 Open Access Article
Open Access ArticleNovel diazenyl chalcones with a 2-arylidene hydrazineylidene thiazole moiety: synthesis, anticancer potential, spectroscopic characterisation, and molecular docking studies
Mariem M. El-Samolya,
Esmail M. El-Fakharany*b,
Yousra A. El-Maradnyb,
Abdallah E. Abdallah c,
F. A. Tahera,
Nadia T. A. Dawoud
c,
F. A. Tahera,
Nadia T. A. Dawoud *a and
Doaa R. Lotfya
*a and
Doaa R. Lotfya
aChemistry Department, Faculty of Science, Girls, Al-Azhar University, Nasr City, Cairo, Egypt
bProtein Research Department, Genetic Engineering and Biotechnology Research Institute, City of Scientific Research and Technological Applications (SRTA-City), New Borg AL-Arab, 21934, Alexandria, Egypt. E-mail: esmailelfakharany@yahoo.co.uk
cPharmaceutical Medicinal Chemistry & Drug Design Department, Faculty of Pharmacy (Boys), Al-Azhar University, Nasr City, Cairo, 11884, Egypt. E-mail: dawoudnadia@yahoo.com
First published on 19th January 2026
Abstract
A series of diazenyl chalcones bearing 2-arylidene-hydrazinylidene thiazole units (4–6a,b) were designed, synthesised, and fully characterised by NMR, IR, MS, and UV spectroscopy. The compounds were evaluated for cytotoxicity against normal skin cells and the MDA and HepG2 cancer cell lines. The results reveal that most synthetic chemicals exhibited limited toxicity on normal cells at concentrations of 100 µg mL−1 or below. Additionally, all pharmaceuticals, except for compound 4a, all pharmaceuticals demonstrated antiproliferation activity at quantities below the safety threshold. MDA and HepG2 cells exhibited no significant differences in cytotoxicity; however, compounds 5b and 6a showed the highest selectivity indices. The new compounds exhibited excellent affinity for the MMP-2 receptor, with values between −32.67 and −14.98 kcal mol−1. Compounds 6a and 6b demonstrated the most advantageous results, with values of −32.67 and −32.27 kcal mol−1, respectively, in comparison to the ligand's value of −33.73 kcal mol−1. Compound 5b attained a third-place ranking with a score of −29.72 kcal mol−1. Compounds 4a and 4b demonstrated relatively lower scores; however, compound 5a revealed markedly worse performance in comparison to other derivatives. Finally, these findings should be regarded as preliminary, indicating promising biological potential but not confirming any specific mechanism of action.
Introduction
Cancer remains a major global health challenge, largely due to the uncontrolled proliferation of abnormal cells. Unlike normal cells, cancer cells bypass regulatory mechanisms governing growth and survival, enabling continuous division, invasion of surrounding tissues, and metastasis to distant organs. These malignant behaviours arise from accumulated genetic mutations and dysregulation of multiple intracellular signalling pathways.1Heterocyclic compounds have played a pivotal role in anticancer drug development. Many clinically used chemotherapeutic agents—such as methotrexate, vinblastine, vincristine, daunorubicin, 5-fluorouracil and doxorubicin—contain heterocyclic scaffolds, reflecting the structural versatility and broad pharmacological potential of these molecules.2 Among heterocycles, Schiff bases (characterised by an imine ![[double bond splayed left]](https://www.rsc.org/images/entities/char_e009.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– group) are particularly prominent due to their π-acceptor properties, strong metal-chelating ability and diverse biological activities, including antibacterial, antioxidant and antiproliferative effects.3–9 Likewise, the 4-thiazolidinone framework represents a privileged scaffold in medicinal chemistry, widely reported for its anti-inflammatory, antibacterial, antidiabetic, antioxidant and anticancer properties.10–15 In particular, 5-arylidenethiazolidin-4-one derivatives have emerged as promising drug-like candidates.16–20 Azo-chalcones constitute another notable class, combining the reactive α,β-unsaturated carbonyl system of chalcones with an azo (–N
N– group) are particularly prominent due to their π-acceptor properties, strong metal-chelating ability and diverse biological activities, including antibacterial, antioxidant and antiproliferative effects.3–9 Likewise, the 4-thiazolidinone framework represents a privileged scaffold in medicinal chemistry, widely reported for its anti-inflammatory, antibacterial, antidiabetic, antioxidant and anticancer properties.10–15 In particular, 5-arylidenethiazolidin-4-one derivatives have emerged as promising drug-like candidates.16–20 Azo-chalcones constitute another notable class, combining the reactive α,β-unsaturated carbonyl system of chalcones with an azo (–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) group, giving rise to photoresponsive molecules with reported antimicrobial, anti-inflammatory and anticancer activities.21–23
N–) group, giving rise to photoresponsive molecules with reported antimicrobial, anti-inflammatory and anticancer activities.21–23
The design of the present diazenyl chalcone series bearing a 2-arylidene hydrazinylidene thiazole moiety was motivated by the pharmacophoric features of these two bioactive scaffolds. Chalcones are well-established Michael acceptors with anticancer, anti-inflammatory and enzyme–inhibitory activities, primarily attributed to their α,β-unsaturated system. Thiazole-based heterocycles, including thiazolidinones and hydrazinylidene derivatives, exhibit potent cytotoxic and matrix metalloproteinase (MMP) inhibitory effects. Integrating these motifs through a diazenyl linker was expected to (i) enhance molecular conjugation and electronic distribution, (ii) improve binding interactions with MMP-2 via additional hydrogen-bonding and π–π stacking sites, and (iii) generate hybrid molecules capable of synergistic anticancer activity. In silico docking supported this design, predicting favourable binding energies.
Thus, the synthetic strategy aimed to develop hybrid molecules with enhanced anticancer potential and high affinity toward MMP targets.
Materials and methods
All melting point ranges were taken on a Gallenkamp electric melting point apparatus using the one-end open capillary method and were uncorrected. The IR spectra of the compounds were recorded on the Bruker ALPHA II Fourier Transform Infrared Spectrometer at a wavenumber of (4000–400 cm−1). The 1H and 13C-NMR spectra were recorded on a Bruker AV 300 and Jeol 500 MHz spectrometer operating at 300 and 500 MHz for 1H and 75, 125 MHz for 13C using dimethyl dimethylsulfoxide as solvent and tetramethylsilane (TMS) as an internal reference. Mass spectra (MS) were recorded on the Thermo Scientific GCMS model (ISQ LT) using Thermo X-Calibur software. Elemental analyses were performed on the elemental analyzer (CHNS) set: Flash 2000 Thermo Scientific. The reactions were followed up, and the purity of products was carried out on silica gel G60F (Merck) plates that were precoated to a thickness of 0.2 mm, visualizing the spots under ultraviolet light and or dichloromethane/PE (l![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), and/or DCM/drop of ethanol as mobile phases. The PerkinElmer Lambda 35 spectrophotometer was used to deliver the visible spectra for all compounds.
3), and/or DCM/drop of ethanol as mobile phases. The PerkinElmer Lambda 35 spectrophotometer was used to deliver the visible spectra for all compounds.
Chemistry and characterization
We confirmed the structures of these newly synthesised compounds using FTIR, 1H-NMR, and 13C-NMR spectroscopy, elemental analysis, and mass spectra.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The products were then filtered and washed many times with hot ethanol and water, and then diethyl ether, resulting in a high yield of thiosemicarbazones (1a–f).
3). The products were then filtered and washed many times with hot ethanol and water, and then diethyl ether, resulting in a high yield of thiosemicarbazones (1a–f).
2-(4-Chlorobenzylidene)hydrazine-1-carbothioamide (1a). White crystals, yield 85%, m.p. 218–220 °C. FTIR (cm−1): 3430(NH), 3279, 3164(NH2), 3070(CH)Ar, 2795(CH)Al, 1284(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1599(C
S), 1599(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1522(C
N), 1522(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 816(C–Cl). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.49 (s, 1H, NH), 8.24 (s, 1H, CH
C), 816(C–Cl). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.49 (s, 1H, NH), 8.24 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.02 (s, 2H, NH2), 7.78–7.35 (m, 4H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 178.0, 141.0, 134.3, 133.0, 128.8(2 × C), 128.7(2 × C). Elemental analysis: calcd: C, 44.97; H, 3.77; Cl, 16.59; N, 19.67; S, 15.00; found: C, 44.64; H, 3.54; Cl, 16.43; N, 19.46; S, 14.65.
N), 8.02 (s, 2H, NH2), 7.78–7.35 (m, 4H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 178.0, 141.0, 134.3, 133.0, 128.8(2 × C), 128.7(2 × C). Elemental analysis: calcd: C, 44.97; H, 3.77; Cl, 16.59; N, 19.67; S, 15.00; found: C, 44.64; H, 3.54; Cl, 16.43; N, 19.46; S, 14.65.
2-Benzylidenehydrazine-1-carbothioamide (1b). Pale yellow crystals, yield 51%, m.p. 157–160 °C. FTIR (cm−1): 3417(NH), 3247, 3143(NH2), 3024(CH)Ar, 2819(CH)Al, 1289(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1588(C
S), 1588(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1534(C
N), 1534(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (500 MHz, DMSO-d6): δ ppm; 12.02, (s, 1H, NH), 8.39 (s, 1H, CH
C). 1H-NMR (500 MHz, DMSO-d6): δ ppm; 12.02, (s, 1H, NH), 8.39 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.75–7.52 (m, 4H, Ar–H + NH2). 13C-NMR (125 MHz, DMSO-d6): δ ppm; 174.2, 155.0, 135.1, 133.1, 129.2(2 × C), 128.9(2 × C). Elemental analysis: calcd: C, 53.61; H, 5.06; N, 23.44; S, 17.89; found: C, 53.33; H, 5.00; N, 23.32; S, 17.67.
N), 7.75–7.52 (m, 4H, Ar–H + NH2). 13C-NMR (125 MHz, DMSO-d6): δ ppm; 174.2, 155.0, 135.1, 133.1, 129.2(2 × C), 128.9(2 × C). Elemental analysis: calcd: C, 53.61; H, 5.06; N, 23.44; S, 17.89; found: C, 53.33; H, 5.00; N, 23.32; S, 17.67.
2-(4-Methoxybenzylidene)hydrazine-1-carbothioamide (1c). Yellow crystals, yield 66%, m.p. 174–176 °C. FTIR (cm−1): 3401(NH), 3285, 3143(NH2), 2970(CH)Ar, 1241(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1596(C
S), 1596(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1172(C–O). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.29 (s, 1H, NH), 8.07 (s, 1H, CH
N), 1172(C–O). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.29 (s, 1H, NH), 8.07 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.00 (s, 2H, NH2), 7.88–6.93 (m, 4H, Ar–H), 3.78 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.5, 160.6, 142.2, 128. 9(2 × C), 126.7, 114.1(2 × C), 55.2. Elemental analysis: calcd: C, 51.66; H, 5.30; N, 20.08; S, 15.32; found: C, 51.45; H, 5.26; N, 20.01; S, 15.10.
N), 8.00 (s, 2H, NH2), 7.88–6.93 (m, 4H, Ar–H), 3.78 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.5, 160.6, 142.2, 128. 9(2 × C), 126.7, 114.1(2 × C), 55.2. Elemental analysis: calcd: C, 51.66; H, 5.30; N, 20.08; S, 15.32; found: C, 51.45; H, 5.26; N, 20.01; S, 15.10.
2-(2-Hydroxybenzylidene)hydrazine-1-carbothioamide (1d). Beige crystals, yield 70%, m.p. 240–242 °C. FTIR (cm−1); 3441(OH), 3314(NH), 3171, 3134(NH2), 3022(CH)Ar, 2985(CH)Al, 1263(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1602(C
S), 1602(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1535(C
N), 1535(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.35 (s, 1H, NH), 9.83 (1H, OH), 8.38 (s, 1H, CH
C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.35 (s, 1H, NH), 9.83 (1H, OH), 8.38 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.07 (s, 2H, NH2), 7.89–6.78 (m, 4H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.5, 156.3, 140.0, 131.2, 126.8, 120.1, 119.3, 116.0. Elemental analysis: calcd: C, 49.22; H, 4.65; N, 21.52; S, 16.42; found: C, 49.02; H, 4.45; N, 21.32; S, 16.22.
N), 8.07 (s, 2H, NH2), 7.89–6.78 (m, 4H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.5, 156.3, 140.0, 131.2, 126.8, 120.1, 119.3, 116.0. Elemental analysis: calcd: C, 49.22; H, 4.65; N, 21.52; S, 16.42; found: C, 49.02; H, 4.45; N, 21.32; S, 16.22.
2-(4-Hydroxybenzylidene)hydrazine-1-carbothioamide (1e). Beige crystals, yield 63.5%, m.p. 258–260 °C. FTIR (cm−1); 3467(OH), 3357(NH), 3185, 3119(NH2), 3030(CH)Ar, 2919 (CH)Al, 1265(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1582(C
S), 1582(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1549(C
N), 1549(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.21 (s, 1H, NH), 9.86 (s, 1H, OH), 8.01 (s, 1H, CH
C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.21 (s, 1H, NH), 9.86 (s, 1H, OH), 8.01 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.79 (s, 2H, NH2), 7.62–6.77 (m, 4H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.4, 159.2, 142.8, 129.1(2 × C), 125.1, 115.6(2 × C). Elemental analysis: calcd: C, 49.22; H, 4.65; N, 21.52; S, 16.42; found: C, 49.02; H, 4.43; N, 21.36; S, 16.26.
N), 7.79 (s, 2H, NH2), 7.62–6.77 (m, 4H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.4, 159.2, 142.8, 129.1(2 × C), 125.1, 115.6(2 × C). Elemental analysis: calcd: C, 49.22; H, 4.65; N, 21.52; S, 16.42; found: C, 49.02; H, 4.43; N, 21.36; S, 16.26.
2-(4-Hydroxy-3-methoxybenzylidene)hydrazine-1-carbothioamide (1f). White crystals, yield 33%, m.p. 200 °C. FTIR (cm−1); 3529(OH), 3437(NH), 3278, 3154(NH2), 3037(CH)Ar, 2938(CH)Al, 1278(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1587(C
S), 1587(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1545(C
N), 1545(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.21 (s, 1H, NH), 9.43 (s, 1H, OH), 8.06 (s, 1H, CH
C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.21 (s, 1H, NH), 9.43 (s, 1H, OH), 8.06 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.96–6.76 (m, 6H, Ar–H + NH2), 3.82 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.1, 148.6, 147.8, 142.7, 125.3, 122.1, 115.0, 109.0, 55.5. Elemental analysis: calcd: C, 47.99; H, 4.92; N, 18.65; S, 14.23; found: C, 47.76; H, 4.83; N, 18.48; S, 14.11.
N), 7.96–6.76 (m, 6H, Ar–H + NH2), 3.82 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 177.1, 148.6, 147.8, 142.7, 125.3, 122.1, 115.0, 109.0, 55.5. Elemental analysis: calcd: C, 47.99; H, 4.92; N, 18.65; S, 14.23; found: C, 47.76; H, 4.83; N, 18.48; S, 14.11.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ethanol–water mixture.
1 ethanol–water mixture.
2(-4-Chlorobenzylidene)hydrazinylidene)thiazolidin-4-one (2a). White crystals, yield 82%, m.p. 310–312 °C. FTIR (cm−1); 3419(NH), 2928(CH)Ar, 2744(CH)Al, 1707(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1640(C
O), 1640(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1540(C
N), 1540(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 829(C–Cl). 1H-NMR (500 MHz, DMSO-d6): δ ppm; 12.03 (s, 1H, NH), 8.39 (s, 1H, CH
C), 829(C–Cl). 1H-NMR (500 MHz, DMSO-d6): δ ppm; 12.03 (s, 1H, NH), 8.39 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.77–7.50 (m, 4H, Ar–H), 3.89 (s, 2H, SCH2). 13C-NMR (125 MHz, DMSO-d6): δ ppm; 174.2, 165.8, 155.0, 135.1, 133.1, 129.2(2 × C), 128.9(2 × C), 33.0. Elemental analysis: calcd: C, 47.34; H, 3.18; Cl, 13.97; N, 16.56; S, 12.64; found: C, 47.22; H, 3.05; Cl, 13.76; N, 16.43; S, 12.52.
N), 7.77–7.50 (m, 4H, Ar–H), 3.89 (s, 2H, SCH2). 13C-NMR (125 MHz, DMSO-d6): δ ppm; 174.2, 165.8, 155.0, 135.1, 133.1, 129.2(2 × C), 128.9(2 × C), 33.0. Elemental analysis: calcd: C, 47.34; H, 3.18; Cl, 13.97; N, 16.56; S, 12.64; found: C, 47.22; H, 3.05; Cl, 13.76; N, 16.43; S, 12.52.
2(Benzylidene)hydrazinylidene)thiazolidin-4-one (2b). Yellow crystals, yield 68%, m.p. 260–263 °C. FTIR (cm−1); 3150(NH), 3056(CH)Ar, 2756(CH)Al, 1709(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1640(C
O), 1640(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1588(C
N), 1588(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.21 (s, 1H, NH), 8.05 (s, 1H, CH
C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.21 (s, 1H, NH), 8.05 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.93–6.76 (m, 5H, Ar–H), 3.82 (s, 2H, SCH2). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 174.3, 165.5, 156.3, 134.2, 130.7, 128.9(2 × C), 127.7(2 × C), 33.1. Elemental analysis: calcd: C, 54.78; H, 4.14; N, 19.16; S, 14.62; found: C, 54.53; H, 4.08; N, 19.13; S, 14.35.
N), 7.93–6.76 (m, 5H, Ar–H), 3.82 (s, 2H, SCH2). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 174.3, 165.5, 156.3, 134.2, 130.7, 128.9(2 × C), 127.7(2 × C), 33.1. Elemental analysis: calcd: C, 54.78; H, 4.14; N, 19.16; S, 14.62; found: C, 54.53; H, 4.08; N, 19.13; S, 14.35.
2(-4-Methoxybenzylidene)hydrazinylidene)thiazolidin-4-one (2c). Yellow crystals, yield 92%, m.p. 260–263 °C. FTIR (cm−1); 3648(OH), 2971(CH)Ar, 2730(CH)Al, 1716(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1629(C
O), 1629(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1596(C
N), 1596(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.88 (1H, s, NH), 8.32 (s, 1H, CH
C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 11.88 (1H, s, NH), 8.32 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.71–7.00 (4H, m, Ar–H), 3.87 (s, 2H, SCH2), 3.80 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 174.2, 161.3, 155.9, 144.8, 129.3(2 × C), 126.8, 114.3(2 × C), 55.3, 33.0. Elemental analysis: calcd: C, 53.00; H, 4.45; N, 16.86; S, 12.86; found: C, 52.83; H, 4.40; N, 16.64; S, 12.67.
N), 7.71–7.00 (4H, m, Ar–H), 3.87 (s, 2H, SCH2), 3.80 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 174.2, 161.3, 155.9, 144.8, 129.3(2 × C), 126.8, 114.3(2 × C), 55.3, 33.0. Elemental analysis: calcd: C, 53.00; H, 4.45; N, 16.86; S, 12.86; found: C, 52.83; H, 4.40; N, 16.64; S, 12.67.
2(-2-Hydroxybenzylidene)hydrazinylidene)thiazolidin-4-one (2d). Simone crystals, yield 90%, m.p. 300–303 °C. FTIR (cm−1); 3419(OH), 3041, 2929(CH)Ar, 2856(CH)Al, 1703(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1596(C
O), 1596(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1493(C
N), 1493(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.06 (s, 1H, NH), 10.86 (s, 1H, OH), 8.63 (s, 1H, CH
C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.06 (s, 1H, NH), 10.86 (s, 1H, OH), 8.63 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.58–6.91 (m, 4H, Ar–H), 3.96 (s, 2H, CH2). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 173.9, 164.6, 158.0, 157.8, 132.2, 130.6, 119.5, 118.4, 116.3, 33.4. Elemental analysis: calcd: C, 53.00; H, 4.45; N, 16.86; S, 12.86. found: C, 5 2.70; H, 4.20; N, 16.66; S, 12.71.
N), 7.58–6.91 (m, 4H, Ar–H), 3.96 (s, 2H, CH2). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 173.9, 164.6, 158.0, 157.8, 132.2, 130.6, 119.5, 118.4, 116.3, 33.4. Elemental analysis: calcd: C, 53.00; H, 4.45; N, 16.86; S, 12.86. found: C, 5 2.70; H, 4.20; N, 16.66; S, 12.71.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of ethanol and water.
1 mixture of ethanol and water.
2-(4-Chlorobenzylidene)hydrazinylidene)-5-(2-hydroxybenzylidene)thiazolidin-4-one (3a). Yellow crystals, yield 23%, m.p. 297–300 °C. FTIR (cm−1); 3744(OH), 3039(CH)Ar, 2753(CH)Al, 1693(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1641(C
O), 1641(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1592(C
N), 1592(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 821(C–Cl). 1HNMR (300 MHz, DMSO-d6): δ ppm; 11.84 (s, H, NH), 10.25 (s, 1H, OH), 8.51 (s, 1H, CH
C), 821(C–Cl). 1HNMR (300 MHz, DMSO-d6): δ ppm; 11.84 (s, H, NH), 10.25 (s, 1H, OH), 8.51 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.89 (s, 1H, CH
N), 7.89 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.84–6.96 (m, 8H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 168.1, 160.8, 157.1, 156.2, 135.5, 132.9, 131.6, 129.6(2 × C), 129.1(2 × C), 128.4, 124.3, 121.6, 120.7, 119.7, 116.1. MS (m/z, %): 357 (M+, 30). Elemental analysis: calcd: C, 57.07; H, 3.38; Cl, 9.91; N, 11.74; S, 8.96; found: C, 57.00; H, 3.20; Cl, 9.65; N, 11.54; S, 8.75.
C), 7.84–6.96 (m, 8H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 168.1, 160.8, 157.1, 156.2, 135.5, 132.9, 131.6, 129.6(2 × C), 129.1(2 × C), 128.4, 124.3, 121.6, 120.7, 119.7, 116.1. MS (m/z, %): 357 (M+, 30). Elemental analysis: calcd: C, 57.07; H, 3.38; Cl, 9.91; N, 11.74; S, 8.96; found: C, 57.00; H, 3.20; Cl, 9.65; N, 11.54; S, 8.75.
2-(Benzylidene)hydrazinylidene)-5-(2-hydroxybenzylidene)thiazolidin-4-one (3b). Yellow crystals, yield 61%, m.p. 298–300 °C. FTIR (cm−1); 3249(OH), 3058(CH)Ar, 2764(CH)Al, 1676(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1641(C
O), 1641(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1590(C
N), 1590(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 465(OH, Enol). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.47 (s, 1H, NHCO/N
C), 465(OH, Enol). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.47 (s, 1H, NHCO/N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH), 10.68 (s, 1H, OH), 8.51 (s, 1H, CH
C–OH), 10.68 (s, 1H, OH), 8.51 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.91 (s, 1H, CH
N), 7.91 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.84–6.99 (m, 9H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.8, 160.0, 157.5, 157.2, 134.0, 131.5, 130.9, 128.9, 128.4, 128.0(2 × C), 124.4, 121.5, 120.7, 119.6, 116.2. MS (m/z, %): 323 (M+, 8). Elemental analysis: calcd: C, 63.14; H, 4.05; N, 12.99; S, 9.91; found: C, 63.01; H, 4.00; N, 12.85; S, 9.67.
C), 7.84–6.99 (m, 9H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.8, 160.0, 157.5, 157.2, 134.0, 131.5, 130.9, 128.9, 128.4, 128.0(2 × C), 124.4, 121.5, 120.7, 119.6, 116.2. MS (m/z, %): 323 (M+, 8). Elemental analysis: calcd: C, 63.14; H, 4.05; N, 12.99; S, 9.91; found: C, 63.01; H, 4.00; N, 12.85; S, 9.67.
5-(2-Hydroxybenzylidene)-2-(4-methoxybenzylidene)hydrazinylidene)thiazolidin-4-one (3c). Yellow crystals, yield 53.19%, m.p. 272–274 °C. FTIR (cm−1); 3400(OH), 3222(NH), 3058(CH)Ar, 2762(CH)Al, 1686(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1636(C
O), 1636(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1598(C
N), 1598(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 468(OH, Enol). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.38 (s, 1H, NHCO/N
C), 468(OH, Enol). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.38 (s, 1H, NHCO/N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH), 10.49 (s, 1H, OH), 8.43 (s, 1H, CH
C–OH), 10.49 (s, 1H, OH), 8.43 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.88 (s, 1H, C
N), 7.88 (s, 1H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.78–6.96 (m, 8H, Ar–H), 3.81 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.8, 161.5, 158.6, 157.1, 157.1, 131.5, 129.7(2× C), 128.3, 126.6, 124.1, 121.6, 120.7, 119.6, 116.1, 114.4(2 × C), 55.4. MS (m/z, %): 353 (M+, 18). Elemental analysis: calcd: C, 61.18; H, 4.28; N, 11.89; S, 9.07; found: C, 61.09; H, 4.15; N, 11.63; S, 9.02.
CH), 7.78–6.96 (m, 8H, Ar–H), 3.81 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.8, 161.5, 158.6, 157.1, 157.1, 131.5, 129.7(2× C), 128.3, 126.6, 124.1, 121.6, 120.7, 119.6, 116.1, 114.4(2 × C), 55.4. MS (m/z, %): 353 (M+, 18). Elemental analysis: calcd: C, 61.18; H, 4.28; N, 11.89; S, 9.07; found: C, 61.09; H, 4.15; N, 11.63; S, 9.02.
5-(2-Hydroxybenzylidene)-2-(2-hydroxybenzylidene)hydrazinylidene)thiazolidin-4-one (3d). Yellow crystals, yield 51%, m.p. 312–314 °C. FTIR (cm−1); 3426(OH), 3154(NH), 2933(CH)Ar, 1677(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1591(C
O), 1591(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1523(C
N), 1523(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.53 (s, 1H, NHCO/N
C). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.53 (s, 1H, NHCO/N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH), 10.68 (s, 1H, OH), 10.46 (s, 1H, OH), 8.72 (s, 1H, CH
C–OH), 10.68 (s, 1H, OH), 10.46 (s, 1H, OH), 8.72 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.91 (s, 1H, C
N), 7.91 (s, 1H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.68–6.98 (m, 8H, H–Ar). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.6, 159.0, 158.0, 157.5, 157.1, 132.5, 132.1, 131.8, 129.9, 128.4, 124.9, 120.9, 120.5, 119.7, 118.8, 116.4, 116.1. MS (m/z, %): 339 (M+, 23). Elemental analysis: calcd: C, 60.17; H, 3.86; N, 12.38; S, 9.45; found: C, 59.87; H, 3.61; N, 12.18; S, 9.19.
CH), 7.68–6.98 (m, 8H, H–Ar). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.6, 159.0, 158.0, 157.5, 157.1, 132.5, 132.1, 131.8, 129.9, 128.4, 124.9, 120.9, 120.5, 119.7, 118.8, 116.4, 116.1. MS (m/z, %): 339 (M+, 23). Elemental analysis: calcd: C, 60.17; H, 3.86; N, 12.38; S, 9.45; found: C, 59.87; H, 3.61; N, 12.18; S, 9.19.
2-(4-Chlorobenzylidene)hydrazinylidene)-5-(5-(4-chlorophenyl)diazenyl)-2-hydroxybenzylidene)thiazolidin-4-one (4a) [Cl/Cl dye]. Red crystals, yield 78%, m.p. 296–298 °C. FTIR (cm−1); 3833(OH), 3236(NH), 3044(CH)Ar, 2750(CH)Al, 1681(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1639(C
O), 1639(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1589(C
N), 1589(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1488(N
C), 1488(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 822(C–Cl), 468(OH, Enol). 1HNMR (300 MHz, DMSO-d6): δ ppm; 11.64 (s, 1H, NH) 10.41 (s, 1H, OH), 8.50 (s, 1H, CH
N), 822(C–Cl), 468(OH, Enol). 1HNMR (300 MHz, DMSO-d6): δ ppm; 11.64 (s, 1H, NH) 10.41 (s, 1H, OH), 8.50 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.07 (s, 1H, CH
N), 8.07 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.88–6.95 (m, 11H, Ar–H). 13C-NMR (100 MHz, DMSO-d6)): δ ppm; 174.4, 161.0, 158.6, 151.3, 145.4(2× C), 145.0, 133.0, 132.8(2× C), 130.2(2× C), 126.5(2 × C), 125.5(2 × C), 124.8, 124.7,123.5, 120.9, 118.4, 118.6, 116.8. MS (m/z, %): 496 (M+, 12). Elemental analysis: calcd: C, 55.66; H, 3.05; Cl, 14.28; N, 14.11; S, 6.46; found: C, 55.53; H, 3.00; Cl, 14.08; N, 14.07; S, 6.33.
C), 7.88–6.95 (m, 11H, Ar–H). 13C-NMR (100 MHz, DMSO-d6)): δ ppm; 174.4, 161.0, 158.6, 151.3, 145.4(2× C), 145.0, 133.0, 132.8(2× C), 130.2(2× C), 126.5(2 × C), 125.5(2 × C), 124.8, 124.7,123.5, 120.9, 118.4, 118.6, 116.8. MS (m/z, %): 496 (M+, 12). Elemental analysis: calcd: C, 55.66; H, 3.05; Cl, 14.28; N, 14.11; S, 6.46; found: C, 55.53; H, 3.00; Cl, 14.08; N, 14.07; S, 6.33.
2-(4-Chlorobenzylidene)hydrazinylidene)-5-(2-hydroxy-5-(p-tolyldiazenyl)benzylidene)thiazolidin-4-one (4b)[Cl/Me dye]. Red crystals, yield 70%, m.p. 277–280 °C. FTIR (cm−1); 3480(OH), 3250(NH), 3030(CH)Ar, 2775(CH)Al, 1682(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1632 (C
O), 1632 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1591(C
N), 1591(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1488(N
C), 1488(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1HNMR (300 MHz, DMSO-d6): δ ppm; 9.92 (s, 1H, NH), 9.15 (s, 1H, OH), 8.50 (s, 1H, CH
N). 1HNMR (300 MHz, DMSO-d6): δ ppm; 9.92 (s, 1H, NH), 9.15 (s, 1H, OH), 8.50 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.10 (s, 1H, CH
N), 8.10 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.85–6.90 (m, 11H, Ar–H), 2.42 (s, 3H, CH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 168.0, 163.6, 160.9, 150.5, 145.7(2× C), 145.3, 141.8, 133.0, 130.4(2× C), 128.8(2× C), 125.5(2× C), 123.1, 122.9(2× C), 121.4, 119.3, 117.0, 116.9, 116.6, 21.0. MS (m/z, %): 475 (M+, 40). Elemental analysis: calcd: C, 60.57; H, 3.81; Cl, 7.45; N, 14.71; S, 6.74; found: C, 60.45; H, 3.66; Cl, 7.32; N, 14.63; S, 6.68.
C), 7.85–6.90 (m, 11H, Ar–H), 2.42 (s, 3H, CH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 168.0, 163.6, 160.9, 150.5, 145.7(2× C), 145.3, 141.8, 133.0, 130.4(2× C), 128.8(2× C), 125.5(2× C), 123.1, 122.9(2× C), 121.4, 119.3, 117.0, 116.9, 116.6, 21.0. MS (m/z, %): 475 (M+, 40). Elemental analysis: calcd: C, 60.57; H, 3.81; Cl, 7.45; N, 14.71; S, 6.74; found: C, 60.45; H, 3.66; Cl, 7.32; N, 14.63; S, 6.68.
2-(Benzylidene)hydrazinylidene)-5-(5-(4-chlorophenyl)diazenyl)-2-hydroxybenzylidene)thiazolidin-4-one (5a)[H/Cl dye]. Red crystals, yield 89%, m.p. 288–290 °C. FTIR (cm−1); 3470(OH), 3236(NH), 3061(CH)Ar, 2777(CH)Al, 1682(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1637(C
O), 1637(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1590(C
N), 1590(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1483(N
C), 1483(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 833(C–Cl). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.19 (s, 1H, NH), 10.36 (s, 1H, OH), 8.53 (s, 1H, CH
N), 833(C–Cl). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.19 (s, 1H, NH), 10.36 (s, 1H, OH), 8.53 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.10 (s, 1H, CH
N), 8.10 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.97–6.96 (m, 12H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.8, 160.5, 159.9, 157.5, 150.5, 144.8, 135.4, 133.9, 131.1, 129.6(2 × C), 128.9(2 × C), 128.3, 127.8(2 × C), 126.8(2 × C), 123.9, 122.9, 122.7, 121.5, 116.7. MS (m/z, %): 461 (M+, 12). Elemental analysis: calcd: C, 59.80; H, 3.49; Cl, 7.67; N, 15.16; S, 6.94; found: C, 59.56; H, 3.42; Cl, 7.15; N, 15.09; S, 6.75.
C), 7.97–6.96 (m, 12H, Ar–H). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.8, 160.5, 159.9, 157.5, 150.5, 144.8, 135.4, 133.9, 131.1, 129.6(2 × C), 128.9(2 × C), 128.3, 127.8(2 × C), 126.8(2 × C), 123.9, 122.9, 122.7, 121.5, 116.7. MS (m/z, %): 461 (M+, 12). Elemental analysis: calcd: C, 59.80; H, 3.49; Cl, 7.67; N, 15.16; S, 6.94; found: C, 59.56; H, 3.42; Cl, 7.15; N, 15.09; S, 6.75.
2-(Benzylidene)hydrazinylidene)-5-(2-hydroxy-5-(p-tolyldiazenyl)benzylidene)thiazolidin-4-one (5b)[H/Me dye]. Red crystals, yield 77%, m.p. 287–290 °C. FTIR (cm−1); 3649(OH), 3160(NH), 3060(CH)Ar, 2920(CH)Al, 1676(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1638(C
O), 1638(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1589(C
N), 1589(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1500(N
C), 1500(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 468(OH, Enol). 1HNMR (300 MHz, DMSO-d6): δ ppm; 12.03 (s, 1H, NH), 9.20 (s, 1H, OH), 8.54 (s, 1H, CH
N), 468(OH, Enol). 1HNMR (300 MHz, DMSO-d6): δ ppm; 12.03 (s, 1H, NH), 9.20 (s, 1H, OH), 8.54 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.09 (s, 1H, CH
N), 8.09 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 7.92–7.14 (m, 12H, Ar–H), 2.42 (s, 3H, CH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.6, 159.9, 157.5, 149.9, 145.0, 141.2, 134.0, 131.1, 130.6, 130.3, 129.9, 128.9, 128.4, 128.3, 127.9, 127.2, 123.1, 122.3, 121.7, 121.3, 120.7, 120.6, 116.5, 21.1. MS (m/z, %): 441 (M+, 46). Elemental analysis: calcd: C, 65.29; H, 4.34; N, 15.86; S, 7.26. Found: C, 65.14; H, 4.22; N, 15.73; S, 7.12.
C) 7.92–7.14 (m, 12H, Ar–H), 2.42 (s, 3H, CH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.6, 159.9, 157.5, 149.9, 145.0, 141.2, 134.0, 131.1, 130.6, 130.3, 129.9, 128.9, 128.4, 128.3, 127.9, 127.2, 123.1, 122.3, 121.7, 121.3, 120.7, 120.6, 116.5, 21.1. MS (m/z, %): 441 (M+, 46). Elemental analysis: calcd: C, 65.29; H, 4.34; N, 15.86; S, 7.26. Found: C, 65.14; H, 4.22; N, 15.73; S, 7.12.
5-(5-(4-Chlorophenyl)diazenyl)-2-hydroxybenzylidene)-2-(4-methoxybenzylidene)hydrazinylidene) thiazolidin-4-one (6a)[MeO/Cl dye]. Yellow crystals, yield 78%, m.p. 302–304 °C. FTIR (cm−1); 3490(OH), 3200(NH), 3066(CH)Ar, 2767(CH)Al, 1681(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1644(C
O), 1644(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1591(C
N), 1591(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1512(N
C), 1512(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 828(C–Cl), 465(OH, Enol). 1HNMR (300 MHz, DMSO-d6): δ ppm; 12.31 (s, 1H, NHCO/N
N), 828(C–Cl), 465(OH, Enol). 1HNMR (300 MHz, DMSO-d6): δ ppm; 12.31 (s, 1H, NHCO/N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH), 10.36 (s, 1H, OH), 8.44 (s, 1H, CH
C–OH), 10.36 (s, 1H, OH), 8.44 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.09(CH
N), 8.09(CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.96–7.01 (m, 11H, Ar–H), 3.82 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 161.6, 160.3, 158.3, 157.8, 157.3, 150.5, 145.7, 136.2, 135.3, 134.4, 131.6, 130.8, 129.6, 128.1, 126.5, 124.0, 123.4, 123.1, 123.0, 122.7, 121.5, 116.7, 114.8, 55.4. MS (m/z, %): 491 (M+, 34). Elemental analysis: calcd: C, 58.60; H, 3.69; Cl, 7.21; N, 14.24; S, 6.52; found: C, 58.43; H, 3.50; Cl, 7.16; N, 14.21; S, 6.35.
C), 7.96–7.01 (m, 11H, Ar–H), 3.82 (s, 3H, OCH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 161.6, 160.3, 158.3, 157.8, 157.3, 150.5, 145.7, 136.2, 135.3, 134.4, 131.6, 130.8, 129.6, 128.1, 126.5, 124.0, 123.4, 123.1, 123.0, 122.7, 121.5, 116.7, 114.8, 55.4. MS (m/z, %): 491 (M+, 34). Elemental analysis: calcd: C, 58.60; H, 3.69; Cl, 7.21; N, 14.24; S, 6.52; found: C, 58.43; H, 3.50; Cl, 7.16; N, 14.21; S, 6.35.
5-(2-Hydroxy-5-(p-tolyldiazenyl)benzylidene)-2-(4-methoxybenzylidene)hydrazinylidene)thiazolidine-4-one (6b)[MeO/Me dye]. Red crystals, yield 77%, m.p. 298–300 °C. FTIR (cm−1); 3400(OH), 3200(NH), 3062(CH)Ar, 2762(CH)Al, 1682(C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1640(C
O), 1640(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1593(C
N), 1593(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1511(N
C), 1511(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 463(OH, Enol). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.54 (s, 1H, NHCO), 10.35 (s, 1H, OH), 8.47 (s, 1H, CH
N), 463(OH, Enol). 1H-NMR (300 MHz, DMSO-d6): δ ppm; 12.54 (s, 1H, NHCO), 10.35 (s, 1H, OH), 8.47 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.09 (s, 1H, CH
N), 8.09 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 7.91–7.03 (m, 11H, Ar–H), 3.84 (s, 3H, OCH3), 2.42 (s, 3H, CH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.3, 161.6, 159.7, 158.1, 157.2, 152.8, 149.9, 145.0, 141.2, 129.9(2× C), 129.6, 126.7, 126.5, 123.2, 122.8, 122.3(2× C), 122.0, 121.3, 116.5, 114.4(2× C), 55.3, 21.0. MS (m/z, %): 471 (M+, 31). Elemental analysis: calcd: C, 63.68; H, 4.49; N, 14.85; S, 6.80; found: C, 63.46; H, 4.43; N, 14.67; S, 6.63.
C), 7.91–7.03 (m, 11H, Ar–H), 3.84 (s, 3H, OCH3), 2.42 (s, 3H, CH3). 13C-NMR (75 MHz, DMSO-d6): δ ppm; 167.3, 161.6, 159.7, 158.1, 157.2, 152.8, 149.9, 145.0, 141.2, 129.9(2× C), 129.6, 126.7, 126.5, 123.2, 122.8, 122.3(2× C), 122.0, 121.3, 116.5, 114.4(2× C), 55.3, 21.0. MS (m/z, %): 471 (M+, 31). Elemental analysis: calcd: C, 63.68; H, 4.49; N, 14.85; S, 6.80; found: C, 63.46; H, 4.43; N, 14.67; S, 6.63.
Biological activity
Docking study
The default docking protocol of MOE 2014 software was applied according to the methodology described in the supplementary data.32,33Statistical analysis
All experiments were performed in triplicate, and the results were statistically analyzed. Data were expressed as mean ± standard error (SEM). Statistical significance was determined using two-way analysis of variance (ANOVA) followed by Sidak's multiple comparisons test, with a significance threshold set at p < 0.05. Additionally, nonlinear regression analysis was performed on the normalized transformed data to fit the dose–response curves.Results and discussion
Chemistry
The development of novel compounds with potential anticancer activity remains a major focus due to the persistent need for selective and effective therapeutic agents. Molecular hybridisation, which combines pharmacologically active fragments from different molecules, offers a strategy for generating hybrid scaffolds with enhanced biological properties compared with their individual precursors.Following this approach, a series of 2-[4-substituted-benzylidene]-hydrazine-1-carbothioamide derivatives (1a–f) was synthesised by condensing various aromatic aldehydes—4-chlorobenzaldehyde, benzaldehyde, 4-methoxybenzaldehyde, 2-hydroxybenzaldehyde, 4-hydroxybenzaldehyde, and 4-hydroxy-3-methoxybenzaldehyde—with thiosemicarbazide in ethanol using a catalytic amount of acetic acid (Scheme 1).34,35 The structures were confirmed by FTIR, 1H NMR, and 13C NMR spectroscopy. In the 1H NMR spectra, N–H protons appeared at 11.21–12.02 ppm, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N protons at 8.01–8.39 ppm, and NH2 protons at 7.79–8.07 ppm. The 13C NMR spectra showed thioxo (C
N protons at 8.01–8.39 ppm, and NH2 protons at 7.79–8.07 ppm. The 13C NMR spectra showed thioxo (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) carbons at 177.16–178.09 ppm, consistent with the proposed structures.
S) carbons at 177.16–178.09 ppm, consistent with the proposed structures.
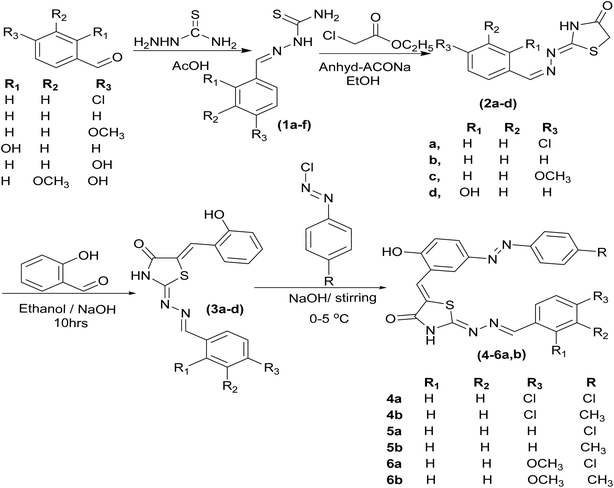 | ||
| Scheme 1 Synthetic routes to thiosemicarbazones (1a–f), thiazolidine derivatives (2a–d), chalcones (3a–c) and diazenyl-chalcones (4–6a,b). | ||
The thiosemicarbazone intermediates 1a–d was cyclised with ethyl chloroacetate in ethanol in the presence of anhydrous sodium acetate under reflux to yield 2-(4-substituted-benzylidene)-hydrazinylidene-thiazolidin-4-one derivatives (2a–d).36–38 This step generated the thiazolidin-4-one ring, a key pharmacophoric element for subsequent hybridisation. Spectroscopic analysis indicated tautomeric mixtures of ketone and enol forms. In 1H NMR spectra, the SCH2 protons of the thiazolidinone ring appeared as singlets at 3.82–3.96 ppm, while CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N protons resonated at 8.39–9.43 ppm. The 13C NMR spectra exhibited characteristic signals for the carbonyl carbon (C
N protons resonated at 8.39–9.43 ppm. The 13C NMR spectra exhibited characteristic signals for the carbonyl carbon (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) at 173.93–174.36 ppm and the methylene carbon (CH2) at 33.03–33.42 ppm.
O) at 173.93–174.36 ppm and the methylene carbon (CH2) at 33.03–33.42 ppm.
Next, a Claisen–Schmidt condensation of 2a–d with salicylaldehyde in ethanol containing NaOH produced the chalcone derivatives 3a–d, forming 2-(4-substituted-benzylidene)hydrazinylidene-5-(2-hydroxybenzylidene)thiazolidin-4-one hybrids.24,30,39,40 These compounds incorporate both thiazolidinone and hydroxybenzylidene motifs, enhancing planarity and conjugation. Spectroscopic characterization confirmed their structures and Z-geometry: the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH methine proton appeared at 7.88–8.51 ppm, and the absence of methylene proton signals from the thiazolidinone ring, with the phenolic OH proton observed as a singlet at 10.25–10.66 ppm. IR, 13C NMR, mass spectrometry, and elemental analysis further supported structural assignment.
CH methine proton appeared at 7.88–8.51 ppm, and the absence of methylene proton signals from the thiazolidinone ring, with the phenolic OH proton observed as a singlet at 10.25–10.66 ppm. IR, 13C NMR, mass spectrometry, and elemental analysis further supported structural assignment.
Finally, diazenyl chalcone derivatives 4–6a,b was obtained by diazotization of the corresponding aromatic amines followed by electrophilic coupling with chalcone intermediates 3a–c. This sequence efficiently introduced the azo (–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) linkage onto the 2-(4-substituted-benzylidene)hydrazinylidene-5-(2-hydroxybenzylidene)thiazolidin-4-one scaffold, affording hybrid molecules with extended π-conjugation and enhanced structural rigidity.
N–) linkage onto the 2-(4-substituted-benzylidene)hydrazinylidene-5-(2-hydroxybenzylidene)thiazolidin-4-one scaffold, affording hybrid molecules with extended π-conjugation and enhanced structural rigidity.
Spectroscopic data confirmed the proposed structures. The IR spectra of compounds 4–6a,b exhibited characteristic absorptions for N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N at 1480–1515 cm−1, C
N at 1480–1515 cm−1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 1676–1682 cm−1, and broad O–H bands at 3300–3649 cm−1. Their 1H NMR spectra showed aromatic proton resonances in the region of 6.96–7.91 ppm. As a representative example, compound 6b displayed IR bands at 1682 cm−1 (C
O at 1676–1682 cm−1, and broad O–H bands at 3300–3649 cm−1. Their 1H NMR spectra showed aromatic proton resonances in the region of 6.96–7.91 ppm. As a representative example, compound 6b displayed IR bands at 1682 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1640 cm−1 (C
O), 1640 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), and 1511 cm−1 (N
N), and 1511 cm−1 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). Its 1H NMR spectrum featured singlets at 10.35, 8.47, 8.09, 3.84, and 2.42 ppm, corresponding to OH, CH
N). Its 1H NMR spectrum featured singlets at 10.35, 8.47, 8.09, 3.84, and 2.42 ppm, corresponding to OH, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH, OCH3, and CH3 protons, respectively, along with a downfield singlet at approximately 12.65 ppm assigned to the NH–CO tautomeric form. The 13C NMR spectrum of 6b further supported the structure, giving prominent resonances at 167.37 ppm (C
CH, OCH3, and CH3 protons, respectively, along with a downfield singlet at approximately 12.65 ppm assigned to the NH–CO tautomeric form. The 13C NMR spectrum of 6b further supported the structure, giving prominent resonances at 167.37 ppm (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 161.60 ppm (C–OH), 55.38 ppm (OCH3), and 21.03 ppm (CH3).
O), 161.60 ppm (C–OH), 55.38 ppm (OCH3), and 21.03 ppm (CH3).
Mass spectral analysis of 6b showed a molecular ion at m/z 471 (M+, 31%) consistent with C25H21N5O3S, along with a base peak at m/z 468 (100%) corresponding to C25H18N5O3S, confirming the proposed molecular framework. A complete set of correlation data is provided in the SI.
Overall, this synthetic strategy [Scheme 1] afforded a series of diazenyl chalcone hybrids incorporating the 2-arylidene hydrazinylidene thiazolidin-4-one core. These molecules integrate multiple pharmacophores—including the chalcone α,β-unsaturated carbonyl system, the thiazolidinone moiety, the hydroxybenzylidene group, and the azo linkage—features that are expected to act synergistically to enhance biological properties, particularly anticancer potential.
Visible spectroscopy
The structures of the synthesised diazenyl chalcones were validated using the UV-Vis absorption spectra (4–6a,b). In Witt's chromophore theory, bathochromic shifts are caused by the modulation of π-conjugation by substituents that donate electrons (such as NHCO CH3, OCH3 or CH3) on one aromatic ring and by substituents that withdraw electrons (such as NO2, CN, or halogens) on the other. To find the maximum absorption wavelengths (λmax), the spectra were recorded in DMSO at room temperature in this investigation. Fig. 1 shows the predicted visible absorption spectra of the six dyes. Dye 4a (Cl/Cl) and dye 4b (Cl/CH3) are highly visible-light absorbers. When compared to dye 4a, dye 4b exhibits a more pronounced red shift. The methyl group on the diazenyl-benzylidene molecule donates electrons, reducing the energy required for the electronic transition. There is a clear absorption band at about 540 nm for dye 5a (H/Cl) and a broader and less intense band for dye 5b (H/CH3), which indicates less conjugation because of the methyl group. There is more complexity in the spectra of dyes 6a (OCH3/Cl) and 6b (OCH3/CH3). The absorption peak at approximately 550 nm is clearly visible in compound 6a, whereas a wider, redder band centred at about 538 nm is seen in dye 6b, suggesting weaker electronic transitions. Generally, changes in the substituents on the benzylidene units greatly impact the complexity, location, and strength of the visible absorption bands. Due to the increased conjugation between the diazenyl group and the thiazolidin-4-one core, the 4-chlorophenyl derivatives (Cl/Cl, H/Cl, OCH3/Cl) exhibit stronger, blue-shifted transitions. Because the electron-donating methyl group reduces conjugation effectiveness, the p-tolyl derivatives (Cl/CH3, H/CH3, OCH3/CH3) show weaker, red-shifted absorptions. These spectral features highlight the synthesised dyes' potential as wavelength-selective spectroscopic probes and dye-sensitised solar cell possibilities. | ||
| Fig. 1 UV-visible absorption spectra for diazenyl chalcones disperse dyes incorporating 2-arylidene hydrazineylidene thiazole. | ||
Biological activity
We assessed the cell viability of synthetic compounds (4–6a,b) using the MTT assay on normal HSF cells, MDA (breast cancer), and HepG2 (liver cancer) cells (Table S1 and Fig. S7). Most compounds exhibited little to no toxicity on normal cells at concentrations of 100 µg mL−1 or lower. Furthermore, except for compound 4a, all compounds demonstrated anticancer activity at concentrations below the safe dose. No significant differences in cytotoxicity were observed between MDA and HepG2 cells, though compounds 5a, 5b, and 6a showed the highest selectivity indices, as presented in Table 1. The selectivity indices for 5a, 5b, and 6a were 30.84, 34.14, and 36.27 against MDA, and 28.89, 34.68, and 38.93 against HepG2 cells, respectively. Interestingly, Table 1 indicates that the compounds 5a, 5b, and 6a exhibited more anticancer activity against HepG2 cells with IC50 values of 27.92 ± 2.33, 24.17 ± 2.67 and 22.26 ± 2.79 µM, respectively, potent than 5-FU (32.14 ± 2.72 µM). Furthermore, compounds 5a, 5b, and 6a showed IC50 value of 26.16 ± 3.81, 24.56 ± 2.28 and 23.89 ± 3.37 µM more than 5-FU (27.91 ± 1.64 µM) as presented in Table 1. Microscopic examination revealed noticeable changes in MDA (Fig. 2) and HepG2 cells (Fig. 3), including cell shrinkage and nuclear condensation, particularly at higher concentrations, indicating apoptosis induction. Fluorescence microscopy using acridine orange/ethidium bromide (AO/EB) staining confirmed apoptosis, with viable cells exhibiting green fluorescence and apoptotic cells displaying greenish-yellow to red fluorescence due to membrane disruption. An ideal anticancer compound exhibits cytotoxic effects on cancer cells at low concentrations (low IC50) while maintaining minimal toxicity to normal cells at higher concentrations (high CC50), resulting in a high selectivity index (SI = CC50/IC50). The selectivity index is a well-established parameter used to assess the therapeutic window and in vitro efficacy of anticancer agents.38| Compound | HSF | HepG2 | MDA | ||
|---|---|---|---|---|---|
| CC50 | IC50 | SI | IC50 | SI | |
| a Statistical analysis was performed using two-way ANOVA followed by Sidak's multiple comparisons test, showing no significant differences (p > 0.05) in selectivity index (SI) between MDA and HepG2 cells across all conditions. | |||||
| 4a | 707.07 ± 7.39 | 574 ± 2.57 | 1.23 | 331.72 ± 4.06 | 2.14 |
| 4b | 794.74 ± 4.66 | 31.66 ± 2.91 | 25.09 | 37.71 ± 3.13 | 21.08 |
| 5a | 806.72 ± 7.04 | 27.92 ± 2.33 | 28.89 | 26.16 ± 3.81 | 30.84 |
| 5b | 611.56 ± 5.46 | 24.17 ± 2.67 | 34.68 | 24.56 ± 2.28 | 34.14 |
| 6a | 866.6 ± 6.61 | 22.26 ± 2.79 | 38.93 | 23.89 ± 3.37 | 36.27 |
| 6b | 973.25 ± 4.83 | 137.71 ± 4.37 | 7.07 | 97.75 ± 4.09 | 9.96 |
| 5-FU | 37.83 ± 3.11 | 32.14 ± 2.72 | 1.18 | 27.91 ± 1.64 | 1.36 |
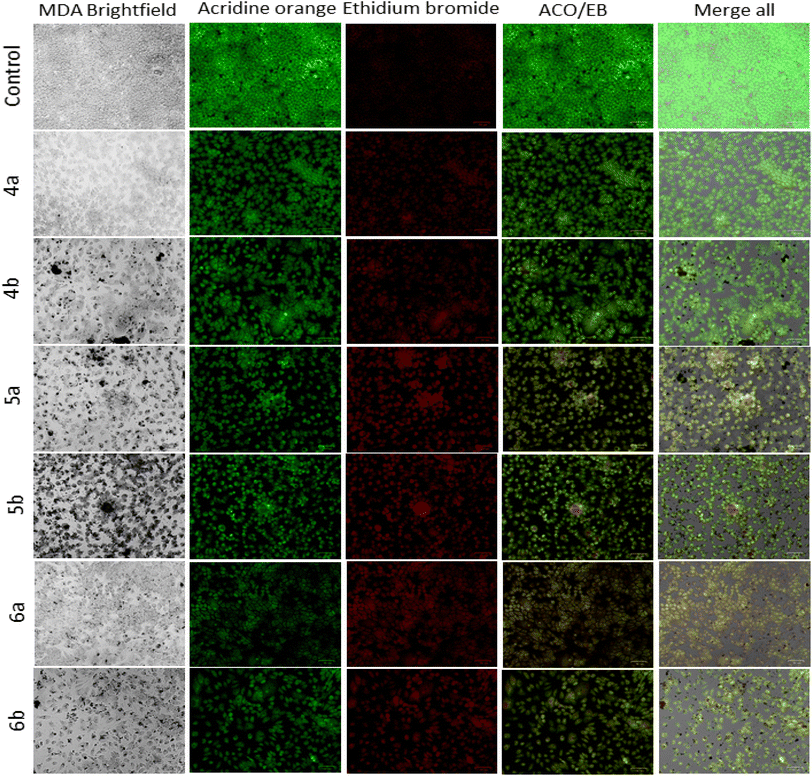 | ||
| Fig. 2 Fluorescent microscopy analysis of MDA cells treated with synthetic compounds (4–6a,b) and stained with acridine orange/ethidium bromide (AO/EB). | ||
 | ||
| Fig. 3 Fluorescent microscopy analysis of HepG2 cells treated with synthetic compounds (4–6a,b) and stained with acridine orange/ethidium bromide (AO/EB). | ||
Bright field images show the viability of MDA cells after treatment with varying concentrations of synthetic compounds (4–6a,b). Fluorescent images display live cells in green (stained with AO) and dead or apoptotic cells in red (stained with EB). The merged AO/EB images distinguish live cells (green), early apoptotic cells (yellow/orange), and late apoptotic cells (red). The final column in the figure represents a merged image combining all fluorescence channels, including acridine orange (green) and ethidium bromide (red), overlaid with the brightfield image.
Bright field images show the viability of HepG2 cells after treatment with varying concentrations of synthetic compounds (4–6a,b). Fluorescent images display live cells in green (stained with AO) and dead or apoptotic cells in red (stained with EB). The merged AO/EB images distinguish live cells (green), early apoptotic cells (yellow/orange), and late apoptotic cells (red). The final column merges all channels, providing a comprehensive overview of cell viability and structural integrity across the different treatments.
Our study evaluated the migration inhibitory effects of synthetic substances on MDA and HepG2 cells by a wound healing experiment. Phase-contrast photographs of the wound area at IC50 doses demonstrated substantial migratory inhibition relative to untreated control cancer cells. The yellow dashed lines denote the wound margins at 0 and 48 hours (Fig. 4A and 5A). Compounds 5a, 5b, and 6a exhibited a pronounced anti-migration effect on MDA cells, whilst compounds 5b and 6a displayed a comparable effect on HepG2 cells, as indicated by the minimal wound closure percentages and reduced migration rates (Fig. 4B and 5B). Additionally, gene expression study revealed that the synthetic compounds (4–6a,b) downregulated the expression of matrix metalloproteinases (MMP-2 and MMP-9), with the exception of compound 4a. These enzymes are crucial facilitators of cancer cell migration and invasion. The expression of MMP-2 was diminished relative to untreated cells, indicating that the compounds exercise their anti-metastatic actions by disrupting essential pathways associated with tumor cell migration and metastasis (Fig. 4C and 5C).
Docking study
We conducted docking study in order to get insights on the affinity and binding patterns of the new designed molecules to some important enzymes highly related to cancer, especially to cell migration process. On the basis of promising biological effects of the new derivatives as inhibitors to cell migration, we suggested that Matrix metalloproteinase-2 (MMP-2) and matrix metalloproteinase-9 (MMP-9) are likely to be suitable proteins for docking study. Especially gene expression study revealed that the synthetic compounds (4–6a,b) downregulated the expression of both MMP-2 and MMP-9. Molecular operating environment software (MOE) was used to make calculations and suggest binding free energies and binding modes according to default docking protocol. The crystal structure of the enzymes (ID: 1HOV and 1GKC for MMP-2 and MMP-9, respectively) were obtained from protein data bank website (PDB). The obtained results revealed that the new derivatives have strong affinities to both MMP-2 and MMP-9. The binding free energies of the new molecules and ligands are outlined in Table 2.| Comp. ID | Ligand | 4a | 4b | 5a | 5b | 6a | 6b |
|---|---|---|---|---|---|---|---|
| 1HOV score (kcal mol−1) | −33.73 | −25.19 | −22.80 | −14.98 | −29.72 | −32.27 | −32.67 |
| 1GKC score (kcal mol−1) | −14.95 | −24.32 | −23.76 | −23.38 | −23.70 | −23.30 | −21.54 |
It can be noticed that the new molecules showed good affinity to MMP-2 receptor ranging from −32.67 to −14.98 kcal mol−1. Compounds 6a and 6b showed the best results with scores (−32.67 and −32.27 kcal mol−1, respectively) comparable to that of the ligand (33.73 kcal mol−1). Compound 5b came in the third order with score of −29.72 kcal mol−1. Compounds 4a and 4b showed relatively weaker scores, while compound 5a was far weaker than other derivatives. With respect to MMP-9 receptor, the new derivatives revealed far better scores (ranging from −24.32 to −21.54 kcal mol−1) than that of the ligand (−14.95 kcal mol−1). It is clear that docking scores of the new candidates are ranged from −24.32 to −21.54 kcal mol−1, indicating comparable binding affinities. Compound 4a came first with a score of −24.32 kcal mol−1. Compound 6b was the last one, while compounds 4b, 5a, 5b, and 6a were comparable to 4a. Regarding the binding modes of the new molecules to MMP-2, different binding interactions to amino acids of the active site were obtained. Fig. 6 illustrates the binding pattern of compound 6b to 1HOV pocket, where it formed three hydrogen bonds with Val117, Ala136, and Thr143. Additionally, it displayed a Pi–Pi interaction with His130 and a Pi–cation interaction with His 120. Moreover, it revealed hydrophobic interactions with Leu150, Val42, Phe148, and Leu137.
Binding mode of compound 6a is illustrated in Fig. 7. It can be noticed that 6a showed hydrogen bonds with Ala136 and Thr143. It also revealed a Pi–Pi interaction with Phe148 and a Pi–cation interaction with His120. In addition to hydrophobic interactions with Arg149, Val42, Ala119, and Leu137.
Meanwhile, the binding mode of the designed compounds to MMP-9 are represented by Fig. 8 and 9, which illustrate the interaction patterns of compounds 4a and 5b, respectively. We can see that compound 4a exhibited hydrogen bonds with Ala189 and Leu188 and Pi–alkyl interactions with Val398, Leu188, Leu187, and Arg424. In addition, it revealed amide–Pi interaction with Leu397 and hydrophobic interaction with His401.
Similarly, compound 5b showed hydrogen bonds with Leu188 and Ala189 and amide Pi stacking with Leu397. Furthermore, it displayed Pi–alkyl interactions with Val398, Leu187, Leu188, Arg424, and Pro421. In addition to hydrophobic interaction with Leu418 (see Fig. 10).
Structure–activity relationship (SAR)
Cytotoxic activity and selectivity index of the synthesised azo derivatives (4–6a,b) were evaluated against MDA-MB-231 and HepG2 cancer cell lines, with selectivity assessed relative to normal WI-38 cells. Table 1 shows that the biological activity of these compounds depends heavily on the nature and position of substituents on the phenyl rings. Among all synthesised derivatives, compound 6a exhibited the highest cytotoxic selectivity, with SI values of 36.27 (MDA-MB-231) and 38.93 (HepG2). The presence of both methoxy (–OCH3) and chloro (–Cl) substituents is what makes this activity so impressive. The chloro group increases molecular hydrophobicity and thereby improves membrane permeability, while the methoxy group contributes additional hydrophobic interactions with Val42, Ala119, and Leu137. As an electron-donating substituent, –OCH3 also enhances π–π stacking with Phe148, collectively strengthening the ligand–protein binding affinity. On the other hand, compound 4a had the lowest selectivity (SI = 2.14 for MDA-MB-231; 1.23 for HepG2). The presence of only a chlorine substituent reduces electronic richness and limits the variety and strength of stabilising interactions within the active site, resulting in weaker cytotoxic performance.Compounds 4b, 5a, and 5b demonstrated consistently high selectivity (SI = 21.08–34.14 for MDA-MB-231; 25.09–34.68 for HepG2). These results can be attributed to the presence of both electron-withdrawing (Cl) and electron-donating (CH3) groups on the aromatic system. The combination of these substituents creates a balanced electronic distribution that enhances intermolecular contacts and promotes stable ligand–protein complexes.
In comparison, compound 6b showed moderate selectivity (SI = 9.96 for MDA-MB-231; 7.07 for HepG2). The reduced activity suggests that symmetrical substitution—whether strongly electron-donating or electron-withdrawing—results in a less favourable electronic arrangement that compromises optimal binding and consequently diminishes anticancer potency.
As illustrated in Fig. 11, the SAR analysis indicates the following trends:
Mixed substituents (EDG + EWG) favour high selectivity and potency, likely due to enhanced molecular polarisation and improved binding stability.
Symmetrical substituents reduce cytotoxicity, implying an electronic imbalance that does not support strong protein interactions.
The azo group (–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) plays a crucial role in biological activity by extending π-conjugation and facilitating additional π–π and hydrophobic interactions.
N–) plays a crucial role in biological activity by extending π-conjugation and facilitating additional π–π and hydrophobic interactions.
Author contributions
N. D.: suggesting points of research [Schemes 1], supervision, methodology development, formal analysis of all the synthesised compounds [Schemes 1], writing–review & editing. D. L.: supervision, and formal analysis of the synthesised compounds. M. M. E.: preparation of all the synthesised compounds. E. M. E. and Y. A. E. are responsible for evaluating the biological activities, conducting formal analyses, performing investigations, visualising results, and writing the original draft. A. E. A.: the molecular docking study, writing, editing and reviewing. F. A. T.: the UV-Vis study, writing, editing and reviewing, and methodology.Conflicts of interest
The authors declare no competing interests.Data availability
All data generated or analysed during this study are included in this published article as supplementary information (SI) files. Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra07769a.Acknowledgements
The authors are grateful and would like to thank the Academy of Scientific Research and Technology, Cairo, Egypt for their financial contribution to this research, provided through the scientist for the Next generation (SNG), (Cycle 8), grant in industry areas, Egyptian Petroleum Research Institute (EPRI), Al-Azhar University/Faculty of Science (girls), and City of Scientific Research and Technological Applications (SRTA-City). This opportunity has been pivotal in my academic and professional growth, allowing me to pursue my research with dedication and focus.References
- R. Rajaraman, D. L. Guernsey, M. M. Rajaraman and S. R. Rajaraman, Cancer Cell Int., 2006, 6, 1–26 CrossRef.
- R. Singh, Key Heterocyclic Cores for Smart Anticancer Drug–Design Part I, Bentham Science Publishers, 2022 Search PubMed.
- J. Ceramella, D. Iacopetta, A. Catalano, F. Cirillo, R. Lappano and M. S. Sinicropi, Antibiotics, 2022, 11(191), 1–23 Search PubMed.
- K. Shahzad Munawar, S. Muhammad Haroon, S. A. Hussain and H. Raza, J. Basic Appl. Sci., 2018, 14, 217–229 CrossRef.
- C. M. da Silva, D. L. da Silva, L. V. Modolo, R. B. Alves, M. A. de Resende, C. V. B. Martins and Â. de Fátima, J. Adv. Res., 2011, 2, 1–8 CrossRef.
- A. Hameed, M. Al-Rashida, M. Uroos, S. Abid Ali and K. M. Khan, Expert Opin. Ther. Pat., 2017, 27, 63–79 CrossRef CAS PubMed.
- D. Iacopetta, R. Lappano, A. Mariconda, J. Ceramella, M. S. Sinicropi, C. Saturnino, M. Talia, F. Cirillo, F. Martinelli, F. Puoci, C. Rosano, P. Longo and M. Maggiolini, Int. J. Mol. Sci., 2020, 21, 7797 CrossRef CAS PubMed.
- A. Pandurangan, N. Singh and A. K. Tiwari, Acad. Sci., 2012, 4, 5–11 Search PubMed.
- A. Kajal, S. Bala, S. Kamboj, N. Sharma and V. Saini, J. Catal., 2013, 2013, 1–14 Search PubMed.
- R. N. Sharma, F. P. Xavier, K. K. Vasu, S. C. Chaturvedi and S. S. Pancholi, J. Enzyme Inhib. Med. Chem., 2009, 24, 890–897 CrossRef CAS PubMed.
- W. D. Alrohily, M. E. Habib, S. M. El-Messery, A. Alqurshi, H. El-Subbagh and E.-S. E. Habib, Microb. Pathog., 2019, 136, 103674 Search PubMed.
- O. Bozdağ-Dündar, M. Ceylan-Ünlüsoy, V. Eugen and R. Ertan, Arzneimittelforschung, 2011, 56, 621–625 Search PubMed.
- T. A. Farghaly, G. S. Masaret, Z. A. Muhammad and M. F. Harras, Bioorg. Chem., 2020, 98, 103761 Search PubMed.
- A. Mohammadi-Farani, A. R. Foroumadi, M. R. Kashani and A. R. Aliabadi, Iran. J. Basic Med. Sci., 2014, 17, 502–508 Search PubMed.
- B. Z. Kurt, I. Gazioglu, F. Sonmez and M. Kucukislamoglu, Bioorg. Chem., 2015, 59, 80–90 Search PubMed.
- R. K. Singh, D. N. Prasad and T. R. Bhardwaj, Med. Chem. Res., 2015, 24, 1534–1545 CrossRef CAS.
- C. Tratrat, A. Petrou, A. Geronikaki, M. Ivanov, M. Kostić, M. Soković, I. S. Vizirianakis, N. F. Theodoroula and M. Haroun, Molecules, 2022, 27, 1930 Search PubMed.
- W. S. Liu, R. R. Wang, H. Yue, Z. H. Zheng, X. H. Lu, S. Q. Wang, W. L. Dong and R. L. Wang, J. Biomol. Struct. Dyn., 2020, 38, 3814–3824 CrossRef CAS PubMed.
- A. D. Patel, T. Y. Pasha, P. Lunagariya, U. Shah, T. Bhambharoliya and R. K. P. Tripathi, ChemMedChem, 2020, 15, 1229–1242 CrossRef CAS.
- D. Kaminskyy, A. Kryshchyshyn and R. Lesyk, Eur. J. Med. Chem., 2017, 140, 542–594 CrossRef CAS.
- R. Morphy and Z. Rankovic, J. Med. Chem., 2005, 48, 6523–6543 CrossRef CAS.
- M. Bhat, B. Poojary, B. S. Kalal, P. M. Gurubasavaraja Swamy, S. Kabilan, V. Kumar, N. Shruthi, S. A. Alias Anand and V. R. Pai, Future Med. Chem., 2018, 10, 1017–1036 CrossRef CAS PubMed.
- S. D. Tupare, S. Digamber and T. K. E. S. Anandibai, Int. J. Sci. Res., 2024, 9, 292–297 Search PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65(1–2), 55–63 CrossRef CAS PubMed.
- E. M. El-Fakharany, W. B. Elsharkawy, Y. A. El-Maradny and H. El-Gendi, J. Food Sci., 2024, 89, 5130–5149 CrossRef CAS.
- N. T. A. Dawoud, E. M. El-Fakharany, A. E. Abdallah, H. El-Gendi and D. R. Lotfy, Sci. Rep., 2022, 12, 1–17 Search PubMed.
- M. A. Ebraheem, E. M. El-Fakharany, S. M. Husseiny and F. A. Mohammed, Microb. Cell Factories, 2024, 23, 200 CrossRef CAS PubMed.
- S. E. El-Didamony, M. H. Kalaba, M. H. Sharaf, E. M. El-Fakharany, A. Osman, M. Sitohy and B. Sitohy, Front. Microbiol., 2024, 15 DOI:10.3389/fmicb.2024.1419917.
- H. M. Habib, E. M. El-Fakharany, U. D. Souka, F. M. Elsebaee, M. G. El-Ziney and W. H. Ibrahim, , DOI: DOI:10.3390/nu14173536.
- H. M. Habib, E. M. El-Fakharany, E. Kheadr and W. H. Ibrahim, Sci. Rep., 2022, 12, 12393 CrossRef CAS PubMed.
- A. E. Abdallah, S. I. Eissa, M. M. S. Al Ward, R. R. Mabrouk, A. B. M. Mehany and M. A. El-Zahabi, Bioorg. Chem., 2021, 109, 104695 CrossRef CAS PubMed.
- A. E. Abdallah, M. S. Alesawy, S. I. Eissa, E. M. El-Fakharany, M. H. Kalaba, M. H. Sharaf, N. M. Abo Shama, S. H. Mahmoud, A. Mostafa, A. A. Al-Karmalawy and H. Elkady, New J. Chem., 2021, 45(36), 16557–1657 RSC.
- M. Donmez, M. Sekerci, R. Adiguzel, E. Oğuz, F. Türkan, U. Yildiko and N. Colak, Mol. Diversity, 2024, 29, 1109–1127 CrossRef.
- M. Islam, A. Khan, M. T. Shehzad, A. Hameed, N. Ahmed, S. A. Halim, M. Khiat, M. U. Anwar, J. Hussain, R. Csuk, Z. Shafiq and A. Al-Harrasi, Bioorg. Chem., 2019, 87, 155–162 CrossRef CAS.
- E. M. El-Fakharany, N. T. A. Dawoud, H. El-Gendi, A. E. Abdallah and D. R. Lotfy, Egypt. J. Chem., 2021, 64, 6565–6576 Search PubMed.
- N. T. A. Dawoud, H. El-Gendi, D. R. Lotfy, E. M. El-Fakharany and M. H. Abdellattif, ChemistrySelect, 2024, 9, e202402828 CrossRef CAS.
- B. N. N. Mallappa, G. Chandra Sharma, M. Jangir, A. Sharma and N. Singh Chauhan, Results Chem., 2024, 10, 101662 CrossRef CAS.
- M. M. El-Samoly, N. T. A. Dawoud, E. M. El-Fakharany, H. M. Mashaly, D. M. Abbas and D. R. Lotfy, Egypt. J. Chem., 2026, 69, 613–622, DOI:10.21608/ejchem.2025.415928.12225.
- Z. Ngaini, M. A. Jefferi and S. Farooq, Nat. Prod. Res., 2024, 1–10 CrossRef.
- G. M. El-Sherbiny, M. H. Kalaba, A. M. Foda, A. S. E. D. Youssef, I. A. Elsehemy, E. E. Farghal and E. M. El-Fakharany, Microb. Pathog., 2024, 192, 106705 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |








