Open Access Article
Open Access ArticleEfficient and specific DNA-targeting using single-stranded LNA/MOE mixmers and chimeric Invader:XenoRNA probes
Michaela E.
Everly
 and
Patrick J.
Hrdlicka
and
Patrick J.
Hrdlicka
 *
*
Department of Chemistry, University of Idaho, Moscow, Idaho 83844-2343, USA. E-mail: hrdlicka@uidaho.edu
First published on 16th October 2025
Abstract
DNA-targeting properties of heteroduplexes between intercalator-functionalized oligonucleotides and various Xeno RNAs (i.e., LNA/MOE mixmers, or fully modified MOE, O2′-Me, or 2′-F strands) are disclosed. Chimeric Invader:LNA/MOE probes and some single-stranded LNA/MOE mixmers enable efficient invasion of double-stranded DNA targets with exceptional binding specificity.
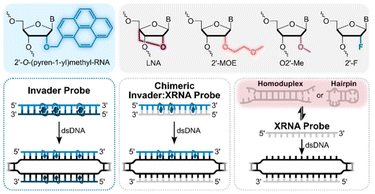
Nucleic acid-based therapeutics are transforming modern medicine. However, while over twenty RNA-targeting modalities have received FDA approval, no DNA-targeting oligonucleotides (ONs) have reached the clinic, as chromosomal DNA is a more challenging target. To realize sequence-specific recognition of double-stranded DNA (dsDNA), probes must invade Watson-Crick base pairs (bps) or bind via the duplex grooves, with polyamides,1 and triplex forming oligonucleotides2 and peptide nucleic acids (PNAs)3 being representative examples of the latter. However, groove binders are limited in targeting scope (e.g., triplex formation is generally restricted to polypurine regions). Strand-invading probes such as single-stranded modified PNAs4–8 and locked nucleic acids (LNAs),9,10 or various double-stranded probes (dsProbes),11–15 can target and unzip Watson-Crick bps of dsDNA targets to form more stable bps between individual probe strands and complementary DNA (cDNA). While capable of targeting mixed-sequence dsDNA regions, single-stranded (ss) strand-invading probes modified with affinity-enhancing modifications often form secondary structures that compromise binding efficiency, whereas dsProbes must be engineered to denature easily whilst maintaining high cDNA affinity.
In pursuit of improved strategies for mixed-sequence dsDNA-recognition, we have explored Invader probes,16,17i.e., dsProbes with +1 interstrand zipper arrangements† of intercalator-functionalized nucleotides like 2′-O-(pyren-1-yl)methyl-RNA (Fig. 1). This arrangement forces pyrene moieties between the π-stacks of neighboring bps, resulting in violation of the nearest neighbor exclusion principle18,19 (NNEP) and duplex destabilization.‡ Conversely, the individual probe strands display high affinity towards cDNA as duplex formation results in pyrene intercalation and strongly stabilizing stacking interactions with flanking bps (NNEP is not violated, Fig. 1). The difference in stability drives dsDNA-invasion, and has been used to enable detection of various biological targets.16,17,20,21 As an extension of our original strategy, we recently introduced chimeric Invader probes, i.e., heteroduplexes between densely modified Invader strands and complementary strands of miniPEG-γPNA (MPγPNA), serine-γPNA (SerγPNA), or LNA.22–24 This strategy was inspired by prior studies on DNA-targeting heteroduplex probes between intercalator-modified ONs and complementary RNA, PNA, or LNA strands,25–27 and relies on the observation that intercalators are often poorly accommodated in PNA/DNA and A-type (RNA-like) duplexes,28,29 but well-tolerated in B-type (DNA-like) duplexes. The chimeric Invader probes were found to be activated for dsDNA-invasion, with more efficient and specific recognition than the corresponding ssProbes.22–24
Here, we describe the dsDNA-recognition properties of new chimeric probe designs consisting of densely modified Invader strands and complementary Xeno RNAs (XRNAs), i.e., fully modified 2′-O-methoxyethyl (MOE), 2′-O-methyl (O2′-Me), 2′-Fluoro (2′-F) or LNA/MOE mixmers.
Fourteen 13-mer probes – seven double-stranded chimeric probes and seven ssXRNAs – were designed to target a model dsDNA target and evaluated against the corresponding conventional Invader and chimeric Invader:LNA probes (Table 1).24 Different LNA/MOE mixmer designs were studied to determine if the relative placement of LNA and MOE monomers vis-à-vis the pyrene-functionalized monomers impacts dsDNA-invasion. Thus, LNA or MOE monomers were incorporated as blocks (“blks”) of 2–4 residues, positioned across from the Invader monomers, or in a systematically alternating (“alt”) fashion with either LNA or MOE monomers across from the Invader monomers (Table 1).
| Name | dsProbe | T m [ΔTm] (°C) | dsProbe | |||
|---|---|---|---|---|---|---|
| ssXRNA Strand | Probe duplex | 5′-Strand vs. cDNA | 3′-Strand vs. cDNA | TA (°C) | ||
a 2′-O-(Pyren-1-yl)methyluridine, LNA, MOE, O2′-Me, 2′-F, and unmodified DNA monomers represented as blue, dark purple, peach, light purple, green, and grey circles, respectively. LNA and MOE “C” = 5-methyl-cytosin-1-yl monomer. ΔTm = change in Tm relative to unmodified DNA duplex 5′-GGTATATATAGGC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3′-CCATATATATCCG (Tm = 37.5 °C). Tms were recorded in medium salt phosphate buffer ([Na+] = 110 mM, [Cl−] = 100 mM, pH 7.0 (NaH2PO4/Na2HPO4), [EDTA] = 0.2 mM) using 1.0 μM of each strand. “n.d.” = not determined.
b Irregular profiles and/or broad transitions (Fig. S1 and S2).
c
T
ms determined from differential thermal denaturation curves (Fig. S2).
d Previously reported in ref. 21 and 24. 3′-CCATATATATCCG (Tm = 37.5 °C). Tms were recorded in medium salt phosphate buffer ([Na+] = 110 mM, [Cl−] = 100 mM, pH 7.0 (NaH2PO4/Na2HPO4), [EDTA] = 0.2 mM) using 1.0 μM of each strand. “n.d.” = not determined.
b Irregular profiles and/or broad transitions (Fig. S1 and S2).
c
T
ms determined from differential thermal denaturation curves (Fig. S2).
d Previously reported in ref. 21 and 24.
|
||||||
| blkMOE1 INVd |

|
>80.0 [>+42.5] | —b | 66.5c [+29.0] | 63.0 [+25.5] | n.d. |
| blkMOE2 INVd |

|
53.0 [+15.5] | 73.0 [+35.5] | 69.5 [+32.0] | 63.0 [+25.5] | 22.0 |
| altMOE3 INVd |

|
73.5 [+36.0] | 74.5b [+37.0] | 72.5c [+35.0] | 63.0 [+25.5] | 23.5 |
| altMOE4 INVd |

|
>80.0 [>+42.5] | —b | ∼77.0c [+39.5] | 63.0 [+25.5] | n.d. |
| fullMOE INVd |

|
34.5 [−3.0] | 53.0 [+15.5] | 52.5 [+15.0] | 63.0 [+25.5] | 25.0 |
| fullOMe INVd |

|
24.0 [−13.5] | 53.0 [+15.5] | 46.0 [+8.5] | 63.0 [+25.5] | 18.5 |
| fullF INVd |

|
28.0 [−9.5] | 59.0 [+21.5] | 54.0 [+16.5] | 63.0 [+25.5] | 20.5 |
| INVu INVd |

|
n.d. | 51.0 [+13.5] | 61.0 [+23.5] | 63.0 [+25.5] | 35.5 |
| LNA1 INVd |

|
>80.0 [>+42.5] | 31.5 [−6.0] | 71.0 [+33.5] | 63.0 [+25.5] | 65.0 |
As expected, individual XRNA and Invader strands form stable duplexes with cDNA (thermal denaturation temperatures (Tms) = 46.0–77.0 °C), with the order of stability decreasing as follows: alt LNA/MOE ≥ LNA > blk LNA/MOE > Invader > 2′-F ∼ MOE > O2′-Me (Table 1). Conversely, the stability of the conventional Invader probe INVu:INVd is lower than the corresponding duplexes between individual Invader strands and cDNA (Tm = 51.0 °C vs. 61.0–63.0 °C). The term thermal advantage (TA), calculated as TA = Tm (upper strand vs. cDNA) + Tm (lower strand vs. cDNA) − Tm (probe duplex) − Tm (dsDNA), is a measure of the available driving force for recognition of complementary dsDNA regions, with more positive TA values indicating a stronger driving force.16 Prior studies have demonstrated that conventional and chimeric Invader probes resulting in efficient dsDNA-recognition display substantially positive TA values. INVu:INVd displays a favorable TA value of 35.5 °C. The chimeric Invader:LNA probe LNA1:INVd displays an even more favorable TA value of 65.0 °C, as the probe is far more labile (Tm = 31.5 °C). The chimeric Invader:LNA/MOE probes display irregular denaturation profiles indicative of perturbed duplex geometries, rendering accurate Tm and TA determination challenging. Thus, blkMOE1:INVd and altMOE4:INVd do not have clear transitions, while blkMOE2:INVd and altMOE3:INVd have high-melting irregular transitions (Tms = 73.0–74.5 °C), resulting in moderate TA values of 22.0–23.5 °C. Chimeric probes entailing the fully modified MOE, O2′-Me, or 2′-F are more stable than the corresponding Invader probes (Tms = 53.0–59.0 °C), and also result in moderate TA values of 18.5–25.0 °C. Analysis of pyrene absorbance trends, suggests that XRNA modifications – contrary to expectations – do not deter pyrene intercalation (Fig. S4, Table S3, and associated discussion), resulting in the high stability of chimeric probes.
Thermal denaturation profiles were also recorded for the ssXRNAs in absence of cDNA to assess if stable secondary structures are formed in the partially self-complementary, AT-rich sequence context, as this could interfere with duplex formation. Indeed, clear transitions were observed in all cases (Table 1, Fig. S1). Fully MOE, O2′-Me, and 2′-F modified XRNAs form weakly stable secondary structures (Tms = 24.0–34.5 °C), whereas three of four LNA/MOE strands form very stable secondary structures (Tms > 73.5 °C); blkMOE2 forms a moderately stable secondary structure (Tm = 53.0 °C; Table 1). Denaturation experiments, in which the XRNA concentration was increased 10-fold, were conducted to gain insight into the type of secondary structure formed. All XRNAs, except for blkMOE1, displayed increased Tms at higher concentrations, suggesting that homoduplexes are formed (Fig. 2, Fig. S3, Table S1).
 | ||
| Fig. 2 Predicted secondary structures formed by single-stranded LNA/MOE mixmers. Color scheme as in Table 1. | ||
The greater stability of the secondary structures observed for altMOE3 and altMOE4vis-à-visblkMOE2 may reflect their greater number of affinity-enhancing LNA monomers in the self-complementary region. blkMOE1 did not result in an increased Tm with higher strand concentrations, pointing to formation of a stable intramolecular hairpin (Fig. 2, Fig. S3, Table S1). The modification pattern of blkMOE1 appears to favor intramolecular interactions. We speculate that this is due to the formation of stable LNA:LNA bps in the center of the hairpin structure (Fig. 2). Hairpin formation by the other LNA/MOE mixmers would likely require formation of two MOE:LNA or MOE:MOE bps (Fig. 2), which might not be sufficiently stable to promote hairpin formation.
The dsDNA-targeting properties of the chimeric Invader:XRNA probes and ssXRNAs were evaluated using an established electrophoretic mobility shift assay (EMSA) that relies on a chemiluminescent readout.30 A 5-fold molar excess of probe was incubated with a digoxigenin (DIG)-labeled DNA hairpin (DH1) comprised of a 13-mer double-stranded target region (complementary to the probes) linked at one end by a T10 loop. The unimolecular nature of DH1 renders it a high-melting target (Tm = 58.5 °C; Table S5). Successful invasion of DH1 by ssProbes or dsProbes is expected to form binary or ternary invasion complexes, respectively, with reduced electrophoretic mobility on non-denaturing polyacrylamide gels relative to unbound DH1 (Fig. 3a).
Conventional Invader probe INVu:INVd and chimeric Invader:LNA probe LNA1:INVd resulted in substantial invasion of DH1 (∼95%; Fig. 3b). Conversely, individual Invader strands only result in trace invasion of DH1,24 while LNA1 resulted in more moderate invasion (∼55%; Fig. 3b). Three of four chimeric Invader:LNA/MOE probes resulted in substantial invasion (>85%; Fig. 3c), while invasion was more moderate with blkMOE2:INVd (∼40%; Fig. 3c), in line with TA (Table 1) and ΔGrec values (Table S2). Unexpectedly, three of four single-stranded LNA/MOE mixmers resulted in efficient invasion of DH1 (>85%; Fig. 3c), while invasion with altMOE4 was more moderate (∼50%; Fig. 3c), likely due to its formation of a stable homoduplex (Tm > 80 °C; Table 1). The chimeric probes entailing the fully MOE, O2′-Me and 2′-F strands resulted in moderate DH1 invasion (30–65%; Fig. 3b) in agreement with their less favorable TA (Table 1) and ΔGrec values (Table S2), whereas the individual probe strands resulted in little to no recognition.
Next, the dsDNA-binding specificities of the chimeric Invader:LNA/MOE probes and the single-stranded LNA/MOE mixmers were evaluated. Thus, a 25-fold molar excess of each probe was incubated with DNA hairpins DH2–DH7, which only differ at position 6 or 9 (a or b, respectively, Fig. 4) of the double-stranded region relative to the probes. Akin to our prior observations with conventional Invader, chimeric Invader:LNA, and chimeric Invader:γPNA probes,22–24 every chimeric Invader:LNA/MOE probes displayed outstanding binding specificity as evidenced by the (near-)complete absence of DH2–DH7 invasion (Fig. 4), at conditions resulting in (near-)complete invasion of DH1 (Fig. 4). In contrast – and similar to our prior observations with single-stranded LNA,24 γPNA, and SerγPNA probes22,23 – three of the four LNA/MOE mixmers result in substantial non-specific recognition of DH2–DH7 (Fig. 4). Interestingly, LNA/MOE mixmer blkMOE1 displayed near-perfect discrimination of DH2–DH7 (Fig. 4). Presumably, the excellent binding specificity of the chimeric Invader:LNA/MOE probes is due to stringency clamping effects that are often observed with structured metastable probes.31 Binding to incorrect dsDNA targets would require (i) denaturation of both the dsProbe and dsDNA target region, and (ii) formation of two less stable, mismatched duplexes in the invasion complexes.32 A similar effect might explain the excellent binding specificity of blkMOE1. Thus, the intramolecular hairpin formed by blkMOE1 is likely more stable than the complexes formed with mismatched DNA hairpins, but less stable than the complex formed with the fully matched DH1, whereas the secondary structures formed by the other single-stranded LNA/MOE mixmers are less stable than the complexes formed with the fully matched and mismatched DNA hairpins, resulting in poor binding specificity.
 | ||
| Fig. 4 Representative electrophoretograms from specificity experiments. DH1–DH7 were incubated with a 25-fold molar probe excess. For sequences and Tms of DH1–DH7, see Table S5. Experimental conditions as in Fig. 3. The electrophoretograms for altMOE3 and altMOE4 are from one gel, with an irrelevant lane excised. | ||
Informed by the results from the initial screen and specificity experiments, the four chimeric Invader:LNA/MOE probes and the single-stranded LNA/MOE mixmer blkMOE1 were selected for dose–response experiments to determine C50 values, i.e., the probe concentration resulting in 50% recognition of DH1 (Fig. 5). The four chimeric Invader:LNA/MOE probes displayed C50 values below 250 nM, with blkMOE1:INVd displaying a comparable C50 value to the corresponding conventional Invader and chimeric Invader:LNA probes (C50 = 55–85 nM). Remarkably, blkMOE1 resulted in near-stoichiometric recognition of DH1 (C50 = 35 nM; Fig. 5).
 | ||
| Fig. 5 (a and b) Dose–response curves for invasion of DH1 by blkMOE1 and chimeric Invader:LNA/MOE probes. For representative electrophoretograms, see Fig. S6; experimental conditions as in Fig. 3. (c) C50 values and percent DH1 invasion when using 5-fold probe excess (Inv5X). For Inv5X data for blkMOE2, altMOE3, altMOE4, and the probes entailing fully modified XRNAs, see Table S4; “±” = standard deviation from at least three trials. Data for LNA1:INVd and INVu:INVd are from ref. 24. | ||
In conclusion, these results suggest that chimeric Invader:LNA/MOE probes have considerable promise for efficient and highly specific recognition of dsDNA targets. Perhaps even more exciting – given their structural simplicity and commercial availability – is the prospect of using single-stranded LNA/MOE mixmers for mixed-sequence dsDNA-invasion. Further studies are needed to determine the robustness of this strategy, but the high binding efficiency and specificity observed with blkMOE1 is promising. Results from such studies will be reported in due course.
Conflicts of interest
PJH is an inventor on patents pertaining to Invader probes, which have been issued to the University Idaho.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ob01570g.Acknowledgements
This publication was made possible by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant #P20GM103408.References
- Y. Kawamoto, T. Bando and H. Sugiyama, Bioorg. Med. Chem., 2018, 26, 1393–1411 CrossRef CAS
.
- Y. Mikame and A. Yamayoshi, Pharmaceutics, 2023, 15, 2515 CrossRef CAS
.
- N. Brodyagin, M. Katkevics, V. Kotikam, C. A. Ryan and E. Rozners, Beilstein J. Org. Chem., 2021, 17, 1641–1688 CrossRef CAS PubMed
.
- A. Dragulescu-Andrasi, S. Rapireddy, B. M. Frezza, C. Gayathri, R. R. Gil and D. H. Ly, J. Am. Chem. Soc., 2006, 128, 10258–10267 CrossRef CAS PubMed
.
- R. Bahal, B. Sahu, S. Rapireddy, C.-M. Lee and D. H. Ly, ChemBioChem, 2012, 13, 56–60 CrossRef CAS PubMed
.
- P. R. Bohländer, T. Vilaivan and H.-A. Wagenknecht, Org. Biomol. Chem., 2015, 13, 9223–9230 RSC
.
- S. A. Thadke, V. M. Hridya, J. D. R. Perera, R. R. Gil, A. Mukherjee and D. H. Ly, Commun. Chem., 2018, 1, 79 CrossRef PubMed
.
- H. Zheng, I. Botos, V. Clausse, H. Nikolayevskiy, E. E. Rastede, M. F. Fouz, S. J. Mazur and D. H. Appella, Nucleic Acids Res., 2021, 49, 713–725 CrossRef CAS PubMed
.
- K. M. L. Hertoghs, J. H. Ellis and I. R. Catchpole, Nucleic Acids Res., 2003, 31, 5817–5830 CrossRef CAS PubMed
.
- E. M. Zaghloul, A. S. Madsen, P. M. D. Moreno, I. I. Oprea, S. El-Andaloussi, B. Bestas, P. Gupta, E. B. Pedersen, K. E. Lundin, J. Wengel and C. I. E. Smith, Nucleic Acids Res., 2011, 39, 1142–1154 CrossRef CAS PubMed
.
- Y. Aiba, M. Shibata and O. Shoji, Appl. Sci., 2022, 12, 3677 CrossRef CAS
.
- J. Lohse, O. Dahl and P. E. Nielsen, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11804–11808 CrossRef CAS PubMed
.
- Y. Aiba, Y. Honda and M. Komiyama, Chem. – Eur. J., 2015, 21, 4021–4026 CrossRef CAS PubMed
.
- M. López-Tena, L. Farrera-Soler, S. Barluenga and N. Winssinger, JACS Au, 2023, 3, 449–458 CrossRef
.
- V. V. Filichev, B. Vester, L. H. Hansen and E. B. Pedersen, Nucleic Acids Res., 2005, 33, 7129–7137 CrossRef CAS
.
- D. C. Guenther, G. H. Anderson, S. Karmakar, B. A. Anderson, B. A. Didion, W. Guo, J. P. Verstegen and P. J. Hrdlicka, Chem. Sci., 2015, 6, 5006–5015 RSC
.
- C. P. Shepard, R. G. Emehiser, S. Karmakar and P. J. Hrdlicka, Molecules, 2023, 28, 127 CrossRef CAS
.
- H. Ihmels and D. Otto, Top. Curr. Chem., 2005, 258, 161204 CrossRef
.
- O. Persil and N. V. Hud, Trends Biotechnol., 2007, 25, 433–436 CrossRef CAS PubMed
.
- B. Denn, S. Karmakar, D. C. Guenther and P. J. Hrdlicka, Chem. Commun., 2013, 49, 9851–9853 RSC
.
- R. Emehiser, E. Hall, D. C. Guenther, S. Karmakar and P. J. Hrdlicka, Org. Biomol. Chem., 2020, 18, 56–65 RSC
.
- R. G. Emehiser and P. J. Hrdlicka, Org. Biomol. Chem., 2020, 18, 1359–1368 RSC
.
- R. G. Emehiser, K. Dhuri, C. Shepard, S. Karmakar, R. Bahal and P. J. Hrdlicka, Org. Biomol. Chem., 2022, 20, 8714–8724 RSC
.
- M. E. Everly, R. G. Emehiser and P. J. Hrdlicka, Org. Biomol. Chem., 2025, 23, 619–628 RSC
.
- T. Bryld, T. Højland and J. Wengel, Chem. Commun., 2004, 1064–1065 RSC
.
- V. V. Filichev, U. B. Christensen, E. B. Pedersen, B. R. Babu and J. Wengel, ChemBioChem, 2004, 5, 1673–1679 CrossRef CAS PubMed
.
- H. Asanuma, R. Niwa, M. Akahane, K. Murayama, H. Kashida and Y. Kamiya, Bioorg. Med. Chem., 2016, 24, 4129–4137 CrossRef CAS PubMed
.
- P. Wittung, S. K. Kim, O. Buchardt, P. Nielsen and B. Norden, Nucleic Acids Res., 1994, 22, 5371–5377 CrossRef CAS PubMed
.
- V. Marin, H. F. Hansen, T. Koch and B. A. Armitage, J. Biomol. Struct. Dyn., 2004, 21, 841–850 CrossRef CAS
.
- M. E. Everly, P. J. Wieber, I. Al Janabi and P. J. Hrdlicka, An electrophoretic mobility shift assay with chemiluminescent readout to evaluate DNA-targeting oligonucleotide-based probes, protocols.io, 2025. DOI:10.17504/protocols.io.4r3l218rxg1y/v1.
- V. V. Demidov and M. D. Frank-Kamenetskii, Trends Biochem. Sci., 2004, 29, 62–71 CrossRef CAS PubMed
.
- S. X. Chen, D. Y. Zhang and G. Seelig, Nat. Chem., 2013, 5, 782–789 CrossRef CAS PubMed
.
Footnotes |
| † For interstrand zipper nomenclature definition, see SI. |
| ‡ For a more detailed explanation of the NNEP, see SI. |
| This journal is © The Royal Society of Chemistry 2026 |