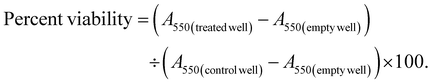Open Access Article
Open Access ArticleBerberine-doped liposomes enhance factor IX mutant mRNA delivery for protein replacement therapy in hemophilia B
Porkizhi
Arjunan
ab,
Gokulnath
Mahalingam
ab,
Harini
Manogaran
a,
Mohankumar
Murugesan
ab,
Saravanabhavan
Thangavel
ab and
Srujan
Marepally
 *ab
*ab
aCentre for Stem Cell Research (CSCR) (a Unit of inStem, Bengaluru), India. E-mail: srujankm@cmcvellore.ac.in
bChristian Medical College Campus, Vellore 632002, Tamil Nadu, India
First published on 15th October 2025
Abstract
Hemophilia B, caused by factor IX (FIX) deficiency, remains the best candidate for mRNA-based gene therapy. However, efficient hepatic delivery of mRNA continues to be a significant challenge. In this study, we developed and evaluated berberine-functionalized cationic liposomes as a novel delivery platform for FIX mutant mRNAs. Berberine incorporation enhanced the liposome's membrane fluidity and fusion potential, facilitating improved intracellular delivery and endosomal escape. Six different FIX variants were designed, transcribed, and screened for optimal expression. Our results demonstrate that berberine liposomes significantly enhance hepatocyte transfection and enable sustained FIX production, particularly with the TmL mutant. These findings suggest that berberine-functionalized liposomes represent a delivery strategy for mRNA therapeutics, leveraging the natural liver tropism common to lipid-based systems while aiming to enhance hepatocyte transfection efficiency.
1. Introduction
Hemophilia B is an X-linked recessive bleeding disorder caused by mutations in the F9 gene, leading to deficient or dysfunctional coagulation factor IX (FIX).1 Patients with hemophilia B experience recurrent bleeding episodes, which can result in joint damage, muscle hematomas, and life-threatening hemorrhages. The standard treatment involves prophylactic or on-demand FIX protein replacement therapy, but frequent infusions, high costs, and the potential for inhibitor development remain significant challenges.2,3 Recently, gene therapy approaches using adeno-associated virus (AAV) vectors have shown promise. However, concerns around issues such as pre-existing immunity, limited vector packaging capacity, and the potential for hepatotoxicity, limit their widespread application.4,5mRNA-based therapeutics have emerged as a non-integrative, transient alternative for gene replacement therapies. mRNA delivery can circumvent issues associated with viral vectors, such as immunogenicity and genomic integration, while enabling controlled and repeatable dosing.6 While lipid-based systems naturally accumulate in the liver, optimizing hepatocyte-specific delivery and endosomal escape remains a challenge for mRNA therapies. As the liver is the primary site of FIX synthesis, lipid carriers are essential to protect unstable mRNA from nuclease degradation and enhance its uptake and translation.7
Wild-type FIX (WT-FIX) serves as a baseline control to compare the activities of engineered gain-of-function mutants. WT-FIX exhibits physiological clotting function, but its natural activity is limited, necessitating frequent dosing in replacement therapy.8–11 WT-FIX serves as a reference for evaluating the performance of engineered gain-of-function mutants, helping to establish a baseline. The limitations are that higher therapeutic doses are required due to moderate clotting activity and short plasma half-life. To enhance the therapeutic potential of FIX mRNA, we employed engineered variants designed to increase the activity, stability, and secretion. Single mutant FIX alanine (Sm A) and leucine (Sm L) introduce specific amino acid substituents to modulate FIX12 activation kinetics and clotting activity while retaining native FIX secretion and stability. Sm A is engineered for enhanced clotting activity while minimizing the risk of spontaneous activation. Sm L is designed to optimize FIX secretion efficiency while maintaining physiological clotting function.13 Sm variants may exhibit improved therapeutic efficiency compared to WT-FIX while maintaining similar secretion profiles.9,14 Double mutant FIX (Dm A and Dm L) introduces two specific mutations, fine-tuning enzymatic activity and protein stability. Dm A (double mutant A) combines activity-enhancing and stability-enhancing mutations, leading to higher clotting efficiency than that of Sm A. Dm L (double mutant L) incorporates modifications to increase FIX half-life and improve secretion, resulting in prolonged therapeutic effects. Dm variants are expected to show higher FIX expression levels and extended circulation times, reducing dosing frequency. Triple mutant FIX Tm A and Tm L are high-performance variants representing the most optimized FIX variants, engineered for maximal clotting efficiency, extended half-life, and enhanced secretion. Tm A (triple mutant A) is specifically designed for superior thrombin activation and clot formation, which could make it an ideal candidate for severe hemophilia B patients.15 Tm L (triple mutant L) exhibits optimal secretion and stability, with potentially the highest therapeutic index among FIX variants.16 Tm variants are likely to outperform WT-FIX and other mutants regarding clotting efficiency, prolonged activity, and requiring lower therapeutic doses.
Lipid nanoparticles (LNPs) have been widely employed for hepatic mRNA delivery, notably in COVID-19 vaccines.7,17 Ionizable lipids in LNPs improve endosomal escape and cytoplasmic delivery, but concerns remain regarding their immunogenicity, toxicity, and suboptimal liver targeting. The development of liver-specific and biocompatible delivery vehicles is critical for advancing mRNA-based therapies for conditions such as hemophilia B.18–21
Berberine, a naturally occurring isoquinoline alkaloid, has been extensively studied for its hepatoprotective, anti-inflammatory, and metabolic regulatory properties.22,23 Berberine may further enhance liver-directed delivery by interacting with hepatocyte transporters, building upon the inherent hepatic tropism of lipid nanoparticles, and has been incorporated into delivery systems to enhance bioavailability. Additionally, berberine incorporation into liposomal bilayers has been associated with increased membrane fluidity and fusion capacity, likely due to its ability to disrupt tight lipid packing.24,25 These biophysical changes may enhance intracellular delivery by promoting liposome–cell membrane interactions and endosomal escape, thereby improving the cytoplasmic release of the mRNA cargo.
Given its hepatoprotective nature and ability to modulate intracellular pathways, berberine is hypothesized to enhance the stability, uptake, and translation of FIX mRNA in hepatocytes. These properties, combined with berberine's potential to modify the liposome structure, make it an ideal candidate for enhancing mRNA delivery and release. This study investigates the possibility of berberine-functionalized cationic liposomes as a novel delivery system for FIX mutant mRNAs. We evaluate whether berberine-functionalized liposomes enhance transfection efficiency and FIX expression in hepatocytes, leveraging both berberine's hepatoprotective properties and the inherent liver tropism characteristic of lipid-based delivery systems.26–29 Additionally, the integration of berberine in the liposomal formulation is expected to modulate the structural characteristics of the liposome itself. Specifically, berberine's presence can induce a repulsion effect within the liposomal hydrophobic core, reducing lipid packing density and creating a more fluidic bilayer structure. This increased fluidity could enhance the membrane's flexibility and reduce entrapment of the mRNA cargo, facilitating more efficient endosomal escape and release of mRNA into the cytoplasm. This effect on the lipid bilayer structure may contribute to an improved balance between membrane stability and fluidity, optimizing the liposome's ability to fuse with endosomal membranes during mRNA delivery.
We hypothesize that berberine-modified liposomes will exhibit superior FIX expression by the TmL mutant variant and functional clotting activity compared to conventional lipid nanoparticles (LNPs). By enhancing the stability and bioavailability of the therapeutic mRNA, our findings could support the development of mRNA therapies for liver-associated disorders, utilizing the inherent biodistribution patterns of lipid-based carriers for hemophilia B and other genetic disorders.
2. Methods
2.1. Lipid synthesis
The DMe16 amide lipid was synthesized following established protocols. DOPE and berberine were obtained from TCI Chemicals, India.2.2. Culture of mammalian cells
The HEK-293T human embryonic kidney, CHO (Chinese hamster ovarian), Hepa 1–6 murine hepatoma, SK-HEP-1 human hepatic adenocarcinoma, and HepG2 human liver cancer cell lines (ATCC, USA) were cultured under optimal conditions using Dulbecco's modified Eagle's medium (DMEM), SK-HEP-1 and HepG2 in Eagle's Minimum Essential Medium (EMEM) (Gibco™, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS) and an antibiotic mix of penicillin (5000 U mL−1) and streptomycin (0.1 mg mL−1) (Gibco™, Thermo Fisher Scientific, USA). The culture medium was refreshed every two days, and the cells were maintained at 37 °C in a humidified incubator with 5% CO2. Once the cells reached 80–90% confluence, they were passaged using 0.25% trypsin-EDTA (Gibco™).2.3. Lipid mixture preparation
Berberine liposomes were prepared by dissolving varying molar ratios of berberine, DME 16, and DOPE in a chloroform–methanol mixture (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in a glass vial while maintaining a constant amount of DOPE lipid (Table S1). The organic solvent evaporated under a stream of nitrogen gas to create a thin lipid film, followed by vacuum drying for 2 hours. The dried film was then hydrated overnight with 1 mL of sterile deionized water. The resulting mixture was vortexed slowly for 2 minutes at room temperature to produce multilamellar vesicles (MLVs). These MLVs were subsequently sonicated in a water bath for 5 minutes and further processed using a probe sonicator for 1 minute with 15-second pulses and 15-second intervals to generate small unilamellar vehicles (SUVs).
1) in a glass vial while maintaining a constant amount of DOPE lipid (Table S1). The organic solvent evaporated under a stream of nitrogen gas to create a thin lipid film, followed by vacuum drying for 2 hours. The dried film was then hydrated overnight with 1 mL of sterile deionized water. The resulting mixture was vortexed slowly for 2 minutes at room temperature to produce multilamellar vesicles (MLVs). These MLVs were subsequently sonicated in a water bath for 5 minutes and further processed using a probe sonicator for 1 minute with 15-second pulses and 15-second intervals to generate small unilamellar vehicles (SUVs).
2.4. Characterization of liposomes
Liposomes were characterized by using Zetasizer equipment to determine their hydrodynamic diameter (HDD), zeta potential, and polydispersity index (PDI) in Milli-Q water. Samples were loaded into folded capillary cells, and each parameter was measured three times.2.5. mRNA binding assay
The electrostatic interactions between cationic liposomes and nucleic acids (mRNA) were evaluated using an agarose gel retardation assay. Lipoplexes, formed by combining liposomes and mRNA, were prepared at different N/P ratios (nitrogen to phosphate) ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 4
1 to 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and incubated at room temperature for 20 minutes. Mixed with 6× loading dye, these lipoplexes were loaded onto a 1% agarose gel, with mRNA alone as a control.
1 and incubated at room temperature for 20 minutes. Mixed with 6× loading dye, these lipoplexes were loaded onto a 1% agarose gel, with mRNA alone as a control.
2.6. Heparin displacement
Lipoplexes were prepared using the same method of retardation assay. Subsequently, 10 μL of heparin (1 mg mL−1), a negatively charged molecule (mRNA), was added to the lipoplexes, and the mixture was incubated for 20 minutes. The resulting samples were then analyzed using 1% agarose gel electrophoresis.2.7. RNase sensitivity assay
The optimized lipoplex formulation was selected for a sensitivity assay. Lipoplex samples of the negative control (RNase−) and positive control (RNase+) were prepared in 50 μL volumes. To each tube, 100 ng of RNase, an enzyme, was added, and the mixtures were incubated at room temperature for 30 minutes. Subsequently, 5 g of proteinase K was added to the samples, which were incubated at 50 °C on a thermomixer for 15 minutes. Following this, 100 μL of a phenol–chloroform mixture was added to each tube, vigorously mixed, and incubated at room temperature for 5 minutes. The tubes were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes, and the aqueous phase was collected. From this, 10 μL was mixed with RNA loading dye and heated to 65 °C for 30 minutes. The samples were then cooled on ice and loaded onto an agarose gel for analysis.
000 rpm for 15 minutes, and the aqueous phase was collected. From this, 10 μL was mixed with RNA loading dye and heated to 65 °C for 30 minutes. The samples were then cooled on ice and loaded onto an agarose gel for analysis.
2.8. In vitro cytotoxicity
Cytotoxicity was evaluated using an MTT-based assay. HEK-293T cells (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per well) were seeded into 96-well plates and incubated at 37 °C with 5% CO2 for 16–20 hours to achieve 70–80% confluency. Lipoplex was then transfected into the cells, followed by a 48-hour incubation under the same conditions. Afterwards, 20 μL of MTT solution (5 mg mL−1 in PBS, pH 7.4) was added, and the cells were incubated for 4 hours. The medium was subsequently removed, and 150 μL of DMSO was added to dissolve the formazan crystals. The absorbance was measured at 570 nm using a Spectra Max i3X microplate reader.
000 per well) were seeded into 96-well plates and incubated at 37 °C with 5% CO2 for 16–20 hours to achieve 70–80% confluency. Lipoplex was then transfected into the cells, followed by a 48-hour incubation under the same conditions. Afterwards, 20 μL of MTT solution (5 mg mL−1 in PBS, pH 7.4) was added, and the cells were incubated for 4 hours. The medium was subsequently removed, and 150 μL of DMSO was added to dissolve the formazan crystals. The absorbance was measured at 570 nm using a Spectra Max i3X microplate reader.2.9. Uptake assay
Red fluorescent liposomes were prepared for cellular uptake studies by incorporating 1% Rho-PE while maintaining the specified lipid ratios (Table S1). Following liposome preparation, transfection experiments were conducted, and cellular uptake was quantified via flow cytometry 4 hours post-transfection.2.10. In vitro transfection for eGFP and FIX
Five distinct cell lines, HEK-293T, CHO, Skhep-1, HepG2, and Hepa R-1–6, were used for transfection studies. The cells were seeded at a density of 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 48-well plates and allowed to grow for 16 hours before transfection. Lipoplexes were formulated with LPX (Lipoplex), Ber. LPX (Berberine. Lipoplex) and con. LPX (Control. Lipoplex) at an mRNA-to-lipid N/P charge ratio of 2
000 cells per well in 48-well plates and allowed to grow for 16 hours before transfection. Lipoplexes were formulated with LPX (Lipoplex), Ber. LPX (Berberine. Lipoplex) and con. LPX (Control. Lipoplex) at an mRNA-to-lipid N/P charge ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in a serum-free medium, followed by a 20-minute incubation at room temperature. The study used 0.2 g of eGFP mRNA to encode a green fluorescent protein (GFP). After incubation, the lipoplexes were transfected into the cells, maintained at 37 °C in a 5% CO2 environment. GFP expression was analyzed 48 hours later using fluorescence microscopy (Leica DM100) and FACS analysis (BD Celesta). The same procedure was followed for FIX mRNA. After 16 hours of post-transfection, the media were changed and supplemented with vitamin K and ITS (insulin-transferring selenite). The cell lysate and media supernatant were collected 72 hours later for western blot analysis.
1 in a serum-free medium, followed by a 20-minute incubation at room temperature. The study used 0.2 g of eGFP mRNA to encode a green fluorescent protein (GFP). After incubation, the lipoplexes were transfected into the cells, maintained at 37 °C in a 5% CO2 environment. GFP expression was analyzed 48 hours later using fluorescence microscopy (Leica DM100) and FACS analysis (BD Celesta). The same procedure was followed for FIX mRNA. After 16 hours of post-transfection, the media were changed and supplemented with vitamin K and ITS (insulin-transferring selenite). The cell lysate and media supernatant were collected 72 hours later for western blot analysis.
2.11. Preparation of variant mutant FIX plasmids by site-directed mutagenesis
The variant mutant FIX plasmids were generated by site-directed mutagenesis using PCR-based primer incorporation and subsequent cloning. The WT-FIX plasmid was used as the template. Two complementary primers containing the desired mutation(s) were designed to introduce specific alterations in the coding sequence of the human factor IX (FIX) gene (Table S2). A PCR was set up using the wild-type plasmid as a template and the mutagenic primers. The reaction conditions were optimized to amplify the desired mutated fragment. Following PCR amplification, the product was treated with DpnI to digest the parental, non-mutated plasmid DNA. The ligation mixture was transformed into a competent E. coli strain, DH5α cells, and individual colonies were screened by colony PCR for confirmation. Positive clones were grown in an appropriate growth medium, and plasmid DNA was extracted using a standard plasmid isolation procedure. The mutation was verified by Sanger sequencing (Fig. S2), and the final mutant plasmids were used for subsequent IVT mRNA preparation experiments.2.12. IVT eGFP and mutant FIX mRNA preparation and purification
The DNA template for eGFP and FIX was generated using a specific primer set. Six distinct mRNA variants for FIX with m1Ψ-modification were synthesized as described in the protocol in ref. 22.2.13. FRET assay
FRET assays were performed to assess molecular interactions between proteins in cells. The cells were transfected with donor and acceptor fluorophore-tagged mRNA using a berberine lipid mixture and allowed to express the constructs for 24–48 hours. Following transfection, the cells were washed with PBS and incubated in the buffer for imaging. FRET measurements were conducted using a microscope equipped with appropriate filter sets for both donor and acceptor fluorophores. The donor was excited at the excitation wavelength, and emission was detected at the donor emission wavelength. The acceptor was excited by the donor's emission and measured at the acceptor emission wavelength. FRET efficiency was calculated based on the difference in acceptor fluorescence with and without donor excitation using the formula:Negative controls included cells expressing only the donor or acceptor, and positive controls used known interacting proteins. Data were analyzed to compare FRET efficiency across experimental conditions.
2.14. Western blot
Factor IX (FIX), a secreted protein, was analyzed from both cell culture media (supernatants: representing secreted/extracellular protein) and cell lysates (representing intracellular protein).
Preparation of the medium supernatant: to quantify secreted FIX, cells were cultivated in either low-serum or serum-free media (to minimize cross-reactivity between FIX and bovine FIX present in FBS). After transfection, the culture media were collected, clarified by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–15
000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10–15 minutes to remove cell debris, and stored at −80 °C until analysis. Preparation of cell lysate: cells were washed twice with cold PBS to remove residual media and detached. Lysis was performed using RIPA buffer (composition: 50 mM Tris-HCl pH7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitors). The lysate was incubated on ice for 30 minutes, followed by centrifugation at 12
000g for 10–15 minutes to remove cell debris, and stored at −80 °C until analysis. Preparation of cell lysate: cells were washed twice with cold PBS to remove residual media and detached. Lysis was performed using RIPA buffer (composition: 50 mM Tris-HCl pH7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitors). The lysate was incubated on ice for 30 minutes, followed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–15
000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 minutes at 4 °C. The supernatant (total protein lysate) was collected for downstream analysis. Quantification: the protein concentration of both cell lysates and media supernatant samples was determined by the Bradford assay using BSA as a standard. For western blotting, 30 μg of total protein (either cell lysate or media protein) was resolved by 10% SDS-PAGE and transferred to a PVDF membrane (WH3135834). The membrane was blocked with 3% BSA in TBST, incubated with a human factor IX antibody (Thermo Scientific, MA5-292541:1000), and then goat anti-rabbit IgG H&L-HRP (Invitrogen, ab205718, 1
000g for 30 minutes at 4 °C. The supernatant (total protein lysate) was collected for downstream analysis. Quantification: the protein concentration of both cell lysates and media supernatant samples was determined by the Bradford assay using BSA as a standard. For western blotting, 30 μg of total protein (either cell lysate or media protein) was resolved by 10% SDS-PAGE and transferred to a PVDF membrane (WH3135834). The membrane was blocked with 3% BSA in TBST, incubated with a human factor IX antibody (Thermo Scientific, MA5-292541:1000), and then goat anti-rabbit IgG H&L-HRP (Invitrogen, ab205718, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). Beta-actin (MA5-15739) with anti-mouse secondary (ab31430) was used as a loading control. Detection was performed using a chemiluminescent substrate and visualized with a Chem-Doc system (Bio-Rad, Hercules, CA, USA).
000). Beta-actin (MA5-15739) with anti-mouse secondary (ab31430) was used as a loading control. Detection was performed using a chemiluminescent substrate and visualized with a Chem-Doc system (Bio-Rad, Hercules, CA, USA).
2.15. Statistical analysis
Ordinary one-way ANOVA and multiple t-tests were used in the statistical analysis, and they were performed with GraphPad Prism (Version 8.02, GraphPad Prism).3. Results and discussion
3.1. Characterization of lipid mixtures
The lipid mixtures exhibit key features, including nanoscale size, uniform dispersion in solution, and a positive surface charge below +75 mV, making them promising candidates for non-viral gene therapy. Liposomes were prepared in various formulations using different lipid and co-lipid concentrations, as detailed in Table S1. The structures of the lipids, co-lipids, and the resulting liposomes are depicted in Fig. 1A. As shown in Fig. 1B, the lipid mixture has an average size within the 160 nm range. The ζ-potential measurements confirm successful quantification and accuracy, with all liposomes maintaining a positive charge below +75 mV except B. LPX, Con. Lipo and con. LPX (Fig. 1C). Furthermore, the lipid mixture demonstrates uniform dispersion in solution, with a polydispersity index (PDI) below 0.3, as illustrated in Fig. 1D.3.2. Lipid mixture complex analysis of mRNA
A series of assays were conducted to evaluate liposome binding efficacy, stability, and cellular interactions. A gel retardation assay demonstrated robust mRNA complexation across all lipid formulations and N/P charge ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 4
1, and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), as shown in Fig. S1a. A heparin displacement assay assessed the lipoplex stability against competitive binding in the bloodstream. Heparin, a highly negatively charged polysaccharide found in the extracellular matrix and on cell surfaces was added to lipoplexes containing mRNA. Despite heparin's presence, the lipoplexes maintained stability, as evidenced in Fig. S1b.
1), as shown in Fig. S1a. A heparin displacement assay assessed the lipoplex stability against competitive binding in the bloodstream. Heparin, a highly negatively charged polysaccharide found in the extracellular matrix and on cell surfaces was added to lipoplexes containing mRNA. Despite heparin's presence, the lipoplexes maintained stability, as evidenced in Fig. S1b.
An RNase I protection assay confirmed liposomes’ ability to protect mRNA from enzymatic degradation. Lipoplexes with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P charge ratio effectively shielded mRNA from RNase I degradation, with intact mRNA bands observed after phenol–chloroform extraction as a negative control (Fig. S1c). In contrast, naked mRNA exposed to RNase I underwent hydrolysis, confirming its susceptibility. Lipoplexes, however, resisted degradation and consistently accumulated in the aqueous phase.
1 N/P charge ratio effectively shielded mRNA from RNase I degradation, with intact mRNA bands observed after phenol–chloroform extraction as a negative control (Fig. S1c). In contrast, naked mRNA exposed to RNase I underwent hydrolysis, confirming its susceptibility. Lipoplexes, however, resisted degradation and consistently accumulated in the aqueous phase.
3.3. In vitro compatibility of lipid mixtures
The cytotoxicity of different lipoplex formulations was evaluated by assessing cell viability. As shown in Fig. 2A all formulations maintained high cell viability (>80%), indicating minimal cytotoxic effects. The darker-coloured bars represent the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratio formulations, while the lighter-coloured bars correspond to the 2
1 charge ratio formulations, while the lighter-coloured bars correspond to the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratio formulations. The LPX group exhibited the highest cell viability, with both 1
1 charge ratio formulations. The LPX group exhibited the highest cell viability, with both 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratios showing no significant toxicity. The B. LPX formulation also demonstrated high cell viability, with only a slight reduction in viability compared to that of LPX. The Con. LPX group showed slightly lower viability than the other groups, but the difference was not substantial. Importantly, no significant difference in cytotoxicity was observed between the 1
1 charge ratios showing no significant toxicity. The B. LPX formulation also demonstrated high cell viability, with only a slight reduction in viability compared to that of LPX. The Con. LPX group showed slightly lower viability than the other groups, but the difference was not substantial. Importantly, no significant difference in cytotoxicity was observed between the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratios within each formulation, suggesting that increasing the ratio does not negatively impact cell viability. The results indicate that all tested LPX formulations exhibit excellent biocompatibility, with cell viability consistently above 80%, suggesting minimal cytotoxic effects. This is crucial for potential in vitro and in vivo applications of these lipoplexes. The slightly reduced viability in Con. LPX could be attributed to formulation-specific properties affecting cellular interactions. The similar cytotoxicity profiles between 1
1 charge ratios within each formulation, suggesting that increasing the ratio does not negatively impact cell viability. The results indicate that all tested LPX formulations exhibit excellent biocompatibility, with cell viability consistently above 80%, suggesting minimal cytotoxic effects. This is crucial for potential in vitro and in vivo applications of these lipoplexes. The slightly reduced viability in Con. LPX could be attributed to formulation-specific properties affecting cellular interactions. The similar cytotoxicity profiles between 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratios suggest that increasing the lipid-to-nucleic acid ratio does not compromise cell viability, optimising transfection efficiency without introducing toxicity. Further studies, such as long-term viability assays and apoptosis analysis, may be required to confirm the safety profile of these formulations (Fig. 2A).
1 charge ratios suggest that increasing the lipid-to-nucleic acid ratio does not compromise cell viability, optimising transfection efficiency without introducing toxicity. Further studies, such as long-term viability assays and apoptosis analysis, may be required to confirm the safety profile of these formulations (Fig. 2A).
The cellular uptake efficiency of different lipoplex (LPX) formulations was assessed, and the results are presented in Fig. 2B. The LPX formulations showed moderate uptake efficiency, with the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio exhibiting slightly higher uptake than the 1
1 ratio exhibiting slightly higher uptake than the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. B. LPX demonstrated the highest uptake among the tested formulations, with both 1
1 ratio. B. LPX demonstrated the highest uptake among the tested formulations, with both 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratios reaching nearly 80–90% uptake. Con. LPX showed significantly lower uptake compared to LPX and B. LPX, with the 1
1 charge ratios reaching nearly 80–90% uptake. Con. LPX showed significantly lower uptake compared to LPX and B. LPX, with the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio exhibiting an uptake of approximately 40%. In contrast, the 2
1 ratio exhibiting an uptake of approximately 40%. In contrast, the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio slightly improved but remained lower than those of the other formulations. The data suggest that B. LPX exhibits the most efficient cellular uptake, indicating an improved formulation for intracellular delivery. The higher uptake observed in the 2
1 ratio slightly improved but remained lower than those of the other formulations. The data suggest that B. LPX exhibits the most efficient cellular uptake, indicating an improved formulation for intracellular delivery. The higher uptake observed in the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratio formulations across all groups suggests that increasing the lipid-to-nucleic acid ratio enhances cellular internalization. The lower uptake of Con. LPX indicates that its composition might not be as effective in facilitating cellular entry, potentially due to differences in lipid composition or charge interactions. There is a significant difference in uptake between B. LPX and Con. LPX, highlighting the importance of formulation optimization for improving intracellular delivery. Further studies, such as endocytic pathway inhibition assays, could help determine the uptake mechanism for these formulations. Additionally, correlating uptake efficiency with transfection efficiency and functional gene expression would provide deeper insights into the effectiveness of these lipoplexes for gene delivery applications.
1 charge ratio formulations across all groups suggests that increasing the lipid-to-nucleic acid ratio enhances cellular internalization. The lower uptake of Con. LPX indicates that its composition might not be as effective in facilitating cellular entry, potentially due to differences in lipid composition or charge interactions. There is a significant difference in uptake between B. LPX and Con. LPX, highlighting the importance of formulation optimization for improving intracellular delivery. Further studies, such as endocytic pathway inhibition assays, could help determine the uptake mechanism for these formulations. Additionally, correlating uptake efficiency with transfection efficiency and functional gene expression would provide deeper insights into the effectiveness of these lipoplexes for gene delivery applications.
3.4. FRET assay
The Förster Resonance Energy Transfer (FRET) assay was conducted to evaluate the membrane fusion and structural dynamics of different liposomal formulations over 30 minutes. As shown in the graph (Fig. 2C), fluorescence recovery (%) increased over time for all three formulations—standard LPX (red circles), berberine-functionalized LPX (B. LPX, green squares), and control LPX (Con. LPX, purple triangles). Among them, B. LPX exhibited the highest fluorescence recovery, reaching approximately 85% at 30 minutes, whereas LPX showed moderate recovery (∼60%), and Con. LPX demonstrated the lowest recovery (∼20%). The fluorescence recovery trend suggests that B. LPX has a significantly higher membrane fluidity and fusion capacity than other formulations (Fig. 2C).The increased fluorescence recovery in B. LPX suggests that the incorporation of berberine enhances the membrane's dynamic properties, potentially due to its ability to modulate lipid packing and introduce repulsive interactions within the hydrophobic core. This agrees with previous studies that indicate berberine can disrupt rigid lipid structures, increasing bilayer fluidity and facilitating molecular rearrangement. The higher FRET recovery in B. LPX implies more efficient liposome–liposome or liposome–cell membrane interactions, which could enhance endosomal escape and intracellular delivery of the encapsulated cargo, such as mRNA.
In contrast, LPX showed moderate fluorescence recovery, indicating a relatively stable but less fluidic membrane environment. The lowest fluorescence recovery in Con. LPX suggests a more rigid and stable bilayer structure, potentially reducing fusion and intracellular delivery efficiency. The observed differences in fluorescence recovery support the hypothesis that berberine-functionalized liposomes improve membrane flexibility, which may enhance the efficiency of gene delivery systems by promoting fusion events necessary for endosomal escape. Overall, these results suggest that B. LPX could serve as a promising platform for enhanced hepatic mRNA delivery by optimizing membrane dynamics and fusion efficiency. Further mechanistic studies are required to correlate these findings with transfection efficiency in hepatocytes.
3.5. Screening of the lipid mixture in hepatic cell lines
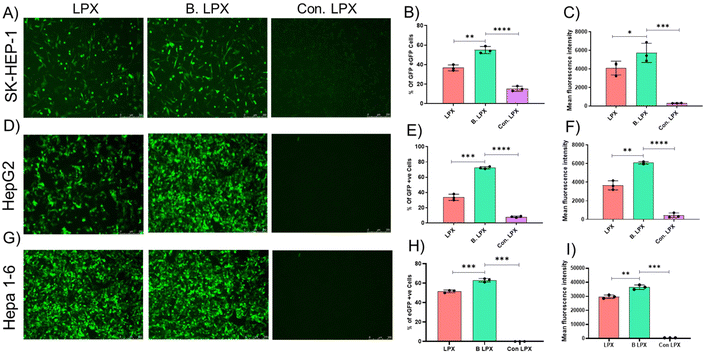
To evaluate the transfection efficiency of different liposome formulations, we transfected SK-HEP-1, HepG2, and Hepa 1–6 cells with chemically modified eGFP mRNA-loaded liposomes. GFP expression was analyzed using fluorescence microscopy and flow cytometry, providing insights into transfection fluorescence imaging, which confirmed eGFP expression in all three hepatic cell lines following transfection. However, the intensity and distribution of GFP signals varied across the formulations. SK-HEP-1cells exhibited moderate GFP expression with B. LPX, while LPX showed lower intensity, indicating reduced transfection efficiency, and with con. LPX, there is minimal transfection, as shown in Fig. 3A–C. HepG2 cells displayed strong GFP fluorescence, especially in B. LPX-treated cells, with improved signal intensity compared to those of Con. LPX and reduced eGFP in LPX (Fig. 3D–F). Hepa 1–6 cells showed the highest GFP fluorescence among the three cell lines, with B. LPX inducing widespread and intense GFP expression and a reduced level in LPX and zero transfection with Con. LPX (Fig. 3G–I). The percentage of GFP-positive cells and mean fluorescence intensity (MFI) were measured to quantify transfection efficiency. In SK-HEP-1 Cells, B. LPX shows 55% GFP-positive cells, a 1.3-fold increase in MFI compared to that of LPX. LPX shows 36% GFP-positive cells, indicating lower transfection efficiency and basal level expression compared to those of Con. LPX (Fig. 3B and C). In HepG2 cells, B. LPX shows 72.3% GFP-positive cells, a 1.66-fold increase in MFI compared to that of LPX. LPX shows 33% GFP-positive cells, confirming successful but less efficient transfection and a low-level expression in Con. LPX (Fig. 3E and F). In Hepa 1–6 cells, B. LPX shows 63% GFP-positive cells, a 1.2-fold increase in MFI compared to that of LPX. LPX shows 51% GFP-positive cells, indicating efficient transfection but lower than those of B. LPX and Con. LPX, pronounced, as usual, as having less or zero transfection efficiency (Fig. 3H and I). The negative control (untreated cells) showed negligible GFP fluorescence, confirming that GFP expression was solely due to liposome-mediated mRNA delivery. The results demonstrate that B. LPX enhances mRNA transfection efficiency in all three hepatic cell lines, with the highest efficiency observed in HepG2 (72.3% GFP+, 1.66-fold MFI increase), followed by Hepa 1–6 (63% GFP+, 1.22-fold MFI increase) and SK-HEP-1 (55% GFP+, 1.3-fold MFI increase). The higher transfection efficiency in Hepa 1–6 and HepG2 cells may be attributed to higher lipid uptake capacity and endosomal escape efficiency, making them more responsive to B. LPX. SK-HEP-1 cells showed lower GFP expression, possibly due to differences in membrane composition, lipid uptake mechanisms, or endosomal processing in this cell line. These findings suggest that B. LPX is a promising lipid formulation for hepatic mRNA delivery, with Hepa 1–6 and HepG2 showing the highest potential for liver-targeted gene therapies; particularly, B. LPX favours FIX production from hepatocytes, whereas SK-HEP-1 is an endothelial cell more responsible for factor VIII production. Further studies exploring endosomal escape mechanisms, in vivo validation, and long-term gene expression are necessary to optimize these formulations for clinical translation.3.6. Screening of the lipid mixture in non-hepatic cell lines
To assess the transfection efficiency of eGFP mRNA using different liposomal formulations in non-hepatic cell lines such as HEK-293T and CHO cells, fluorescence microscopy was performed, and the images revealed distinct differences in eGFP expression among the three formulations. B. LPX-treated cells exhibited the highest fluorescence intensity and the most widespread eGFP expression in both cell lines. LPX-treated cells displayed moderate fluorescence signals, suggesting effective but slightly lower transfection efficiency. Con. LPX-treated cells showed weak fluorescence, indicating poor transfection efficiency compared to those of LPX and B. LPX. Quantification of eGFP-positive cells (%) and mean fluorescence intensity (MFI) provided a more detailed comparison among the formulations: in HEK-293T cells, B. LPX showed 94% eGFP-positive cells and LPX showed 85% eGFP-positive cells (Fig. 4A–C). In CHO cells, B. LPX showed 40% eGFP-positive cells, and LPX showed 22% eGFP-positive cells. Fold increase analysis revealed that, compared to the fluorescence intensity (MFI) in Con. LPX-treated cells, those in LPX- and B. LPX-treated cells showed significant improvements (Fig. 4D–F). In HEK-293T cells, LPX exhibited a ∼8-fold increase over Con. LPX and B. LPX showed a ∼19-fold increase over Con. LPX. In CHO cells, LPX displayed a ∼1.3-fold increase over Con. LPX and B. LPX, resulting in a ∼1.7-fold rise over Con. LPX.The results demonstrate that B. LPX is the most efficient liposomal formulation for mRNA delivery, showing the highest eGFP expression levels and fluorescence intensity in both HEK-293T and CHO cells. The enhanced performance of B. LPX may be attributed to improved lipid composition, higher endosomal escape efficiency, and better mRNA protection. LPX also showed good transfection efficiency but with slightly lower fluorescence intensity, suggesting that while effective, it may have reduced the mRNA uptake or translation efficiency compared to B. LPX. In contrast, Con. LPX exhibited poor transfection efficiency, highlighting the necessity of optimized lipid formulations for effective mRNA delivery. Across both cell lines, HEK-293T cells consistently exhibited higher transfection efficiency than CHO cells. This trend aligns with previous studies, as HEK-293T cells are highly permissive to transfection, whereas CHO cells require more optimized conditions. However, the significant improvement of B. LPX demonstrates its potential for mRNA delivery even in less transfectable cell lines. These results underscore the importance of lipid composition in mRNA delivery and suggest that B. LPX could be a promising formulation for gene therapy applications requiring high transfection efficiency and protein expression levels.
3.7. Creating site-directed mutagenesis of variant mutant FIX mRNA
FIX mutations have an impact on expression and activity; FIX plays a crucial role in the coagulation cascade, and its functional enhancement can significantly benefit hemophilia B gene therapy. Our study introduced a series of mutations in the epidermal growth factor-like domain and peptidase FIX gene through site-directed mutagenesis, confirmed via sequencing. These designed mutations could help improve FIX expression, secretion, and enzymatic activity.The rationale for mutations and their functional implications of the Padua mutation (R338L) in FIX-Padua I is a well-characterized gain-of-function mutation that enhances FIX activity. R338L demonstrated an 8-fold increase in enzymatic activity, enabling near-curative clotting factor levels. Mechanistically, R338L enhances FIXa's affinity for factor X in a factor VIIIa (FVIIIa) dependent manner, accelerating coagulation.9,14,30 FIX-Padua II R338A demonstrated a 3-fold increase in clotting activity compared to WT-FIX in activated partial thromboplastin assays. The mutation did not abolish binding to factor VIII (FVIIIa) but altered heparin affinity. The substitution enhanced catalytic activity (kcat/Km) for factor X activation in the presence of FVIIIa. The R338A variant is distinct from R338L but shares mechanistic insights into how residue 338 modulates FIXa activity.31 The FIX inclusion of this mutation in our study aims to assess its expression in hepatic cell lines and its potential for improving clotting efficacy. In addition to the Padua mutation, we introduced Ala substitutions in the FIX catalytic domain and linker regions, leading to TripleA (A86:A227:A338), TripleAL (A86:A227:L338), and DoubleA (A227:A338) variants. These substitutions were hypothesized to enhance FIX folding and secretion. Incorporating Ala at positions 86 and 227 could reduce steric hindrance, promoting a more stable protein conformation. A properly folded protein is often more efficiently trafficked through the endoplasmic reticulum (ER) and Golgi apparatus, leading to increased secretion. To improve enzymatic efficiency, residue 227 (Glu → Ala) is in the activation peptide region of FIX, which plays a role in enzyme activation. The substitution may reduce potential steric clashes during activation, enhancing FIXa function. By increasing stability and half-life, the Leu substitution (R338L) in FIX-TripleAL and FIX-DoubleL may alter the electrostatic interactions within the serine protease domain, potentially stabilizing the active conformation and prolonging FIXa half-life. The recombinant gain of functional mutated FIX with its modified amino acid residue and modified codon are mentioned in Fig. 5A. The detailed amino acid residue modification with the specific codon is shown in Fig. 5B.
These structural modifications could enhance the overall activity of FIX while simultaneously increasing expression levels through improved protein folding and secretion efficiency. For potential applications in gene therapy, our findings suggest that FIX mutants with TripleA, TripleAL, and DoubleL modifications may be promising candidates for gene therapy due to their increased expression potential and functional enhancement. If these mutations contribute to both higher protein yields and increased activity, they could reduce the therapeutic dosing requirements, making them ideal for liver-directed mRNA or gene therapy approaches. Future studies need to focus on characterizing the functional kinetics of these mutants, including factor X activation assays and thrombin generation studies, to validate their enhanced coagulation potential.
3.8. Mutant mRNA FIX protein expression
4. Conclusions
This study highlights the potential of berberine-functionalized cationic liposomes as a novel platform for hepatic delivery of gain-of-function FIX mutant mRNAs, with the TmL mutant demonstrating superior expression compared to the widely studied SmL variant. The enhanced FIX protein production observed suggests that the additional mutations in TmL improve protein stability, folding, or secretion efficiency, positioning TmL as a promising candidate for next-generation mRNA-based therapies for hemophilia B.4.1 Limitations
Despite the promising results of this study, several limitations should be acknowledged. Firstly, the biological activity of the expressed FIX variants was not directly validated using functional clotting assays such as activated partial thromboplastin time (aPTT), factor X (FX) activation, or thrombin generation assays. Our assessment of FIX protein relied primarily on western blot analysis, and thus, the pro-coagulant function of the delivered FIX remains to be confirmed in future studies. Secondly, our experiments were conducted in hepatoma cell lines (Hepa 1–6 and HepG2), and we did not evaluate delivery efficiency or FIX expression in primary hepatocytes. Since primary hepatocytes may differ from immortalized cell lines in terms of uptake, metabolism, and gene expression, future studies using primary hepatocytes or in vivo models are necessary to validate the translational potential and therapeutic efficacy of berberine-functionalized liposomes. Western blot quantification (Fig. 6) was performed with only two biological replicates, limiting the statistical power of the analysis. Regarding scalability, although our liposome synthesis and characterization were performed at laboratory scale, further work is needed to determine the feasibility of large-scale production, batch consistency, and long-term stability to support clinical translation.Overall, these limitations highlight important directions for future research to fully establish the functional efficacy, mechanistic basis, and clinical relevance of this delivery platform for hemophilia B therapy.
Author contributions
Srujan Marepally: conceptualization, formal analysis, funding acquisition, investigation, project administration, resources, supervision, and writing, review & editing. Porkizhi Arjunan: data curation, conceptualization, formal analysis, methodology, software, validation, visualization, writing – original draft, and review & editing. Gokulnath Mahalingam: conceptualization, formal analysis, methodology and software. Harini: methodology. Saravanabhavan Thangavel: writing, review, and editing. Mohan Kumar Murugesan: writing, reviewing, and editing.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article are included as part of the supplementary information (SI). The author will also provide raw data upon request. The Supplementary Information file includes additional figures supporting the main text (Fig. S1–S2), complexation studies, and sequence confirmation data. Tables S1–S3 provide details of lipid mixtures, primer sets, and fold changes in FIX expression relative to WT and SmL (Padua L) in cell lysates and media supernatants.Supplementary information is available. See DOI: https://doi.org/10.1039/d5pm00135h.
Acknowledgements
We want to thank Durga Kathirvelu and Priyanka Sankar for their initial experimental help.References
- K. Lee, J. Q. Chau and Y. B. Suber, Enhanced procoagulant activity of select hemophilia B causing factor IX variants with emicizumab, Blood, 2024, 144(11), 1230–1235 CrossRef PubMed.
- P. M. Mannucci and E. G. Tuddenham, The hemophilias–from royal genes to gene therapy, N. Engl. J. Med., 2001, 344(23), 1773–1779 CrossRef PubMed.
- S. W. Pipe, G. Gonen-Yaacovi and O. G. Segurado, Hemophilia A gene therapy: current and next-generation approaches, Expert Opin. Biol. Ther., 2022, 22(9), 1099–1115 CrossRef PubMed.
- A. C. Nathwani, U. M. Reiss, E. G. Tuddenham, C. Rosales, P. Chowdary and J. McIntosh, et al., Long-term safety and efficacy of factor IX gene therapy in hemophilia B, N. Engl. J. Med., 2014, 371(21), 1994–2004 CrossRef PubMed.
- L. A. George, S. K. Sullivan, A. Giermasz, J. E. J. Rasko, B. J. Samelson-Jones and J. Ducore, et al., Hemophilia B Gene Therapy with a High-Specific-Activity Factor IX Variant, N. Engl. J. Med., 2017, 377(23), 2215–2227 CrossRef PubMed.
- U. Sahin, K. Karikó and Ö Türeci, mRNA-based therapeutics–developing a new class of drugs, Nat. Rev. Drug Discovery, 2014, 13(10), 759–780 CrossRef PubMed.
- X. Hou, T. Zaks and R. Langer, Lipid nanoparticles for mRNA delivery, Nat Rev Mater., 2021, 6(12), 1078–1094 CrossRef PubMed.
- W. Wu, L. Xiao, X. Wu, X. Xie, P. Li and C. Chen, et al., Factor IX alteration p.Arg338Gln (FIX Shanghai) potentiates FIX clotting activity and causes thrombosis, Haematologica, 2021, 106(1), 264–268 CrossRef PubMed.
- B. J. Samelson-Jones, J. D. Finn, L. J. Raffini, E. P. Merricks, R. M. Camire and T. C. Nichols, et al., Evolutionary insights into coagulation factor IX Padua and other high-specific-activity variants, Blood Adv., 2021, 5(5), 1324–1332 CrossRef PubMed.
- F. A. Al-Allaf, M. M. Taher, Z. Abduljaleel, A. Bouazzaoui, M. Athar and N. M. Bogari, et al., Molecular Analysis of Factor VIII and Factor IX Genes in Hemophilia Patients: Identification of Novel Mutations and Molecular Dynamics Studies, J. Clin. Med. Res., 2017, 9(4), 317–331 CrossRef PubMed.
- L. Gerasimavicius and B. J. Livesey, Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure, Nat Commun., 2022, 13(1), 3895 CrossRef CAS PubMed.
- P. E. Monahan, J. Sun, T. Gui, G. Hu, W. B. Hannah and D. G. Wichlan, et al., Employing a gain-of-function factor IX variant R338L to advance the efficacy and safety of hemophilia B human gene therapy: preclinical evaluation supporting an ongoing adeno-associated virus clinical trial, Hum. Gene Ther., 2015, 26(2), 69–81 CrossRef CAS PubMed.
- T. VandenDriessche and M. K. Chuah, Hyperactive Factor IX Padua: A Game-Changer for Hemophilia Gene Therapy, Mol. Ther., 2018, 26(1), 14–16 CrossRef CAS PubMed.
- P. Simioni, D. Tormene, G. Tognin, S. Gavasso, C. Bulato and N. P. Iacobelli, et al., X-linked thrombophilia with a mutant factor IX (factor IX Padua), N. Engl. J. Med., 2009, 361(17), 1671–1675 CrossRef CAS PubMed.
- C. Y. Kao, C. N. Lin, I. S. Yu, M. H. Tao, H. L. Wu and G. Y. Shi, et al., FIX-Triple, a gain-of-function factor IX variant, improves haemostasis in mouse models without increased risk of thrombosis, Thromb. Haemostasis, 2010, 104(2), 355–365 CrossRef CAS PubMed.
- C. Y. Kao, S. J. Yang, M. H. Tao, Y. M. Jeng, I. S. Yu and S. W. Lin, Incorporation of the factor IX Padua mutation into FIX-Triple improves clotting activity in vitro and in vivo, Thromb. Haemostasis, 2013, 110(2), 244–256 CrossRef CAS PubMed.
- G. Mahalingam, H. K. Rachamalla, P. Arjunan, K. V. Karuppusamy, Y. Periyasami and A. Mohan, et al., SMART-lipid nanoparticles enabled mRNA vaccine elicits cross-reactive humoral responses against the omicron sub-variants, Mol. Ther., 2024, 32(5), 1284–1297 CrossRef PubMed.
- X. Xu and T. Xia, Recent Advances in Site-Specific Lipid Nanoparticles for mRNA Delivery, 2023, 3(3), 192–203 Search PubMed.
- B. Lohchania, P. Arjunan, G. Mahalingam, A. Dandapani, P. Taneja and S. Marepally, Lipid Nanoparticle-Mediated Liver-Specific Gene Therapy for Hemophilia B, Pharmaceutics, 2024, 16(11), 1427 CrossRef PubMed.
- P. Arjunan, D. Kathirvelu, G. Mahalingam, A. K. Goel, U. G. Zacharaiah and A. Srivastava, et al., Lipid-nanoparticle-enabled nucleic acid therapeutics for liver disorders, Acta Pharm. Sin. B, 2024, 14(7), 2885–2900 CrossRef PubMed.
- P. Arjunan, G. Mahalingam and P. Sankar, Base-modified factor VIII mRNA delivery with galactosylated lipid nanoparticles as a protein replacement therapy for haemophilia A, Biomater. Sci., 2024, 12(19), 5052–5062 RSC.
- M. Zhou, Y. Deng, M. Liu, L. Liao, X. Dai and C. Guo, et al., The pharmacological activity of berberine, a review for liver protection, Eur. J. Pharmacol., 2021, 890, 173655 CrossRef PubMed.
- R. Domitrović, H. Jakovac and G. Blagojević, Hepatoprotective activity of berberine is mediated by inhibition of TNF-α, COX-2, and iNOS expression in CCl(4)-intoxicated mice, Toxicology, 2011, 280(1–2), 33–43 CrossRef PubMed.
- X. Chang, Z. Wang, J. Zhang, H. Yan, H. Bian and M. Xia, et al., Lipid profiling of the therapeutic effects of berberine in patients with nonalcoholic fatty liver disease, J. Transl. Med., 2016, 14, 266 CrossRef PubMed.
- Q. Rong, B. Han, Y. Li, H. Yin, J. Li and Y. Hou, Berberine Reduces Lipid Accumulation by Promoting Fatty Acid Oxidation in Renal Tubular Epithelial Cells of the Diabetic Kidney, Front. Pharmacol., 2021, 12, 729384 CrossRef PubMed.
- S. Sardana, R. Gupta, K. Madan, D. Bisht, V. S. Rana and S. Bhargava, et al., Advance drug delivery and combinational drug approaches for hepatoprotective action of berberine: a progressive overview with underlying mechanism, RPS Pharm. Pharmacol. Rep., 2023, 2, 1–23 Search PubMed.
- Y. Lu, N. S. Mohammad, J. Lee, A. M. Aranyos, K. A. Serban and M. L. Brantly, Berberine potentiates liver inflammation and fibrosis in the PI*Z hAAT transgenic murine model, PLoS One, 2024, 19(9), e0310524 CrossRef PubMed.
- Q. Nie, M. Li, C. Huang, Y. Yuan, Q. Liang and X. Ma, et al., The clinical efficacy and safety of berberine in the treatment of non-alcoholic fatty liver disease: a meta-analysis and systematic review, J. Transl. Med., 2024, 22(1), 225 CrossRef PubMed.
- X. Liu, Q. Liang, Y. Wang, S. Xiong and R. Yue, Advances in the pharmacological mechanisms of berberine in the treatment of fibrosis, Front. Pharmacol., 2024, 15, 1455058 CrossRef PubMed.
- G. Almeida-Porada, A new “FIX” for hemophilia B gene therapy, Blood, 2021, 137(21), 2860–2861 CrossRef PubMed.
- B. J. Samelson-Jones, J. D. Finn, L. A. George, R. M. Camire and V. R. Arruda, Hyperactivity of factor IX Padua (R338L) depends on factor VIIIa cofactor activity, JCI Insight, 2019, 5(14), e128683 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |