Polyimidazolium amphiphilic dendrimers on thiacalix[4]arene and gallic acid platforms via copper-free click chemistry: synthesis, self-assembly and DNA binding
Ilshat M.
Bogdanov
a,
Angelina A.
Fedoseeva
a,
Anastasiya A.
Glukhova
a,
Elza D.
Sultanova
 a,
Timur A.
Mukhametzyanov
a,
Timur A.
Mukhametzyanov
 a,
Vladimir G.
Evtugyn
b,
Svetlana E.
Solovieva
b,
Vladimir A.
Burilov
a,
Vladimir G.
Evtugyn
b,
Svetlana E.
Solovieva
b,
Vladimir A.
Burilov
 *a and
Igor S.
Antipin
*a and
Igor S.
Antipin
 a
a
aAlexander Butlerov Institute of Chemistry, Kazan Federal University, 18 Kremlevskaya Str., 420008 Kazan, Russia. E-mail: ultrav@bk.ru
bArbuzov Institute of Organic and Physical Chemistry, FRC Kazan Scientific Center of RAS, 8 Arbuzov Str., 420088 Kazan, Russia
First published on 19th November 2025
Abstract
A divergent synthetic scheme based on copper-free click chemistry was used to obtain two types of bromine-containing dendrimers with a thiacalix[4]arene or gallic acid core. These dendrimers were then terminated with cationic imidazolium groups via the Menshutkin reaction to obtain a series of amphiphilic dendrimers. The critical aggregation concentrations of the dendrimers were determined using three different probes. It was demonstrated that the solubilization capacity with respect to the hydrophobic Orange OT substrate logically increases with the transition from the second to the third generation, whereas the first generation does not solubilize the dye. In the case of the thiacalix[4]arene core, the most compact aggregates (approximately 80 nm) were obtained in the second generation. In the case of gallic acid, the most compact aggregates (approximately 30 nm) were obtained in the second and third generations, with the third generation forming the most monodisperse particles. Steady-state and time-resolved fluorescence spectroscopy revealed that the third generation on both the gallic acid and thiacalix[4]arene platforms interacts most effectively with calf thymus DNA, displacing ethidium bromide. The addition of third-generation dendrimers causes DNA compaction, forming particles measuring approximately 70–120 nm, and results in surface recharging to +40 mV. Since the CD data show that the addition of dendrimers results in DNA ordering (transition to the C-form), the obtained second- and third-generation dendrimers are promising as potential matrices for stabilization, storage, or delivery of nucleic acids in future studies.
Introduction
The development of dendrimers has greatly advanced supramolecular chemistry, bio/nanotechnology and materials science.1–5 Amphiphilic dendrimers (ADs), which combine hydrophilic and hydrophobic dendritic domains into a single macromolecule, can self-assemble into vesicle-like dendrimerosomes,6,7 an ability that gives them an advantage over conventional symmetric dendrimers. The size of dendrimerosomes can be controlled by adjusting the volumes of the hydrophilic and hydrophobic fragments (influencing the amphiphile packing parameter). Any groups can be introduced into the polar part of ADs, including biocompatible ligands, targeting fragments and probes for visualization. The combination of internal ‘pockets’ and hydrophobic fragments enables the encapsulation of drugs. When equipped with cationic head groups, dendrimerosomes can effectively interact with nucleic acids to form compact lipoplexes (DNA and RNA). Therefore, ADs show great promise in biotechnology for encapsulating and delivering both drugs and nucleic acids for selective gene therapy.8–12 ADs also demonstrate superiority over other systems in catalysis. The numerous cavities of dendrimers, which are rich in sigma-donor atoms, stabilize metal particles.13,14 Their ability to self-assemble and encapsulate hydrophobic reagents also allows metal complex catalysis to be supplemented by micellar catalysis.15,16The copper-catalysed azide–alkyne cycloaddition (CuAAC) reaction is one of the most convenient tools for constructing and functionalizing dendrimers.17–19 It is also one of the most common click chemistry reactions and the 2022 Nobel Prize was awarded for it.20 However, despite its many advantages, the CuAAC reaction has one significant disadvantage: residual copper impurities are difficult to remove from the final dendrimers because the metal is trapped in chelated polythiazole ‘pockets’.21 When considering dendrimers for use in biotechnology, the issues of purity and toxic copper content become acute; copper ions can easily bind with amino acid residues, damaging the structure and function of proteins and causing the formation of reactive oxygen species.22 The copper-free Huisgen reaction (cycloaddition of azides with activated alkynes or AAC) does not have these drawbacks. The AAC reaction is underutilized in click chemistry, despite the fact that the rate, atom economy, and ease of isolation and purification of AAC reaction products are often advantageous compared to those of the CuAAC reaction.
There are also just a few examples of copper-free AAC chemistry being used in dendrimer synthesis.23–25
Thanks to its ability to selectively introduce lipophilic and hydrophilic moieties onto the lower rim, the thiacalixarene platform enables the production of a variety of AD structures.15,16,26 Combining the thiacalixarene platform with the copper-free click chemistry approach will facilitate the production of a broad spectrum of ADs with potential biomedical applications.
Although the thiacalix[4]arene core provides effective spatial separation of lipophilic and hydrophilic fragments while preserving the hydrophobic cavity, simpler molecular platforms that provide amphiphilic architectures can also be used to create ADs. In this regard, particular attention in dendrimer's chemistry is paid to gallic acid derivatives. Gallic acid derivatives are most commonly used as a branching point in the construction of dendrimers.27,28 However, they can also act as a dendrimer core. Necessary polar dendron groups can be introduced into the O-alkylated gallic acid core using CuAAC chemistry, which has been successfully demonstrated for the synthesis of self-assembling Janus dendrimers29,30 and switchable amphiphilic rotaxanes,31 and as a hydrophobic fragment for amphiphilic proteins using micelle-assisted activity-based protein-labeling technology.32
We herein propose a divergent approach for the construction of ADs up to the third generation, based on the AAC reaction, involving two cores: gallic acid and p-tert-butylthiacalix[4]arene. The aggregation characteristics of the obtained dendrimers and their use in interaction with the model DNA will be demonstrated.
Results and discussion
Synthesis
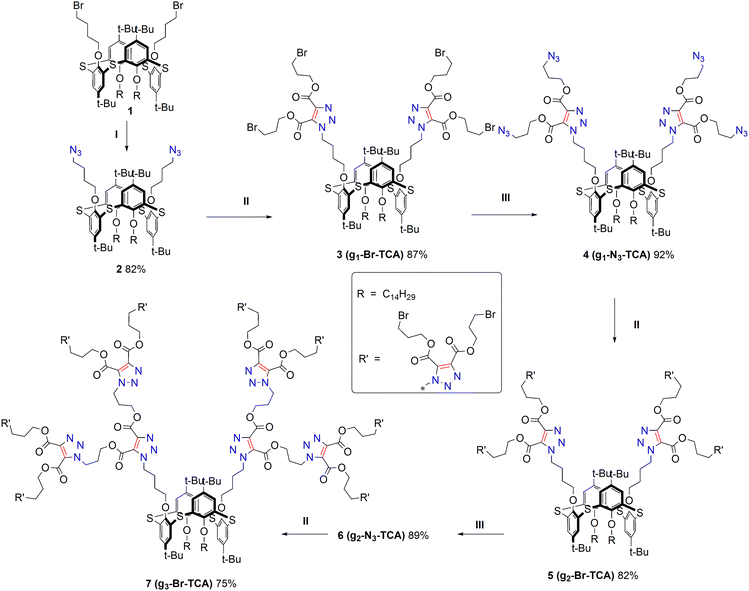
A divergent scheme involving 1,4-bis(3-bromopropyl)-2-butynedioate as a key focal module was chosen for the copper-free AAC reaction to construct dendrimers up to the third generation. To this end, a core consisting of thiacalix[4]arene 2, with tetradecyl fragments on one side of the macrocyclic platform and azidobutyl fragments on the other side, was synthesized from the corresponding bromide33 derivative 1 using a 10-fold excess of sodium azide in DMF at 120 °C under microwave heating (400 W). The structure of the target azide 2 was confirmed by a set of physical methods. In the 1H NMR spectrum of the product, a signal from the –CH2N3 protons undergoes an upfield shift of 0.72 ppm compared to the parent bromide 1.In the IR spectrum, the most characteristic feature is the appearance of an intense absorption band in the region of 2100 cm−1, corresponding to the stretching asymmetric vibrations of the azide group. In the high-resolution mass spectrum with electrospray ionization (HRESI MS), a peak of the cationized molecule [M + NH4]+ was detected (Fig. S1). Core 2 was subjected to an AAC click reaction with 2.2 equiv. of 1,4-bis(3-bromopropyl)-2-butynedioate in toluene at 65 °C (Scheme 1). According to TLC, complete conversion of the initial substrate was achieved within 6 hours. The target product was isolated in 87% yield. In the 1H NMR spectrum of compound 3, the proton signal from the –CH2Trz fragments shifts downfield (Δ 1.36 ppm), with new multiplets appearing at δ 4.54–4.47, 3.58–3.51 and 2.38–2.23 ppm (Fig. 1). These correspond to the proton signals from the ester (–CH2–CH2–CH2–Br) methylene groups. The most notable feature of the 13C NMR spectrum is the appearance of four new signals in the range of 130–170 ppm. Thus, the signals at 160.3 and 158.3 ppm correspond to the signals of the non-equivalent carbon atoms of carbonyl groups, and the signals at 140.3 and 129.4 ppm correspond to the signals of triazole ring carbons. The IR spectrum of compound 3 shows a new band at 1735 cm−1, characteristic of the stretching vibrations of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds in esters. The HRESI mass spectrum revealed peaks of the cationized molecules [M + H]+, [M + NH4]+ and [M + K]+ (Fig. S2).
O bonds in esters. The HRESI mass spectrum revealed peaks of the cationized molecules [M + H]+, [M + NH4]+ and [M + K]+ (Fig. S2).
For the subsequent generation growth, dendrimer 3 was further introduced into the activation stage of peripheral groups by substitution with sodium azide in DMF. The reaction lasted for 6 hours at 65 °C. The yield of azide 4 after isolation was 92%. In the 1H NMR spectrum of azide G1 4, replacing the bromide with azide results in a shift of 0.05 ppm and 0.3 ppm in the signals of the protons of the α- and β-methylene fragments, respectively.
An absorption band from the azide group at 2100 cm−1 is observed in the IR spectrum of product 6. A series of cationized molecule ions were detected in the high-resolution mass spectrum.
Thus, dendrimers of the second and third generations containing 8 (compound 5) and 16 (compound 7) bromide fragments at the periphery were synthesized by repeating the AAC reaction and substitution with sodium azide sequentially. The 1H NMR spectra show (Fig. 1) that the AAC reaction of compounds 4 and 6 with 1,4-bis(3-bromopropyl)-2-butynedioate results in a significant downfield shift (Δ 1.32 ppm) of the proton signal from the –CH2–N3 methylene groups, as well as a smaller shift (Δ 0.24 ppm) of the β-methylene proton signals. Changes in the NMR spectra going from the first to the third generation can clearly be traced by the –CH2–Br proton signals at 3.1–3.6 ppm, as these do not overlap with any other signals.
Thus, the integrated intensity increases from 8 to 32 protons from the first to the third generation (compounds 3, 5, and 7), which is in full agreement with the structure. Additionally, from the second generation onwards, a signal from the –CH2-Trz protons appears at 4.7–4.9 ppm, corresponding to an integrated intensity of 8 protons for compounds 6 and 7, or 24 protons for compound 7. Moving from the first to the third generation in the 13C NMR spectra reveals an increase in the number of signals from the carbon atoms of the triazole fragments, which correspond to the C4 and C5 carbon atoms of the triazole ring at 1.40 and 1.29 ppm, respectively (Fig. S7). Thus, the spectrum of the first-generation dendrimer 3 shows two signals, whereas the spectrum of the second-generation dendrimer 5 shows six signals.
A similar increase in signals is observed in the 158–160 ppm region, which corresponds to the signals of the carbon atoms of the carboxyl groups. HRESI mass spectrometry revealed the peaks of cationized molecule ions formed by the addition of H+, NH4+, Na+ and K+, which confirmed the composition of the dendrimers.
In the course of the work, dendrimers with bromide groups based on a non-macrocyclic gallic acid platform were also obtained (Scheme 2). An azide derivative of tris(tetradecyl)methyl gallate 8 was chosen as the core. This compound was obtained according to the literature method.34 Similarly, by repeating the sequence of the click reaction and substitution with sodium azide, dendrimers 9–13 from the first to third generations were obtained with yields of 86%, 81% and 75%, respectively (Scheme 3). The structures of compounds 9–13 were proven and characterized by a set of physical research methods. 1H NMR analysis (see Fig. S8–S12) of the first-generation dendrimer 9 reveals two triplets of proton signals corresponding to the –CH2–O– fragment in the 4.5 ppm region and two triplets corresponding to the –CH2–Br fragment in the 3.5 ppm region. The Ar-CH2-Trz protons appear as a singlet at 5.69 ppm. When transitioning to the second generation (compound 11), two triplets of CH2-Trz protons appear at 4.79 and 4.75 ppm. The –CH2–O– protons of the inner layer appear as two triplets at 4.43 and 4.38 ppm. The proton signals of the outer layer –CH2–O– and –CH2–Br fragments appear at 4.50 and 3.53 ppm, with an integral intensity corresponding to eight protons. In the third-generation dendrimer, the success of the modification can be judged by the appearance of a signal from the –CH2–Br protons, which has an intensity corresponding to 16 protons. As an example, Fig. S19 shows fragments of the 13C NMR spectra of compounds 8, 9, 11 and 13. Just as in the case of dendrimers based on the thiacalixarene core, the appearance of signals from the nonequivalent carbon atoms of the triazole ring (C4 and C5 atoms) and carboxyl carbon atoms is observed. For compound 11, 6 signals of the 6 magnetically non-equivalent triazole carbons appear. For compound 13, 12 signals out of the 14 signals appear due to their overlap. The IR spectra of the dendrimers show an intense band at 1735 cm−1, corresponding to the stretching vibrations of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds in esters. Peaks of cationized molecule ions formed by the addition of H+, NH4+, Na+ and K+ were observed in the high-resolution mass spectra of all dendrimers.
O bonds in esters. Peaks of cationized molecule ions formed by the addition of H+, NH4+, Na+ and K+ were observed in the high-resolution mass spectra of all dendrimers.
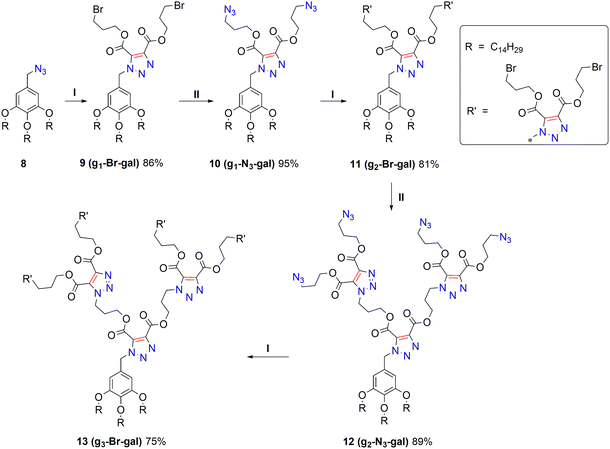 | ||
| Scheme 2 Syntheses of I–III generation dendrimers based on the gallic acid core. (I) 1,4-Bis(3-bromopropyl)-2-butynedioate, toluene, 65 °C, 6 h; (II) NaN3, DMF, 65 °C, 6 h. | ||
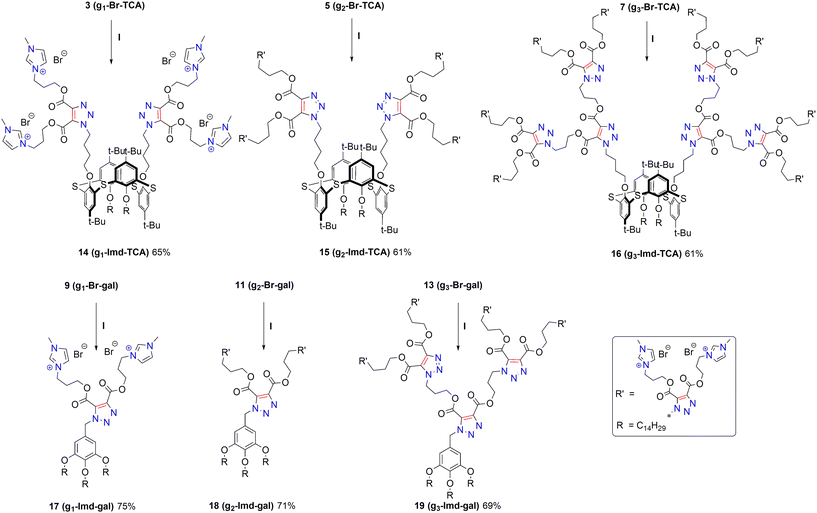 | ||
| Scheme 3 Synthesis of imidazolium dendrimers based on thiacalixarene (14–16) and methyl gallate (17–19). (I) N-Methylimidazole, acetonitrile, 80 °C, 10 h. | ||
The bromine-containing dendrimers were then subjected to a quaternization reaction with N-methylimidazole (Scheme 3). N-Methylimidazole was used at a ratio of 1.2 equivalents per bromide group. The reaction was carried out in acetonitrile at 80 °C. Complete conversion of the initial compounds was achieved after 10 hours. The yields of the final products were 61–75%. The structure of the obtained compounds was proven by a set of physical research methods. Fig. S20 shows the 1H NMR spectra of the starting bromide 9 and its quaternization product 17 as an example. The most characteristic feature in the spectra is the appearance of new signals of the imidazolium ring protons at δ 9.22 ppm, 9.16 ppm, 7.80 ppm, 7.74 ppm, 7.71 ppm and 7.68 ppm, as well as the signals of the methyl group protons at δ 3.90–3.82 ppm, which overlap with the signals of the –CH2OAr fragment protons. The signals of –CH2–Br in the initial bromide 9 are shifted downfield by Δ 0.81 ppm as a result of quaternization. Cationized molecule ions [M − nBr]+n were observed in the HRESI mass spectra for all dendrimers except thiacalixarene 16 due to the difficulty of ionizing a molecule with a charge of +16 (Fig. S13–S18).
Aggregation studies
The 1st–3rd generation dendrimers obtained from the thiacalix[4]arene (TCA-G1–3) and gallic acid (Gal-G1–3) cores have an amphiphilic structure due to the presence of positively charged imidazolium groups and 2 or 3 hydrophobic tetradecyl fragments. Their aggregation properties were studied using fluorescence and spectrophotometric methods, as well as dynamic light scattering (DLS) (Table 1). To determine the critical aggregation concentration (CAC), a fluorescence method was used with a pyrene probe, which is known for its ability to change the ratio of the emission intensity of the first and third emission bands (I1 and I3, Fig. 2) when it enters the hydrophobic layer of aggregates.35 In this way, it is possible to determine the CACpyrene from the plot of I1/I3 pyrene emission vs. the concentration of dendrimers using the Boltzmann equation (see SI Table S1). The formation of pyrene excimers is also observed when transitioning to the micellar cavity (Fig. 2A). Next, we examined the ability of the formed complexes to solubilize the water-insoluble dye Orange OT. Orange OT is a hydrophobic probe, completely insoluble in water; therefore, it exhibits no absorption at 495 nm without the presence of a surfactant. The addition of a surfactant results in the appearance of absorption at 495 nm due to encapsulation of the dye within the non-polar core of aggregates.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 cm−1); 25 °C
400 M−1 cm−1); 25 °C
| System | CACpyrene, mM | CACsol, mM | CACEosin, mM | S, OT, mM | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D D |
|---|---|---|---|---|---|
| Gal-G1 | 1.05 | — | 0.091 | 10.20 | |
| Gal-G2 | 0.09 | 0.03 | 0.011 | 66 | 3.08 |
| Gal-G3 | 0.07 | 0.015 | 0.004 | 103 | −11.16 |
| TCA-G1 | 0.04 | — | 0.016 | — | 12.38 |
| TCA-G2 | 0.02 | 0.024 | 0.007 | 87 | −1.86 |
| TCA-G3 | 0.05 | 0.038 | 0.003 | 108 | −30.34 |
This can be used to determine the solubilization CAC constant (CACsol),36 while demonstrating the potential of using dendrimers as nanocontainers for water-insoluble substances. The solubilizing ability of the aggregates (S), which corresponds to the number of moles of Orange OT solubilized by one mole of the dendrimer, is determined according to eqn (1):37
| S = B/(εext × l) | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 cm−1). Interestingly, both the first-generation amphiphilic molecules do not solubilize Orange OT within the 0–0.5 mM concentration range, whereas their second- and third-generation analogues do, with the solubilizing ability increasing with each generation (Table 1). The thiacalixarene-based dendrimers exhibit greater solubilizing ability; it appears that the macrocyclic cavity also contributes to solubilization.38
400 M−1 cm−1). Interestingly, both the first-generation amphiphilic molecules do not solubilize Orange OT within the 0–0.5 mM concentration range, whereas their second- and third-generation analogues do, with the solubilizing ability increasing with each generation (Table 1). The thiacalixarene-based dendrimers exhibit greater solubilizing ability; it appears that the macrocyclic cavity also contributes to solubilization.38
Solubilization only occurs after the formation of associates, which enables the CACsol to be determined (Table 1). If the absence of solubilization in this concentration range (CACpyrene was 1 mM) for Gal-G1 is logical, then for TCA-G1, it is possibly due to the formation of loose associates without a pronounced hydrophobic core. Notably, for both pyrene and Orange OT, the CAC increases when transitioning from the second to the third generation of dendrimers on the thiacalixarene platform. It appears that the optimal hydrophilic–lipophilic balance is achieved in the second generation of the macrocycle containing eight charged groups and that the hydrophobicity of the two tetradecyl fragments is clearly insufficient when transitioning to TCA-G3 with sixteen charged imidazolium groups.
The interaction between the dendrimers and a negatively charged dye Eosin Y was studied (Fig. S21 and Table S2). This interaction occurs due to electrostatic forces and the hydrophobic effect. Quenching and a subsequent bathochromic shift are associated with Eosin Y entering a non-polar environment due to its solubilisation in micelles.39 This dye transition can be used to determine the CACEosin of the dendrimers (Table 1). Furthermore, in contrast to the CAC values determined using uncharged probes (pyrene and Orange OT), partial charge compensation occurs in the case of Eosin Y, leading to a decrease in CAC values.40 Due to compensation for the dendrimer's positive charge, electrostatic repulsion decreases, reflected in a decrease in CAC values. Thus, for the TCA-G3 macrocycle, which has bulky charged fragments, a CACEosin decrease of one order of magnitude is observed.
The average size and polydispersity index of the dendrimers were determined before and after the CAC using the DLS method (Table 2). All dendrimers exhibit a polymodal intensity distribution with the presence of both small nanoparticles and submicron/micron aggregates, likely formed by micelle aggregation (Table S3). The largest aggregates are formed by the first-generation dendrimers; in the case of Gal-G1, micron-sized particles are formed, while in the case of TCA-G1, the sizes approach the micron scale. After the CAC, all dendrimers form aggregates smaller than those in the pre-micellar region.
| System | C, mM | PDI | Z, nm |
|---|---|---|---|
| 20 mM Tris with pH 7.4, 25 °C. | |||
| Gal-G1 | 0.025–0.1 | — | — |
| 1 | 1 | 7935 ± 1500 | |
| 1.5 | 0.541 ± 0.057 | 1423 | |
| Gal-G2 | 0.025 | 0.520 ± 0.017 | 36 ± 1 |
| 0.05 | 0.436 ± 0.053 | 32 ± 1 | |
| 0.1 | 0.545 ± 0.096 | 35 ± 1 | |
| 0.25 | 0.454 ± 0.017 | 33 ± 1 | |
| Gal-G3 | 0.025 | 0.310 ± 0.015 | 46 ± 13 |
| 0.05 | 0.302 ± 0.018 | 28 ± 2 | |
| 0.1 | 0.215 ± 0.001 | 20 ± 0.5 | |
| 0.25 | 0.482 ± 0.048 | 51 ± 0.5 | |
| TCA-G1 | 0.005 | — | — |
| 0.05 | 0.408 ± 0.064 | 535 ± 11 | |
| 0.08 | 0.481 ± 0.008 | 561 ± 2 | |
| TCA-G2 | 0.005 | 0.925 ± 0.029 | 66 ± 1 |
| 0.03 | 0.579 ± 0.004 | 80 ± 1 | |
| 0.06 | 0.626 ± 0.013 | 73 ± 1 | |
| 0.1 | 0.512 ± 0.003 | 226 ± 2 | |
| TCA-G3 | 0.005 | 0.545 ± 0.008 | 711 ± 5 |
| 0.06 | 0.584 ± 0.122 | 356 ± 52 | |
| 0.08 | 0.575 ± 0.021 | 349 ± 100 | |
| 0.1 | 0.561 ± 0.144 | 367 ± 102 | |
At the same time, dendrimers based on thiacalix[4]arene form larger aggregates compared to the gallic dendrimers. For thiacalix[4]arene dendrimers, the most compact particles are formed by the TCA-G2 dendrimers. It appears that in this case, the optimal amphiphile packing parameter41 (the optimal ratio of the volume of the polar head group to the hydrophobic part) is achieved. Conversely, the Gal-G2 and Gal-G3 dendrimers are characterized by the most compact sizes and a lower polydispersity index. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D (ref. 42) values for the synthesized dendrimers were also calculated. According to the data obtained (Table 1), the Gal-G1 and TCA-G1 dendrimers have an extremely high positive log
D (ref. 42) values for the synthesized dendrimers were also calculated. According to the data obtained (Table 1), the Gal-G1 and TCA-G1 dendrimers have an extremely high positive log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D value, indicating their high lipophilicity, which appears to be the driving force behind the formation of micron-sized aggregate associates. In the case of the 2nd generation dendrimers, the log
D value, indicating their high lipophilicity, which appears to be the driving force behind the formation of micron-sized aggregate associates. In the case of the 2nd generation dendrimers, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D value differs significantly between the two different cores. Thus, in the case of Gal-G2, lipophilicity prevails, while in the case of TCA-G2, hydrophilicity prevails, which is associated with the twofold higher number of polar groups in the latter case. Strong hydrophobic interactions in the case of the Gal-G2 dendrimer contribute to the formation of more compact aggregates. Finally, for the 3rd generation dendrimers, there is also a huge difference in log
D value differs significantly between the two different cores. Thus, in the case of Gal-G2, lipophilicity prevails, while in the case of TCA-G2, hydrophilicity prevails, which is associated with the twofold higher number of polar groups in the latter case. Strong hydrophobic interactions in the case of the Gal-G2 dendrimer contribute to the formation of more compact aggregates. Finally, for the 3rd generation dendrimers, there is also a huge difference in log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D values: the Gal-G3 dendrimer remains more hydrophobic, while the TCA-G3 dendrimer has the highest negative log
D values: the Gal-G3 dendrimer remains more hydrophobic, while the TCA-G3 dendrimer has the highest negative log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D, which is due to 16 ionized imidazolium groups. In this case, as in the case of the second generation dendrimers, the greater lipophilicity of the Gal-G3 dendrimer leads to the formation of more compact aggregates. The tendency to form aggregate associates is clearly visible for both the gallic and thiacalix[4]arene dendrimers. Thus, with a further increase in the concentration (0.25 mM for Gal-G3 and 0.1 mM for TCA-G2), the size and polydispersity index increase significantly, indicating the coalescence of aggregates into larger ones.
D, which is due to 16 ionized imidazolium groups. In this case, as in the case of the second generation dendrimers, the greater lipophilicity of the Gal-G3 dendrimer leads to the formation of more compact aggregates. The tendency to form aggregate associates is clearly visible for both the gallic and thiacalix[4]arene dendrimers. Thus, with a further increase in the concentration (0.25 mM for Gal-G3 and 0.1 mM for TCA-G2), the size and polydispersity index increase significantly, indicating the coalescence of aggregates into larger ones.
Thus, from the viewpoint of aggregation behaviour, it cannot be said that the dendrimer platform has a dramatic effect on the CAC values. The CAC values determined using three independent dyes are similar for the second- and third-generation dendrimers on the gallic acid and thiacalix[4]arene platforms, as is their solubilization capacity. In terms of the size and dispersion of the aggregates formed, the most compact aggregates of 20–50 nm with a monomodal distribution are formed by the second- and third-generation gallic acid derivatives, which is apparently due to their greater lipophilicity, while the thiacalix[4]arene dendrimers form polymodal aggregates of about 70–80 nm in the case of the second generation dendrimers and submicron aggregates of about 350–700 nm in the case of the third generation dendrimers. This behaviour may be associated with both insufficient lipophilicity and the influence of the bulkier three-dimensional architecture of the macrocycle, which may hinder self-organization into stable and compact aggregates, in contrast to the flat platform of gallic acid.
DNA binding abilities
Nucleic acids play a vital role in life processes as they carry hereditary information and control the biological synthesis of proteins and enzymes. DNA and mRNA are actively used in the creation of gene therapy products, particularly vaccines against various diseases, including cancer.43,44Various methods are employed to deliver DNA into the cell nucleus to minimise immunogenicity; the most prevalent of these is liposomal delivery.45–47 Amphiphilic dendrimers, which have branched positively charged head groups and the ability to self-assemble, are capable of forming stable lipoplexes with DNA and exhibit high delivery efficiency in this regard.45–47 A convenient method for studying the interaction of molecules with DNA is the use of competitive displacement of the ethidium bromide (EthBr) dye. EthBr is known to coexist in both the intercalation and electrostatic modes of binding with the nitrogenous bases of DNA and in the minor groove, respectively.48 Binding to DNA leads to a significant increase in fluorescence intensity and lifetime.49 In competitive binding of DNA with a guest molecule, EthBr is displaced from the DNA. This is evident from the quenching of fluorescence,50 which allows the binding constant of the dendrimer with the DNA to be calculated. The graphs (Fig. 3) showing fluorescence quenching in Stern–Volmer coordinates reveal that quenching occurs linearly for the 3rd generation dendrimers, indicating the predominance of a single type of quenching mechanism. In the case of the 2nd generation dendrimers, quenching of the dye–DNA complex occurs non-linearly, which indicates a mixed interaction. In the case of the 1st generation dendrimers, no quenching occurs at the concentrations studied, which indicates the absence of interaction. For the 3rd generation dendrimers, Stern–Volmer binding constants were calculated.51Table 3 shows the Stern–Volmer quenching constants (KSV) and bimolecular quenching constants (Kq), which were calculated for dendrimer–ctDNA systems using eqn (2):
| F0/F = 1 + KSV[ctDNA] | (2) |
| Kq = KSV/τ0 | (3) |
lg[(F0 − F)/F] = lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb + n Kb + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg[dendrimer]. lg[dendrimer]. | (4) |
| Dendrimers | T, K | K SV × 105 (M−1) | K q × 1013 (M−1 s−1) | K b × 106 (M−1) | n | ΔH0, kJ mol−1 | ΔS0, J mol−1 K−1 | ΔG0, kJ mol−1 |
|---|---|---|---|---|---|---|---|---|
| Gal-G3 | 293 | 3.7 | 3.7 | 1.3 | 1.2 | 93.30 | 432.33 | −34.84 |
| 298 | 2.5 | 2.5 | 2.6 | 1.4 | −36.58 | |||
| 308 | 4.9 | 4.9 | 2.3 | 1.4 | −39.49 | |||
| TCA-G3 | 293 | 3.2 | 3.2 | 1.3 | 1.4 | 42.73 | 262.47 | −34.80 |
| 298 | 3.1 | 3.1 | 1.6 | 1.3 | −35.42 | |||
| 308 | 6.7 | 6.7 | 2.9 | 1.3 | −36.90 |
The calculated value of Kb suggested that the 3rd generation dendrimers have strong binding affinity towards ctDNA. For the 3rd generation dendrimers, fluorescence titration was performed at different temperatures (Table 3 and Fig. S22) to calculate the thermodynamic parameters using eqn (SI1) and (SI2). It is well known52,53 that, depending on the trends in enthalpy ΔH0 and entropy ΔS0 accompanying the formation of dendrimer–ctDNA complexes, various intermolecular interactions can dominate: (1) if ΔH0 < 0 and ΔS0 < 0, hydrogen bonding and van der Waals forces are preferable; (2) if ΔH0 > 0 and ΔS0 > 0, hydrophobic interactions are favorable; and (3) if ΔH0 < 0 and ΔS0 > 0, electrostatic interactions are beneficial. A negative change in Gibbs free energy (ΔG < 0) indicates (Table 3) that complex formation is thermodynamically favorable. Based on the enthalpy and entropy data obtained, the main contribution to the 3rd generation DNA–dendrimer interaction is due to hydrophobic interaction.
The binding interaction of dendrimers with EthBr–ctDNA was further confirmed by measuring the fluorescence lifetime. The fluorescence decay profiles of EB–DNA in the presence of dendrimers (0.01 mM) are presented in Fig. 4. The calculated decay parameters obtained using single- and double-exponential approximations (DAS-6) for the analyzed systems are listed in Table 4. The free EthBr in an aqueous solution shows single-exponential decay with a lifetime of 1.56 ± 0.2 ns, and for the binary EthBr–ctDNA system, biexponential decay is observed, corresponding to lifetimes of 26.6 ± 3 (54%) and 1.3 ± 0.4 (46%) ns (mean lifetime: 14.4 ns). These results are consistent with the literature data.54 The data obtained from lifetime measurements are consistent with the data obtained from fluorescence titration. Thus, when the 1st generation dendrimers are added, the lifetime of EthBr does not differ from the lifetime in a double EthBr–ctDNA system (mean lifetime: 14.4 ns).
 | ||
| Fig. 4 Fluorescence decay curves of EthBr–ctDNA and EthBr–DNA in the presence of dendrimers. [ctDNA] = 0.05 mM, [EthBr] = 0.01 mM, [dendrimers] = 0.012 mM, 20 mM Tris with pH 7.4. | ||
| Sample | τ 1, ns | α 1 | τ 2, ns | α 2 |
|---|---|---|---|---|
| [ctDNA] = 0.05 mM, [EthBr] = 0.01 mM, [dendrimers] = 0.012 mM, 20 mM Tris with pH 7.4. | ||||
| EthBr | 1.56 ± 0.2 | 100 | — | — |
| EthBr–ctDNA | 1.3 ± 0.4 | 46 | 26.6 ± 3 | 54 |
| +Gal-G1 | 1.3 ± 0.4 | 47 | 26.2 ± 3 | 53 |
| +Gal-G2 | 1.8 ± 0.3 | 69 | 23.8 ± 2 | 31 |
| +Gal-G3 | 2.5 ± 0.2 | 100 | — | — |
| +TCA-G1 | 1.3 ± 0.4 | 47 | 26.1 ± 3 | 53 |
| +TCA-G2 | 2.0 ± 0.2 | 78 | 22.9 ± 3 | 22 |
| +TCA-G3 | 2.4 ± 0.2 | 100 | — | — |
That is, EthBr is not displaced at all in the presence of 1st generation dendrimers. When switching to the 2nd generation, the mean lifetime for Gal-G2 is 8.6 nanoseconds, and for TCA-G2, it is 6.5 ns, i.e., the proportion of free EthBr increases significantly. Only in the case of the 3rd generation is complete EthBr release achieved (lifetime: 2.4 ns). As can be seen from the DNA titration graphs (Fig. 5A), the addition of 2nd and 3rd generation dendrimers results in ctDNA compaction. The addition of 1st generation dendrimers, in contrast, leads to strong aggregation of the system and the formation of micron-sized particles.
 | ||
| Fig. 5 Z-Average (A) and zeta potential (B) of ctDNA in the presence of dendrimers. [ctDNA] = 0.05 mM, [dendrimer] = 0–0.015 mM, 20 mM Tris with pH 7.4, 25 °C. | ||
When using the 2nd and 3rd generation dendrimers, upon reaching a concentration of 0.015 mM, the system is recharged to +30 mV (Fig. 5B), which is enough to ensure colloidal stability.55 The recharging of the system passes through a minimum charge of approximately −40 mV, which may be associated with the unwinding of ctDNA due to its winding around liposomes. A subsequent increase in the concentration of the dendrimer carrying cationic groups then leads to a complete recharging of the system. It is noteworthy that compaction occurs in the presence of dendrimers at concentrations as low as ∼0.005 mM. This corresponds to the CACEosin values obtained with the anionic dye Eosin Y, which, like ctDNA, leads to partial charge compensation of dendrimers.
Transmission electron microscopy (Fig. 6) showed that the dendrimers themselves form aggregates with a size of about 50 nm in the case of Gal-G3, while in the case of TCA-G3, there is a clear tendency to form large associates (about 200 nm) formed from aggregates of 25–30 nm. When dendrimers are added to ctDNA, the typical fibril-like structures of free ctDNA56 disappear, which is consistent with the data obtained from dynamic and electrophoretic light scattering in solution (Fig. 5). The formation of monodisperse systems with an average size of about 70–120 nm is observed. At the same time, the TCA-G3 dendrimer forms looser aggregates with uneven edges compared to the more hydrophobic Gal-G3 dendrimer (Fig. 7).
 | ||
| Fig. 6 TEM microphotographs of ctDNA (A), Gal-G3 (B), ctDNA–Gal-G3 (C), TCA-G3 (D) and ctDNA–TCA-G3 (E). [ctDNA] = 0.05 mM, [dendrimer] = 0.01 mM. | ||
UV-visible spectroscopy can detect conformational changes in the DNA helix when it interacts with ligands, as evidenced by red/blue shifts and hyperchromic/hypochromic effects. This technique is often employed to evaluate substance–DNA interactions, and ctDNA typically exhibits an absorption spectrum with a maximum at 260 nm (Fig. 8). Changes in the absorption spectra are observed when the dendrimers are added in all cases. Furthermore, a hypochromic effect is evident for TCA-G1 and Gal-G1. This effect is most pronounced with TCA-G1 and may be associated with the greater hydrophobicity of 1st generation amphiphilic dendrimers. Binding due to hydrophobic interactions leads to base pair ordering and strengthening of stacking interactions. In the case of 2nd and 3rd generation dendrimers, a hyperchromic effect accompanied by a bathochromic shift is observed. A bathochromic shift of the DNA absorption maximum is often observed during both intercalation and binding to the minor groove.57 However, in true intercalation, the shift is usually greater than 14 nm and is accompanied by a hypochromic effect.58 The hyperchromic effect may be associated with disruption to stacking interactions between base pairs, indicating partial unwinding of the DNA helix and strong electrostatic interactions. This is consistent with an increase in ionized dendrimer fragments during the transition from the 1st to the 3rd generation.
 | ||
| Fig. 8 Circular dichroism (CD) spectra of ctDNA in the presence and absence of dendrimers. [ctDNA] = 0.05 mM, [dendrimer] = 0.02 mM, 20 mM Tris with pH 7.4, 25 °C. | ||
In the circular dichroism (CD, Fig. 8) spectra, the positive band at 275 nm is associated with π–π* transitions of bases and the right-handed B-form of DNA. The negative band at 245 nm reflects the helical structure of DNA and its chirality.59 When the dendrimers are added to the ctDNA solution, a slight hypochromic signal is observed at 275 nm, accompanied by a bathochromic shift. This shift increases with the transition from the 1st to 3rd generation dendrimers, reaching 5 nm. There is also a decrease in intensity at 245 nm, also accompanied by a bathochromic shift of 5 nm. Similar changes in the spectrum are often observed during the transition from the B-form to the C-form of DNA, which is often achieved through electrostatic interaction with cationic liposomes.60 The cationic groups of the dendrimer neutralize the negative charge of the phosphate fragments, decreasing repulsion between the chains and making the DNA helix denser, stiffer and more stable.
Thus, the 2nd and especially 3rd generation dendrimers bind most effectively to ctDNA, causing its compaction. The greatest compaction is achieved in the case of the 2nd generation dendrimers on the gallic acid core (120 nm), which can be attributed to its greater lipophilicity in the 2nd and 3rd dendrimer series and is consistent with the data on its self-aggregation. Spectrophotometry data indicate that the spectral characteristics of systems containing 2nd and 3rd generation dendrimers correspond to their interaction with the minor groove of DNA. According to the CD data, the addition of dendrimers causes a transition from the B-form to the more stable C-form.
Conclusions
A series of amphiphilic dendrimers with a thiacalix[4]arene or gallic acid core and imidazolium terminal groups were synthesized using copper-free click chemistry. The aggregation ability and solubilization capacity of imidazolium dendrimers in relation to a hydrophobic dye have been studied in detail. The second- and third-generation dendrimers on both platforms have been shown to interact effectively with the model calf thymus DNA, forming compact nanoparticles with a high positive zeta potential. According to the circular dichroism data, the addition of dendrimers does not lead to significant conformational changes in the DNA helix. This study opens up new possibilities for the easy synthesis of cationic dendrimers of various generations without copper catalyst impurities and demonstrates the potential for using amphiphilic imidazolium dendrimers to bind, store, and stabilize nucleic acids.Experimental section
Sample preparation
All solutions were prepared using Adrona Crystal E30 MilliQ water with a resistivity of 0.055 microSiemens (SIA “Adrona”, Latvia, Riga). The pH levels of the solutions were maintained at 7.4 using 20 mM TRIS-HCl (tris(hydroxymethyl)aminomethane) buffer. Various amounts of ctDNA were dissolved in 20 mM Tris-HCl buffer (pH 7.4) and kept at 4 °C for 24 h. The concentration of calf thymus DNA was evaluated by using a previously described approach.61Steady-state fluorescence study
The emission spectra were recorded using a Jobin Yvon Horiba Fluorolog-3 (HORIBA Jobin Yvon SAS, France) spectrofluorometer. Fluorescence emissions were recorded in the 340–430 nm range with λex = 335 nm and a 2.5 nm slit (for pyrene) and in the 535–750 nm range with λex = 520 nm and a 3 nm slit (for EthBr) using a 10 mm quartz cuvette.Time-resolved fluorescence study
Time-resolved fluorescence was obtained using a Horiba Jobin Yvon time-correlated single-photon counting (TCSPC) spectrometer (HORIBA Jobin Yvon SAS, France). A 454 nm nano-LED light source (pulse duration = 1.3 ns) was used for the measurements. The instrument response function was recorded using diluted Ludox AS-40 colloidal silica as a prompt. The decay data were analyzed using DAS-6 software, and an acceptable χ2 value was kept below 1.2 for a good fit.Dynamic and electrophoretic light scattering study
DLS and ELS measurements were carried out using a Zetasizer Nano ZS instrument (Malvern Panalytical, UK) with a 4 mW 633 nm He–Ne laser light source and a light scattering angle of 173 degrees. The solutions were filtered through a 0.8 µm filter before the measurements to remove dust. The experiments were carried out in DTS 0012 disposable plastic cells (size) or in DTS 1070 disposable folded capillary cells (zeta potential) (Sigma-Aldrich, USA) at 298 K with at least three experiments for each system.TEM
TEM was performed using a Hitachi HT7700 ExaLens (Hitachi High-Tech Corporation, Tokyo, Japan) in the Interdisciplinary Center for Analytical Microscopy of Kazan Federal University. The images were acquired at an accelerating voltage of 100 kV. The samples were ultrasonicated in water for 10 min, dispersed on 200 mesh copper grids with continuous formvar support films and then dried for 3 hours. Energy dispersive X-ray spectroscopy was performed using an Oxford Instruments X-MaxN 80T detector.UV absorption spectroscopy study
The UV-Vis spectra were recorded using a Shimadzu UV-2600 (Shimadzu, Kyoto, Japan) spectrophotometer. The spectra of ctDNA were recorded between 210 and 350 nm using a quartz cuvette of 10 mm path length. Solubilization of the dye (Orange OT) was performed by adding an excess amount of crystalline Orange OT to the solutions of dendrimers. These solutions were allowed to equilibrate for about 48 h at a constant temperature (25 °C), followed by filtration. Then the absorbance at 495 nm was measured.Circular dichroism study
The CD spectra were recorded using a J-1500 CD (Jasco, Tokyo, Japan) spectrophotometer equipped with a Peltier temperature controller maintained at 25 °C, using a quartz cuvette of path length 10 mm. All the CD spectra of different samples were recorded in the wavelength range of 230–320 nm at 25 °C by taking points at 0.5 nm. Each spectrum was the average of three runs at 25 °C and a five minute equilibration before each scan.Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) D calculation
D calculation
The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D calculations were performed using the Chemaxon online tool (https://playground.calculators.cxn.io/).
D calculations were performed using the Chemaxon online tool (https://playground.calculators.cxn.io/).
Author contributions
Conceptualization: V. B., I. A. and S. S.; methodology: I. B., V. B. and E. S.; investigation: I. B., A. F., A. G., E. S., T. M., and V. E.; resources: I. A.; data curation: E. S., V. B. and I. B.; writing – original draft preparation: I. B., E. S., and V. B.; writing – review and editing: V. B.; visualization: V. B., E. S. and I. B.; supervision: V. B.; project administration: V. B.; funding acquisition: I. A. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5py00984g.Acknowledgements
This research was funded by the Russian Science Foundation (22-13-00304-П, https://rscf.ru/en/project/22-13-00304/).References
- S. Zawadzki, Á. Martín-Serrano, E. Okła, M. Kędzierska, S. Garcia-Gallego, P. O. López, F. J. de la Mata, S. Michlewska, T. Makowski, M. Ionov, E. Pędziwiatr-Werbicka, M. Bryszewska and K. Miłowska, Sci. Rep., 2024, 14, 1–19 CrossRef.
- E. Karakhanov, A. Maximov and A. Zolotukhina, Polymers, 2022, 14, 981 CrossRef CAS PubMed.
- J. Wang, Z. Wang, G. Zhang, J. Rodrigues, H. Tomás, X. Shi and M. Shen, Biomater. Sci., 2024, 12, 1346–1356 RSC.
- M. Vonlanthen, F. Cuétara-Guadarrama, K. Sorroza-Martínez, I. González-Méndez, A. S. Estrada-Montaño and E. Rivera, Dyes Pigm., 2024, 222, 111873 CrossRef CAS.
- M. Pérez-Ferreiro, A. M. Abelairas, A. Criado, I. J. Gómez and J. Mosquera, Polymers, 2023, 15, 4369 CrossRef.
- B. Klajnert-Maculewicz, A. Janaszewska and A. Majecka, Chem. Commun., 2023, 59, 14611–14625 RSC.
- M. Filippi, J. Martinelli, G. Mulas, M. Ferraretto, E. Teirlinck, M. Botta, L. Tei and E. Terreno, Chem. Commun., 2014, 50, 3453–3456 RSC.
- S. Nummelin, V. Liljeström, E. Saarikoski, J. Ropponen, A. Nykänen, V. Linko, J. Seppälä, J. Hirvonen, O. Ikkala, L. M. Bimbo and M. A. Kostiainen, Chem. – Eur. J., 2015, 21, 14433–14439 CrossRef CAS.
- M. Gomes, I. Resende, Y. Zamoshchak, D. Araújo, J. Castro, D. Dhumal, L. Peng, R. S. Santos and N. F. Azevedo, J. Controlled Release, 2025, 384, 113850 CrossRef CAS PubMed.
- J. Chen, A. Ellert-Miklaszewska, S. Garofalo, A. K. Dey, J. Tang, Y. Jiang, F. Clément, P. N. Marche, X. Liu, B. Kaminska, A. Santoni, C. Limatola, J. J. Rossi, J. Zhou and L. Peng, Nat. Protoc., 2021, 16, 327–351 CrossRef CAS PubMed.
- J. Chen, D. Zhu, X. Liu and L. Peng, Acc. Mater. Res., 2022, 3, 484–497 CrossRef CAS PubMed.
- C. Galanakou, D. Dhumal and L. Peng, Biomater. Sci., 2023, 11, 3379–3393 RSC.
- K. Yamamoto, T. Imaoka, M. Tanabe and T. Kambe, Chem. Rev., 2020, 120, 1397–1437 CrossRef CAS PubMed.
- V. Maraval, A. Hameau, R. Laurent and A.-M. Caminade, Coord. Chem. Rev., 2025, 542, 216788 CrossRef CAS.
- E. A. Ocherednyuk, E. D. Sultanova, E. G. Makarov, A. A. Fedoseeva, A. A. Khannanov, V. G. Evtugyn, S. E. Solovieva, V. A. Burilov and I. S. Antipin, New J. Chem., 2024, 48, 13999–14012 RSC.
- A. M. Fatykhova, E. D. Sultanova, V. A. Burilov, B. K. Gafiatullin, A. A. Fedoseeva, T. A. Veshta, M. A. Ziganshin, S. A. Ziganshina, V. G. Evtugyn, D. R. Islamov, K. S. Usachev, S. E. Solovieva and I. S. Antipin, New J. Chem., 2023, 47, 19223–19234 RSC.
- K. Sorroza-Martínez, I. González-Méndez, R. D. Martínez-Serrano, J. D. Solano, A. Ruiu, J. Illescas, X. X. Zhu and E. Rivera, RSC Adv., 2020, 10, 25557–25566 RSC.
- N. El Brahmi, S. El Kazzouli, S. Mignani, M. Bousmina and J. P. Majoral, Tetrahedron, 2013, 69, 3103–3133 CrossRef CAS.
- D. Astruc, L. Liang, A. Rapakousiou and J. Ruiz, Acc. Chem. Res., 2012, 45, 630–640 CrossRef CAS.
- P. Wu, ACS Chem. Biol., 2022, 17, 2959–2961 CrossRef CAS PubMed.
- S. Neumann, M. Biewend, S. Rana and W. H. Binder, Macromol. Rapid Commun., 2020, 41, 1900359 CrossRef CAS.
- G. J. Brewer, Chem. Res. Toxicol., 2010, 23, 319–326 Search PubMed.
- S. P. Amaral, J. Correa and E. Fernandez-Megia, Green Chem., 2022, 24, 4897–4901 RSC.
- M. Arseneault, I. Levesque and J.-F. Morin, Macromolecules, 2012, 45, 3687–3694 CrossRef CAS.
- S. Parcero-Bouzas, J. Correa, C. Jimenez-Lopez, B. Delgado Gonzalez and E. Fernandez-Megia, Biomacromolecules, 2024, 25, 2780–2791 CrossRef CAS PubMed.
- I. I. Stoikov, I. S. Antipin, V. A. Burilov, A. R. Kurbangalieva, N. V. Rostovskii, A. S. Pankova, I. A. Balova, Y. O. Remizov, L. M. Pevzner, M. L. Petrov, A. V. Vasilyev, A. D. Averin, I. P. Beletskaya, V. G. Nenajdenko, E. K. Beloglazkina, S. P. Gromov, S. S. Karlov, T. V. Magdesieva, A. A. Prishchenko, S. V. Popkov, A. O. Terent'ev, G. V. Tsaplin, T. P. Kustova, L. B. Kochetova, N. A. Magdalinova, E. A. Krasnokutskaya, A. V. Nyuchev, Y. L. Kuznetsova, A. Y. Fedorov, A. Y. Egorova, V. S. Grinev, V. V. Sorokin, K. L. Ovchinnikov, E. R. Kofanov, A. V. Kolobov, V. L. Rusinov, G. V. Zyryanov, E. V. Nosov, V. A. Bakulev, N. P. Belskaya, T. V. Berezkina, D. L. Obydennov, V. Y. Sosnovskikh, S. G. Bakhtin, O. V. Baranova, V. S. Doroshkevich, G. Z. Raskildina, R. M. Sultanova, S. S. Zlotskii, V. D. Dyachenko, I. V. Dyachenko, A. S. Fisyuk, V. V. Konshin, V. V. Dotsenko, E. A. Ivleva, A. N. Reznikov, Y. N. Klimochkin, D. A. Aksenov, N. A. Aksenov, A. V. Aksenov, V. V. Burmistrov, G. M. Butov, I. A. Novakov, K. S. Shikhaliev, N. V. Stolpovskaya, S. M. Medvedev, N. V. Kandalintseva, O. I. Prosenko, E. B. Menshchikova, A. A. Golovanov and S. Y. Khashirova, Russ. J. Org. Chem., 2024, 60, 1361–1584 CrossRef CAS.
- A. R. Araújo, J. Correa, V. Dominguez-Arca, R. L. Reis, E. Fernandez-Megia and R. A. Pires, ACS Appl. Mater. Interfaces, 2021, 13, 59673–59682 CrossRef.
- S. P. Amaral, M. Fernandez-Villamarin, J. Correa, R. Riguera and E. Fernandez-Megia, Org. Lett., 2011, 13, 4522–4525 CrossRef CAS PubMed.
- S. Nummelin, V. Liljeström, E. Saarikoski, J. Ropponen, A. Nykänen, V. Linko, J. Seppälä, J. Hirvonen, O. Ikkala, L. M. Bimbo and M. A. Kostiainen, Chem. – Eur. J., 2015, 21, 14433–14439 CrossRef CAS.
- S. Nummelin, M. Selin, S. Legrand, J. Ropponen, J. Seitsonen, A. Nykänen, J. Koivisto, J. Hirvonen, M. A. Kostiainen and L. M. Bimbo, Nanoscale, 2017, 9, 7189–7198 RSC.
- Z.-Q. Cao, Y.-C. Wang, A.-H. Zou, G. London, Q. Zhang, C. Gao and D.-H. Qu, Chem. Commun., 2017, 53, 8683–8686 RSC.
- B. S. Sandanaraj, M. M. Reddy, P. J. Bhandari, S. Kumar and V. K. Aswal, Chem. – Eur. J., 2018, 24, 16085–16096 CrossRef CAS PubMed.
- V. A. Burilov, M. B. K. Gafiatullin, D. A. Mironova, E. D. Sultanova, V. G. Evtugyn, Y. N. Osin, D. R. Islamov, K. S. Usachev, S. E. Solovieva and I. S. Antipin, Eur. J. Org. Chem., 2020, 2180–2189 CrossRef CAS.
- H. Cheng, R. Zhang, T. Li, X. Peng, M. Xia, Y. Xiao and X. Cheng, New J. Chem., 2017, 41, 8443–8450 RSC.
- M. Yan, B. Li and X. Zhao, Food Chem., 2010, 122, 1333–1337 CrossRef CAS.
- A. B. Mirgorodskaya, R. A. Kushnazarova, S. S. Lukashenko, A. D. Voloshina, O. A. Lenina, L. Ya and O. G. Sinyashin, J. Mol. Liq., 2018, 269, 203–210 CrossRef CAS.
- G. Gaynanova, L. Vasileva, R. Kashapov, D. Kuznetsova, R. Kushnazarova, A. Tyryshkina, E. Vasilieva, K. Petrov, L. Zakharova and O. Sinyashin, Molecules, 2021, 26(22), 6786 CrossRef CAS PubMed.
- G. A. Gaynanova, A. M. Bekmukhametova, R. R. Kashapov, A. Y. Ziganshina and L. Y. Zakharova, Chem. Phys. Lett., 2016, 652, 190–194 CrossRef CAS.
- N. O. Mchedlov-Petrossyan and V. N. Kleshchevnikova, J. Chem. Soc., Faraday Trans., 1994, 90, 629–640 RSC.
- V. A. Burilov, G. A. Fatikhova, M. N. Dokuchaeva, R. I. Nugmanov, D. A. Mironova, P. V. Dorovatovskii, V. N. Khrustalev, S. E. Solovieva and I. S. Antipin, Beilstein J. Org. Chem., 2018, 14, 1980–1993 CrossRef CAS PubMed.
- R. Nagarajan, Langmuir, 2002, 18, 31–38 CrossRef CAS.
- M. L. Landry and J. J. Crawford, ACS Med. Chem. Lett., 2020, 11, 9–13 CrossRef PubMed.
- V. P. Chavda, R. Pandya and V. Apostolopoulos, Expert Rev. Vaccines, 2021, 20, 1549–1560 CrossRef CAS.
- B. Lu, J. M. Lim, B. Yu, S. Song, P. Neeli, N. Sobhani, P. K, S. R. Bonam, R. Kurapati, J. Zheng and D. Chai, Front. Immunol., 2024, 15, 1–24 CAS.
- G. Y. Tseu and K. A. Kamaruzaman, Molecules, 2023, 28(3), 1498 CrossRef CAS PubMed.
- R. Chelliah, M. Rubab, S. Vijayalakshmi, M. Karuvelan, K. Barathikannan and D.-H. Oh, Next Nanotechnol., 2025, 8, 100209 CrossRef.
- J. S. Son, R. Chow, H. Kim, T. Lieu, M. Xiao, S. Kim, K. Matuszewska, M. Pereira, D. Le Nguyen and J. Petrik, Reprod. Biol. Endocrinol., 2023, 21, 75 CrossRef CAS.
- C. G. Reinhardt and T. R. Krugh, Biochemistry, 1978, 17, 4845–4854 CrossRef CAS.
- D. P. Heller and C. L. Greenstock, Biophys. Chem., 1994, 50, 305–312 CrossRef CAS PubMed.
- P. O. Vardevanyan, A. P. Antonyan, M. A. Parsadanyan, M. A. Shahinyan and G. A. Melkonyan, Int. J. Spectrosc., 2015, 2015, 586231 Search PubMed.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, 3rd Principles of fluorescence spectroscopy, Springer, New York, USA, 3rd edn, 2006 Search PubMed.
- M. Akram, H. Lal and Kabir-ud-Din, Bioorg. Chem., 2022, 119, 105555 CrossRef CAS PubMed.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- K. K. Athira and R. L. Gardas, ACS Phys. Chem. Au, 2025, 5, 387–397 CrossRef CAS.
- H. Elsana, T. O. B. Olusanya, J. Carr-wilkinson, S. Darby, A. Faheem and A. A. Elkordy, Sci. Rep., 2019, 9, 15120 CrossRef PubMed.
- Y. Fan, H. Wang, C. He, F. Qiao, S. Wang and Y. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 23333–23341 CrossRef CAS.
- M. Sirajuddin, S. Ali and A. Badshah, J. Photochem. Photobiol., B, 2013, 124, 1–19 CrossRef CAS PubMed.
- Y. Sun, S. Bi, D. Song, C. Qiao, D. Mu and H. Zhang, Sens. Actuators, B, 2008, 129, 799–810 CrossRef CAS.
- J. Kypr, I. Kejnovská, D. Renčiuk and M. Vorlíčková, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS.
- N. J. Zuidam, Y. Barenholz and A. Minsky, FEBS Lett., 1999, 457, 419–422 CrossRef CAS PubMed.
- R. B. Inman and D. O. Jordan, Biochim. Biophys. Acta, 1960, 42, 530–532 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |