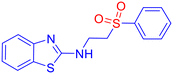
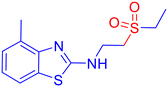
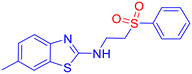
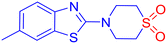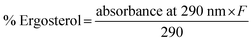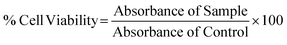Open Access Article
Open Access ArticleMicrowave-assisted single-step synthesis of cyclic and acyclic β-aminosulfones and evaluation of their antifungal activity targeting CYP51
Abigail Bibiana
Pinheiro
a,
Soumik
Saha
a,
Madhuri
Madduri
b,
Utpal
Roy
b,
Lakshmi Sudhir
Menon
b,
Sumit
Biswas
b,
Amrita
Chatterjee
 *a and
Mainak
Banerjee
*a and
Mainak
Banerjee
 *a
*a
aDepartment of Chemistry, Birla Institute of Technology and Science, Pilani, KK Birla Goa Campus, Zuarinagar, Sancoale, Goa 403726, India. E-mail: amrita@goa.bits-pilani.ac.in; mainak@goa.bits-pilani.ac.in
bDepartment of Biological Sciences, Birla Institute of Technology and Science, Pilani, KK Birla Goa Campus, Zuarinagar, Sancoale, Goa 403726, India
First published on 9th January 2026
Abstract
The alarming rise of drug resistance has created an urgent need for novel antifungal agents. On the other hand, growing environmental concerns necessitate the development of sustainable alternatives to conventional synthetic methods for active pharmaceutical ingredients (APIs). β-Aminosulfones represent a potent yet underexplored functionality in biologically active compounds. In this work, a series of β-aminosulfone derivatives, designed as potential antifungal agents, were synthesized via a one-step, reagent-free, catalyst-free double aza-Michael addition of 2-aminobenzothiazoles, biogenic amines, or aromatic amines with different vinyl sulfones. The reactions were carried out in water under microwave irradiation (150 °C, 10 min), affording β-aminosulfones in excellent yields by simple filtration, without the need for further work-up or purification. This cost-effective process makes the production cost comparable to raw materials (e.g., compound 3d at $3.43 per g). Molecular docking, performed using the Glide module of the Schrödinger suite, revealed potential inhibitory activity against the fungal target CYP51 (PDB ID: 5V5Z), with reasonably good docking scores ranging from −5.69 to −8.25 kcal mol−1 for benzothiazole derivatives and −5.73 to −7.05 kcal mol−1 for biogenic amines. In silico ADMET profiling of selected compounds indicated promising drug-like attributes, satisfying Lipinski's rule. β-Aminosulfones were screened for their in vitro antifungal activity against various Candida species, and they exhibited MIC values ranging from 16 to 64 μg mL−1. Notably, one compound (3d, 0.051 μM) showed comparable potency to fluconazole (0.052 μM) against Candida glabrata. Through ergosterol depletion assays, the mechanism of antifungal activity could be linked to the CYP51 inhibition pathway. Although these compounds exhibited moderate docking scores against 1KZN for antibacterial activity (−3.96 to −5.63 kcal mol−1), the spot test revealed insignificant inhibition. Cytotoxicity studies with selected molecules revealed that these aza-sulfones are non-toxic to human cells, encouraging further studies with β-aminosulfone scaffolds.
Introduction
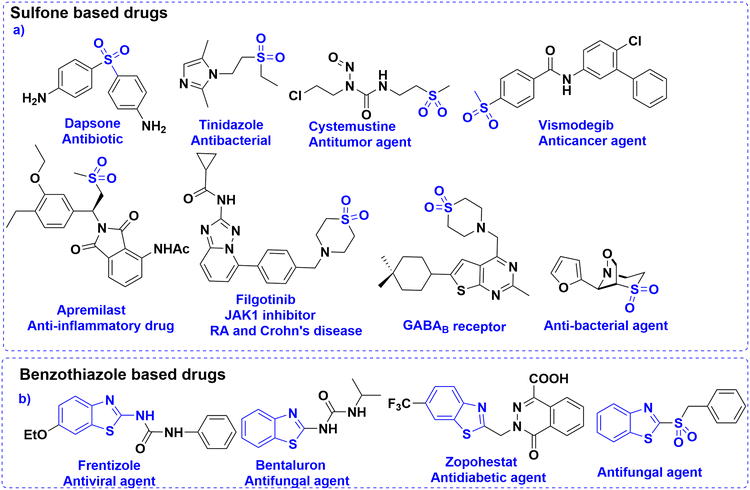
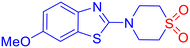
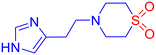
Fungal infections pose a significant public health concern, particularly in individuals with compromised immune systems.1 The most common pathogens include Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus. Among these, C. albicans is a major cause of invasive fungal infections (IFIs). It ranks as the fourth leading cause of hospital-acquired bloodstream infections, with a mortality rate of nearly 40%.2 The increasing prevalence of opportunistic fungal infections, combined with the rise of resistance to current antifungal agents such as azoles, echinocandins, and polyenes, highlights the urgent need for novel antifungal compounds with new mechanisms of action and improved resistance profiles.3–5 Lanosterol 14α-demethylase (CYP51), a cytochrome P450 enzyme essential in ergosterol biosynthesis, remains a critical target in antifungal drug discovery.6 Ergosterol is a vital component of fungal cell membranes, and inhibition of this disrupts membrane integrity, leading to cell death.7 However, resistance to existing azole inhibitors has emerged due to active site mutations and efflux pump upregulation.8,9 This underscores the urgent need for novel scaffolds targeting the enzyme with high selectivity and potency.Sulfones are recognized as highly versatile scaffolds in the development of biologically active compounds and pharmacophores.10–13 Sulfone functionality is present in several marketed drugs like tinidazole (to treat infections caused by protozoa),14 dapsone (to help control dermatitis herpetiformis),15etc. (Fig. 1a). From a chemical perspective, the sulfonyl group is a well-established activating functionality that facilitates the formation of carbon–carbon and carbon–heteroatom bonds in various organic transformations.16 To the context of study, β-aminosulfones are a chemically versatile class of molecules, and they are omnipresent in various bioactive compounds and drug molecules as well.11 In particular, cyclic β-amino sulfones, viz., thiomorpholine-1,1-dioxides, show promising pharmaceutical activities toward Chagas disease,17 filgotinib inhibitor,18 as γ-aminobutyric acid type B receptor,19etc. (Fig. 1a). Although sulfone possesses immense potential as a “drug-like” scaffold, due to its polarity and redox behaviour, it is a less explored pharmacophore.
While azole-based antifungals, such as fluconazole, remain a cornerstone in the treatment of fungal infections,20 their prolonged use has led to the emergence of drug-resistant fungal strains. On the other hand, benzothiazoles (BTAs) are well-established heterocycles in drug discovery.21 Their planar, electron-rich structure allows favourable interactions with diverse biomolecular targets, and their adaptability supports functional modifications for enhanced pharmacokinetic and pharmacodynamic profiles.22 Numerous BTA-based compounds are used clinically to treat various conditions, including cancers, bacterial and fungal infections, inflammatory diseases, etc. (Fig. 1b).23–25 Even sulfone-containing benzothiazole showed promising bioactivities.26
Biogenic amines, produced as metabolites of amino acids in the human body via enzymatic processes, play crucial roles as regulators in various physiological systems, particularly within the central nervous and cardiovascular systems.27–29 Given their biological relevance, these amines are occasionally explored in drug development.30–33 Incorporating biogenic amine moieties into drug scaffolds may enhance biological compatibility and target engagement.
Therefore, a simple combination of sulfone functionality with biogenic amines appears to be a rational strategy for developing effective antifungal agents.
Although “computation” has significantly reduced the synthetic activities from random synthesis and screening of “hits” to only the potential “leads”, their synthesis is often a rate-limiting factor in drug-discovery projects.34,35 While it is often assumed that the impact of medicinal chemistry is trivial compared to the impact of development and manufacturing activities, it has been estimated that drug discovery is responsible for generating 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 2 million kg of waste, with a further 150
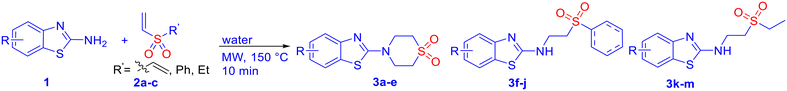
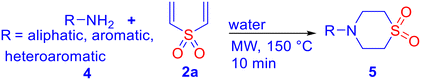
000 to 2 million kg of waste, with a further 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 1.5 million kg during the preclinical process.36 In the long run, various regulatory measures are expected to be implemented on industrial production. The replacement of conventional solution-based chemistry with green and sustainable methods is envisaged to adhere to good laboratory practice (GLP) as per the needs of the time, while maintaining cost-efficacy. The prospective APIs derived from β-aminosulfones are cost-effective and benign, as they can be synthesized in a single step by aza-Michael addition of amines to vinyl sulfones, commonly in a solution phase in the presence of catalysts, such as toxic Lewis acids (e.g., AlCl3).37,38 A limited number of solvent-free methods for β-aminosulfones are also available.39 Notably, microwave-assisted organic synthesis (MAOS) is a well-validated sustainable technology that provides significant advantages over conventional heating, including shorter reaction times, higher yields, and improved selectivities.40 Recently, our group developed a microwave-assisted catalyst- and organic-solvent-free green protocol for the synthesis of cyclic and acyclic β-aminosulfones via aza-Michael addition,41 which opens a new avenue for API syntheses with β-aminosulfones as the key structural motif. In view of our focused efforts on developing biologically active agents,39,42 we report herein the design, green and cost-effective synthesis, and in vitro antifungal assay of β-aminosulfones as potent antifungal agents. The mode of action of these compounds with the potential receptor cytochrome P450 14α-sterol demethylase was investigated through molecular docking studies within the active site of fungal CYP51 and ergosterol assay.
000 to 1.5 million kg during the preclinical process.36 In the long run, various regulatory measures are expected to be implemented on industrial production. The replacement of conventional solution-based chemistry with green and sustainable methods is envisaged to adhere to good laboratory practice (GLP) as per the needs of the time, while maintaining cost-efficacy. The prospective APIs derived from β-aminosulfones are cost-effective and benign, as they can be synthesized in a single step by aza-Michael addition of amines to vinyl sulfones, commonly in a solution phase in the presence of catalysts, such as toxic Lewis acids (e.g., AlCl3).37,38 A limited number of solvent-free methods for β-aminosulfones are also available.39 Notably, microwave-assisted organic synthesis (MAOS) is a well-validated sustainable technology that provides significant advantages over conventional heating, including shorter reaction times, higher yields, and improved selectivities.40 Recently, our group developed a microwave-assisted catalyst- and organic-solvent-free green protocol for the synthesis of cyclic and acyclic β-aminosulfones via aza-Michael addition,41 which opens a new avenue for API syntheses with β-aminosulfones as the key structural motif. In view of our focused efforts on developing biologically active agents,39,42 we report herein the design, green and cost-effective synthesis, and in vitro antifungal assay of β-aminosulfones as potent antifungal agents. The mode of action of these compounds with the potential receptor cytochrome P450 14α-sterol demethylase was investigated through molecular docking studies within the active site of fungal CYP51 and ergosterol assay.
Results and discussion
Chemistry
Notably, the current protocol for the syntheses of the potential antifungal agents (3 or 5) is significantly cost-effective and “greener”. A quick cost estimation of the prospective antifungal agents, based on the cost of raw materials only, revealed that these scaffolds can be synthesized at approximately the same cost as the starting materials for this high-yielding, work-up and purification-free process. For example, 2-aminonitrobenzothiazole and tyramine-derived products, 3d and 5e, cost approximately $3.43 per g and $6.09 per g, respectively, with respect to only the raw materials' cost from the Merck catalogue. Notably, the synthesis of fluconazole, a widely used antifungal drug, involves at least a three-step synthetic pathway,43 and incurs a high production cost [approximately $130.5 per g], making it significantly more expensive compared to our single-step synthetic approach.
In silico antifungal studies by molecular docking
Assuming the antimicrobial potential of β-aminosulfone derivatives, we aimed to assess the binding affinity and interaction profiles of a variety of β-aminosulfones, including those previously reported by our group.41The sulfone derivatives were docked into the active site pockets (MET508, PHE 228, SER507, SER506, CYS470, PHE233, MET306, THR311, LEU376, PRO230, ILE231, GLY307) of the crystal structure of ergosterol (lanosterol 1,4α-demethylase) (CYP51) from C. albicans (PDB ID: 5V5Z) to better understand the binding interactions between these ligands and the target protein. The CYP51 family is an intriguing subject for fundamental P450 structure–function studies and is also a key clinical drug target.44 Once the protein was prepared, a grid box (−45, −15, 29) for PDB ID: 5V5Z and (25, 25, 40) for PDB ID: 1KZN was generated around the protein using the Receptor Grid Generation tool of Glide in Maestro. The docking simulations revealed that several compounds demonstrated strong binding affinities, with docking scores ranging from −5.69 to −8.25 kcal mol−1, indicating favourable interactions within the active site of CYP51 [PDB ID: 5V5Z] as shown in Table 3. Benzothiazole derivatives have demonstrated significant potential in molecular docking studies due to their heterocyclic scaffold, which enables strong binding interactions such as hydrogen bonding and π–π stacking, with key amino acid residues in biological targets, contributing to their promising bioactivity profiles against CYP51.
| Compounds | Binding affinities | Compounds | Binding affinities |
|---|---|---|---|
| PDB ID: 5V5Z | PDB ID: 5V5Z | ||
| (kcal mol−1) | (kcal mol−1) | ||
| 3a | −6.58 | 5a | −6.24 |
| 3b | −8.25 | 5b | −5.74 |
| 3c | −6.85 | 5c | −5.73 |
| 3d | −6.61 | 5d | −5.85 |
| 3e | −6.54 | 5e | −6.15 |
| 3f | −6.53 | 5f | −6.77 |
| 3g | −7.30 | 5g | −7.05 |
| 3h | −6.79 | ||
| 3i | −6.9 | ||
| 3j | −6.61 | ||
| 3k | −5.98 | ||
| 3l | −5.69 | ||
| 3m | −7.24 |
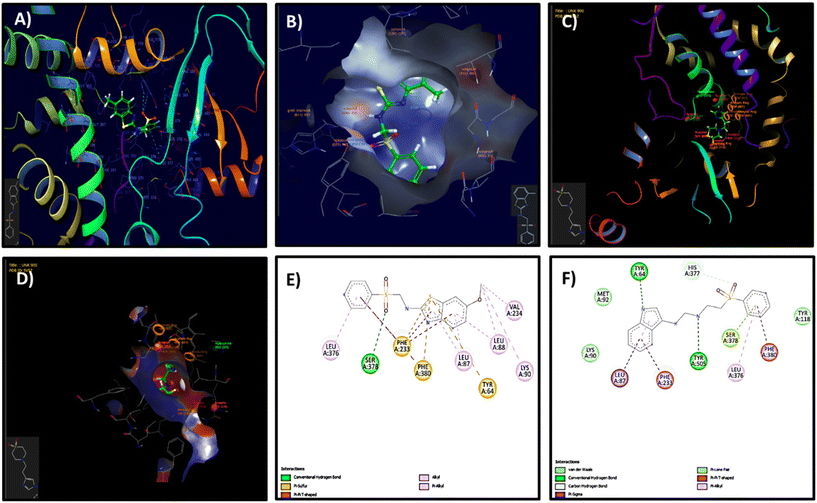
The molecular docking study revealed that the benzothiazole sulfone derivatives (3a–3m) formed a diverse array of stabilizing interactions within the active site of the target protein (Table S1 and S2 of SI). Notably, the nitro derivative (3d) exhibited multiple conventional hydrogen bonds with residues such as ASP-73, VAL-71, VAL-167, and ARG-76, indicating strong polar interactions. Additionally, an attractive charge interaction was observed with GLU-50, which enhanced binding affinity. The compound also engaged in various π interactions, including amide-π stacking with ASN-46 and ALA-47, π–sigma interactions with VAL-43 and THR-165, and π–alkyl interaction with ILE-78. The wide range of interactions, complemented by van der Waals contacts, suggests a well-anchored and stable binding conformation (Fig. 2A and B). In contrast, the 6-methyl derivative (3b), also maintains hydrogen bonding with HIS377 and SER378, but lacks the stronger polar contacts observed in the first structure. Instead, it relies more heavily on π–π stacking and van der Waals interactions with residues like PHE380, PHE233, and TYR64, suggesting that its binding is primarily stabilized by hydrophobic and aromatic interactions. Overall, while both ligands occupy a similar binding pocket and share key interactions, the compound, 3d exhibits a more balanced polar–hydrophobic interaction profile, which could translate to enhanced binding affinity and specificity over the second, more π-stacking-dominated structure.
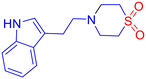
Biogenic amines generally dock in a similar way to benzothiazoles because they form stronger hydrogen bonds, offer ionic interactions, are more flexible, fit better in diverse binding sites, and often mimic natural bioactive compounds (Tables S1 and S2 of SI). For example, the docking interaction profiles of the tyramine-based and tryptamine-based sulfone compounds (5b and 5c) show both structural and interactional differences that may influence their binding affinities and biological activities. The tyramine divinyl sulfone (5b), featuring a phenolic group, engages in strong hydrogen bonding with active site residues such as ARG469 and HIS468, and its –OH group may further stabilize binding through additional polar contacts. The benzene ring of tyramine allows for moderate π–π interactions with nearby aromatic residues like TYR118 and PHE105 (Fig. 2C and D). Whereas the tryptamine-divinyl sulfone (5c), containing an indole ring, presents a more extended conjugated π-system, enhancing aromatic stacking interactions within the binding pocket. Although it lacks the –OH group present in tyramine, the indole nitrogen can participate in weaker hydrogen bonding or dipolar interactions, and its planarity allows for snug fitting and deeper penetration into the hydrophobic pocket. Overall, while the tyramine scaffold offers strong hydrogen bonding through its hydroxyl group, the tryptamine scaffold provides superior aromatic interactions and structural rigidity, potentially contributing to improved binding stability and bioactivity. The primary molecular interactions observed for the compounds highlight distinct binding profiles, which include π–π T-shaped interactions with aromatic residues such as PHE-233, PHE-380, and TYR-64. π–Alkyl interactions with LEU-87, LEU-88, and VAL-234, contributing to hydrophobic stabilization. A single conventional hydrogen bond with SER-378. Overall, interactions were primarily hydrophobic/aromatic (Fig. 2E). This implies that the compound depends heavily on aromatic stacking and hydrophobic contacts, with limited polar interactions. Such an interaction profile may result in weaker binding strength and reduced specificity within the active site. However, the tyramine–phenyl vinyl sulfone derivative exhibited multiple hydrogen bonds with residues TYR-64, SER-378, and TYR-505, strengthening polar interactions within the binding pocket. Stabilization was further supported by carbon–hydrogen bonding and π–π T-shaped interactions with PHE-233 and PHE-380. In addition, π–lone pair interactions and extensive van der Waals contacts were observed, including interactions with HIS-377, a residue often implicated in catalytic or anchoring functions. Collectively, these interactions with both aromatic and polar residues highlight a well-balanced and stable binding profile (Fig. 2F). This suggests that the compound is formed from a combination of polar and hydrophobic interactions, contributing to a more stable and specific binding within the active site. This suggests a potentially stronger binding affinity compared to biogenic amines and possibly better biological activity.
As mentioned previously, heterocycle-based sulfones (5h–5l) and functionalized carbocycles (5m–5y) were docked against the CYP51 target, demonstrating favourable docking scores (see Table S3, SI). Once again, the docking scores are similar to those of benzothiazole and biogenic amine-derived sulfones, encouraging in vitro antifungal activity studies. In contrast, docking of the compounds (3 and 5) against DNA gyrase (PDB ID: 1KZN) obtained comparatively lower scores (see Tables S3 and S4, SI).
Evaluation of in vitro antifungal activity
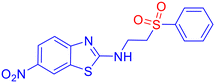
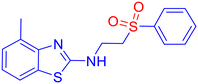
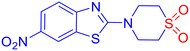
To evaluate the antifungal activity of the newly synthesized compounds, 3a–m and 5a–y, a preliminary spot test assay was performed against a panel of clinically relevant fungal pathogens, including Candida albicans (SC 5314), Candida glabrata (Cg ATCC 2001/-), and Candida tropicalis (Ct ATCC 750/-) (Fig. S1, SI). Considering the significant inhibitory activity shown by the compounds, the minimum inhibitory concentration (MIC) was determined using the broth microdilution method, following the guidelines established by the Clinical and Laboratory Standards Institute (CLSI) (M27-A3 and M38-A2).45,46 Fluconazole (Flc) and amphotericin B (AmB) were included as reference standards. The concentration of the compounds was set at 2000 μg mL−1 using the well diffusion method. The compounds were found to be active against fungal species. Several compounds exhibited satisfactory antifungal activity, with MICs in the range of 16–64 μg mL−1 (Table 4). Notably, derivatives containing electron-withdrawing groups (e.g., nitro) and electron-donating groups (e.g., methyl) on the aromatic ring displayed superior potency, suggesting that electronic modulation plays a significant role in antifungal efficacy. Compound 3d [6-nitro-N-(2-(phenylsulfonyl)ethyl)benzo[d]thiazol-2-amine sulfone derivative] showed the most potent activity, with an MIC value of 16 μg mL−1 (0.051 μM), although its docking value is −6.61 kcal mol−1 in comparison to −8.25 kcal mol−1 with an MIC of 64 μg mL−1 (0.226 μM) for the compound bearing methyl (3b) against Candida species. However, it is difficult to draw any specific inference unless the inhibition mechanism is thoroughly understood. Notably, when compared to fluconazole, a well-established antifungal medication with an MIC value of 0.052 μM against Candida glabrata, our newly synthesized β-aminosulfone nitro derivative (3d) showed an MIC value of 0.051 μM, indicating similar potency. Considering the favourable docking scores against DNA gyrase (Table S4, SI), we proceeded with a preliminary antimicrobial evaluation using the spot test method on bacterial strains S. aureus and E. coli. However, the compounds exhibited insignificant inhibition. Therefore, further studies were not carried out to determine MICs against bacteria.| Compound | C. alb. | C. gla. | C. tro. | Compound | C. alb. | C. gla. | C. tro. |
|---|---|---|---|---|---|---|---|
| 3a | 0.238 | 0.238 | 0.238 | 5a | 0.139 | 0.139 | 0.139 |
| 3b | 0.226 | 0.113 | 0.226 | 5b | 0.125 | 0.250 | 0.125 |
| 3c | 0.107 | 0.107 | 0.107 | 5c | 0.114 | 0.229 | 0.114 |
| 3d | 0.051 | 0.051 | 0.051 | 5d | 0.094 | 0.189 | 0.094 |
| 3e | 0.226 | 0.226 | 0.226 | 5e | 0.083 | 0.083 | 0.083 |
| 3f | 0.20 | 0.20 | 0.20 | 5f | 0.072 | 0.072 | 0.072 |
| 3g | 0.192 | 0.096 | 0.192 | 5g | 0.088 | 0.088 | 0.088 |
| 3h | 0.091 | 0.091 | 0.091 | ||||
| 3i | 0.088 | 0.088 | 0.088 | ||||
| 3j | 0.192 | 0.096 | 0.192 | ||||
| 3k | 0.236 | 0.236 | 0.236 | ||||
| 3l | 0.225 | 0.112 | 0.225 | ||||
| 3m | 0.225 | 0.225 | 0.225 |
ADMET prediction of sulfone derivatives
Next, ADMET-based in silico evaluation of pharmacokinetic properties was conducted for selected compounds that showed good MIC values (viz.3b, 3d, 3h, 5e, and 5h) using the ADMETlab 2.0 online platform to assess drug likeness (Table 5). Computational ADME predictions offer reasonably good insights into pharmacokinetics and help assess oral bioavailability as the most suitable route of administration.47 A radar map was used to assess each compound's bioavailability and physicochemical properties, which provide further information about the drug likeness of β-aminosulfone derivatives (Fig. S2, SI). Firstly, all the compounds under investigation showed acceptance of Lipinski's rule of 5, indicating they are more likely to be orally active. Their predicted solubilities are high, indicating aqueous compatibility that favours oral absorption. Compounds 3b, 3h, and 5e showed ADMET properties more or less comparable to fluconazole.48 Compound 5e closely matched in HBD/HBA (1/7) and TPSA (86.79 Å2), though its lower log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (−2.17) suggested reduced lipophilicity. Compound 3h exhibited a similar drug-likeness score (0.74) and log
P (−2.17) suggested reduced lipophilicity. Compound 3h exhibited a similar drug-likeness score (0.74) and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (3.16), indicating balanced hydrophilic–lipophilic characteristics favorable for absorption. Compound 3b also displayed good drug-likeness (0.80) and moderate physicochemical properties. In contrast, 5g, with higher MW (362.09), HBD/HBA (3/10), and TPSA (142.19 Å2), showed reduced membrane permeability and oral absorption. Notably, the nitro-derivative, 3d, that showed comparable MICs to fluconazole has a marginally lower drug likeness score (0.62) and a slightly higher log
P (3.16), indicating balanced hydrophilic–lipophilic characteristics favorable for absorption. Compound 3b also displayed good drug-likeness (0.80) and moderate physicochemical properties. In contrast, 5g, with higher MW (362.09), HBD/HBA (3/10), and TPSA (142.19 Å2), showed reduced membrane permeability and oral absorption. Notably, the nitro-derivative, 3d, that showed comparable MICs to fluconazole has a marginally lower drug likeness score (0.62) and a slightly higher log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S value than the acceptable range. However, the ADME provides only a qualitative idea about a drug candidate, and based on the experimental observations, 3d can still be a promising candidate. It satisfies Pfizer's rule and GSK's rule. Overall, ADMET profiles of selected compounds provide a fair idea about the drug likeness of β-aminosulfone derivatives, as they satisfy Lipinski's rule (Table 5).
S value than the acceptable range. However, the ADME provides only a qualitative idea about a drug candidate, and based on the experimental observations, 3d can still be a promising candidate. It satisfies Pfizer's rule and GSK's rule. Overall, ADMET profiles of selected compounds provide a fair idea about the drug likeness of β-aminosulfone derivatives, as they satisfy Lipinski's rule (Table 5).
| Entry | MW | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
HBD | HBA | TPSA (Å2) | LR | Drug likeliness |
|---|---|---|---|---|---|---|---|---|
LR: Lipinski's rule; log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P: log of the octanol/water partition coefficient; log P: log of the octanol/water partition coefficient; log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S: log of aqueous solubility; HBD: number of hydrogen bond donors; HBA: number of hydrogen bond acceptors; TPSA: topological polar surface area. S: log of aqueous solubility; HBD: number of hydrogen bond donors; HBA: number of hydrogen bond acceptors; TPSA: topological polar surface area. |
||||||||
| 3b | 282.05 | 2.20 | −3.73 | 0 | 4 | 50.27 | Accepted | 0.80 |
| 3d | 312.02 | 2.02 | −4.61 | 0 | 6 | 90.17 | Accepted | 0.62 |
| 3h | 348.06 | 3.16 | −5.32 | 1 | 5 | 68.29 | Accepted | 0.74 |
| 5e | 381.18 | −2.17 | −1.13 | 1 | 7 | 86.79 | Accepted | 0.52 |
| 5g | 361.09 | −1.97 | −1.00 | 3 | 10 | 142.19 | Accepted | 0.51 |
| Fluconazole | 306.10 | 0.40 | −1.72 | 1 | 7 | 81.65 | Accepted | 0.75 |
| Acceptable range | <500 | <5 | −4 to 0.5 | 0–7 | 0–12 | <140 | — | >0.67 |
SEM analysis
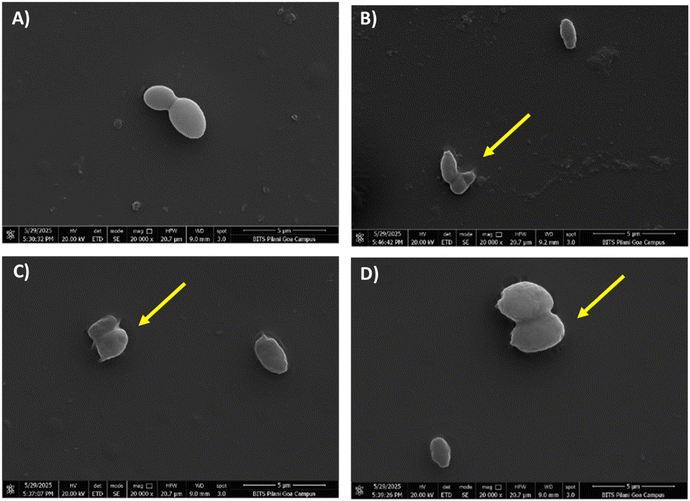
A scanning electron microscopy (SEM) study was conducted on selected candidates to support and confirm the MICs of 3 and 5, obtained from the broth microdilution method. SEM was conducted to visualize the 24 h untreated control samples, serving as a negative control for comparison with treated benzothiazole-based scaffolds (3d, 3h) and biogenic amine-based derivatives (5e, 5g), and fluconazole as a positive control (Fig. 3 and S3, SI). In SEM images, healthy, oval-shaped morphological features were observed for 24 h-old C. glabrata (Fig. 3A). Cells treated with fluconazole showed disrupted membranes (Fig. S3E and F, SI). Similar morphological disruptions were also observed in the fungal spores that were treated with synthesized compounds 3d, (16 μg mL−1 or 0.051 μM), 3h (32 μg mL−1 or 0.091 μM), 5e (32 μg mL−1 or 0.083 μM), and 5g (32 μg mL−1 or 0.083 μM), respectively, as shown in Fig. 3B–D and S3G–J, SI. These findings suggest a mechanism of action comparable to that of fluconazole, involving the complete disruption of the fungal cell membrane through CYP51 inhibition.Ergosterol assay
In the synthetic pathway of ergosterol, inhibition of CYP51 activity can cause the aggregation of the upstream composition, and ergosterol depletion is connected to antifungal activity through the CYP51 inhibition mechanism. An ergosterol depletion assay was conducted to gain insight into the inhibition mechanism of β-aminosulfone derivatives.49,50 Representative molecules 3c, 3d, 5e were selected based on their MIC values, fluconazole was used as the control, and ergosterol depletion at concentrations corresponding to ½ MIC, MIC, and 2× MIC was analyzed. A UV spectrophotometer was used to detect changes in absorption at 290 nm, a signature UV-vis absorption peak of ergosterol, in the corresponding composition of Candida glabrata, Candida tropicalis, and Candida albicans. The ergosterol depletion assay50 allowed a direct comparison of the inhibitory effects of β-aminosulfones 3c, 3d, and 5e across three Candida species (Tables 6, S6 and S7 and Fig. S4 of SI).| Compd | 3c | 3d | 5e | Fluconazole | ||||
|---|---|---|---|---|---|---|---|---|
| μg mL−1 | Mean ± SD | % ergosterol | Mean ± SD | % ergosterol | Mean ± SD | % ergosterol | Mean ± SD | % ergosterol |
| a The absorbance data represent the mean value of three independent replicas. b The absorbance of our standard ergosterol was 0.137 at 290 nm, which corresponds to 100% of ergosterol. | ||||||||
| 8 | — | — | 0.0177 ± 0.00208 | 12.88 | — | — | 0.0029 ± 0.0001 | 2.11 |
| 16 | 0.0027 ± 0.0001 | 1.94 | 0.0033 ± 0.002098 | 2.41 | 0.0053 ± 0.0015 | 3.88 | 0.0027 ± 0.0001 | 1.99 |
| 32 | 0.0013 ± 0.0006 | 0.97 | 0.0017 ± 0.003055 | 1.24 | 0.0040 ± 0.0026 | 2.92 | 0.0019 ± 0.0009 | 1.38 |
| 64 | 0.0008 ± 0.0001 | 0.56 | — | — | 0.0013 ± 0.0006 | 0.97 | — | — |
As shown in Table 6, across all tested species, the compounds exhibited a progressive reduction in ergosterol content with increasing concentration, confirming a dose-dependent inhibition of ergosterol biosynthesis. To elaborate, for Candida tropicalis, compound 3d showed a marked decline in ergosterol content from 12.88% at ½ MIC to 2.41% at MIC to 1.24% at 2× MIC; similarly, 3c and 5e reduced ergosterol from 1.94% and 3.88% to 0.56% and 0.97%, respectively across the same range. A similar trend was observed in Candida glabrata (Table S6, SI). At ½ MIC, ergosterol levels were reduced to 4.52%, 11.3%, and 4.74% for 3c, 3d, and 5e, respectively. At MIC, the ergosterol content decreased further to approximately 2.6–2.9%, comparable to that of fluconazole (1.99%), confirming consistent dose-dependent inhibition. Notably, at 2× MIC, 3c and 5e achieved the lowest ergosterol values (<2.0%). These findings indicate that the tested compounds effectively impair ergosterol synthesis in C. glabrata. A similar trend was observed for Candida albicans as well (Table S7, SI). In this case, ergosterol synthesis inhibition was less significant, but the same trend of dose-dependent depletion was followed (approximately 65% of the control at ½ MIC to 15–20% at MIC and to <10% at 2× MIC). The strong dose-dependent depletion of the absorbance of the ergosterol peak at 290 nm for these compounds indicates highly effective disruption of ergosterol biosynthesis, similar to fluconazole's activity under identical conditions. Overall, two trends emerge: (i) species-specific susceptibility follows C. tropicalis > C. glabrata > C. albicans, and (ii) compound-specific potency highlights 3d as slightly superior across all strains, correlating with MIC data and docking results.
Inhibition mechanism
The molecular docking, ergosterol depletion assay, and SEM analysis offered a preliminary understanding of the antifungal activity of β-aminosulfones through the CYP51 inhibition pathway. Firstly, molecular docking through the Glide module of the Schrödinger suite against the active site of CYP51 revealed favourable hydrogen-bonded and hydrophobic interactions with docking scores in excess of −6.0 kcal mol−1, suggesting plausible inhibition of ergosterol biosynthesis. The SEM images showed clear membrane disruption in treated cells, similar to what is observed for fluconazole, which is known to operate through CYP51 inhibition. Furthermore, ergosterol depletion assays demonstrated a clear dose-dependent reduction in ergosterol content, providing further mechanistic evidence that the β-aminosulfones act through CYP51 inhibition. Together, these studies establish inhibition of ergosterol biosynthesis and consequent membrane damage as key mechanistic insights underlying the antifungal activity of the synthesized β-aminosulfones.Cytotoxicity
To evaluate the compatibility of β-aminosulfone-derived antifungal agents against human cells, selected compounds from both series (3 and 5) were tested for cytotoxicity against HUH7 immortalized hepatocyte-derived carcinoma cell lines using the MTT assay. These epithelial-like cells were cultured for 24 h in complete media (DMEM, 15% FBS, 1% antimycotic, antibiotic solution) after being seeded into a 96-well plate at a density of 2 × 104 cells per mL for a total volume of 100 μL each. After treatment with varying concentrations of the compounds in DMSO, the cell was incubated for 24 and 48 h, after which MTT reagent was added. The optical density of the colored formazan crystals helped to quantify the % of viable cells. Generally, the cell viability was approximately 80% for all tested compounds at 128 μg mL−1 after 24 h, indicating that the new antifungal agents are sufficiently benign to human cells, well above their MICs (Fig. 4). Further, the cell viability was plotted against the logarithm of concentration (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C) and the IC50 values of 3d and 5e were obtained as 1.088 μM (339.6 μg mL−1) and 0.457 μM (174.1 μg mL−1) after 24 h of incubation, respectively from which the selectivity index (SI = IC50/MIC) were calculated as 21.2 for 3d and 5.5 for 5e when calculated with the MICs against C. albicans. Notably, the SI of 3d, which exhibits comparable antifungal potential to fluconazole, is significantly high, indicating the good pharmacological potential of this lead. Overall, the cytotoxicity assay supports the therapeutic potential of the β-aminosulfone scaffold.
C) and the IC50 values of 3d and 5e were obtained as 1.088 μM (339.6 μg mL−1) and 0.457 μM (174.1 μg mL−1) after 24 h of incubation, respectively from which the selectivity index (SI = IC50/MIC) were calculated as 21.2 for 3d and 5.5 for 5e when calculated with the MICs against C. albicans. Notably, the SI of 3d, which exhibits comparable antifungal potential to fluconazole, is significantly high, indicating the good pharmacological potential of this lead. Overall, the cytotoxicity assay supports the therapeutic potential of the β-aminosulfone scaffold.
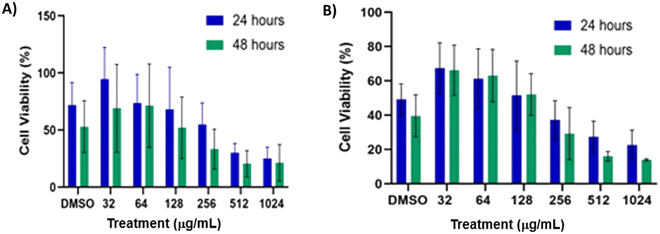 | ||
| Fig. 4 Representative examples for (A) benzothiazoles (3d) and (B) biogenic amine-derived β-amino sulfones (5e), showing reasonable cell viability even up to the concentration of 128 μg mL−1. | ||
Conclusion
In summary, we have successfully synthesized a novel series of β-aminosulfone derivatives incorporating benzothiazole and biogenic amine motifs through a simple, efficient, and environmentally friendly synthetic protocol. This method utilizes commercially available amines and vinyl sulfones under microwave-assisted, reagent- and catalyst-free conditions. The approach offers notable advantages, including rapid reaction times, wide substrate scope, and the absence of tedious work-up procedures, aligning with the principles of green chemistry. The newly synthesized compounds were evaluated for their antifungal activity against clinically relevant fungal pathogens, including C. albicans, C. glabrata, and C. tropicalis. Many derivatives exhibited significant antifungal activity, with MIC values ranging from 16 to 64 μg mL−1. Notably, the β-aminosulfone nitro derivative (3d) showed similar activity to fluconazole against C. glabrata (MIC 0.051 μM). Molecular docking studies against CYP51 revealed favorable binding interactions, primarily through hydrogen bonding and hydrophobic contacts, suggesting that inhibition of ergosterol biosynthesis is a plausible mechanism of action. Ergosterol depletion assays further confirmed concentration-dependent inhibition, with compound 3d showing the most potent depletion, particularly in C. tropicalis. While in silico ADMET profiling highlighted the best candidate, 3d as having optimal solubility, BBB penetration, and low hepatotoxicity, consistent with favorable drug-like properties. Importantly, cytotoxicity assays demonstrated that the active compounds were non-toxic to mammalian cells at relevant concentrations. Together, these findings position the benzothiazole/biogenic amine-based β-aminosulfones as promising lead scaffolds for antifungal drug development.Experimental
Chemicals and equipment
All chemicals were obtained from commercial suppliers and used as received, without any additional purification. LR-grade solvents were sourced from local vendors. Microwave-assisted reactions were performed using a CEM Discover microwave synthesizer. The progress of these reactions was tracked using thin-layer chromatography (TLC) on silica gel plates (0.25 mm, aluminium-backed, 60F-254), with visualization under UV light at wavelengths of 254 or 365 nm. 1H NMR and carbon 13C NMR spectra were recorded on Bruker Avance spectrometers operating at 500 MHz, using tetramethylsilane (TMS) as the internal standard. Chemical shifts are given in parts per million (δ), and standard nomenclature was used for peak multiplicities. Infrared (IR) spectra were collected on a Shimadzu IR Affinity-1 FTIR spectrometer. Liquid chromatography mass spectrometry (LC-MS) data were acquired using an Agilent 6460 Triple Quadrupole LC-MS system with electrospray ionization (ESI) as the ion source. CHNS data were recorded with a Vario MICRO elementar CHNS analyser. Molecular docking simulations were carried out using the Glide module in Schrödinger's Maestro software (version 13.9). Theoretical ADMET studies were conducted using the ADMETlab 2.0 online platform (https://admetmesh.scbdd.com/service/evaluation/cal) for in silico evaluation of pharmacokinetic and toxicity properties. Absorbance measurements for biological studies were done using a Shimadzu UV-1800 UV-vis spectrophotometer. The optical density was measured at 570 nm in an ELIZA Plate Reader (Multiskan GO, Thermo). Field emission scanning electron microscopy (FESEM) images were obtained with a Quanta 250 FEG microscope (FEI).General procedure for the synthesis of β-amino sulfones (3–5)
A 10 mL microwave reaction vessel was charged with an amine (compound 1, 0.5 mmol) and a vinyl sulfone (0.5 mmol) in 3 mL of water. A magnetic stir bar was placed, and the vessel was sealed with a septum. The mixture was then subjected to microwave irradiation at 150 °C and 200 W for 10 minutes, during which a pressure of 250 psi developed. After completion, the reaction mixture was allowed to cool down to room temperature, the resulting sufficiently pure solid product was collected by filtration, dried under vacuum, and characterized.![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2998, 2938, 1548, 1531, 1488, 1445, 1397, 1376, 1329, 1263, 1174, 1116 cm−1; LCMS (ESI): m/z 269.04 [M + H]+; CHNS analysis calcd (%) for C11H12N2O2S2: C, 49.23; H, 4.51; N, 10.64; S, 23.89; found: C, 49.21; H, 4.40; N, 10.82; S, 23.76.
2998, 2938, 1548, 1531, 1488, 1445, 1397, 1376, 1329, 1263, 1174, 1116 cm−1; LCMS (ESI): m/z 269.04 [M + H]+; CHNS analysis calcd (%) for C11H12N2O2S2: C, 49.23; H, 4.51; N, 10.64; S, 23.89; found: C, 49.21; H, 4.40; N, 10.82; S, 23.76.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2986, 2914, 2896, 2702, 1611, 1528, 1427, 1300, 1144, 1055, 1000 cm−1; LCMS (ESI): m/z 283.06 [M + H]+; CHNS analysis calcd (%) for C12H14N2O2S2: C, 51.04; H, 5.00; N, 9.92; S, 22.71; found: C, 51.23; H, 4.87; N, 9.65; S, 22.49.
2986, 2914, 2896, 2702, 1611, 1528, 1427, 1300, 1144, 1055, 1000 cm−1; LCMS (ESI): m/z 283.06 [M + H]+; CHNS analysis calcd (%) for C12H14N2O2S2: C, 51.04; H, 5.00; N, 9.92; S, 22.71; found: C, 51.23; H, 4.87; N, 9.65; S, 22.49.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2985, 2916, 2899, 2704, 1622, 1556, 1433, 1288, 1128, 1078 cm−1; LCMS (ESI): m/z 299.1 [M + H]+; CHNS analysis calcd (%) for C12H14N2O3S2: C, 48.31; H, 4.73; N, 9.39; S, 21.49; found: C, 48.11; H, 4.71; N, 9.37; S, 21.47.
2985, 2916, 2899, 2704, 1622, 1556, 1433, 1288, 1128, 1078 cm−1; LCMS (ESI): m/z 299.1 [M + H]+; CHNS analysis calcd (%) for C12H14N2O3S2: C, 48.31; H, 4.73; N, 9.39; S, 21.49; found: C, 48.11; H, 4.71; N, 9.37; S, 21.47.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2995, 2916, 1664, 1556, 1433, 1288, 1128, 1078 cm−1; LCMS (ESI): m/z 314.03 [M + H]+; CHNS analysis calcd (%) for C11H11N3O4S2: C, 42.16; H, 3.54; N, 13.41; S, 20.46; found: C, 42.21; H, 3.62; N, 13.51; S, 20.41.
2995, 2916, 1664, 1556, 1433, 1288, 1128, 1078 cm−1; LCMS (ESI): m/z 314.03 [M + H]+; CHNS analysis calcd (%) for C11H11N3O4S2: C, 42.16; H, 3.54; N, 13.41; S, 20.46; found: C, 42.21; H, 3.62; N, 13.51; S, 20.41.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 3004, 2965, 2941, 1542, 1521, 1496, 1415, 1392, 1359, 1325, 1286, 1137, 1048 cm−1; LCMS (ESI): m/z 283.06 [M + H]+; CHNS analysis calcd (%) for C12H14N2O2S2: C, 51.04; H, 5.01; N, 9.92; S, 22.71; found: C, 50.96; H, 4.98; N, 9.35; S, 22.08.
3004, 2965, 2941, 1542, 1521, 1496, 1415, 1392, 1359, 1325, 1286, 1137, 1048 cm−1; LCMS (ESI): m/z 283.06 [M + H]+; CHNS analysis calcd (%) for C12H14N2O2S2: C, 51.04; H, 5.01; N, 9.92; S, 22.71; found: C, 50.96; H, 4.98; N, 9.35; S, 22.08.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2991, 2933, 2344, 1527, 1439, 1288, 1139, 1072 cm−1; LCMS (ESI): m/z 319.06 [M + H]+; CHNS calcd (%) for C15H14N2O2S2: C, 56.58; H, 4.43; N, 8.80; S, 20.14; found: C, 56.64; H, 4.63; N, 9.12; S, 20.27.
2991, 2933, 2344, 1527, 1439, 1288, 1139, 1072 cm−1; LCMS (ESI): m/z 319.06 [M + H]+; CHNS calcd (%) for C15H14N2O2S2: C, 56.58; H, 4.43; N, 8.80; S, 20.14; found: C, 56.64; H, 4.63; N, 9.12; S, 20.27.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2964, 2827, 2354, 1462, 1320, 1119, 1030, 865, 750 cm−1; LCMS (ESI): m/z 333.07 [M + H]+; CHNS analysis calcd (%) for C16H16N2O2S2: C, 57.81; H, 4.85; N, 8.43; S, 19.29; found: C, 57.95; H, 4.82; N, 8.56; S, 19.23.
2964, 2827, 2354, 1462, 1320, 1119, 1030, 865, 750 cm−1; LCMS (ESI): m/z 333.07 [M + H]+; CHNS analysis calcd (%) for C16H16N2O2S2: C, 57.81; H, 4.85; N, 8.43; S, 19.29; found: C, 57.95; H, 4.82; N, 8.56; S, 19.23.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2994, 2914, 1594, 1533, 1455, 1294, 1116, 1000, 789 cm−1; LCMS (ESI): m/z 283.06 [M + H]+; CHNS analysis calcd (%) for C16H16N2O3S2: C, 55.15; H, 4.63; N, 8.04; S, 18.40; found: C, 55.35; H, 4.71; N, 7.97; S, 17.98.
2994, 2914, 1594, 1533, 1455, 1294, 1116, 1000, 789 cm−1; LCMS (ESI): m/z 283.06 [M + H]+; CHNS analysis calcd (%) for C16H16N2O3S2: C, 55.15; H, 4.63; N, 8.04; S, 18.40; found: C, 55.35; H, 4.71; N, 7.97; S, 17.98.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 3355, 2916, 2338, 1556, 1433, 1288, 1128, 1078, 822, 733 cm−1; LCMS (ESI): m/z 364.04 [M + H]+; CHNS analysis calcd (%) for C15H16N3O4S2: C, 49.17; H, 4.40; N, 11.47; S, 17.50; found: C, 49.34; H, 4.29; N, 12.02; S, 17.63.
3355, 2916, 2338, 1556, 1433, 1288, 1128, 1078, 822, 733 cm−1; LCMS (ESI): m/z 364.04 [M + H]+; CHNS analysis calcd (%) for C15H16N3O4S2: C, 49.17; H, 4.40; N, 11.47; S, 17.50; found: C, 49.34; H, 4.29; N, 12.02; S, 17.63.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 3360, 2916, 2839, 2350, 1617, 1572, 1527, 1455, 1299, 1144, 1072, 1006, 800, 733, 666 cm−1; LCMS (ESI): m/z 333.07 [M + H]+; CHNS analysis calcd (%) for C16H16N2O2S2: C, 57.81; H, 4.85; N, 8.43; S, 19.29; found: C, 58.06; H, 4.89; N, 8.62; S, 19.03.
3360, 2916, 2839, 2350, 1617, 1572, 1527, 1455, 1299, 1144, 1072, 1006, 800, 733, 666 cm−1; LCMS (ESI): m/z 333.07 [M + H]+; CHNS analysis calcd (%) for C16H16N2O2S2: C, 57.81; H, 4.85; N, 8.43; S, 19.29; found: C, 58.06; H, 4.89; N, 8.62; S, 19.03.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2985, 2850, 2811, 1564, 1527, 1456, 1294, 1116, 1044, 794, 752 cm−1; LCMS (ESI): m/z 271.06 [M + H]+; CHNS analysis calcd (%) for C11H14N2O2S2: C, 48.67; H, 5.32; N, 10.36; S, 23.72; found: C, 48.38; H, 5.58; N, 10.74; S, 23.96.
2985, 2850, 2811, 1564, 1527, 1456, 1294, 1116, 1044, 794, 752 cm−1; LCMS (ESI): m/z 271.06 [M + H]+; CHNS analysis calcd (%) for C11H14N2O2S2: C, 48.67; H, 5.32; N, 10.36; S, 23.72; found: C, 48.38; H, 5.58; N, 10.74; S, 23.96.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2944, 2827, 2333, 1461, 1282, 1122, 1049, 983, 850 cm−1; LCMS (ESI): m/z 285.07 [M + H]+; CHNS analysis calcd (%) for C12H16N2O2S2: C, 50.68; H, 5.67; N, 9.85; S, 22.55; found: C, 50.65; H, 5.62; N, 9.81; S, 22.72.
2944, 2827, 2333, 1461, 1282, 1122, 1049, 983, 850 cm−1; LCMS (ESI): m/z 285.07 [M + H]+; CHNS analysis calcd (%) for C12H16N2O2S2: C, 50.68; H, 5.67; N, 9.85; S, 22.55; found: C, 50.65; H, 5.62; N, 9.81; S, 22.72.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 3355, 2905, 2350, 1561, 1527, 1466, 1283, 1116, 1044, 794, 722 cm−1; LCMS (ESI): m/z 285.39 [M + H]+; CHNS analysis calcd (%) for C12H16N2O2S2: C, 50.68; H, 5.67; N, 9.85, S, 22.55; found: C, 51.09; H, 5.82; N, 9.85; S, 22.86.
3355, 2905, 2350, 1561, 1527, 1466, 1283, 1116, 1044, 794, 722 cm−1; LCMS (ESI): m/z 285.39 [M + H]+; CHNS analysis calcd (%) for C12H16N2O2S2: C, 50.68; H, 5.67; N, 9.85, S, 22.55; found: C, 51.09; H, 5.82; N, 9.85; S, 22.86.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 3004, 2933, 2827, 1500, 1299, 1105, 1355, 1250, 1083 cm−1; LCMS (ESI): m/z 230.09 [M + H]+; CHNS analysis calcd (%) for C9H15N3O2S: C, 47.14; H, 6.59; N, 18.33; S, 13.98; found: C, 47.31; H, 6.70; N, 18.49; S, 13.67.
3004, 2933, 2827, 1500, 1299, 1105, 1355, 1250, 1083 cm−1; LCMS (ESI): m/z 230.09 [M + H]+; CHNS analysis calcd (%) for C9H15N3O2S: C, 47.14; H, 6.59; N, 18.33; S, 13.98; found: C, 47.31; H, 6.70; N, 18.49; S, 13.67.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2992, 2929, 2745, 2222, 1565, 1400, 1254, 1122, 1002, 752 cm−1; LCMS (ESI): m/z 256.1 [M + H]+; CHNS analysis calcd (%) for C12H17NO3S: C, 47.14; H, 6.59; N, 18.33; S, 13.98; found: C, 47.37; H, 6.74; N, 18.49; S, 13.59.
2992, 2929, 2745, 2222, 1565, 1400, 1254, 1122, 1002, 752 cm−1; LCMS (ESI): m/z 256.1 [M + H]+; CHNS analysis calcd (%) for C12H17NO3S: C, 47.14; H, 6.59; N, 18.33; S, 13.98; found: C, 47.37; H, 6.74; N, 18.49; S, 13.59.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 3014, 2995, 2921, 1584, 1428, 1319, 1284, 1028, 722 cm−1; LCMS (ESI): m/z 279.12 [M + H]+; CHNS analysis calcd (%) for C14H18N2O2S: C, 47.14; H, 6.59; N, 18.33; S, 13.98; found: C, 47.11; H, 6.28; N, 18.49; S, 13.69.
3014, 2995, 2921, 1584, 1428, 1319, 1284, 1028, 722 cm−1; LCMS (ESI): m/z 279.12 [M + H]+; CHNS analysis calcd (%) for C14H18N2O2S: C, 47.14; H, 6.59; N, 18.33; S, 13.98; found: C, 47.11; H, 6.28; N, 18.49; S, 13.69.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2985, 2927, 2911, 2874, 2854, 2785, 1581, 1471, 1379, 1339, 1291, 1276, 1192, 1040 cm−1; LCMS (ESI): m/z 279.12 [M + H]+; CHNS analysis calcd (%) for C13H26N2O4S2: C, 46.13; H, 7.74; N, 8.28; S, 18.94; found: C 46.12; H 7.71; N 8.49; S 18.69.
2985, 2927, 2911, 2874, 2854, 2785, 1581, 1471, 1379, 1339, 1291, 1276, 1192, 1040 cm−1; LCMS (ESI): m/z 279.12 [M + H]+; CHNS analysis calcd (%) for C13H26N2O4S2: C, 46.13; H, 7.74; N, 8.28; S, 18.94; found: C 46.12; H 7.71; N 8.49; S 18.69.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2987, 2916, 2910, 2873, 2846, 2789, 1585, 1470, 1382, 1346, 1290, 1279, 1184, 1046 cm−1; LCMS (ESI): m/z 382.20 [M + H]+; CHNS analysis calcd (%) for C15H31N3O4S2: C, 47.22; H, 8.19; N, 11.01; S, 16.81; found: C, 46.92; H, 8.14; N, 10.96; S, 16.69.
2987, 2916, 2910, 2873, 2846, 2789, 1585, 1470, 1382, 1346, 1290, 1279, 1184, 1046 cm−1; LCMS (ESI): m/z 382.20 [M + H]+; CHNS analysis calcd (%) for C15H31N3O4S2: C, 47.22; H, 8.19; N, 11.01; S, 16.81; found: C, 46.92; H, 8.14; N, 10.96; S, 16.69.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2976, 2922, 2916, 2877, 2849, 2792, 1591, 1476, 1381, 1349, 1296, 1266, 1172, 1051 cm−1; LCMS (ESI): m/z 439.2 [M + H]+; CHNS analysis calcd (%) for C18H38N4O4S2: C, 49.29; H, 8.73; N, 12.77; S, 14.62; found: C, 49.30; H, 8.72; N, 12.79; S, 14.67.
2976, 2922, 2916, 2877, 2849, 2792, 1591, 1476, 1381, 1349, 1296, 1266, 1172, 1051 cm−1; LCMS (ESI): m/z 439.2 [M + H]+; CHNS analysis calcd (%) for C18H38N4O4S2: C, 49.29; H, 8.73; N, 12.77; S, 14.62; found: C, 49.30; H, 8.72; N, 12.79; S, 14.67.
![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) 2997, 2916, 2855, 1733, 1711, 1450, 1355, 1250, 1083 cm−1; LCMS (ESI): m/z 362.1 [M + H]+; CHNS analysis calcd (%) for C13H19N3O7S: C, 43.21; H, 5.30; N, 11.63; S, 8.87; found: C, 43.11; H, 5.24; N, 11.76; S, 8.86.
2997, 2916, 2855, 1733, 1711, 1450, 1355, 1250, 1083 cm−1; LCMS (ESI): m/z 362.1 [M + H]+; CHNS analysis calcd (%) for C13H19N3O7S: C, 43.21; H, 5.30; N, 11.63; S, 8.87; found: C, 43.11; H, 5.24; N, 11.76; S, 8.86.
Molecular docking studies
The molecular docking studies were conducted using the Glide module of the Schrödinger suite Maestro 13.9 program. The crystal structure of ergosterol (lanosterol 14-alpha demethylase) (CYP51) from C. albicans (PDB ID: 5V5Z) at 2.00 Å resolution was imported from the protein data bank (https://www.rcsb.org/). Protein preparation was done using the Protein Preparation Wizard, this process is divided into three parts: the first part being the pre-processing, where hydrogen was added to the protein, converting selenomethionine to methionine, filling missing side chains, and filling missing loops using Prime. The second part involves assigning H-bonds using PROPKA at pH 7.0 to optimize the protein, followed by minimization of the protein using the OPLS4 force field in the final step. Once the protein was prepared, a grid box (−45, −15, 29) was generated around the protein using the Receptor Grid Generation tool of Glide in Maestro. Subsequently, the ligands were imported to Schrödinger Maestro in SDF format to be prepared using the LigPrep module. While preparing ligands for docking, hydrogens were added to all ligands, bond orders were assigned, bond lengths and angles were adjusted, stereochemistry was determined, and ring conformations were determined, followed by setting the ionization state at pH 7.0. Additionally, partial charges were applied using the OPLS4 force field, followed by an energy minimization process. For the docking of the molecules, Glide's Standard Precision (SP) mode with default settings was used along with a post-docking minimization step. Both docked ligands were imported and evaluated in Maestro 13.9. A program to explore the hydrogen bonds and hydrophobic interactions of the functional groups of ligands with amino acids present in the binding site of the protein.ADMET prediction of sulfone derivatives
ADMET properties were predicted using ADMETlab 2.0 (https://admetmesh.scbdd.com), a free web-based platform for evaluating small molecule absorption, distribution, metabolism, excretion, and toxicity. Built with cheminformatics tools (RDKit, Scopy) and deep learning (PyTorch, DGL), it applies a Multi-task Graph Attention (MGA) framework trained on large curated datasets to predict over 50 ADMET endpoints. The tool provides comprehensive profiles including physicochemical properties, drug-likeness, substructure alerts (e.g., PAINS), and probability-based classification outputs. Batch predictions were performed via SMILES input with results in CSV format. With accuracy comparable to or better than SwissADME, admetSAR 2.0, and pkCSM, ADMETlab 2.0 offers a robust platform for early-stage in silico drug discovery.In vitro biological activity study: strains and growth conditions
The fungal strains Candida glabrata ATCC 2001, Candida tropicalis ATCC 750, and C. albicans SC5314 were maintained as 20% glycerol stock at −80 °C. Bacterial strains E. coli and S. aureus were also included as test strains. Antimicrobial activity was evaluated using Sabouraud dextrose broth (SD Broth) with fungal strains and nutrient broth used for bacterial strains.Preliminary biological activity
All β-aminosulfone derivatives were dissolved in pure DMSO in measured quantities and tested for their antifungal activity against Gram-positive Candida species. The respective selective media, dextrose agar broth (Hi-Media) for Candida species. Sabouraud dextrose broth (SDB), or nutrient medium, was mixed with 1.5% agar powder and autoclaved at 121 °C/15 lbs. Wells were created on the Sabouraud dextrose agar (SDA) plates for fungal strains or nutrient agar plates for bacterial strains, using a sterile well borer (5 mm). Then the plates were cultured with actively grown cultures using the spread plate technique, and selected dilutions of the synthesized derivatives with respective concentrations were inoculated. After inoculation, the cultured plates were incubated at 37 °C for 24 h, and the antifungal activity was evaluated based on the zone of inhibition (mm) around the well. The tested plates were incubated at 37 °C for 24 h for Candida species. Subsequently, these plates were checked for the zone of inhibition, if any, around the spot. DMSO was also screened for antifungal effect in the same way as a negative control.Inoculum preparation
The inocula of yeast cultures were prepared according to CLSI (M27-A3) guidelines. The organisms were grown on SDA plates at 37 °C for 24 h. A few colonies of test strains were resuspended in 1 mL of 0.85% sterile saline. The optical density (OD) of the cell suspension at 530 nm was adjusted to an absorbance of 0.08–0.1, corresponding to a suspension of 1 × 106 to 5 × 106 cells per mL. The cell suspension was then diluted with RPMI 1640 medium to obtain a working suspension of 5 × 102 to 2.5 × 103 cells per mL. Minimum inhibitory concentrations (MICs) were determined using the broth microdilution protocol, as per the CLSI recommendations (CLSI, 2017), against the C. albicans and non-C. albicans strains used in the present study.Determination of minimum inhibitory concentrations (MICs)
All the synthesized compounds were further screened for their antifungal activity by the broth microdilution method as per CLSI guidelines.45,46 Experimental procedures were carried out in RPMI-1640 medium supplemented with L-glutamine, phenol red, 0.2% glucose, and 0.165 M MOPS buffer (pH 7.0 ± 0.2), and excluded sodium bicarbonate. A 96-well sterile microtiter plate was used, with one column reserved as a negative control (broth only). Two-fold serial dilutions of each drug candidate were prepared in RPMI-1640 (pH 7.0 ± 0.1). 100 μL of diluted cell suspension of 1 × 103 to 5 × 103 cells per mL was added to the 100 μL of 2–512 μg mL−1 dilutions of antifungal compounds. The plates were incubated at 37 °C for 24–48 h. The obtained results were analyzed by visual optical density for the determination of MIC. The standard antifungal drugs, fluconazole and amphotericin B (AMB), were employed as standards. All the experiments were repeated three times.Scanning electron microscopy (SEM)
SEM analysis was used to analyze the effect of sulfones against C. glabrata as previously described.46 In brief, the antifungal test compounds at concentrations of 16 or 32 μg mL−1 were added with fresh RPMI medium to treat the cells. After the 24 h post-treatment, samples were fixed overnight with 2.5% glutaraldehyde at 4 °C, washed twice with 0.1 M sodium cacodylate buffer, and then incubated with osmium tetroxide for 30 min, followed by a wash with 0.1 M sodium cacodylate buffer. The dehydration process was performed with a gradual dilution of ethanol (30, 40, 50, 60, 70, 80, 90, and 100% for 10 min each). Then, the samples were fixed on aluminium stubs, sputter-coated with gold, and observed using a scanning electron microscope (FEI, Quanta 250 FEG 30 kV), at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 magnification.
000 magnification.
Method for ergosterol assay
In this assay, Candida species, Candida glabrata (ATCC Cg 2001/-), Candida tropicalis (ATCC Cg 750/-), and Candida albicans (SC 5314), were used as test strains, with fluconazole serving as the positive control and ergosterol as a reference. Test drug concentrations were 8, 16, and 32 μg mL−1. After 24 h of incubation, cultures were harvested, washed with sterile distilled water, and the wet weight of each cell pellet was recorded. To each pellet, 3 mL of 25% alcoholic potassium hydroxide (prepared by dissolving 25 g of KOH in 35 mL of sterile water and bringing the total volume to 100 mL with absolute ethanol) was added, and vortexed for 1 minute. The suspensions were transferred to sterile borosilicate glass screw-cap tubes and incubated at 85 °C for 1 h. Sterols were extracted using petroleum ether, and the petroleum ether phase was collected into clean screw-cap tubes and stored at −20 °C for up to 24 h. Before analysis, a 20 μL aliquot of the extract was diluted fivefold in absolute ethanol and scanned spectrophotometrically from 240 to 300 nm. The presence of ergosterol and its late biosynthetic intermediate 24(28)-DHE produced a characteristic four-peak UV spectrum, while a flat line indicated the absence of detectable ergosterol. The study was repeated three times, and the mean values are presented in Table 6.Cytotoxicity
The cytotoxicity analysis for compounds was performed on HUH7 immortalized hepatocyte-derived carcinoma cell lines. The cells were cultured for 24 h in complete media (DMEM, 15% FBS, 1% antimycotic, antibiotic solution) after being seeded into a 96-well plate at a density of 2 × 104 cells per mL (as measured by a hemocytometer) for a total volume of 100 μL each. Throughout the assay, the 96-well plates were incubated in a 5% CO2 incubator at 37 °C. After 24 h of sub-culturing into a 96-well plate, the cell lines were checked for attachment under the microscope and subjected to treatment at varying concentrations of 32–1024 μg mL−1 for all samples. The samples are dissolved in DMSO with a stock concentration of 2 mg mL−1. DMSO at a concentration of 256 μg mL−1 was kept as a positive control. Following treatment, the cells were incubated for 24 and 48 h, after which 20 μL of MTT reagent was added to all wells and incubated again for 3.5–4 h. The formazan crystals formed by the addition of MTT reagent (RM1131, Himedia) were dissolved in DMSO and incubated in the dark for 15 minutes for color formation. The optical density was measured at 570 nm in an ELIZA Plate Reader (Multiskan GO, Thermo).The OD570 values obtained were further calculated as per the formula given below and plotted against their concentration in Graph Pad Prism 9.3.1.
Author contributions
The manuscript was written through the contributions of all authors. ABP carried out docking studies and biological evaluation of the synthesized compounds; SS was engaged in the synthesis of the β-aminosulfone derivatives; MM helped with the antifungal evaluation study; UR contributed with designing antifungal activity study and revision of bioassay part; LRM and SB carried out cytotoxicity study; AC contributed with supervision and revision of the manuscript; MB was engaged in the design of the project, investigation, and oversight of the entire project, supervision, manuscript writing, and reviewing. All authors have approved the final version of the manuscript.Conflicts of interest
The authors confirm that there is no conflict of interest in any sort.Data availability
The data for this work will be available on request from the authors.Supplementary information (SI): it contains binding interactions; in vitro antifungal activities of compounds 5h–5y; SEM images; ergosterol depletion graphs; copies of NMR spectra etc. See DOI: https://doi.org/10.1039/d5md00907c.
Acknowledgements
M. B. is thankful to DBT-BUILDER (project no. BT/INF/22/SP42543/2021) for financial support. A. B. P. is indebted to BITS Pilani, KK Birla Goa campus for the JRFship. S. S. is grateful to BITS Pilani, KK Birla Goa campus for the SRFship. Sania Srivastava is duly acknowledged for some help in cytotoxicity studies. We acknowledge the central sophisticated instrumentation facilities (CSIFs) of BITS Pilani, KK Birla Goa campus, for NMR, LCMS, and SEM analysis.References
- Y. Lee, E. Puumala, N. Robbins and L. E. Cowen, Antifungal Drug Resistance: Molecular Mechanisms in Candida albicans and Beyond, Chem. Rev., 2020, 121, 3390–3411 CrossRef PubMed.
- C. C. Lai, C. K. Tan, Y. T. Huang, P. L. Shao and P. R. Hsueh, Current Challenges in the Management of Invasive Fungal Infections, J. Infect. Chemother., 2008, 14, 77–85 CrossRef CAS.
- J. Perlroth, B. Choi and B. Spellberg, Nosocomial Fungal Infections: Epidemiology, Diagnosis, and Treatment, Med. Mycol., 2007, 45, 321–346 CrossRef.
- M. J. Rueping, J. J. Vehreschild and O. A. Cornely, Invasive Candidiasis and Candidemia: From Current Opinions to Future Perspectives, Expert Opin. Invest. Drugs, 2009, 18, 735–748 CrossRef CAS.
- N. Liu, J. Tu, G. Dong, Y. Wang and C. Sheng, Emerging New Targets for the Treatment of Resistant Fungal Infections, J. Med. Chem., 2018, 61, 5484–5511 CrossRef CAS PubMed.
- J. Zhang, L. Li, Q. Lv, L. Yan, Y. Wang and Y. Jiang, The Fungal CYP51s: Their Functions, Structures, Related Drug Resistance, and Inhibitors, Front. Microbiol., 2019, 10, 691 CrossRef PubMed.
- D. G. Sant, S. G. Tupe, C. V. Ramana and M. V. Deshpande, Fungal Cell Membrane—Promising Drug Target for Antifungal Therapy, J. Appl. Microbiol., 2016, 121, 1498–1510 Search PubMed.
- R. Rajendran, E. Mowat, E. McCulloch, D. F. Lappin, B. Jones, S. Lang and G. Ramage, Azole Resistance of Aspergillus Fumigatus Biofilms is Partly Associated with Efflux Pump Activity, Antimicrob. Agents Chemother., 2011, 55, 2092–2097 CrossRef CAS.
- J. E. Parker, A. G. Warrilow, C. L. Price, J. G. Mullins, D. E. Kelly and S. L. Kelly, Resistance to Antifungals that Target CYP51, ChemBioChem, 2014, 7, 143–161 Search PubMed.
- M. Feng, B. H. Tang, S. Liang and X. Jiang, Sulfur-Containing Scaffolds in Drugs: Synthesis and Application in Medicinal Chemistry, Curr. Top. Med. Chem., 2016, 16, 1200–1216 CrossRef CAS.
- R. Ahmadi and S. Emami, Recent Applications of Vinyl Sulfone Motif in Drug Design and Discovery, Eur. J. Med. Chem., 2022, 234, 114255 CrossRef CAS.
- D. C. Meadows and J. Gervay-Hague, Vinyl Sulfones: Synthetic Preparations and Medicinal Chemistry Applications, Med. Res. Rev., 2006, 26, 793–814 CrossRef CAS.
- N. K. Konduru, S. Dey, M. Sajid, M. Owais and N. Ahmed, Synthesis and Antibacterial and Antifungal Evaluation of Some Chalcone-Based Sulfones and Bisulfones, Eur. J. Med. Chem., 2013, 59, 23–30 CrossRef CAS PubMed.
- A. A. Carmine, R. N. Brogden, R. C. Heel, T. M. Speight and G. S. Avery, Tinidazole in Anaerobic Infections: A Review of its Antibacterial Activity, Pharmacological Properties and Therapeutic Efficacy, Drugs, 1982, 24, 85–117 CrossRef CAS PubMed.
- G. Wozel and C. Blasum, Dapsone in Dermatology and Beyond, Arch. Dermatol. Res., 2014, 306, 103–124 CrossRef CAS PubMed.
- J. Morales-Sanfrutos, J. Lopez-Jaramillo, M. Ortega-Muñoz, A. Megia-Fernandez, F. Perez-Balderas, F. Hernandez-Mateo and F. Santoyo-Gonzalez, Vinyl Sulfone: A Versatile Function for Simple Bioconjugation and Immobilization, Org. Biomol. Chem., 2010, 8, 667–675 RSC.
- J. Franco, L. Scarone and M. A. Comini, Drugs and Drug Resistance in African and American Trypanosomiasis, Annu. Rep. Med. Chem., 2018, 51, 97–133 CAS.
- F. Vanhoutte, M. Mazur, O. Voloshyn, M. Stanislavchuk, A. Van Der Aa, F. Namour, R. Galien, L. Meuleners and G. Van't Klooster, Efficacy, Safety, Pharmacokinetics, and Pharmacodynamics of Filgotinib, a Selective JAK-1 Inhibitor, After Short-Term Treatment of Rheumatoid Arthritis: Results of Two Randomized Phase IIA Trials, Arthritis Rheumatol., 2017, 69, 1949–1959 CrossRef CAS PubMed.
- N. Murai, Y. Kondo, S. Akuzawa, T. Mihara, N. Shiraishi, S. Kakimoto and M. A. Matsumoto, Novel GABAB Receptor Positive Allosteric Modulator, ASP8062, Exerts Analgesic Effects in a Rat Model of Fibromyalgia, Eur. J. Pharmacol., 2019, 865, 172750 CrossRef CAS.
- A. Vena, G. Tiseo, M. Falcone, C. Bartalucci, C. Marelli, M. Cesaretti and M. Bassetti, Impact of Fluconazole Resistance on the Outcomes of Patients with Candida Parapsilosis Bloodstream Infections: A Retrospective Multicenter Study, Clin. Infect. Dis., 2025, 80, 540–550 CrossRef CAS.
- A. Kamal, M. A. H. Syed and S. M. Mohammed, Therapeutic Potential of Benzothiazoles: A Patent Review (2010–2014), Expert Opin. Ther. Pat., 2015, 25, 335–349 CrossRef CAS PubMed.
- R. S. Keri, M. R. Patil, S. A. Patil and S. Budagumpi, A Comprehensive Review in Current Developments of Benzothiazole-Based Molecules in Medicinal Chemistry, Eur. J. Med. Chem., 2015, 89, 207–251 CrossRef CAS PubMed.
- V. F. Carvalho, E. O. Barreto and M. F. Serra, Aldose Reductase Inhibitor Zopolrestat Restores Allergic Hyporesponsiveness in Alloxan-Diabetic Rats, Eur. J. Pharmacol., 2006, 549, 173–178 CrossRef CAS PubMed.
- I. Hutchinson, S. A. Jennings, B. R. Vishnuvajjala, A. D. Westwell and M. F. Stevens, Antitumor Benzothiazoles Synthesis and Pharmaceutical Properties of Antitumor 2-(4-aminophenyl) Benzothiazole Amino Acid Prodrugs, J. Med. Chem., 2002, 45, 744–747 CrossRef CAS.
- N. Herrera Cano, M. S. Ballari, A. G. López and A. N. Santiago, New Synthesis and Biological Evaluation of Benzothiazole Derivatives as Antifungal Agents, J. Agric. Food Chem., 2015, 63, 3681–3686 CrossRef CAS PubMed.
- M. S. Ballari, N. H. Cano, D. A. Wunderlin, G. E. Feresin and A. N. Santiago, One-pot Sequential Synthesis and Antifungal Activity of 2-(Benzylsulfonyl) Benzothiazole Derivatives, RSC Adv., 2019, 9, 29405–29413 RSC.
- P. Celano, S. B. Baylin and R. A. Casero, Polyamines Differentially Modulate the Transcription of Growth-Associated Genes in Human Colon Carcinoma, J. Biol. Chem., 1989, 264, 8922–8927 CrossRef CAS.
- N. Seiler, A Guide to Polyamines, in The Quarterly Review of Biology, ed. S. S. Cohen, The University of Chicago Press, Chicago, IL, 1999, vol. 74, pp. 342–343 Search PubMed.
- U. Bachrach, Polyamines and Cancer: Minireview Article, Amino Acids, 2004, 26, 307–309 CrossRef CAS.
- S. Pockes, D. Wifling, M. Keller, A. Buschauer and S. Elz, Highly Potent, Stable, and Selective Dimeric Hetarylpropylguanidine-Type Histamine H2 Receptor Agonists, ACS Omega, 2018, 3, 2865–2882 CrossRef CAS.
- S. Babić, J. S. Marjanović, V. M. Divac, O. R. Klisurić, D. Milivojević, J. V. Bogojeski and M. D. Kostić, Molecular Docking Study and In Vitro Evaluation of Apoptotic Effect of Biogenic-Amine-Based N,O-Cu(II) Complexes as Potent Antitumor Agents, J. Coord. Chem., 2025, 78, 1007–1026 CrossRef.
- P. Tata, A. Ghosh, T. Jamma, O. Kulkarni, R. Ganesan and J. Ray Dutta, Caffeic Acid–Biogenic Amine Complexes Outperform Standard Drugs in Reducing Toxicity: Insights from In Vivo Iron Chelation Studies, Mol. Pharmaceutics, 2025, 22, 2985–2996 CrossRef CAS PubMed.
- B. J. McCabe-Sellers, C. G. Staggs and M. L. Bogle, Tyramine in Foods and Monoamine Oxidase Inhibitor Drugs: A Crossroad Where Medicine, Nutrition, Pharmacy, and Food Industry Converge, J. Food Compos. Anal., 2006, 19, S58–S65 CrossRef CAS.
- X. Y. Meng, H. X. Zhang, M. Mezei and M. Cui, Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery, Curr. Comput.-Aided Drug Des., 2011, 7, 146–157 CrossRef CAS.
- D. C. Blakemore, L. Castro, I. Churcher, D. C. Rees, A. W. Thomas, D. M. Wilson and A. Wood, Organic Synthesis Provides Opportunities to Transform Drug Discovery, Nat. Chem., 2018, 10, 383–394 CrossRef CAS PubMed.
- I. Aliagas, R. Berger, K. Goldberg, R. T. Nishimura, J. Reilly, P. Richardson and M. C. Bryan, Sustainable Practices in Medicinal Chemistry Part 2: Green by Design: Miniperspective, J. Med. Chem., 2017, 60, 5955–5968 CrossRef CAS PubMed.
- J. J. Chen, C. V. Lu and R. N. Brockman, Catalyzed Double Michael Addition of Anilines to Vinyl Sulfone, Tetrahedron Lett., 2003, 44, 3459–3462 CrossRef CAS.
- S. Kim, S. Kang, G. Kim and Y. Lee, Copper-Catalyzed Aza-Michael Addition of Aromatic Amines or Aromatic Aza-Heterocycles to α,β-Unsaturated Olefins, J. Org. Chem., 2016, 81, 4048–4057 CrossRef PubMed.
- Z. T. Bhutia, A. Das, M. Biswas, A. Chatterjee and M. Banerjee, 7-Oxa-4-Thia-1-Aza-Bicyclo[3.2.1]Octane4,4-Dioxides: Mechanochemical Synthesis by Tandem Michael Addition–1,3-Dipolar Cycloaddition of Aldoximes and Evaluation of Antibacterial Activities, Eur. J. Org. Chem., 2018, 4, 506–514 CrossRef.
- A. De La Hoz, A. Díaz-Ortiz and P. Prieto, in Alternative Energy Sources for Green Chemistry, ed. G. Stefanidis and A. Stankiewicz, The Royal Society of Chemistry, Cambridge, UK, 2016, pp. 1–33 Search PubMed.
- S. Saha, A. Chatterjee and M. Banerjee, Reagentless Chemistry “On-Water”: An Atom-Efficient and “Green” Route to Cyclic and Acyclic β-Amino Sulfones via Aza-Michael Addition Using Microwave Irradiation, J. Org. Chem., 2023, 88, 15358–15366 Search PubMed.
- Z. T. Bhutia, G. Prasannakumar, A. Das, M. Biswas, A. Chatterjee and M. Banerjee, A Facile, Catalyst-Free Mechano-Synthesis of Quinoxalines and their In-Vitro Antibacterial Activity Study, ChemistrySelect, 2017, 2, 1183–1187 CrossRef.
- S. Korwar, S. Amir, P. N. Tosso, B. K. Desai, C. J. Kong, S. Fadnis and B. F. Gupton, The Application of a Continuous Grignard Reaction in the Preparation of Fluconazole, Eur. J. Org. Chem., 2017, 44, 6495–6498 CrossRef.
- G. I. Lepesheva and M. R. Waterman, Sterol 14α-Demethylase Cytochrome P450 (CYP51), a P450 in All Biological Kingdoms, Biochim. Biophys. Acta, Biomembr., 2007, 1770, 467–477 CrossRef PubMed.
- Clinical and Laboratory Standards Institute (CLSI) Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved Standard-M27, 4th edn, CLSI, Wayne, PA, 2017.
- M. Madhuri, S. M. Rudramurthy and U. Roy, Two Promising Bacillus-derived Antifungal Lipopeptide Leads AF4 and AF5 and Their Combined Effect with Fluconazole on the In Vitro Candida Glabrata Biofilms, Front. Pharmacol., 2024, 15, 1334419 CrossRef PubMed.
- R. Hamdy, A. M. Hamoda, M. Al-Khalifa, V. Menon, R. El-Awady and S. S. Soliman, Efficient Selective Targeting of Candida CYP51 by Oxadiazole Derivatives Designed from Plant Cuminaldehyde, RSC Med. Chem., 2022, 13, 1322–1340 RSC.
- N. Tlapale-Lara, J. López, E. Gómez, L. Villa-Tanaca, E. Barrera, C. H. Escalante and O. Gómez-García, Synthesis, In Silico Study, and In Vitro Antifungal Activity of New 5-(1,3-Diphenyl-1H-Pyrazol-4-yl)-4-Tosyl-4,5-Dihydrooxazoles, Int. J. Mol. Sci., 2024, 25, 5091 CrossRef PubMed.
- O. N. Breivik and J. L. Owades, Yeast Analysis, Spectrophotometric Semimicrodetermination of Ergosterol in Yeast, J. Agric. Food Chem., 1957, 5, 360–363 Search PubMed.
- B. A. Arthington-Skaggs, H. Jradi, T. Desai and C. J. Morrison, Quantitation of Ergosterol Content: Novel Method for Determination of Fluconazole Susceptibility of Candida albicans, J. Clin. Microbiol., 1999, 37, 3332–3337 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |