 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The paradigm shift in therapeutics: a comprehensive review of artificial intelligence in drug delivery systems
Shivam Singh and
Anindita De
and
Anindita De *
*
Department of Pharmaceutics, College of Pharmacy JSS University, Noida-201301, India. E-mail: Shivam.jssuni@gmail.com; aninditanirupa@gmail.com
First published on 12th January 2026
Abstract
The integration of Artificial Intelligence (AI) into the pharmaceutical landscape is heralding a new era of precision medicine, particularly in the domain of drug delivery. Traditional drug development is notoriously time-consuming, expensive, and prone to high attrition rates. AI, with its subfields of machine learning (ML) and deep learning (DL), is poised to de-bottleneck this process by enabling the rational design of intelligent, targeted, and responsive drug delivery systems (DDS). This review meticulously outlines the transformative role of AI across the entire spectrum of advanced drug delivery. We explore how AI algorithms leverage vast chemical and biological datasets to design novel nanocarriers, predict their physicochemical properties, and optimize their formulation for enhanced efficacy and safety. A significant focus is placed on AI-driven targeted and stimuli-responsive DDS for oncology, neurological, and inflammatory diseases. Furthermore, we delve into the emergence of AI-powered closed-loop systems for autonomous drug release. The review is supplemented with detailed tables summarizing key algorithms, recent clinical trials, and a landscape analysis of patents, highlighting the intense commercial and academic interest. Finally, we address the current challenges—including data quality, regulatory hurdles, and model interpretability—and propose future directions for the clinical translation of AI-engineered therapeutics. This synthesis underscores AI not merely as a tool but as a disruptive force, poised to unlock personalized, predictive, and precise drug delivery paradigms.
1. Introduction
The ultimate goal of drug delivery is to transport a therapeutic agent to its site of action at the right time and concentration, maximizing efficacy while minimizing off-target effects.1 Despite decades of research, achieving this remains a formidable challenge. Conventional drug delivery often suffers from poor bioavailability, non-specific distribution, and suboptimal pharmacokinetics, leading to reduced therapeutic indices and adverse side effects.2The advent of nanotechnology provided the first major leap forward, enabling the development of sophisticated Drug Delivery Systems (DDS) like liposomes, polymeric nanoparticles, and dendrimers.3 These systems can improve solubility, extend circulation time, and facilitate passive targeting through the Enhanced Permeability and Retention (EPR) effect, particularly in oncology. However, the rational design of these complex systems is non-trivial.4 Their behaviour in vivo is governed by a multitude of interconnected parameters—size, shape, surface charge, hydrophobicity, and ligand density—making traditional trial-and-error approaches inefficient and costly.5
Artificial Intelligence (AI) – AI refers to the capability of a machine to imitate intelligent human behaviour.6 Machine Learning (ML), a subset of AI, allows systems to learn patterns from data without being explicitly programmed. Deep Learning (DL), a further subset of ML using multi-layered neural networks, excels at identifying complex, non-linear relationships in high-dimensional data.7 The pharmaceutical industry, generating immense volumes of data from high-throughput screening, omics technologies, and medical imaging, is an ideal domain for AI application.8
This review articulates the profound paradigm shift underway in pharmaceutical sciences, where artificial intelligence is revolutionizing the field of drug delivery by transforming it from a largely empirical discipline, reliant on trial-and-error experimentation, into a sophisticated predictive science.9 We will explore the extensive application of AI across the entire development pipeline, beginning with the de novo design of novel drug carriers and intelligent biomaterials with bespoke properties. This extends to the optimization of complex formulation parameters and the streamlining of manufacturing processes to ensure robustness and scalability.10 Furthermore, AI is instrumental in enabling precision targeting strategies and engineering sophisticated stimuli-responsive behaviours for site-specific release. It is also the core engine powering the next generation of closed-loop systems capable of autonomous therapy by continuously adapting to patient-specific physiological signals.11 Critically, AI's predictive power is being harnessed to model intricate pharmacokinetic and pharmacodynamic (PK/PD) profiles, thereby accelerating the selection of optimal candidates. To ground this technological overview in reality, the review will also present a critical analysis of the current landscape through an examination of ongoing clinical trials and global patent activities, ultimately concluding with a discussion of the significant translational challenges and future directions that will shape this rapidly evolving field.12
2. AI-driven design and formulation of nanocarriers
The design of nanocarriers involves selecting from a vast chemical space of materials, surfactants, stabilizers, and drugs. AI models can navigate this complexity to propose optimal formulations with desired properties.2.1. Predictive modeling of nanoparticle properties
A cornerstone of AI's impact in formulation science is the predictive modeling of nanoparticle properties, where machine learning (ML) models are trained on extensive historical datasets to accurately forecast the Critical Quality Attributes (CQAs) of nanocarriers based solely on their initial formulation parameters.13 This approach leverages a suite of powerful algorithms, including Support Vector Regression (SVR) for handling high-dimensional spaces, Random Forest for robust ensemble predictions, Gradient Boosting methods like XGBoost for maximizing predictive accuracy, and complex Artificial Neural Networks (ANNs) capable of deciphering intricate non-linear relationships.14 These models are applied to predict fundamental characteristics such as particle size, polydispersity index (PDI) which is a key indicator of homogeneity, zeta potential governing colloidal stability, and drug loading efficiency crucial for therapeutic efficacy.15 The input variables for these predictions encompass critical formulation factors like polymer concentration, drug-polymer ratio, and solvent choice. A prime example of this capability is the use of an ANN to model the complex, non-linear relationship between the energy input during sonication and the resulting liposome size distribution, a task that is exceptionally challenging for traditional empirical methods.8 This data-driven paradigm allows researchers to bypass extensive experimental screening and rationally design nanoparticles with desired properties from the outset.162.2. Inverse design of nanocarriers
Building upon predictive modeling, a more advanced and powerful application is the inverse design of nanocarriers. This paradigm flips the traditional discovery process on its head: instead of creating a particle and then testing its performance, researchers specify a desired in vivo outcome such as long circulation time or high tumour accumulation and the AI model proactively proposes the optimal nanoparticle design to achieve that precise biological goal.17 This ambitious approach is powered primarily by generative models, which excel at creating novel, high-dimensional data. Key algorithms enabling this include Generative Adversarial Networks (GANs), which pit two neural networks against each other to generate highly realistic candidate designs; Variational Autoencoders (VAEs), which learn the latent, fundamental structure of existing data to produce new, plausible variations; and reinforcement learning, which iteratively improves designs based on a reward function tied to the desired outcome.18 A transformative application of this is the de novo molecular design of entirely novel biodegradable polymers or lipid-like materials for next-generation lipid nanoparticles (LNPs). For instance, a GAN can be trained on a vast database of known biomaterials and their biological properties, learning the complex relationships between chemical structure and function.19 It can then generate a library of new, virtual candidate molecules predicted to exhibit optimized properties for specific tasks, such as the highly efficient and stable delivery of mRNA therapeutics (Fig. 1). These AI-proposed candidates are subsequently prioritized for synthesis and biological testing, dramatically accelerating the discovery of advanced delivery systems (Table 1).20| Algorithm | Type | Application in drug delivery | Example |
|---|---|---|---|
| Random forest | Ensemble ML | Predicting nanoparticle properties, classifying PK profiles | Predicting PDI based on formulation parameters.13 |
| Artificial neural network (ANN) | Deep learning | Modeling complex non-linear relationships in formulation | Predicting drug release kinetics from a hydrogel.21 |
| Convolutional neural network (CNN) | Deep learning | Image analysis for characterization | Analyzing TEM images to quantify nanoparticle aggregation.22 |
| Generative adversarial network (GAN) | Deep learning | Inverse design of novel materials and carriers | Generating novel polymer structures for gene delivery.23 |
| Reinforcement learning | ML | Optimizing drug dosing schedules | Personalizing chemotherapy regimens in closed-loop systems.24 |
3. AI for targeted and stimuli-responsive drug delivery
Moving beyond passive targeting, active targeting using ligands (antibodies, peptides) and stimuli-responsiveness (pH, enzyme, redox) is crucial for precision. AI accelerates the discovery of targeting moieties and predicts responsive behaviours.3.1. Ligand discovery and selection
The quest to identify a high-affinity, highly specific ligand such as a peptide, antibody fragment, or small molecule that targets receptors uniquely overexpressed on diseased cells is a challenge perfectly suited for artificial intelligence, given the astronomically vast combinatorial molecular space and the complex, non-linear nature of molecular recognition. AI transcends traditional brute-force screening by integrating a predictive, generative, and knowledge-driven approach.6 Through advanced virtual screening, models trained on multi-modal datasets—including structural data from the PDB, biophysical binding affinities from sources like Binding DB, and genomic-proteomic expression profiles—leverage sophisticated algorithms such as Graph Neural Networks (GNNs) and Convolutional Neural Networks (CNNs) to perform in-silico evaluations of millions of candidates. These models predict not only binding affinity but also critical specificity, quantifying off-target risks to ensure therapeutic safety.25 Furthermore, AI initiates discovery even before ligand design through knowledge mining: Natural Language Processing (NLP) models like Bio BERT and SciBERT analyze vast scientific literature and patents to perform advanced named entity recognition and relationship extraction, identifying novel, high-potential target-receptor pairs—for instance, a receptor frequently associated with aggressive metastasis but not yet explored therapeutically.26 This end-to-end capability is exemplified by Insilco Medicine's fully integrated AI pipeline, which used its Panda Omics platform for target identification in idiopathic pulmonary fibrosis (IPF) and its Chemistry42 platform, employing a Generative Adversarial Network (GAN), for de novo generation of the novel kinase inhibitor ISM001-055.27 The generator creates new molecular structures, while the discriminator evaluates them based on drug-likeness, synthesizability, and binding criteria—a process directly applicable to designing targeted ligands by training the GAN on ligand databases and specifying desired properties such as stability, size, affinity, and specificity, thereby producing optimized candidates ready for experimental validation.283.2. Optimizing stimuli-responsive systems
The next frontier in precision medicine involves the creation of “smart” drug delivery systems that function like miniature robots, releasing their therapeutic payload only in response to specific biological cues. For systems triggered by internal stimuli, such as the acidic pH of a tumour, high cytoplasmic glutathione, or elevated enzyme levels in diseased tissue—the design challenge is to engineer materials that remain stable in healthy tissue but undergo a precise conformational change at the target site. This constitutes a multi-variable optimization problem of immense complexity, as the formulation must respond to a specific biochemical threshold without premature release. Traditional empirical methods struggle to navigate this complexity, as they can only explore a minuscule fraction of the vast chemical design space and fail to capture the non-linear interactions between polymer chemistry, nanoparticle architecture, and the dynamic physiological environment. This challenge is ideally addressed by artificial intelligence, which enables a predictive simulation approach. Machine learning models, such as Random Forest and XGBoost, are trained on datasets linking formulation parameters (e.g., polymer pKa, cross-linking density, linker chemistry) to release kinetics. These models uncover hidden structure–activity relationships, allowing for the inverse design of components like pH-sensitive shells for endosomal escape or enzyme-specific peptide linkers that minimize off-target cleavage.29,30,33For external stimuli—such as light, ultrasound, or magnetic fields—the design imperative shifts to maximizing the efficiency of energy conversion into a triggered release event. This requires finely tuned material characteristics, such as the phase-transition temperature of liposomes or the adsorption wavelength of a photolabile cage, which are difficult to optimize through iterative experimentation alone. Here, AI-driven generative models excel by proposing novel material combinations and nano-architectures that satisfy a predefined release profile. For instance, researchers can specify a requirement for minimal drug leakage at baseline but rapid, complete payload discharge upon exposure to a specific ultrasound frequency or wavelength of light (Fig. 2). The AI algorithm can then propose ideal candidate formulations, such as liposome compositions with tailored bilayer rigidity or nanoparticle composites with optimized photothermal efficiency, dramatically accelerating the development of systems with unprecedented spatiotemporal control (Table 2).30–33
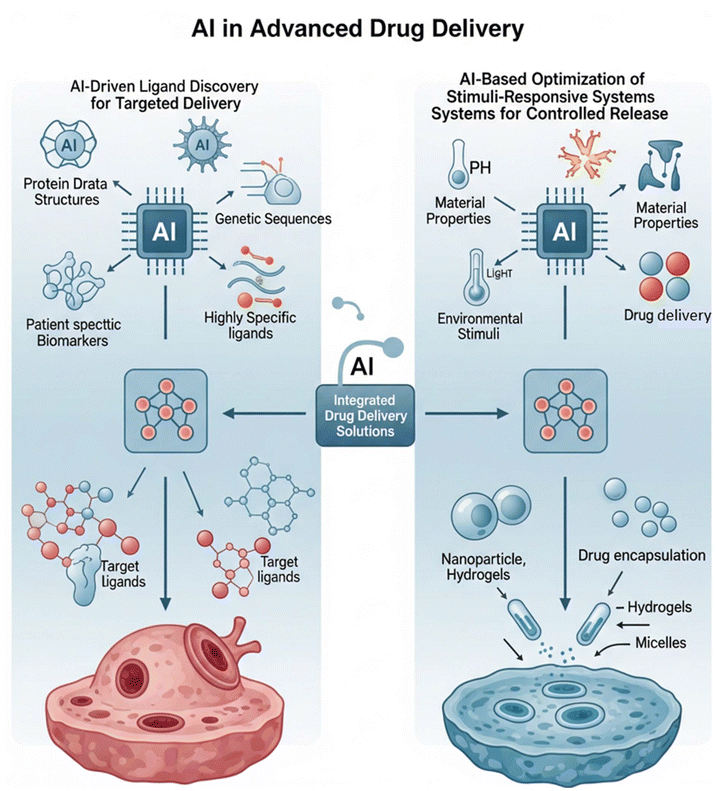 | ||
| Fig. 2 This figure illustrates a conceptual framework for leveraging artificial intelligence to advance precision medicine through intelligent drug delivery systems. | ||
| Application area | Case study/company | AI technology | Key outcome | Ref. |
|---|---|---|---|---|
| mRNA LNP formulation | Acuitas Therapeutics (partnered with Pfizer/BioNTech for COVID-19 vaccine) | ML-based optimization | Rapid optimization of LNP formulations for stability and efficacy, crucial for vaccine development speed. | 34 |
| Novel drug design & delivery | Insilco Medicine | Generative adversarial networks (GANs) | AI-generated novel molecule (ISM001-055) for fibrosis, entering clinical trials. Demonstrates integrated AI approach. | 35 |
| Predicting nanomedicine efficacy | University of Toronto Research | Random forest model | AI-generated novel molecule (ISM001-055) for fibrosis, entering clinical trials. Demonstrates integrated AI approach. | 36 |
| Closed-loop insulin delivery | Doctrine Med | Reinforcement learning | Developed an ML model that accurately predicted the tumour accumulation of nanoparticles based on their physicochemical properties. | 37 |
4. AI in pharmacokinetics and pharmacodynamics (PK/PD) modeling
Predicting the absorption, distribution, metabolism, excretion, and toxicity (ADMET) of a drug, especially when encapsulated in a complex DDS, is critical. AI enhances traditional PK/PD modeling.4.1. PBPK modeling enhancement
Physiologically-Based Pharmacokinetic (PBPK) models are complex mathematical models. AI can be used to optimize the thousands of parameters within these models for specific subpopulations or individual patients, moving towards personalized dosing.38AI significantly enhances Physiologically-Based Pharmacokinetic (PBPK) models by personalizing them for individual patients. Traditional PBPK models rely on average population data, which can be inaccurate for a specific person. AI and machine learning overcome this by analyzing a patient's unique data—such as their genetics, age, organ function, and other health conditions—to predict their personal physiological parameters.39 These AI-predicted parameters are then fed into the sophisticated PBPK model, which acts as a digital twin of the patient. This allows for the simulation of a highly accurate, personalized drug concentration-time profile, ultimately moving beyond one-size-fits-all dosing to truly optimized, individual treatment plans.40
4.2. Predicting drug–drug interactions
AI is crucial for predicting dangerous or ineffective combinations between nanocarrier-based drugs and a patient's other medications.40 These interactions are complex; a second drug might, for instance, disrupt the nanocarrier's shell, release its payload too early, or block the cellular pathways it uses to enter tissues. Machine learning models can be trained on vast datasets of chemical properties, biological targets, and historical interaction reports to identify subtle patterns that humans would miss.41 By analyzing the molecular features of both the nanocarrier system and the co-administered drugs, AI can flag potential risks for altered efficacy or increased toxicity before the treatment is even administered, enabling clinicians to adjust the therapy or choose a safer alternative.425. Clinical translation and patent landscape
The promise of AI is now being tested in clinical settings, and the intellectual property landscape is evolving rapidly.5.1. Clinical trials
While many applications are in pre-clinical stages, several AI-informed therapies are undergoing clinical evaluation. Most are focused on treatment personalization rather than the DDS itself (Table 3).| Trial identifier | Title | Condition | AI component | Status | Sponsor |
|---|---|---|---|---|---|
| NCT04095127 | A study to evaluate the efficacy and safety of ISM001-055 in participants with idiopathic pulmonary fibrosis | Idiopathic pulmonary fibrosis | Treatment designed by insilco medicine's AI platform | Phase II | Insilco Medicine43 |
| NCT05304624 | Personalising insulin dosing using an automated system (PiDaaS) | Type 1 diabetes | AI algorithm for personalized insulin dose calculation | Recruiting | University of Cambridge44 |
| NCT03671083 | A study of LY3298176 (Tirzepatide) in participants with Type 2 diabetes not controlled with diet and exercise alone | Type 2 diabetes | Use of AI for analyzing patient data to predict response (ancillary) | Completed | Eli Lilly and Company45 |
| NCT04293679 | An investigational drug delivery system – DUROS® subcutaneous delivery of fulvestrant in women with advanced breast cancer | Breast cancer | Device-focused, but data analyzed with ML for PK insights | Active, not recruiting | AstraZeneca46 |
| NCT06335095 | A study of QTY-platform designed proteins for opioid overdose reversal | Opioid overdose | AI-driven protein design. Uses AI to deimmunize and optimize the design of novel biologic scavengers (QTY code proteins) that bind to opioids, creating a new drug delivery mechanism for toxins. | Recruiting (2024) | QTY Therapeutics, Inc. |
5.2. Patent landscape
The patent activity reflects the strategic importance of AI in drug delivery. Key players include both established pharma giants and agile AI-focused startups (Table 4).| Patent/publication number | Title | Assignee | Key innovation |
|---|---|---|---|
| WO2020250182A1 | Systems and methods for designing lipid nanoparticles using machine learning | Pfizer | Using ML models to predict LNP formulation parameters for nucleic acid delivery based on desired properties.47 |
| US20220028583A1 | Methods for generating novel therapeutics using generative models | Insilco Medicine | Using GANs for the de novo design of drug molecules and potentially their delivery modalities.48 |
| EP3890701A1 | An apparatus and method for controlling drug delivery | Medtronic | AI-driven closed-loop system for autonomous drug delivery (e.g., insulin, pain relief) based on real-time biosensor data.49 |
| WO2021191376A1 | Machine learning for characterization of nanomedicines | The University of Texas | Using ML (e.g., CNNs) to analyze microscopic images for automated characterization of nanoparticle size and shape.50 |
6. Challenges and future perspectives
Despite the immense potential of AI in revolutionizing drug delivery systems, several significant challenges must be overcome for its widespread clinical adoption. A primary bottleneck is the issue of Data Quality and Availability, as AI models are fundamentally “garbage in, garbage out”; the current lack of large, standardized, and high-quality datasets on nano-formulations and their in vivo performance severely limits model training, making initiatives to create open-source, collaborative databases absolutely crucial. Compounding this is the Model Interpretability or “Black Box” Problem, where complex deep learning algorithms can deliver accurate predictions but fail to provide a understandable rationale for their decisions, which is essential for both regulatory approval and building scientific trust—a gap that the growing field of Explainable AI (XAI) seeks to bridge. Furthermore, there is a substantial Regulatory Science Lag, as bodies like the FDA and EMA, while progressing on frameworks for AI in medical devices, are still evolving specific guidelines for AI-driven drug products, leaving a novel challenge in defining the validation criteria for an AI-generated formulation or therapy. Finally, successful Integration and Workflow requires more than just technology; it necessitates a cultural shift within pharmaceutical companies and the upskilling of scientists to seamlessly incorporate these new tools into existing development pipelines. Looking to the future, overcoming these hurdles could unlock transformative perspectives, including AI-powered “Pharma-factories” capable of the continuous, automated manufacturing of personalized drug delivery systems, the development of patient-specific Digital Twins to simulate and optimize therapies in a virtual environment before administration, and the rise of Multi-modal AI that integrates genomics, proteomics, medical imaging, and data from real-time biosensors to design dynamic, holistic, and truly personalized treatment strategies.7. Conclusion
The integration of Artificial Intelligence into drug delivery represents a foundational paradigm shift from empirical methods to predictive, precision science. AI's value extends systematically across the entire development pipeline, enhancing material design, formulation optimization, and therapeutic personalization. Machine learning algorithms now accurately predict critical quality attributes of complex formulations, while generative models enable inverse design of tailored drug delivery systems. These AI-driven approaches facilitate creation of sophisticated, stimuli-responsive systems that function as “intelligent” therapeutic robots, releasing payloads in response to specific biological cues. The field's advancement depends on addressing three critical challenges. First, establishing robust, standardized datasets through industry-wide collaboration is essential for developing trustworthy models. Second, bridging the gap between computational predictions and clinical efficacy requires advanced multi-scale modeling that integrates AI with physiological-based pharmacokinetic frameworks. Finally, achieving widespread adoption necessitates developing interpretable AI and user-friendly interfaces to build trust among researchers and regulators while democratizing access for non-specialists. By prioritizing these strategic areas data infrastructure, biologically-relevant modeling, and translational usability the scientific community can solidify AI's role as an indispensable partner in therapeutic development. This concerted effort will unlock the technology's full potential, enabling smarter, safer, and highly personalized therapeutics that redefine patient care standards through precise, predictable drug delivery solutions.Conflicts of interest
There are no conflicts to declare.Data availability
This article is a comprehensive review of the existing scientific literature. No new primary data were created or analysed in this study. All data supporting this review, including references to previously reported studies and datasets, are available within the article and its reference list.References
- M. A. Karsdal, S. H. Nielsen, D. J. Leeming, L. L. Langholm, M. J. Nielsen, T. Manon-Jensen, A. Siebuhr, N. S. Gudmann, S. Rønnow, J. M. Sand and S. J. Daniels, The good and the bad collagens of fibrosis–their role in signalling and organ function, Adv. Drug Delivery Rev., 2017, 121, 43–56 CrossRef CAS PubMed.
- E. Blanco, H. Shen and M. Ferrari, Principles of nanoparticle design for overcoming biological barriers to drug delivery, Nat. Biotechnol., 2015, 33(9), 941–951 CrossRef CAS.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet and H. F. Dvorak, et al. Analysis of nanoparticle delivery to tumours, Nat. Rev. Mater., 2016, 1(5), 16014 CrossRef CAS.
- K. Park, The beginning of the end of the nanomedicine hype, J. Controlled Release, 2019, 305, 221–222 CrossRef CAS.
- J. I. Hare, T. Lammers, M. B. Ashford, S. Puri, G. Storm and S. T. Barry, Challenges and strategies in anti-cancer nanomedicine development: An industry perspective, Adv. Drug Delivery Rev., 2017, 108, 25–38 CrossRef CAS PubMed.
- J. Vamathevan, D. Clark, P. Czodrowski, I. Dunham, E. Ferran, G. Lee, B. Li, A. Madabhushi, P. Shah, M. Spitzer and S. Zhao, Applications of machine learning in drug discovery and development, Nat. Rev. Drug Discovery, 2019, 18(6), 463–477 Search PubMed.
- J. M. Stokes, K. Yang, K. Swanson, J. W. Cubillos-Ruiz, A. Donghia, N. M. MacNair, C. R. French, S. Carfrae, L. A. Bloom-Ackermann, Z. Tran and V. M, A deep learning approach to antibiotic discovery, Cell, 2020, 180(4), 688–702 CrossRef CAS PubMed.
- A. Zhavoronkov, Y. A. Ivanenkov, A. Aliper, M. S. Veselov, V. A. Aladinskiy, A. V. Aladinskaya, V. A. Terentiev, D. A. Polykovskiy, M. D. Kuznetsov, A. Asadulaev and Y. Volkov, Deep learning enables rapid identification of potent DDR1 kinase inhibitors, Nat. Biotechnol., 2019, 37(9), 1038–1040 CrossRef CAS PubMed.
- D. Paul, G. Sanap, S. Shenoy, D. Kalyane, K. Kalia and R. K. Tekade, Artificial intelligence in drug discovery and development, Drug Discovery Today, 2021, 26(1), 80–93 Search PubMed.
- M. Al-Mohaya, B. Mesut, A. Kurt and Y. S. Çelik, In silico approaches which are used in pharmacy, J. Appl. Pharm. Sci., 2024, 14(4), 239–253 Search PubMed.
- P. Bannigan, Z. Bao, R. J. Hickman, M. Aldeghi, F. Häse, A. Aspuru-Guzik and C. Allen, Machine learning models to accelerate the design of polymeric long-acting injectables, Nat. Commun., 2023, 14(1), 35 CrossRef CAS PubMed.
- A. Kiani, J. Wang, X. Chen, Y. Liu, Q. Wang and G. Zhou, et al., Smart materials and technologies for precision medicine, Adv. Mater., 2023, 35(43), e2205112 Search PubMed.
- P. Bannigan, Z. Bao, R. J. Hickman, M. Aldeghi, F. Häse, A. Aspuru-Guzik and C. Allen, Machine learning models to accelerate the design of polymeric long-acting injectables, Nat. Commun., 2023, 14(1), 35 Search PubMed.
- H. Narayanan, M. F. Luna, M. von Stosch, M. N. Cruz Bournazou, G. Polotti, M. Morbidelli, A. Butté and M. Sokolov, Bioprocessing in the digital age: the role of process models, Biotechnol. J., 2020, 15(1), 1900172 CrossRef CAS PubMed.
- A. Dey, A. Mitra, S. Pathak, S. Prasad, A. S. Zhang, H. Zhang, X. F. Sun and A. Banerjee, Recent advancements, limitations, and future perspectives of the use of personalized medicine in treatment of colon cancer, Technol. Cancer Res. Treat., 2023, 22, 15330338231178403 CrossRef PubMed.
- Y. H. Cheng, C. He, J. E. Riviere, N. A. Monteiro-Riviere and Z. Lin, Meta-analysis of nanoparticle delivery to tumours using a physiologically based pharmacokinetic modeling and simulation approach, ACS Nano, 2020, 14(3), 3075–3095 Search PubMed.
- S. Li and A. S. Barnard, Inverse Design of Nanoparticles Using Multi–Target Machine Learning, Adv. Theory Simul., 2022, 5(2), 2100414 CrossRef.
- B. Sanchez-Lengeling and A. Aspuru-Guzik, Inverse molecular design using machine learning: Generative models for matter engineering, Science, 2018, 361(6400), 360–365 Search PubMed.
- O. Méndez-Lucio, B. Baillif, D. A. Clevert, D. Rouquié and J. Wichard, De novo generation of hit-like molecules from gene expression signatures using artificial intelligence, Nat. Commun., 2020, 11(1), 10 CrossRef PubMed.
- S. Fan, W. Wang, W. Che, Y. Xu, C. Jin, L. Dong and Q. Xia, Nanomedicines targeting metabolic pathways in the tumour microenvironment: future perspectives and the role of AI, Metabolites, 2025, 15(3), 201 CrossRef CAS.
- H. Narayanan, M. F. Luna, M. von Stosch, M. N. Cruz Bournazou, G. Polotti, M. Morbidelli, A. Butté and M. Sokolov, Bioprocessing in the digital age: the role of process models, Biotechnol. J., 2020, 15(1), 1900172 CrossRef CAS.
- N. Creange, O. Dyck, R. K. Vasudevan, M. Ziatdinov and S. V. Kalinin, Towards automating structural discovery in scanning transmission electron microscopy, Mach. Learn.: Sci. Technol, 2022, 3(1), 015024 Search PubMed.
- A. Zhavoronkov, Y. A. Ivanenkov, A. Aliper, M. S. Veselov, V. A. Aladinskiy, A. V. Aladinskaya, V. A. Terentiev, D. A. Polykovskiy, M. D. Kuznetsov, A. Asadulaev and Y. Volkov, Deep learning enables rapid identification of potent DDR1 kinase inhibitors, Nat. Biotechnol., 2019, 37(9), 1038–1040 CrossRef CAS PubMed.
- R. Padmanabhan, N. Meskin and W. M. Haddad, Reinforcement learning-based control of drug dosing for cancer chemotherapy treatment, Math. Biosci., 2017, 293, 11–20 CrossRef CAS.
- J. M. Stokes, K. Yang, K. Swanson, J. W. Cubillos-Ruiz, A. Donghia, N. M. MacNair, C. R. French, S. Carfrae, L. A. Bloom-Ackermann, Z. Tran and V. M, A deep learning approach to antibiotic discovery, Cell, 2020, 180(4), 688–702 CrossRef CAS PubMed.
- J. Lee, W. Yoon, S. Kim, D. Kim, S. Kim, C. H. So and J. Kang, BioBERT: a pre-trained biomedical language representation model for biomedical text mining, Bioinformatics, 2020, 36(4), 1234–1240 CrossRef CAS PubMed.
- B. Sanchez-Lengeling and A. Aspuru-Guzik, Inverse molecular design using machine learning: Generative models for matter engineering, Science, 2018, 361(6400), 360–365 CrossRef CAS.
- W. Gao, T. Fu, J. Sun and C. Coley, Sample efficiency matters: a benchmark for practical molecular optimization, Adv. Neural Inf. Process. Syst., 2022, 35, 21342–21357 Search PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discovery, 2021, 20(2), 101–124 CrossRef CAS.
- D. Banerjee, D. Rajput, S. Banerjee and V. A. Saharan, Artificial intelligence and its applications in drug discovery, formulation development, and healthcare, in Computer Aided Pharmaceutics and Drug Delivery: An Application Guide for Students and Researchers of Pharmaceutical Sciences, Springer Nature Singapore, Singapore, 2022, pp. 309–380 Search PubMed.
- L. K. Vora, A. D. Gholap, K. Jetha, R. R. Thakur, H. K. Solanki and V. P. Chavda, Artificial intelligence in pharmaceutical technology and drug delivery design, Pharmaceutics, 2023, 15(7), 1916 CrossRef CAS.
- D. Wu, M. Si, H. Y. Xue and H. L. Wong, Nanomedicine applications in the treatment of breast cancer: current state of the art, Int. J. Nanomed., 2017, 5879–5892 Search PubMed.
- T. Ching, D. S. Himmelstein, B. K. Beaulieu-Jones, A. A. Kalinin, B. T. Do, G. P. Way, E. Ferrero, P. M. Agapow, M. Zietz, M. M. Hoffman and W. Xie, Opportunities and obstacles for deep learning in biology and medicine, J. R. Soc., Interface, 2018, 15(141), 20170387 CrossRef PubMed.
- P. R. Cullis and M. J. Hope, Lipid nanoparticle systems for enabling gene therapies, Mol. Ther., 2017, 25(7), 1467–1475 CrossRef CAS PubMed.
- S. Levy and H. Ruohola-Baker, AI-driven designed protein epigenomics, Clin. Res. Oncol., 2023, 1(1), 1 Search PubMed.
- Y. H. Cheng, C. He, J. E. Riviere, N. A. Monteiro-Riviere and Z. Lin, Meta-analysis of nanoparticle delivery to tumors using a physiologically based pharmacokinetic modeling and simulation approach, ACS Nano, 2020, 14(3), 3075–3095 CrossRef CAS PubMed.
- M. K. Bothe, L. Dickens, K. Reichel, A. Tellmann, B. Ellger, M. Westphal and A. A. Faisal, The use of reinforcement learning algorithms to meet the challenges of an artificial pancreas, Expert Rev. Med. Devices, 2013, 10(5), 661–673 CrossRef CAS PubMed.
- M. Zhang, L. Van and M. M. Amiji, Pharmacometric Modeling of Lipid Nanoparticle-Encapsulated mRNA Therapeutics and Vaccines: A Systematic Review, Mol. Ther. – Nucleic Acids, 2025, 36, 7–8 Search PubMed.
- N. A. Miller, M. B. Reddy, A. T. Heikkinen, V. Lukacova and N. Parrott, Physiologically based pharmacokinetic modelling for first-in-human predictions: an updated model building strategy illustrated with challenging industry case studies, Clin. Pharmacokinet., 2019, 58(6), 727–746 CrossRef CAS.
- K. W. Cheung, B. D. van Groen, G. J. Burckart, L. Zhang, S. N. de Wildt and S. M. Huang, Incorporating ontogeny in physiologically based pharmacokinetic modeling to improve pediatric drug development: what we know about developmental changes in membrane transporters, J. Clin. Pharmacol., 2019, 59, S56–S69 CrossRef CAS.
- T. Takeda, M. Hao, T. Cheng, S. H. Bryant and Y. Wang, Predicting drug–drug interactions through drug structural similarities and interaction networks incorporating pharmacokinetics and pharmacodynamics knowledge, J. Cheminf., 2017, 9(1), 16 Search PubMed.
- M. Zitnik, M. Agrawal and J. Leskovec, Modeling polypharmacy side effects with graph convolutional networks, Bioinformatics, 2018, 34(13), i457–i466 CrossRef CAS PubMed.
- Insilco Medicine, Insilico Medicine announces first-in-human dose of ISM001-055, the first AI-discovered novel molecule for idiopathic pulmonary fibrosis [Internet], Bus. Wire, 2021, 1–3 Search PubMed . 2021 Feb 24 [cited 2024 May 10]. Available from: https://www.businesswire.com/news/home/20210224005122/en/.
- L. Bally, H. Thabit and R. Hovorka, Closed-loop for type 1 diabetes – an introduction and appraisal for the generalist, BMC Med., 2017, 15(1), 14, DOI:10.1186/s12916-016-0772-6.
- Eli Lilly Company. Lilly Announces Tirzepatide Delivered Superior Weight Loss Compared to Placebo in Adults with Obesity or Overweight in SURMOUNT-1 [press release on the Internet], Eli Lilly and Company, Indianapolis (IN), 2022, 2022 Apr 28 [cited 2024 May 10]. Available from: https://investor.lilly.com/news-releases/news-release-details/lilly-announces-tirzepatide-delivered-superior-weight-loss Search PubMed.
- L. A. Medina, A. Lo, J. Wu, Y. Wang, G. Klinzing and H. Patel, et al., Population Pharmacokinetics of Subcutaneous Fulvestrant Delivery in Women with ER+ Advanced Breast Cancer, Clin. Pharmacol. Ther., 2023, 113(5), 1121–1132, DOI:10.1002/cpt.2862.
- N. Pardi, M. J. Hogan, P. R. Cullis, M. J. Hope, Y. K. Tam and T. D. Madden, et al., Systems and Methods for Designing Lipid Nanoparticles Using Machine Learning, Patent WO2020250182A1, 2020, World Intellectual Property Organization (WIPO) Search PubMed.
- A. Zhavoronkov, Y. A. Ivanenkov, A. Aliper and M. S. Veselov, Methods for Generating Novel Therapeutics Using Generative Models, Patent US20220028583A1, 2022, United States Patent and Trademark Office (USPTO) Search PubMed.
- D. Brancazio, R. Coles, F. J. Doyle, E. Dassau and J. E. Pinsker, An Apparatus and Method for Controlling Drug Delivery, Patent EP3890701A1, 2021, European Patent Office (EPO) Search PubMed.
- K. P. Johnston, R. O. Williams, A. B. Watts and Z. Cui, Machine Learning for Characterization of Nanomedicines, Patent WO2021191376A1, 2021, World Intellectual Property Organization (WIPO) Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |

