS spectrum of the [M + H]+ ion (m/z 443.1560) exhibited characteristic product ions at m/z = 223.1244, 167.0725, 196.1118, 203.0277, and 205.0438…, which are consistent with the proposed structure of compound 4l (Fig. S106).
2.2.3.16 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(2-fluorophenyl)thiazole (4m). Chemical formula: C24H19FN4S; tortila powder (0.1967 g); yield of 70.33%; melting point: 242–243 °C; Rf = 0.67 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3401 (–CH stretching vibration); 3059 (–CH3); 2976 (–CH2–); 1619 (C
3); IR (λmax, cm−1, KBr): 3401 (–CH stretching vibration); 3059 (–CH3); 2976 (–CH2–); 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1486 (C
N thiazole); 1486 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1338, 1274, 1235, 1125, 1084, 1021 (Ar–F, C–N,
C aromatic); 1338, 1274, 1235, 1125, 1084, 1021 (Ar–F, C–N, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 872, 804, 752 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.23–7.25 (m, 2H, H–C5-thiazole; H–C5-phenyl); 7.28–7.31 (m, 2H); 7.35–7.39 (m, 1H); 7.47–7.50 (m, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.0 Hz, 1H); 7.87 (dd, J1 = 6.5 Hz, J2 = 1.5 Hz, 1H); 8.01–8.04 (m, 1H, –CH
N–NH–); 872, 804, 752 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.23–7.25 (m, 2H, H–C5-thiazole; H–C5-phenyl); 7.28–7.31 (m, 2H); 7.35–7.39 (m, 1H); 7.47–7.50 (m, 1H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.0 Hz, 1H); 7.87 (dd, J1 = 6.5 Hz, J2 = 1.5 Hz, 1H); 8.01–8.04 (m, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.38 (d, J = 1.0 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 167.7 (quanternary carbon, C2-thiazole); 160.3 (quanternary carbon, C–F); 158.7 (quanternary carbon, C4-thiazole); 143.3; 140.2 (–CH
N–); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.38 (d, J = 1.0 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 167.7 (quanternary carbon, C2-thiazole); 160.3 (quanternary carbon, C–F); 158.7 (quanternary carbon, C4-thiazole); 143.3; 140.2 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 139.9 (quanternary carbon, C9a-carbazole); 129.3, 126.1 (quanternary carbon, C4a, C8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole); 124.7 (quanternary carbon, C3-carbazole); 123.6, 122.3 (quanternary carbon, C1-phenyl); 122.1; 120.6; 119.7; 119.3; 116.1; 115.9; 109.7; 109.4; 108.0; 107.9 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SF: 415.1392, found: 415.1392.
N–); 139.9 (quanternary carbon, C9a-carbazole); 129.3, 126.1 (quanternary carbon, C4a, C8a-carbazole); 125.3 (quanternary carbon, C4b-carbazole); 124.7 (quanternary carbon, C3-carbazole); 123.6, 122.3 (quanternary carbon, C1-phenyl); 122.1; 120.6; 119.7; 119.3; 116.1; 115.9; 109.7; 109.4; 108.0; 107.9 (C5-thiazole); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H19N4SF: 415.1392, found: 415.1392.
2.2.3.17 4-(3,4-Dichlorophenyl)-2-(2-((9-ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazole (4n). Chemical formula: C24H18Cl2N4S; colorless amorphous powder (0.2591 g); yield of 82.63%; melting point: 240–241 °C; Rf = 0.68 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3237 (–CH stretching vibration); 3050 (–CH3); 2977 (–CH2–); 1619 (C
3); IR (λmax, cm−1, KBr): 3237 (–CH stretching vibration); 3050 (–CH3); 2977 (–CH2–); 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1489 (C
N thiazole); 1489 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1233 (C–N aromatic); 1136 (C–N); 934, 752 (aromatic ring bearing substituents); 633 (Ar–Cl). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.3 Hz, 1H); 7.49 (t, J = 6.5 Hz, 1H); 7.54 (s, 1H, H–C5-thiazole); 7.64 (d, J = 6.5 Hz, 1H); 7.63–7.68 (m, 2H); 7.84–7.87 (m, 2H); 8.09 (d, J = 2.0 Hz, 1H, –CH
C aromatic); 1233 (C–N aromatic); 1136 (C–N); 934, 752 (aromatic ring bearing substituents); 633 (Ar–Cl). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.33 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.3 Hz, 1H); 7.49 (t, J = 6.5 Hz, 1H); 7.54 (s, 1H, H–C5-thiazole); 7.64 (d, J = 6.5 Hz, 1H); 7.63–7.68 (m, 2H); 7.84–7.87 (m, 2H); 8.09 (d, J = 2.0 Hz, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 8.22 (d, J = 6.5 Hz, 1H); 8.23 (s, 1H); 8.38 (d, J = 1.0 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 147.8 (quanternary carbon, C4-thiazole); 143.3 (–CH
N–); 8.22 (d, J = 6.5 Hz, 1H); 8.23 (s, 1H); 8.38 (d, J = 1.0 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 147.8 (quanternary carbon, C4-thiazole); 143.3 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 135.2 (quanternary carbon, C4-phenyl); 131.4 (quanternary carbon, C3-phenyl); 130.8 (quanternary carbon, C1-phenyl); 129.6; 127.1, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.5 (quanternary carbon, C4b-carbazole); 125.3 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.2; 109.7; 109.4; 105.5 (C5-thiazole); 56.0; 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H18N4SCl2: 465.0707, found: 465.0702.
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 135.2 (quanternary carbon, C4-phenyl); 131.4 (quanternary carbon, C3-phenyl); 130.8 (quanternary carbon, C1-phenyl); 129.6; 127.1, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.5 (quanternary carbon, C4b-carbazole); 125.3 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6; 119.7; 119.2; 109.7; 109.4; 105.5 (C5-thiazole); 56.0; 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H18N4SCl2: 465.0707, found: 465.0702.
2.2.3.18 4-(3-Chloro-4-fluorophenyl)-2-(2-((9-ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazole (4o). Chemical formula: C24H18ClFN4S; light brown powder (0.2532 g); yield of 83.64%; melting point: 257–258 °C; Rf = 0.63 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3050 (–CH3); 2977 (–CH2–); 1613 (C
3); IR (λmax, cm−1, KBr): 3050 (–CH3); 2977 (–CH2–); 1613 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1482 (C
N thiazole); 1482 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1234, 1135 (C–N); 1087 (Ar–F); 1022 (
C aromatic); 1234, 1135 (C–N); 1087 (Ar–F); 1022 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 880, 814, 750 (aromatic ring bearing substituents); 676, 614 (Ar–Cl). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.45–7.50 (m, 3H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.85–7.89 (m, 2H); 8.05 (dd, J1 = 6.0 Hz, J2 = 1.5 Hz, 1H, –CH
N–NH–); 880, 814, 750 (aromatic ring bearing substituents); 676, 614 (Ar–Cl). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.24 (t, J = 6.0 Hz, 1H); 7.45–7.50 (m, 3H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.85–7.89 (m, 2H); 8.05 (dd, J1 = 6.0 Hz, J2 = 1.5 Hz, 1H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 8.22 (d, J = 7 Hz, 1H); 8.23 (s, 1H); 8.38 (s, 1H); 12.05 (s, 1H,
N–); 8.22 (d, J = 7 Hz, 1H); 8.23 (s, 1H); 8.38 (s, 1H); 12.05 (s, 1H, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 157.4; 155.5 (quanternary carbon, C–F); 148.1 (quanternary carbon, C4-thiazole); 143.0 (–CH
N–NH–). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.6 (quanternary carbon, C2-thiazole); 157.4; 155.5 (quanternary carbon, C–F); 148.1 (quanternary carbon, C4-thiazole); 143.0 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 132.7 (quanternary carbon, C1-phenyl); 127.3, 126.1 (quanternary carbon, C4a, 8a-carbazole); 126.0; 125.9; 125.3 (quanternary carbon, C4b-carbazole); 123.5 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.5 (quanternary carbon, C3-phenyl); 119.8; 119.6; 119.2; 117.2; 117.0; 109.6; 109.4; 104.4 (C5-thiazole); 37.1 (–CH2–CH3); 13.70 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H18N4SClF: 449.1003, found: 449.1002.
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9; 132.7 (quanternary carbon, C1-phenyl); 127.3, 126.1 (quanternary carbon, C4a, 8a-carbazole); 126.0; 125.9; 125.3 (quanternary carbon, C4b-carbazole); 123.5 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.5 (quanternary carbon, C3-phenyl); 119.8; 119.6; 119.2; 117.2; 117.0; 109.6; 109.4; 104.4 (C5-thiazole); 37.1 (–CH2–CH3); 13.70 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C24H18N4SClF: 449.1003, found: 449.1002.
2.2.3.19 4-(Benzo[d][1,3]dioxol-5-yl)-2-(2-((9-ethyl-9H-carbazol-3-yl)methylene)hydrazone)thiazole (4p). Chemical formula: C25H20N4O2S; brown powder (0.2167 g); yield of 72.88%; melting point: 232–233 °C; Rf = 0.56 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3294 (–CH stretching vibration); 3045 (–CH3); 2977 (–CH2–); 1626 (C
3); IR (λmax, cm−1, KBr): 3294 (–CH stretching vibration); 3045 (–CH3); 2977 (–CH2–); 1626 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1478 (C
N thiazole); 1478 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1272 (C–N aromatic); 1237 (C–N aromatic); 1116 (Ar–O–CH2–); 1046 (
C aromatic); 1272 (C–N aromatic); 1237 (C–N aromatic); 1116 (Ar–O–CH2–); 1046 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 927 (benzo[d][1,3]dioxole); 876, 819 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 6.01 (s, 2H, –O–CH2–O–); 6.95–6.97 (m, 1H); 7.19 (s, 1H, H–C5-thiazole); 7.25 (t, J = 6.3 Hz, 1H); 7.40–7.41 (m, 2H); 7.49 (t, J = 6.3 Hz, 1H); 7.64 (d, J = 6.5 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H), 8.25 (s, 1H); 8.38 (d, J = 1.0 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.2 (quanternary carbon, C2-thiazole); 147.6 (quanternary carbon, C3a–O–); 146.8 (quanternary carbon, C7a–O–), 143.3 (–CH
N–NH–); 927 (benzo[d][1,3]dioxole); 876, 819 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 6.01 (s, 2H, –O–CH2–O–); 6.95–6.97 (m, 1H); 7.19 (s, 1H, H–C5-thiazole); 7.25 (t, J = 6.3 Hz, 1H); 7.40–7.41 (m, 2H); 7.49 (t, J = 6.3 Hz, 1H); 7.64 (d, J = 6.5 Hz, 1H); 7.68 (d, J = 7.5 Hz, 1H); 7.87 (dd, J1 = 7.3 Hz, J2 = 1.3 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H), 8.25 (s, 1H); 8.38 (d, J = 1.0 Hz, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 168.2 (quanternary carbon, C2-thiazole); 147.6 (quanternary carbon, C3a–O–); 146.8 (quanternary carbon, C7a–O–), 143.3 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9, 126.1 (quanternary carbon, C4a, 8a-carbazole; C5-phenyl); 125.3 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.0; 120.6; 119.7; 119.4; 119.2; 109.7; 109.4, 108.3; 105.9 (C5-thiazole); 101.9; 101.4 (–O–CH2–O–); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H20N4SO2: 441.1385, found: 441.1388.
N–); 140.2 (quanternary carbon, C9a-carbazole); 139.9, 126.1 (quanternary carbon, C4a, 8a-carbazole; C5-phenyl); 125.3 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.0; 120.6; 119.7; 119.4; 119.2; 109.7; 109.4, 108.3; 105.9 (C5-thiazole); 101.9; 101.4 (–O–CH2–O–); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H20N4SO2: 441.1385, found: 441.1388.
2.2.3.20 2-(2-((9-Ethyl-9H-carbazol-3-yl)methylene)hydrazone)-4-(5-fluoro-2-methoxyphenyl)thiazole (4q). Chemical formula: C25H21FN4OS; light brown powder (0.1896 g); yield of 63.19%; melting point: 261–262 °C; Rf = 0.60 (Vhexane
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Vethyl acetate = 7
Vethyl acetate = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3); IR (λmax, cm−1, KBr): 3110 (–CH stretching vibration); 3035 (–CH3); 2972, 2886 (–CH2–); 1611 (C
3); IR (λmax, cm−1, KBr): 3110 (–CH stretching vibration); 3035 (–CH3); 2972, 2886 (–CH2–); 1611 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N azomethine, C
N azomethine, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N thiazole); 1499, 1480 (C
N thiazole); 1499, 1480 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic); 1242 (Ar–O–CH3) 1242 (C–N); 1088 (Ar–F); 1033 (
C aromatic); 1242 (Ar–O–CH3) 1242 (C–N); 1088 (Ar–F); 1033 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH–); 860, 826, 768 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 3.92 (s, 3H, –O–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.12–7.14 (m, 2H, H–C5-thiazole; H–C3-phenyl); 7.24 (t, J1 = 6.1 Hz, 1H); 7.47–7.50 (m, 2H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.0 Hz, 1H); 7.77–7.79 (m, 1H); 7.87 (dd, J1 = 6.5 Hz, J2 = 1.5 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.38 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 166.9 (quanternary carbon, C2-thiazole); 155.4 (quanternary carbon, C5-phenyl); 153.0 (quanternary carbon, C2-phenyl); 143.0 (–CH
N–NH–); 860, 826, 768 (aromatic ring bearing substituents). 1H-NMR (500 MHz, DMSO-d6, 25 °C) δ (ppm): 1.34 (t, J = 6.0 Hz, 3H, –CH2–CH3); 3.92 (s, 3H, –O–CH3); 4.47 (q, J = 6.0 Hz, 2H, –CH2–CH3); 7.12–7.14 (m, 2H, H–C5-thiazole; H–C3-phenyl); 7.24 (t, J1 = 6.1 Hz, 1H); 7.47–7.50 (m, 2H); 7.64 (d, J = 7.0 Hz, 1H); 7.68 (d, J = 7.0 Hz, 1H); 7.77–7.79 (m, 1H); 7.87 (dd, J1 = 6.5 Hz, J2 = 1.5 Hz, 1H); 8.22 (d, J = 6.5 Hz, 1H); 8.25 (s, 1H); 8.38 (s, 1H). 13C-NMR (125 MHz, DMSO-d6, 25 °C) δ (ppm): 166.9 (quanternary carbon, C2-thiazole); 155.4 (quanternary carbon, C5-phenyl); 153.0 (quanternary carbon, C2-phenyl); 143.0 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–); 140.2 (quanternary carbon, C9a-carbazole); 140.0, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.4 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6 (quanternary carbon, C1-phenyl); 119.6; 119.3; 114.9; 114.7; 113.0; 109.7; 109.4; 108.7 (C5-thiazole); 56.0 (–O–CH3); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H21N4SOF: 445.1498, found: 445.1504.
N–); 140.2 (quanternary carbon, C9a-carbazole); 140.0, 126.1 (quanternary carbon, C4a, 8a-carbazole); 125.4 (quanternary carbon, C4b-carbazole); 123.6 (quanternary carbon, C3-carbazole); 122.3; 122.1; 120.6 (quanternary carbon, C1-phenyl); 119.6; 119.3; 114.9; 114.7; 113.0; 109.7; 109.4; 108.7 (C5-thiazole); 56.0 (–O–CH3); 37.1 (–CH2–CH3); 13.7 (–CH2–CH3). HR-MS (ESI+): m/z [M + H]+ calcd for C25H21N4SOF: 445.1498, found: 445.1504.The thiazole derivatives 4a–q were synthesised via a cyclisation reaction between compound (3) and various α-bromoketones in ethanol, yielding pure powders characterised by 1H-NMR and 13C-NMR spectra consistent with the expected substituents. All derivatives exhibited a characteristic triplet signal of one proton at around δ ∼ 7.24 ppm corresponding to the CH-thiazole moiety, confirming successful thiazole ring closure. The products were obtained in high purity with small melting point variations. However, in several derivatives, the 1H-NMR signal of the ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–NH– proton (δ ∼ 12 ppm) disappeared due to its high mobility and possible exchange with protons from the NMR solvent, especially in the presence of trace water. Therefore, future studies on similar derivatives should take note of this proton's instability. The spectral data and yields of each compound are described in the following section.
N–NH– proton (δ ∼ 12 ppm) disappeared due to its high mobility and possible exchange with protons from the NMR solvent, especially in the presence of trace water. Therefore, future studies on similar derivatives should take note of this proton's instability. The spectral data and yields of each compound are described in the following section.
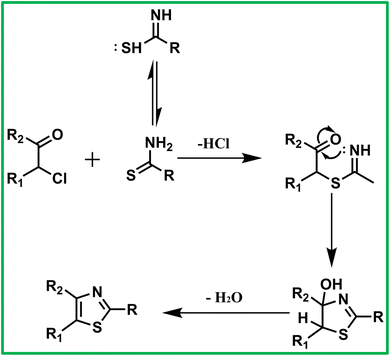
In principle, reactions between sulfur-containing nucleophiles and α-bromoketones may proceed via two competing pathways corresponding to nucleophilic attack by either sulfur or nitrogen at the activated α-carbon center. However, under the present reaction conditions, the sulfur-alkylation pathway is strongly favored. The α-carbon of the α-bromoketone is a soft and highly activated electrophilic site due to the adjacent carbonyl group, making it preferentially susceptible to attack by sulfur, a soft and highly polarizable nucleophile. Moreover, the use of ethanol as a protic solvent significantly reduces the nucleophilicity of nitrogen through hydrogen-bonding interactions, thereby suppressing N-alkylation. The absence of strong bases further limits nitrogen activation while having minimal impact on sulfur reactivity. As a result, the reaction proceeds regioselectively via initial S-alkylation followed by intramolecular cyclization to afford the thermodynamically favored aromatic 1,3-thiazole, consistent with the experimental observation of a single product by TLC and clean 1H and 13C NMR spectra.
The chemical purity of representative compounds 4e, 4f, 4g, and 4l was evaluated by analytical HPLC prior to biological studies. The HPLC chromatograms (Fig. S102–S105) revealed a single predominant peak for each compound, with relative peak areas of 98.64% (4e), 98.66% (4f), 95.64% (4g), and 94.37% (4l), respectively. These results demonstrate that the synthesized compounds possess high chemical purity and confirm that the applied synthetic and purification procedures consistently afford compounds suitable for reliable in vitro and in silico biological evaluation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well for MCF-7, HepG2, and HeLa, and 7500 cells per well for Human non-small cell lung carcinoma cell line (NCI-H460). After incubation for 24 h, cells were treated with different concentrations of the test compounds and incubated for a further 48 h. The cells were then fixed with cold 50% (w/v) trichloroacetic acid (Merck) for 1–3 h, rinsed with distilled water, and stained with 0.2% (w/v) SRB (Sigma) for 20 min. Excess dye was removed by washing five times with 1% acetic acid (Merck), and the bound dye was solubilised in 10 mM Tris base (Promega).30 Absorbance was measured at 492 and 620 nm using a Synergy HT microplate reader (BioTek Instruments). The percentage of growth inhibition (Inh %) was calculated using the equation: Inh % = (1 − [Odt/Odc]) × 100, where Odt and Odc represent the optical density values of the test and control samples, respectively. Ellipticine served as the positive control. Cytotoxicity assays were performed in three independent experiments, each conducted with three technical replicates per concentration. IC50 values were calculated from the averaged data and are reported as mean ± SD.
000 cells per well for MCF-7, HepG2, and HeLa, and 7500 cells per well for Human non-small cell lung carcinoma cell line (NCI-H460). After incubation for 24 h, cells were treated with different concentrations of the test compounds and incubated for a further 48 h. The cells were then fixed with cold 50% (w/v) trichloroacetic acid (Merck) for 1–3 h, rinsed with distilled water, and stained with 0.2% (w/v) SRB (Sigma) for 20 min. Excess dye was removed by washing five times with 1% acetic acid (Merck), and the bound dye was solubilised in 10 mM Tris base (Promega).30 Absorbance was measured at 492 and 620 nm using a Synergy HT microplate reader (BioTek Instruments). The percentage of growth inhibition (Inh %) was calculated using the equation: Inh % = (1 − [Odt/Odc]) × 100, where Odt and Odc represent the optical density values of the test and control samples, respectively. Ellipticine served as the positive control. Cytotoxicity assays were performed in three independent experiments, each conducted with three technical replicates per concentration. IC50 values were calculated from the averaged data and are reported as mean ± SD.2.2.8.1 Molecular docking. The five candidated 4a, 4e, 4f, 4g, 4l have conducted in silico to 4 file target enzymes: such as 4J5T, 2VF5, 4WCU and 1T8I to explain why compound has exposed inhibits diabetes (4J5T), inhibits cancer (1T8I), inhibits anti-inflammatory (4WCU), antibacterial (2VF5) in silico. For docking parameters such as 4J5T.pdb (spacing: 0.600, the numbers of elements (50, 50, 50), and grid box (−18.417, −20.925, 8.009)); 2VF5.pdb: pdb (spacing: 0.500, the numbers of elements (50, 50, 50), and grid box (26.577, 22.697, 8.113)); 4WCU.pdb: (spacing: 0.600, the numbers of elements (50, 50, 50), and grid box (29.286, −59.727, −26.674)); 1T8I.pdb: spacing: 0.600, the numbers of elements (60, 60, 60), and grid box (20.968, 0.667, 41.024)); procedure docking of conformation of compounds: 4a, 4e, 4f, 4g, 4l has followed Scheme S1. The coordinations of active center of enzyme has bene detected by Discovery Studio Visualizer software via article. The target enzymes before processing docking has been checked the residual amino acid on chains of enzyme by Swiss-pdb viewer software. For one target ezyme has removed the water molecules and heteroatoms and saves as by targer.pdb. For structures of ligands, it is optimized by Avogadro software by MMFF94 force field and saved by *.pdb format before reading to Autodock tool. The parameters of docking processing are examinated and selected depend on size of ligand molecule and saved in dock.gpf file. The docking parameters are selected by the numbers of running of 1000 times, docking method of Genetic Algorithm and out put in file dock.dpf.37–44
2.2.8.2 Prediction of drug-likeness and ADMET analysis. The drug-likeness properties and absorption, distribution, metabolism, excretion, and toxicity (ADMET) profiles of the synthesised compounds were evaluated using the online platform ADMETlab2.0. Based on Lipinski's Rule of Five, which serves as a common guideline for predicting oral bioavailability, key physicochemical parameters, such as molecular weight, log
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, and the number of hydrogen bond donors and acceptors, were analysed. These computational tools offer a preliminary yet dependable understanding of the pharmacokinetic properties and safety of prospective drug candidates, markedly decreasing both time and expense relative to conventional methods.45
P, and the number of hydrogen bond donors and acceptors, were analysed. These computational tools offer a preliminary yet dependable understanding of the pharmacokinetic properties and safety of prospective drug candidates, markedly decreasing both time and expense relative to conventional methods.45
2.2.8.3 Molecular dynamics (MD) simulation. To assess the stability and long-term interactions between the investigated compounds and their target enzymes, molecular dynamics (MD) simulations were performed using Desmond (Schrödinger LLC) on a Linux environment (Scheme S2). The optimal ligand–protein complex obtained from docking studies was used as the initial input structure. The system was solvated in a TIP3P water model and enclosed in an orthorhombic box with a 10 Å buffer around the complex. It was then neutralised by adding Na+ or Cl− ions and adjusted to a physiological salt concentration of 0.15 M. Energy minimisation was performed to remove steric clashes, followed by equilibration under the NVT (constant volume and temperature) and NPT (constant pressure and temperature) ensembles at 300 K and 1.01325 bar, using the RESPA integrator with a 2 fs time step.46 The MD simulation was run for 100 nanoseconds (ns) and comprised approximately 63
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649 atoms, including the protein, ligand, counter ions, and 16
649 atoms, including the protein, ligand, counter ions, and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 882 water molecules. The resulting trajectories were analysed to calculate parameters such as root mean square deviation (RMSD), radius of gyration (Rg), and the number of hydrogen bonds between the ligand and protein over time, which provided information about the stability and binding persistence of the ligand within the active site of the target protein.
882 water molecules. The resulting trajectories were analysed to calculate parameters such as root mean square deviation (RMSD), radius of gyration (Rg), and the number of hydrogen bonds between the ligand and protein over time, which provided information about the stability and binding persistence of the ligand within the active site of the target protein.
3. Results and discussion
3.1. Antibacterial activity
The derivatives 4a–q were evaluated for their inhibitory activity against bacterial and fungal strains, including Escherichia coli ATCC 8739, Staphylococcus aureus ATCC 6538, Salmonella typhimurium ATCC 14028, and Candida albicans ATCC 10231. The results showed that compounds 4a, 4c, 4e, 4h, 4k, 4n, 4o, and 4p formed clear inhibition zones on agar plates (Fig. S82–S85). In the sterile ring radius measurement test on agar plates, it was found that for Escherichia coli, Staphylococcus aureus, and Candida albicans, compound 4o at a concentration of 250 µg per well showed the best results with diameters of 8.5 and 10.5 mm, respectively, among the derivatives, while chloramphenicol showed diameters of 22, 26, and 18.8 mm, respectively. For Salmonella typhimurium, compounds 4a, 4e, 4h, and 4n all showed a radius of 8 mm, while chloramphenicol showed a radius of 26 mm. The antibacterial ability, based on the MIC value (Fig. 1), can be compared as follows: 4h < 4p < 4o < 4a < 4k < 4c < 4n < 4e < positive control. Compound 4e, bearing a chloro substituent, demonstrated the highest antibacterial activity within the series, with an MIC = 83.5 µM against the tested bacterial and fungal strains. Although this activity is moderate, it suggests a beneficial contribution of the chlorine atom, likely enhancing membrane interaction and inhibitory behaviour.47 The antibacterial activity of compound 4e, when compared with recently reported thiazole derivatives, remains relatively modest. For example, Sateesh Kuna et al. (2023) synthesized a series of coumarin-containing thiazole derivatives that showed strong antibacterial activity, with MIC values ranging from 7.69 to 22.76 µM.48 The lower activity observed for 4e may reflect limited membrane permeability, indicating that halogenation remains a promising structure–activity relationship optimisation (SAR) direction for designing analogues with improved antibacterial performance. | ||
| Fig. 1 MIC values of substances exhibiting antimicrobial activity: (a) Escherichia coli, (b) Staphylococcus aureus, (c) Salmonella typhimurium, (d) Candida albicans. | ||
3.2. Cytotoxic activity
The anticancer activity of the compounds 4a–q was evaluated against A549, HepG2, and MCF-7 cell lines using the SRB assay, with ellipticine as the reference drug (Fig. S90–S92). Five compounds (4a, 4e, 4g, 4i, 4j) shown significant cytotoxic activity against HepG2 and MCF-7 cell lines. In terms of activity, a comparison based on IC50 values can be made as follows: for MCF-7: 4i < 4j < 4e < 4a < 4g < ellipticine; for HepG2: 4i < 4e < 4j < 4a < 4g < ellipticine; and for A549: 4a < 4g < ellipticine (Fig. 2). Compound 4g showed the most potent cytotoxic activity, with IC50 values of 2.6 ± 0.2 µM for HepG2, 6.4 ± 0.7 µM for MCF-7, and 6.6 ± 0.6 µM for A549, although still less active than Ellipticine (IC50: 0.3–0.4 µM). These findings suggest that the nitro substituent present in 4g may play a critical role in enhancing its affinity toward molecular targets involved in cancer cell proliferation and survival, thereby improving its cytotoxic efficacy.49 In comparison to prior publications on thiazole-based anticancer medicines, the cytotoxic efficacy of compound 4g was deemed promising. Hassan et al. (2023) reported thiazole derivatives with IC50 values of 34–92 µM against A549 and 37–80 µM against MCF-7, relative to erlotinib (IC50 = 30 and 40 µM, respectively).50 Likewise, Kuzu et al. (2025) described a structurally distinct thiazole library exhibiting a broad potency range, with IC50 values of 9.2–182.2 µM (MCF-7), 25.2–195.7 µM (A549), and 7.2–168.0 µM (HepG2), compared to doxorubicin (IC50 = 0.6–1.3 µM).51 In comparison to the reference data, compound 4g demonstrated considerable potency, attaining IC50 values in the low micromolar range across all examined cell lines. The results indicate that 4g is a promising lead compound for subsequent optimization studies, especially with adjustments to solubility and cell membrane permeability. | ||
| Fig. 2 The IC50 values of the derivatives suggest potential toxicity for the three cancer cell lines: MCF-7, HepG2, and A549 analysed. | ||
3.3. Anti-inflammatory activity
The anti-inflammatory activity of the compounds 4a–q was assessed via their ability to suppress nitric oxide (NO) production in LPS-stimulated RAW 264.7 macrophages, using dexamethasone as the reference drug. All compounds exhibited dose-dependent NO inhibition, with seven derivatives (4a, 4d, 4e, 4f, 4g, 4o, and 4p) for adequate inhibitory capacity (Fig. S93). The IC50 values ranged from 20.5 to 15.4 µM (Fig. 3), while dexamethasone displayed an IC50 value of 13.7 ± 1.2 µM. Comparing the anti-inflammatory activity of compounds based on IC50 values reveals the following order: 4o < 4d < 4p < 4a < 4e < 4f < dexamethasone. Notably, compound 4f demonstrated the most potent activity (IC50 = 15.4 ± 1.0 µM), comparable to dexamethasone, while maintaining excellent cell viability (99.8% – Fig. S94). From a SAR perspective, the enhanced potency of 4f can be attributed to the presence of a bromine substituent, which facilitates σ-hole/halogen bonding interactions and improves lipophilicity, thereby supporting membrane penetration and efficient engagement with the iNOS active site.52 | ||
| Fig. 3 The IC50 values of the derivatives indicate strong inhibition of the inflammatory mediator nitric oxide (NO). | ||
In comparison to previously published thiazole derivatives, the performance of 4f is favourable. For instance, Fang et al. (2024) reported compound 12b with an IC50 of 2.6 µM, which surpasses that of methylprednisolone.53 While Lin et al. (2024) documented thiazole derivatives exhibiting IC50 values of 11.0–61.5 µM, compared to dexamethasone (IC50 = 21.2 µM).54 Compound 4f was recognised as a promising derivative for the development of anti-inflammatory medications, possessing the capacity to refine its structure to augment desired efficacy and solubility.
3.4. α-Glucosidase inhibitory activity
The antidiabetic potential of the synthesised derivatives was evaluated based on their α-glucosidase inhibitory activity using p-nitrophenyl-α-D-glucopyranoside (pNPG) as the substrate. All compounds demonstrated concentration-dependent inhibitory activity (Fig. S95), with the inhibitory levels ranked by IC50 in the following order: 4e < 4j < 4o < 4c < 4h < 4m < 4k < 4n < 4q < 4i < acarbose < 4a < 4l, with values ranging from 436 to 41 µM. Remarkably, compounds 4a and 4l, both bearing phenolic hydroxyl groups, exhibited IC50 values of 46 ± 2 µM and 41 ± 2 µM, three times more effective than acarbose (117 ± 8 µM). These findings underscore the key contribution of –OH groups in active-site binding, with the additional –CH2OH moiety in 4l likely providing an extra hydrogen-bonding interaction, accounting for its enhanced affinity.55,56Comparison with recent studies on thiazole derivatives indicates that the inhibitory potential of compounds 4l and 4a is substantial (Fig. 4). Zahra et al. (2023) synthesised various thiazole compounds and documented IC50 values ranging from 99 to 39 µM, whereas acarbose showed an IC50 of 1.21 µM.57 Khan et al. (2023) assembled a variety of thiazole compounds, with IC50 values ranging from 18 to 3 µM. Although some derivatives in Khan's series surpassed acarbose, compound 4l demonstrated threefold more potent inhibition than acarbose under identical assay conditions.58 Likewise, Seliem et al. (2025) reported IC50 values of 112–183 µM, compared with acarbose (IC50 = 394 µM).59 However, their work lacked ADMET and MD validation, limiting drug-development insight. The results indicate that compound 4l represents a promising structure for future antidiabetic drug development targeting type II diabetes.
 | ||
| Fig. 4 The IC50 values of the derivatives demonstrate significant inhibition of the enzyme α-glucosidase. | ||
3.5. Structure–activity relationship (SAR) analysis
Substituents on the phenyl ring at the 4-position of the thiazole core played a pivotal role in modulating the biological activities of the compounds—notably, halogen substituents (Cl, Br) at the para position significantly enhanced activity. Compound 4e (4-Cl) demonstrated the most potent antibacterial effect in the series (MIC = 83.5 µM), while 4n (3,4-Cl) also exhibited favourable antibacterial properties. These findings suggest that chlorine atoms may enhance membrane interactions and bacterial inhibition by increasing lipophilicity and participating in halogen bonding. Similarly, 4f (4-Br) showed the most pronounced anti-inflammatory activity (IC50 ≈ 15.4 µM), comparable to dexamethasone, implying that the bulky bromine substituent may enhance hydrophobic interactions and potentially engage in halogen bonding (σ-hole interactions) with the active site of iNOS.53 These observations align with previous reports indicating that halogenated derivatives (Cl, Br, I) tend to enhance lipid solubility and facilitate halogen bonding with enzyme carbonyl groups, thereby promoting cell membrane penetration and inhibiting microbial growth.60The nitro group (–NO2) also proved highly effective, particularly at the para position. Compound 4g (para-NO2) exhibited the lowest IC50 values across MCF-7, HepG2, and A549 cell lines, significantly outperforming most other derivatives and closely approaching the potency of ellipticine. This enhancement may stem from the strong electron-withdrawing character of the nitro group, which increases the electrophilicity of the conjugated system and favours interactions with biological targets in cancer cells. In contrast, 4h (meta-NO2) displayed the weakest antibacterial activity, underscoring the importance of precise substituent positioning to achieve optimal electronic effects.
Hydroxyl-containing derivatives (–OH), particularly those with phenolic or benzylic hydroxyl groups, showed superior α-glucosidase inhibitory activity. Compounds 4a (para-OH) and 4l (ortho-CH2OH–OH) exhibited IC50 values of 46 µM and 41 µM, respectively, markedly outperforming acarbose (117 µM). The phenolic –OH group provides strong hydrogen bonding capability at the enzyme's active site, while the benzylic –CH2OH moiety in 4l adds additional hydrogen-bonding interactions with surrounding residues, enhancing complex stability.
Other substituents, including –OCH3, –CH3, polycyclic aromatics, –CF3, and –OCF3, generally conferred moderate to weak activity. For instance, compounds 4c (4-OCH3) and 4k (4-CH3) displayed only modest antibacterial or anticancer effects, possibly due to their electron-donating nature, which diminishes the electrophilicity of the conjugated system. Similarly, derivatives with weakly electron-withdrawing groups (e.g., 4b: 4-CF3 and 4d: 4-OCF3) did not show superior activity in any of the biological models tested.
Overall, the SAR trend reveals that electron-withdrawing groups, such as Cl, Br, and NO2, generally enhance antibacterial, anti-inflammatory, and anticancer activity by promoting strong target binding or favorable pharmacokinetic profiles, while electron-donating groups, especially hydroxyls, are more beneficial for α-glucosidase inhibition due to their capacity for hydrogen bonding. These insights provide a solid rationale for future optimisation, recommending halogenation or nitro substitution for antimicrobial and anticancer development, and retention or enhancement of hydroxyl groups for antidiabetic applications.
3.6. In silico docking model
| Entry | Pose | Free energy of bindingb | Kic | The number of hydrogen bondsd | The property and bond lengthd |
|---|---|---|---|---|---|
| a Protein data bank file downloaded from Protein Data Bank.b Calculated using AutoDockTools-1.5.6rc3 and reported in units of kcal mol−1.c Inhibition constants, Ki, in units of µM, derived from free energy of binding (ΔG°).d Based on the Discovery Studio (DSC) 2025 software. | |||||
| 4a | 283 | −8.14 | 1.08 | 0 | — |
| 4l | 612 | −8.56 | 0.53 | 4 | A:Gly312:N-612:O (2.65 Å) |
| A:Lys324:N-612:O (3.11 Å) | |||||
| A:Arg799:N-612:S (3.13 Å) | |||||
| 612:H-A:Leu797:O (2.02 Å) | |||||
| Small ligand in 4J5T | 586 | −4.49 | 513.52 | 3 | 586:H-A:Glu374:O (2.13 Å) |
| 586:H-A:Glu374:O (2.45 Å) | |||||
| Acarbose | 926 | −0.32 | 583.93 × 106 | 3 | A:Thr281:O-926:N (3.18 Å) |
| 926:H-A:Thr281:O (2.37 Å) | |||||
| 926:H-A:Thr 281:O (2.26 Å) | |||||
| 4g | 714 | −7.99 | 1.38 | 2 | A:Arg749:N-714:O (2.90 Å) |
| A:Arg749:N-714:O (3.04 Å) | |||||
| Small ligand in 1T8I | 831 | −7.41 | 3.69 | 2 | 831:H-A:Asp301:O (1.74 Å) |
| 831:H-A:Leu 297:O (1.96 Å) | |||||
| Ellipticine | 585 | −6.98 | 7.71 | 0 | — |
| 4f | 637 | −9.97 | 0.05 | 0 | — |
| Small ligand in 4WCU | 3 | −7.64 | 4.76 | 0 | — |
| Dexamethasone | 410 | −7.18 | 5.46 | 1 | 410:H-B:Pro356:O (1.91 Å) |
| 4e | 674 | −9.54 | 0.10 | 1 | X:Ser349:O-674:Cl (3.10 Å) |
| Small ligand in 2VF5 | 455 | −3.60 | 2.3 × 103 | 0 | — |
| Chloramphenicol | 512 | −4.91 | 251.55 | 3 | X:Gly301:N-512:O (2.81 Å) |
| X:Lys487:N-512:O (2.81 Å) | |||||
| A:512:H-Gly301:O (2.19 Å) | |||||
At the structural level, the functional groups of 4e played different roles in keeping the stable shape. The hydrogen-bonding moiety that directly interacted with Ser349 served as the primary anchoring site. The peripheral aromatic capping group formed amide-π stacking with Lys487, π-alkyl interactions with Leu601 and Arg599, and a bridging van der Waals connection through the connecting unit with Leu601. Additionally, a π-anion hydrophilic interaction with Glu488 contributed to the overall structural stabilisation of the ligand–protein complex. The in silico docking results indicate that compound 4e exhibits a favourable binding affinity toward the target enzyme 2VF5, reflecting its potential for effective enzyme–ligand interactions at the molecular level. However, the in vitro results reveal that the MIC value of 4e remains higher than that of chloramphenicol. This discrepancy suggests that a strong binding affinity predicted by docking does not necessarily translate directly into optimal antibacterial efficacy under biological conditions, most likely due to extra-enzymatic factors such as limited bacterial membrane permeability or insufficient intracellular transport efficiency. Therefore, in this study, molecular docking is primarily employed to elucidate trends in molecular interactions and to support SAR analysis, while also indicating that the structural scaffold of 4e represents a promising framework that requires further optimisation, particularly aimed at improving membrane permeability, to enhance its in vitro antibacterial activity.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (5.9) that can be mitigated through structural modification or formulation strategies. Its drug-likeness risk score (RSK) composite and synthetic accessibility (SAscore = 2.5) indicate practical synthetic feasibility and the potential for scaffold optimisation.61,62 Regarding the ADMET properties, 4l shows moderate absorption, with predicted Caco-2 permeability and low human intestinal absorption (HIA), attributed mainly to its high lipophilicity and poor solubility (log
P (5.9) that can be mitigated through structural modification or formulation strategies. Its drug-likeness risk score (RSK) composite and synthetic accessibility (SAscore = 2.5) indicate practical synthetic feasibility and the potential for scaffold optimisation.61,62 Regarding the ADMET properties, 4l shows moderate absorption, with predicted Caco-2 permeability and low human intestinal absorption (HIA), attributed mainly to its high lipophilicity and poor solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = −7.6). These limitations are, however, addressable through salt formation or prodrug approaches. The distribution profile indicates strong plasma protein binding (PPB = 100%) and a moderate volume of distribution, consistent with peripheral drug behavior. The low blood–brain barrier (BBB) permeability is advantageous for non-central nervous system (CNS) targets. In metabolic predictions, 4l is anticipated to interact with specific CYP isoforms (CYP1A2, 2C9, 2D6), a characteristic prevalent in thiazole scaffolds that can be mitigated through substituent optimization. The excretion rate appears moderate, suggesting no rapid clearance or abnormal accumulation. Toxicologically, we predict potential drug-induced liver injury (DILI) and Ames-positive alerts. Still, the absence of pan-assay interference compounds (PAINS) and low hERG inhibition probability suggests minimal cardiotoxic liability. Overall, 4l can be considered a promising and synthetically accessible scaffold that complies with most classical drug-likeness rules. With rational optimization of lipophilicity, sulfur reactivity, and solubility, it holds potential for advancement toward a viable drug candidate63–66
S = −7.6). These limitations are, however, addressable through salt formation or prodrug approaches. The distribution profile indicates strong plasma protein binding (PPB = 100%) and a moderate volume of distribution, consistent with peripheral drug behavior. The low blood–brain barrier (BBB) permeability is advantageous for non-central nervous system (CNS) targets. In metabolic predictions, 4l is anticipated to interact with specific CYP isoforms (CYP1A2, 2C9, 2D6), a characteristic prevalent in thiazole scaffolds that can be mitigated through substituent optimization. The excretion rate appears moderate, suggesting no rapid clearance or abnormal accumulation. Toxicologically, we predict potential drug-induced liver injury (DILI) and Ames-positive alerts. Still, the absence of pan-assay interference compounds (PAINS) and low hERG inhibition probability suggests minimal cardiotoxic liability. Overall, 4l can be considered a promising and synthetically accessible scaffold that complies with most classical drug-likeness rules. With rational optimization of lipophilicity, sulfur reactivity, and solubility, it holds potential for advancement toward a viable drug candidate63–664. Conclusion
This study reports the successful synthesis and structural confirmation of a series of thiazole derivatives, characterised by NMR, FT-IR, and HR-MS analyses. Among them, compound 4e showed the most potent antibacterial activity (MIC = 83.5 µM), while 4g exhibited potent cytotoxicity (IC50 = 2.6–6.6 µM). Compound 4f displayed notable anti-inflammatory effects (IC50 = 13.4 µM), comparable to dexamethasone. Remarkably, 4a and 4l (–OH, –CH2OH) showed superior α-glucosidase inhibition (IC50 = 45 and 41 µM) to acarbose (117 µM). The molecular docking studies revealed that the ligands exhibited favorable interactions with the respective enzymes. This indicates that the favourable interactions observed in silico align with the positive activities seen in vitro. The molecular dynamic simulations, spanning 100 ns, demonstrated structural stability, with critical residues such as Glu428 and Asn452 exhibiting hydrogen bond occupancy exceeding 80%. The ADMET profile of compound 4l adheres to the principles of drug-likeness. Subsequent research endeavors could focus on the structural optimization and augmentation of hydrophilicity to enhance the bioavailability of the synthesized compounds, alongside in vivo assessments of compound 4l in relation to α-glucosidase activity.Author contributions
Nguyen Duy Khanh: conceptualization, formal analysis, investigation, data curation, writing – original draft, visualization, funding acquisition. Nguyen Thi Hong Anh: formal analysis, investigation, data curation, visualization. Nguyen Thi Mai Tho: methodology, software, validation, writing – review & editing, supervision. Tran Nguyen Minh An: methodology, software, validation, formal analysis, resources, writing – review & editing, supervision, project administration. Nguyen Van Son, Nguyen Thi Nhat Thang: visualization, funding acquisition, data curation. Tran Thuy Quyen: methodology.Conflicts of interest
There are no conflicts to declare.Data availability
All experimental and computational data supporting the findings of this study, including in vitro bioassay results, molecular docking data, molecular dynamics simulations, ADMET screening, and 1H-NMR, 13C-NMR, FT-IR, HPLC, and HR-MS spectra for structural confirmation, are provided in the manuscript and the supplementary information (SI). Additional raw data are available from the corresponding author upon reasonable request. Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra08472e.Acknowledgements
This research is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 104.01-2023.78. The authors also express their sincere appreciation to the Laboratory of the Faculty of Chemical Engineering (FCE), Industrial University of Ho Chi Minh City (IUH), Ho Chi Minh City, Vietnam, for providing essential facilities and technical assistance during the research.References
- D. Ungureanu, B. Tiperciuc, C. Nastasă, I. Ionuţ, G. Marc, I. Oniga and O. Oniga, Pharmaceutics, 2024, 16, 89 Search PubMed.
- K. H. Narasimhamurthy, T. R. Swaroop and K. S. Rangappa, Eur. J. Med. Chem. Rep., 2024, 12, 100225 CAS.
- A. F. Kassem, R. H. Althomali, M. M. Anwar and W. I. El-Sofany, J. Mol. Struct., 2024, 1303, 137510 CrossRef CAS.
- W. A. A. Fadaly, A. Elshewy, M. T. M. Nemr, K. Abdou, A. M. Sayed and N. M. Kahk, Bioorg. Chem., 2024, 152, 107760 CrossRef CAS.
- A. Singh, D. Malhotra, K. Singh, R. Chadha and P. M. S. Bedi, J. Mol. Struct., 2022, 1266, 133479 CrossRef CAS.
- H. Aziz, J. Mol. Liq., 2025, 424, 127064 CrossRef CAS.
- S. Basak, A. Murmu, B. W. Matore, P. P. Roy and J. Singh, Eur. J. Med. Chem. Rep., 2024, 11, 100160 Search PubMed.
- Salahuddin, A. Mazumder, M. J. Ahsan, R. Kumar, Z. Ullah, M. Shahar Yar and K. Shabana, Bioengineering, 2025, 12, 1024 Search PubMed.
- J. Liu and L.-N. Wang, Cochrane Database Syst. Rev., 2023, 2023(1) DOI:10.1002/14651858.CD010693.pub6.
- L. Maram, J. M. Michael, H. Politte, V. S. Srirama, A. Hadji, M. Habibi, M. O. Kelly, R. T. Brookheart, B. N. Finck, L. Hegazy, K. S. McCommis and B. Elgendy, Eur. J. Med. Chem., 2025, 283, 117150 Search PubMed.
- Z. Xu, D. Wu, C. Fang and Y. Li, Des. Monomers Polym., 2023, 26, 90–105 Search PubMed.
- S. A. Patil, S. A. Patil, E. A. Ble-González, S. R. Isbel, S. M. Hampton and A. Bugarin, Molecules, 2022, 27, 6575 Search PubMed.
- G. Wang, S. Sun and H. Guo, Eur. J. Med. Chem., 2022, 229, 113999 Search PubMed.
- F. Grande, G. Ioele, A. Caruso, M. A. Occhiuzzi, H. El-Kashef, C. Saturnino and M. S. Sinicropi, Appl. Sci., 2022, 13, 349 CrossRef.
- C.-H. Chen, C.-S. Liu, X.-M. Guo, J.-P. Tong, J. Huang, T.-T. Shi and J. Sun, J. Mol. Struct., 2024, 1306, 137891 CrossRef CAS.
- D. Ling, C. Xiang, H. Guolin, S. Huisheng and N. Xiaohua, 3 Biotech, 2025, 15, 111 CrossRef.
- S. Drygała, M. Radzikowski and M. Maciejczyk, Front. Pharmacol., 2024, 15, 1489657 CrossRef.
- H. Elimam, Z. Hassan, N. A. A. Alhamshry, K. M. El-Say and A. M. A. Akabawy, Naunyn-Schmiedeberg’s Arch. Pharmacol., 2025, 15 DOI:10.1007/s00210-025-04356-9.
- H. Şahin, A. Arslantürk Bingül, İ. Şengül and M. Bingül, Istanb. J. Pharm., 2023, 53, 39–44 Search PubMed.
- F. Grande, G. Ioele, A. Caruso, M. A. Occhiuzzi, H. El-Kashef, C. Saturnino and M. S. Sinicropi, Appl. Sci., 2022, 13, 349 CrossRef.
- M. I. Abdjan, N. S. Aminah, A. N. Kristanti, I. Siswanto, A. L. L. Windah, T. M. Thant, R. Ramadhan, Y. Takaya, Z. Ul-Haq and M. I. Choudhary, Eng. Sci., 2024, 31 DOI:10.30919/es1286.
- İ. Çapan, M. Hawash, M. T. Qaoud and N. Jaradat, ACS Omega, 2025, 10, 848–861 CrossRef.
- B. Donarska, K. Seklecka, J. Cytarska, K. Piechowska, P. Ledwon, S. Kula, P. Krawczyk, A. Baranowska-Łączkowska and K. Z. Łączkowski, Int. J. Mol. Sci., 2025, 26, 7945 CrossRef CAS.
- S. S. Marufa, M. M. Rahman, M. M. Rahman, J. R. Debnath, M. A. Mim, R. Jahan, H. Nishino, M. S. Alam and M. A. Haque, J. Mol. Struct., 2025, 1321, 139861 CrossRef CAS.
- P. Krawczyk, B. Jędrzejewska, J. Cytarska, K. Seklecka and K. Z. Łączkowski, Sensors, 2024, 24 DOI:10.3390/s24196368.
- S. Verma, P. Gupta, S. Lal, R. Narang, S. Mujwar, S. Saini and J. Sharma, Eur. J. Med. Chem., 2025, 298, 117995 CrossRef CAS PubMed.
- N. M. Hassanin, T. E. Ali, M. A. Assiri and S. M. Abdel-Kariem, RSC Adv., 2024, 14, 17245–17260 RSC.
- C. Maccallini, R. Budriesi, B. De Filippis and R. Amoroso, Int. J. Mol. Sci., 2024, 25, 8486 CrossRef CAS.
- A. Mushtaq, U. Azam, S. Mehreen and M. M. Naseer, Eur. J. Med. Chem., 2023, 249, 115119 CrossRef CAS.
- J. Hughes, S. Rees, S. Kalindjian and K. Philpott, Br. J. Pharmacol., 2011, 162, 1239–1249 CrossRef CAS PubMed.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, J. Pharm. Anal., 2016, 6, 71–79 CrossRef.
- M. A. Kumar, T. N. Minh An, I. J. Lee, S. Park and K. D. Lee, Phosphorus Sulfur Silicon Relat. Elem., 2015, 190, 1160–1168 CrossRef CAS.
- S. H. Ali and A. R. Sayed, Synth. Commun., 2021, 51, 670–700 CrossRef CAS.
- M. A. T. Nguyen, A. K. Mungara, J. A. Kim, K. D. Lee and S. Park, Phosphorus Sulfur Silicon Relat. Elem., 2015, 190, 191–199 CrossRef CAS.
- S. Cheenpracha, E.-J. Park, B. Rostama, J. M. Pezzuto and L. C. Chang, Mar. Drugs, 2010, 8, 429–437 CrossRef CAS.
- V. Kumar, O. Prakash, S. Kumar and S. Narwal, Pharmacogn. Rev., 2011, 5, 19 CrossRef.
- N.-H. Nguyen, N.-M.-A. Tran, T.-H. Duong and G. Van Vo, RSC Adv., 2023, 13, 8190–8201 RSC.
- H. H. Nguyen, N. M. A. Tran, T. H. T. Nguyen, H. C. Vo, C. H. Nguyen, T. H. A. Nguyen, N. H. Nguyen and T. H. Duong, J. Saudi Chem. Soc., 2022, 26, 101489 CrossRef CAS.
- G. Van Vo, T. H. T. Nguyen, T. P. Nguyen, T. H. T. Do, N. M. A. Tran, H. T. Nguyen and T. T. Nguyen, Saudi Pharm. J., 2022, 30, 1301–1314 CrossRef.
- N. D. T. Ha, T. Phuong and V. C. Nguyen, ChemistrySelect, 2023, 8(17), e202300246 CrossRef.
- G. Van Vo, T. H. T. Nguyen, T. P. Nguyen, T. H. T. Do, N. M. A. Tran, H. T. Nguyen and T. T. Nguyen, Saudi Pharm. J., 2022, 30(9) DOI:10.1016/j.jsps.2022.06.018.
- T. B. N. Dao, T. M. T. Nguyen, V. Q. Nguyen, T. M. D. Tran, N. M. A. Tran, C. H. Nguyen, T. H. T. Nguyen, H. H. Nguyen, J. Sichaem, C. L. Tran and T. H. Duong, Molecules, 2021, 26, 1–15 Search PubMed.
- T. T. Ngoc Mai, P. N. Minh, N. T. Phat, T. H. Duong, T. N. Minh An, V. S. Dang, N. Van Hue and M. D. Tri, RSC Adv., 2024, 14, 9326–9338 Search PubMed.
- T. N. M. An, N. Van Cuong, N. M. Quang, T. V. Thanh and M. Alam, ChemistrySelect, 2020, 5, 6339–6349 Search PubMed.
- H. Yang, L. Sun, Z. Wang, W. Li, G. Liu and Y. Tang, J. Chem. Inf. Model., 2018, 58, 2051–2056 CrossRef CAS PubMed.
- S. A. Hollingsworth and R. O. Dror, Neuron, 2018, 99, 1129–1143 CrossRef CAS PubMed.
- O. S. Faleye, B. R. Boya, J.-H. Lee, I. Choi and J. Lee, Pharmacol. Rev., 2024, 76, 90–141 CrossRef CAS.
- M. T. Sayed, S. A. Elsharabasy and A. Abdel-Aziem, Sci. Rep., 2023, 13(1) DOI:10.1038/s41598-023-36705-0.
- S. S. Borisevich, T. E. Aksinina, M. G. Ilyina, V. O. Shender, K. S. Anufrieva, G. P. Arapidi, N. V. Antipova, F. Anizon, Y. J. Esvan, F. Giraud, V. V. Tatarskiy, P. Moreau, M. I. Shakhparonov, M. S. Pavlyukov and A. A. Shtil, Cancers, 2024, 16, 834 CrossRef CAS PubMed.
- A. A. Hassan, N. K. Mohamed, A. A. Aly, M. Ramadan, H. A. M. Gomaa, A. T. Abdel-Aziz, B. G. M. Youssif, S. Bräse and O. Fuhr, Molecules, 2023, 28, 7951 Search PubMed.
- E. Kuzu, E. Arzuk, F. Karakuş, B. Kuzu and H. Genç, Turk. J. Chem., 2025, 49, 215–227 CrossRef PubMed.
- L. T. H. Nguyen, D. H. Vu, M. Q. Pham, Q. A. Ngo and N. B. Vo, RSC Adv., 2025, 15, 2850–2861 Search PubMed.
- S. Fang, X. Huang, F. Cai, G. Qiu, F. Lin and X. Cai, J. Steroid Biochem. Mol. Biol., 2024, 240, 106478 Search PubMed.
- H.-F. Lin, Y.-C. Jiang, Z.-W. Chen and L.-L. Zheng, RSC Adv., 2024, 14, 16349–16357 RSC.
- F. Esposito, N. Pala, M. Carcelli, S. T. Boateng, P. S. D’Aquila, A. Mariani, S. Satta, J. C. Chamcheu, M. Sechi and V. Sanna, J. Enzyme Inhib. Med. Chem., 2023, 38(1) DOI:10.1080/14756366.2023.2236802.
- Z. Soleimani, M. Mohammadi, M. Halimi, S. Safapoor, N. Dastyafteh, E. Safaie, S. Mojtabavi, M. A. Faramarzi, M. Bozorgi-Koushalshahi, B. Larijani, M. Mohammadi-Khanaposhtani and M. Mahdavi, Sci. Rep., 2024, 14, 18693 CrossRef CAS PubMed.
- F. T. Zahra, A. Saeed, A. Ahmed, H. Ismail, M. U. Ijaz and F. Albericio, RSC Adv., 2023, 13, 24988–25001 RSC.
- Y. Khan, S. Khan, R. Hussain, A. Maalik, W. Rehman, M. W. Attwa, R. Masood, H. W. Darwish and H. A. Ghabbour, Pharmaceuticals, 2023, 16, 1650 CrossRef CAS.
- I. A. Seliem, RSC Adv., 2025, 15, 32309–32327 Search PubMed.
- A. Kolarič, M. Kokot, M. Hrast, M. Weiss, I. Zdovc, J. Trontelj, S. Žakelj, M. Anderluh and N. Minovski, Antibiotics, 2021, 10, 862 Search PubMed.
- R. J. Young, Expet Opin. Drug Discov., 2023, 18, 965–972 CrossRef CAS.
- S. Q. Pantaleão, P. O. Fernandes, J. E. Gonçalves, V. G. Maltarollo and K. M. Honorio, ChemMedChem, 2021, 17(1) DOI:10.1002/cmdc.202100542.
- F. Wu, Y. Zhou, L. Li, X. Shen, G. Chen, X. Wang, X. Liang, M. Tan and Z. Huang, Front. Chem., 2020, 8 DOI:10.3389/fchem.2020.00726.
- J. Zhang, H. Li, Y. Zhang, J. Huang, L. Ren, C. Zhang, Q. Zou and Y. Zhang, Briefings Bioinf., 2025, 26(5) DOI:10.1093/bib/bbaf533.
- Y. Zhang, G. Zhang, T. Wang, Y. Chen, J. Wang, P. Li, R. Wang and J. Su, ChemBioChem, 2025, 25(22) DOI:10.1002/cbic.202400297.
- H. Komura, R. Watanabe and K. Mizuguchi, Pharmaceutics, 2023, 15, 2619 CrossRef CAS PubMed.
- A. M. Hassan, H. S. Gattan, A. A. Faizo, M. H. Alruhaili, A. S. Alharbi, L. H. Bajrai, I. A. AL-Zahrani, V. D. Dwivedi and E. I. Azhar, Pharmaceuticals, 2024, 17, 1617 CrossRef CAS PubMed.
- W. Lu and M. J. Buehler, Mater. Adv., 2025, 6, 4267–4285 RSC.
| This journal is © The Royal Society of Chemistry 2026 |