Open Access Article
Open Access ArticleMitochondria-targeting symmetric diiminoguanidines: potent and selective anticancer agents against pancreatic tumors
Sigrid
Lacaille
* and
Andreea R.
Schmitzer
 *
*
Département de chimie- Faculté des arts et des sciences, Université de Montréal. Campus MIL, 1375, Ave. Thérèse Lavoie-Roux, Montréal, Québec H2V 0B3, Canada. E-mail: ar.schmitzer@umontreal.ca
First published on 29th December 2025
Abstract
Pancreatic cancer remains one of the deadliest malignancies of the 21st century, with a five-year survival rate below 12%. Its growing incidence is strongly linked to modern lifestyles, marked by obesity, diabetes, overnutrition, and physical inactivity. Current chemotherapies offer limited success and are often burdened by severe side effects, highlighting the urgent need for more effective and selective treatments. In response, we have developed a new class of easily synthesized, amphiphilic symmetric diiminoguanidines and evaluated their antiproliferative activity against pancreatic cancer cell lines. Several compounds demonstrated remarkable efficacy and selectivity, positioning them as strong candidates for further in vivo evaluation. Fluorescence microscopy revealed that these molecules rapidly localize into mitochondria. Preliminary mechanistic studies suggest their primary target is the mitochondrial respiratory chain. These findings support the potential of diiminoguanidines as affordable, mitochondria-targeting alternatives to existing pancreatic cancer therapies.
Introduction
Pancreatic cancer stands as one of the most aggressive and fatal malignancies in modern oncology.1–3 With a dismal five-year survival rate of less than 12%, it is projected to become the second leading cause of cancer-related deaths in the United States by 2040, surpassing colorectal cancer and second only to lung cancer.3,4 This alarming trend reflects both the increasing incidence of the disease and the lack of effective therapeutic interventions.5,6The rising prevalence of pancreatic cancer in Western societies is closely tied to metabolic and lifestyle-associated risk factors. Type 2 diabetes, obesity, overnutrition, and physical inactivity have been consistently identified as key contributors to pancreatic tumorigenesis.7–11 These conditions, highly prevalent in North America and Europe, create a metabolic and inflammatory environment that fosters cancer development and progression. One of the principal challenges in treating pancreatic cancer is the late-stage diagnosis in most patients. Early clinical signs are either vague or absent, often mistaken for benign gastrointestinal disorders. By the time a definitive diagnosis is made, the disease is frequently at an advanced or metastatic stage, leaving few curative options. Surgical resection, while potentially curative, is only feasible in a minority of cases due to the pancreas's deep retroperitoneal location, rich vascular supply, and the high metastatic potential of pancreatic tumors.11
Chemotherapy remains the cornerstone of treatment for unresectable or metastatic pancreatic cancer. However, current regimens offer only modest improvements in survival and are associated with significant toxicity. Antimetabolites such as 5-fluorouracil (5-FU) and capecitabine disrupt RNA and DNA synthesis but are burdened with adverse effects including myelosuppression, neurotoxicity, and gastrointestinal damage.12–17 Combination therapies like FOLFIRINOX (5-FU, folinic acid, irinotecan, and oxaliplatin) improve efficacy but also amplify systemic toxicity, often limiting their use to patients with good performance status.18–20 Gemcitabine, a nucleoside analog, remains the standard of care in many settings, yet typically extends survival by only a few months.21–25 These clinical limitations underscore the urgent need for novel therapeutic strategies that are both efficacious and well-tolerated.
Mitochondria, the powerhouses of the cell, have emerged as promising targets in cancer therapy due to their essential roles in bioenergetics, apoptosis regulation, and reactive oxygen species (ROS) generation.26–28 Compounds that selectively disrupt mitochondrial function in cancer cells could represent a powerful new avenue for treatment, particularly in metabolically active and chemoresistant cancers such as pancreatic adenocarcinoma.29–32
Over the past decade, our group has concentrated on the development of guanidine-based molecules, particularly biguanides, as promising agents for targeting pancreatic cancer cells (Fig. 1). We previously demonstrated that amphiphilic biguanide salts, most notably a triflate salt of phenylethynylbenzyl (PEB)-substituted biguanide, exhibited potent antiproliferative activity, with IC50 values in the low micromolar range against pancreatic cancer cell lines.33,34 Building on these findings, we further explored bifunctional constructs, including biguanide-based PROTACs and AUTACs, which induced mitochondrial accumulation and led to marked growth inhibition.35,36 These efforts validated the guanidine scaffold as a versatile and bioactive platform in anticancer drug discovery.
 | ||
| Fig. 1 Structure of antiproliferative PEB-biguanide, biguanide-PROTAC and biguanide-AUTAC synthesized in our group. | ||
Encouraged by these results, we hypothesized that another class of guanidine derivatives, iminoguanidines, could serve as valuable building blocks for the design of new amphiphilic molecules with anticancer potential. Like biguanides, iminoguanidines possess basic nitrogen atoms capable of protonation and hydrogen bonding, key features for mitochondrial targeting and intracellular accumulation.37 Moreover, their synthetic accessibility enables rapid generation of structural analogs, facilitating structure–activity relationship (SAR) exploration. In this study, we report the synthesis of a library of amphiphilic symmetric diiminoguanidines, prepared as chloride salts, bearing a range of aromatic, polycyclic aromatic, and heterocyclic substituents. These compounds were evaluated for their antiproliferative activity on pancreatic cancer cell lines. Selectivity toward cancer versus normal cells was assessed, and mechanistic studies were conducted to elucidate their mode of action, with a particular focus on mitochondrial targeting.
Results and discussion
Design and synthesis of amphiphilic symmetric diiminoguanidines as chloride salts
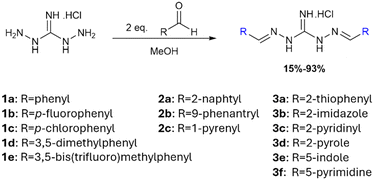
A series of 14 symmetric diiminoguanidines were synthesized as chloride salts (Fig. 2) in order to explore the relationship between molecular structure, specifically side-chain identity and antiproliferative activity against pancreatic cancer cells. Amphiphilicity was a critical design parameter, as it strongly influences a molecule's ability to traverse cell membranes and reach intracellular organelles, such as mitochondria. To modulate amphiphilicity and molecular recognition features, we introduced aromatic rings (both substituted and unsubstituted), polycyclic aromatic systems, and heterocycles as side chains. Aromatic moieties are prevalent in medicinal chemistry due to their capacity for π–π stacking interactions, while polycyclic aromatics offer advantageous photophysical properties for intracellular tracking. Heterocycles, on the other hand, are biocompatible and capable of hydrogen bonding, enhancing their potential to interact with biological targets. The diiminoguanidines were obtained via double condensation of N,N′-diaminoguanidinium hydrochloride with two equivalents of the corresponding aldehydes in methanol, under mild conditions (room temperature to 80 °C, ≤24 h) (Fig. 2). It should be noted that certain compounds were obtained with low yields. In the case of the polycyclic aromatic compounds, the main reason was the low solubility of the final substituted diiminoguanidine hydrochloride in methanol. In the case of the heterocyclic compounds, even higher temperatures or longer reaction times did not allow the increase of the yields.Generally, this synthetic route is operationally simple, efficient, and adheres to green chemistry principles, an important consideration in the development of sustainable therapeutic candidates.
Antiproliferative activity of the diiminoguanidine hydrochlorides
The antiproliferative properties of the synthesized diiminoguanidines were evaluated on two human pancreatic cancer cell lines: KP4, which is characterized by a high metastatic potential, and PANC1, associated with a comparatively more moderate phenotype. As shown in Table 1, all compounds exhibited IC50 (concentration that inhibit cancer cell growth by 50%) in the micromolar range, validating the iminoguanidine scaffold as a promising starting point for the development of anticancer agents. Remarkably, compound 3b, bearing imidazole side chains, demonstrated the most potent activity with an IC50 of 280 nM, while 2c, bearing pyrene groups, was the least active (IC50 = 163 μM).| Compound | Substituent R | IC50 cancer cells (μM) | IC50 healthy cells (μM) | SI IMR90/PANC1 | SI hTERT-HPNE/PANC1 | clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pa Pa |
||
|---|---|---|---|---|---|---|---|---|
| KP4 | PANC1 | IMR90 | hTERT-HPNE | |||||
| a Calculated with Swiss ADME consensus. | ||||||||
| 1a | Phenyl | 9.3 ± 1.4 | 29.6 ± 2.9 | 23.6 ± 0.9 | 62.9 ± 7.5 | 0.8 | 2.1 | 2.24 |
| 1b | p-Fluorophenyl | 9.4 ± 1.3 | 28.6 ± 4.3 | 16.3 ± 2.9 | 50.0 ± 5.4 | 0.6 | 1.7 | 2.83 |
| 1c | p-Chlorophenyl | 36.6 ± 5.5 | 59.1 ± 4.1 | 43.5 ± 5.2 | 48.5 ± 9.7 | 0.7 | 0.8 | 3.29 |
| 1d | 3,5-Dimethylphenyl | 29.1 ± 3.5 | 56.2 ± 3.4 | 89.1 ± 5.3 | 140.7 ± 21.1 | 3.4 | 6.4 | 3.57 |
| 1e | 3,5-Bis(trifluoro)methylphenyl | 134.0 ± 10.7 | 58.2 ± 8.4 | 197.0 ± 27.6 | 374.9 ± 30.0 | 1.6 | 2.5 | 6.4 |
| 2a | 2-Naphtyl | 122.2 ± 21.9 | 104.7 ± 7.3 | 119.2 ± 27.4 | 276.0 ± 55.2 | 1.1 | 2.6 | 4.05 |
| 2b | 9-Phenantryl | 138.6 ± 16.6 | 151.4 ± 13.6 | 190.5 ± 24.7 | 130.6 ± 10.4 | 1.3 | 0.9 | 5.83 |
| 2c | 1-Pyrenyl | 163.2 ± 24.5 | 240.3 ± 28.3 | 280.9 ± 25.3 | 284.3 ± 54 | 1.2 | 1.2 | 6.76 |
| 3a | 2-Thiophenyl | 10.8 ± 1.7 | 50.1 ± 6.0 | 47.9 ± 7.7 | 120.8 ± 20.5 | 1 | 2.4 | 2.26 |
| 3b | 2-Imidazole | 0.28 ± 0.02 | 9.7 ± 1.3 | 180.1 ± 10.8 | 257.9 ± 33.5 | 18.6 | 26.6 | −0.63 |
| 3c | 2-Pyridinyl | 5.4 ± 1.0 | 4.8 ± 0.3 | 95.0 ± 4.6 | 198.8 ± 23.8 | 19.8 | 41.4 | 0.78 |
| 3d | 2-Pyrole | 0.52 ± 0.03 | 4.1 ± 0.3 | 21.6 ± 2.6 | 74.3 ± 7.4 | 5.3 | 18.1 | 0.62 |
| 3e | 2-Indole | 4.9 ± 0.4 | 7.01 ± 1.2 | 5.9 ± 1.0 | 8.8 ± 0.8 | 0.8 | 1.3 | 2.47 |
| 3f | 5-Pyrimidine | 118.9 ± 14.3 | 113.5 ± 22.7 | 95.6 ± 17.2 | 398.1 ± 55.7 | 0.8 | 3.5 | −0.42 |
SAR analysis revealed several trends:
• Heterocyclic side chains (3a–3f) generally conferred higher potency. Imidazole- and oxazole-bearing compounds showed IC50 values well below 5 μM. The exception was 3f (pyrimidine), which showed only modest activity (IC50 = 119 μM), suggesting that not all heterocycles contribute equally to activity.
• Polycyclic aromatic derivatives (2a–2c), such as those containing pyrene (2c) or phenanthracene (2b) moieties, were the least active. Their high lipophilicity may hinder cell membrane penetration or intracellular distribution.
• Phenylic derivatives (1a–1e) showed variable activity with no clear correlation to electronic effects. For example, the unsubstituted phenyl derivative 1a and its para-fluorinated analog 1b exhibited similar activity. Interestingly, a meta-substituted derivative bearing electron-donating methyl groups (1d) yielded an IC50 of 29 μM, whereas its electron-withdrawing trifluoromethylphenyl counterpart (1e) had a significantly reduced activity (IC50 = 134 μM). This observation points to a complex interplay of steric and electronic factors not solely explained by traditional SAR paradigms.
As imine bonds are known to have the ability to be hydrolyzed in acidic aqueous media, we investigated the stability of our compounds. To do so, we mimicked the biological experimental conditions, namely PBS buffer with 4% DMSO at 37 °C for 3 days, and recorded the UV-Vis spectrum every 24 hours. All the recorded spectra were identical, confirming no deterioration of the diiminoguanidine structure for the duration of treatment (see SI).
Role of amphiphilicity (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) P) in antiproliferative activity
P) in antiproliferative activity
To further investigate the impact of amphiphilicity on biological performance, we analyzed the correlation between the calculated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (clog
P (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) values and IC50 data (Fig. 3). The results revealed a compelling inverse relationship between amphiphilicity and antiproliferative efficacy. Compounds with clog
P) values and IC50 data (Fig. 3). The results revealed a compelling inverse relationship between amphiphilicity and antiproliferative efficacy. Compounds with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values between 0.5 and 4 consistently showed strong activity (IC50 < 50 μM), indicating an optimal amphiphilic window that supports both membrane permeability and intracellular distribution. In contrast, compounds with clog
P values between 0.5 and 4 consistently showed strong activity (IC50 < 50 μM), indicating an optimal amphiphilic window that supports both membrane permeability and intracellular distribution. In contrast, compounds with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values > 4 exhibited significantly reduced potency (IC50 > 100 μM), likely due to excessive lipophilicity that hinders effective cellular uptake. Compound 3f, with a clog
P values > 4 exhibited significantly reduced potency (IC50 > 100 μM), likely due to excessive lipophilicity that hinders effective cellular uptake. Compound 3f, with a clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of −0.42, displayed weak activity (IC50 = 119 μM), suggesting that higher hydrophilicity may also be detrimental. However, the high activity of 3b despite its higher hydrophilicity suggests that this is not the only parameter that can explain the cytotoxicity of the studied diiminoguanidines. Overall, these results emphasize the necessity of fine-tuning amphiphilicity of the compounds to achieve an optimal balance for membrane penetration, intracellular targeting, and pharmacological activity. Nevertheless, only a deep study of the mechanism of action and the determination of the precise target of diiminoguanidines in the cells would allow to establish the relationship between the nature of the substituents and their cytotoxic activity.
P of −0.42, displayed weak activity (IC50 = 119 μM), suggesting that higher hydrophilicity may also be detrimental. However, the high activity of 3b despite its higher hydrophilicity suggests that this is not the only parameter that can explain the cytotoxicity of the studied diiminoguanidines. Overall, these results emphasize the necessity of fine-tuning amphiphilicity of the compounds to achieve an optimal balance for membrane penetration, intracellular targeting, and pharmacological activity. Nevertheless, only a deep study of the mechanism of action and the determination of the precise target of diiminoguanidines in the cells would allow to establish the relationship between the nature of the substituents and their cytotoxic activity.
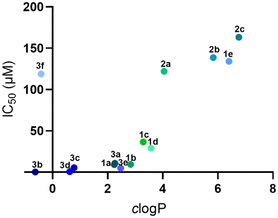 | ||
Fig. 3 Correlation between clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and IC50 values of diiminoguanidines on KP4 pancreatic cancer cells. P and IC50 values of diiminoguanidines on KP4 pancreatic cancer cells. | ||
Selectivity toward cancer versus normal cells
To evaluate the selectivity of our diiminoguanidines for pancreatic cancer cells over non-cancerous cells, we assessed their antiproliferative activity on two healthy human cell lines: hTERT-HPNE (immortalized pancreatic epithelial cells) and IMR-90 (normal human lung fibroblasts). hTERT-HPNE serves as a physiological control representative of the targeted organ, while IMR-90 represents a non-tumorigenic, proliferative cell line that is often sensitive to chemotherapeutic toxicity, providing a stringent measure of off-target effects. Selectivity indices (SI), calculated as the ratio of IC50 in healthy cells to that in cancer cells, revealed important trends. Compounds bearing phenylic (1a–1e) and polycyclic aromatic (2a–2c) side chains exhibited indices low selectivity (SI < 10), suggesting a narrow therapeutic window and potential for toxicity in non-cancerous tissues. In contrast, some heterocyclic derivatives (3a–3d) showed higher selectivity. Notably, compounds 3b–3d, which featured imidazole (3b), pyridine (3c), and pyrrole (3d) side chains, respectively, stood out as the most selective candidates. Compound 3b, in particular, achieved a selectivity index greater than 600 for KP4 cells relative to both hTERT-HPNE and IMR-90 lines, indicating strong preferential activity against malignant cells with minimal cytotoxicity toward normal tissue. Given their potent antiproliferative activity and favorable selectivity profiles, these heterocycle-functionalized diiminoguanidines, especially 3b, represent promising leads for further in vivo evaluation. Their strong discrimination between cancerous and normal cells positions them as attractive candidates for the development of targeted pancreatic cancer therapies with reduced systemic toxicity.Selectivity indexes were calculated for PANC1 cancer cells as well, and can be found in SI. Overall, the selectivity indexes on PANC1 cancer cells were in the same range as the one for KP4, except for compound 3b, suggesting no preference for a particular pancreatic cancer cell line.
Mechanism of action
In light of the potent antiproliferative activity demonstrated by our diiminoguanidines, we next investigated their cellular mechanism of action. Given the amphiphilic and cationic nature of these compounds, characteristics known to facilitate mitochondrial targeting, particularly among guanidinium-containing structures, we hypothesized that mitochondria might be their primary intracellular site of action. To test this, we exploited the inherent fluorescence properties of several of our diiminoguanidine derivatives and performed confocal fluorescence microscopy on live pancreatic cancer cells. Cells were co-stained with MitoTracker™ Deep Red (MTDR), a well-established mitochondrial marker, allowing us to assess potential colocalization. The resulting images revealed a strong spatial overlap between the fluorescent signal of our compounds and that of MTDR. This colocalization was visually confirmed by the emergence of yellow signals in merged images (Fig. 4 and SI), and quantitatively supported by Pearson correlation coefficients exceeding 0.89 for all tested compounds—indicative of a robust mitochondrial localization.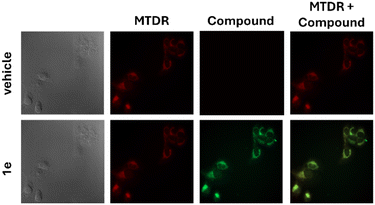 | ||
| Fig. 4 Confocal microscopy images of KP4 pancreatic cancer cells treated with 100 nM diiminoguanidine hydrochloride 1e or vehicle (4% DMSO) and colocalization with MitoTracker deep red (zoom × 63). | ||
To further characterize this localization, we conducted time-course experiments to evaluate the persistence of mitochondrial targeting. Cells treated with diiminoguanidines were imaged at 30 minutes and 24 hours post-incubation. Remarkably, mitochondrial accumulation was evident as early as 30 minutes and remained clearly detectable after 24 hours, indicating that these compounds are not rapidly cleared and instead exhibit sustained mitochondrial residency. Taken together, these findings confirm that our amphiphilic symmetric diiminoguanidines rapidly and durably accumulate in mitochondria. This prolonged retention is likely critical for their antiproliferative efficacy, enabling them to engage mitochondrial targets involved in energy production or apoptosis regulation over an extended period. The observed mitochondrial tropism not only highlights a key feature of the compounds' mechanism of action but also strengthens their candidacy as selective anticancer agents.
Action on the mitochondrial respiratory chain
Mitochondrial dysfunction is a common consequence of therapeutic agents that accumulate within these organelles. When the mitochondrial respiratory chain (MRC) is impaired, cancer cells often respond by shifting their energy production toward aerobic glycolysis, a metabolic reprogramming known as the Warburg effect. To determine whether our diiminoguanidines interfere with mitochondrial respiration, we employed a glycolytic suppression assay using 2-deoxyglucose (2-DG), a well-characterized glycolysis inhibitor. 2-DG is a glucose analog in which the hydroxyl group at the 2-position is absent. This structural modification prevents its metabolism during glycolysis, effectively blocking the glycolytic pathway. By co-administering 2-DG with our compounds, we can test whether the inhibition of glycolysis unmasks a dependence on mitochondrial respiration—a dependence that would become fatal in the presence of mitochondrial impairment. To this end, we treated KP4 pancreatic cancer cells with a non-toxic concentration of 2-DG (1 mM) in combination with each diiminoguanidine at 0.5 × IC50 (Fig. 5). A simple 0.5 × IC50 treatment served as the baseline for partial growth inhibition. If the addition of 2-DG had no further effect, this would suggest the mitochondrial function remains intact, and cells are not compensating via glycolysis. In contrast, a further reduction in cellular proliferation upon co-treatment would indicate that our compounds impair mitochondrial function and that cells, unable to compensate via glycolysis, undergo energy crisis-induced death.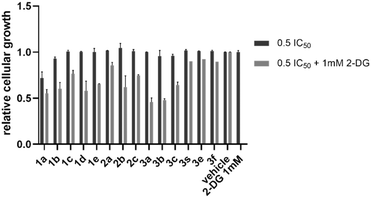 | ||
| Fig. 5 Relative cellular growth of KP4 cancel cells with 0.5 × IC50 of diiminoguanidines and a combination of 0.5 × IC50 of diiminoguanidines and 1 mm 2-DG. | ||
The results were unambiguous: all diiminoguanidines tested exhibited enhanced antiproliferative activity when combined with 2-DG, strongly supporting their inhibitory effect on the mitochondrial respiratory chain. Notably, compound 3b, already identified as the most potent and selective molecule in the series, produced a ∼50% further reduction in cellular viability when used in combination with 2-DG, compared to treatment with 3b alone. This synergistic effect underscores a robust mitochondrial mechanism of action, reinforcing our earlier microscopy-based conclusions.
Together, these findings provide compelling evidence that diiminoguanidines, particularly compound 3b, disrupt mitochondrial energy metabolism, and that this disruption is critical to their antiproliferative efficacy. These results also suggest the potential of using glycolytic inhibition as a co-therapeutic strategy to potentiate the effects of mitochondria-targeting agents in pancreatic cancer.
Conclusions
In this study, we successfully synthesized a series of 14 amphiphilic symmetric diiminoguanidinium hydrochloride salts through straightforward, green condensation reactions between N,N′-diaminoguanidine hydrochloride and a diverse set of aldehydes. By systematically varying the side chains—aromatic, polycyclic aromatic, and heteroaromatic—we modulated both the amphiphilicity and biological activity of the resulting compounds. Biological evaluation on pancreatic cancer cell lines revealed potent antiproliferative activity, with most compounds exhibiting IC50 values below 100 μM. Among them, compound 3b, bearing imidazole side chains, emerged as the lead candidate, with submicromolar IC50 values and an excellent selectivity index exceeding 600 when compared to healthy pancreatic and lung fibroblast cell lines. This high efficacy–selectivity profile positions 3b as a strong candidate for in vivo validation.We identified molecular amphiphilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) as a key determinant of activity: compounds with clog
P) as a key determinant of activity: compounds with clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P between 0.5 and 4 showed significantly improved potency, suggesting optimal membrane permeability is essential for mitochondrial access. Confocal microscopy confirmed that diiminoguanidines rapidly accumulate in mitochondria, where they persist for at least 24 h—indicating prolonged mitochondrial engagement. Furthermore, combination assays with 2-deoxyglucose provided mechanistic evidence that the mitochondrial respiratory chain is a critical functional target, particularly for compound 3b.
P between 0.5 and 4 showed significantly improved potency, suggesting optimal membrane permeability is essential for mitochondrial access. Confocal microscopy confirmed that diiminoguanidines rapidly accumulate in mitochondria, where they persist for at least 24 h—indicating prolonged mitochondrial engagement. Furthermore, combination assays with 2-deoxyglucose provided mechanistic evidence that the mitochondrial respiratory chain is a critical functional target, particularly for compound 3b.
Taken together, this work introduces a new class of synthetically accessible, mitochondria-targeting anticancer agents with a favorable pharmacological profile for pancreatic cancer. The modular synthesis and tunable side chains offer a promising platform for future optimization toward enhanced selectivity, potency, and translational potential.
Materials and methods
General information
All chemicals were purchased from Sigma Aldrich, Oakwood Chemicals, and Combi-Blocks at their highest purity and were used without further purification. NMR spectra were recorded on AVANCE II 400, Bruker AVANCE NEO 400, Bruker AVANCE 500, and Bruker AVANCE 700 spectrometers. High-resolution mass spectra (HRMS) and LCMS purity analysis were performed on Agilent TOF instruments by the regional mass spectrometry center (University of Montreal).Synthesis and characterization of compounds
Synthesis of N,N′-di(phenylmethanimino)guanidine hydrochloride (1a). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and benzaldehyde (2 eq.) yielded compound 1a as a white powder with a 53% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.32 (s, 2H), 8.54 (s, 2H), 8.46 (s, 2H), 7.99–7.90 (m, 4H), 7.55–7.46 (m, 6H). 13C NMR (101 MHz, DMSO-d6) δ 153.3, 149.3, 133.7, 131.2, 129.2, 128.3. HRMS: m/z [M + H]+ calculated for C15H16N5, 266.1400; found 266.1396.
Synthesis of N,N′-di(4-fluorophenylmethanimino)guanidine hydrochloride (1b). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and 4-fluorobenzaldehyde (2 eq.) yielded compound 1b as a white powder with a 61% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.36 (s, 2H), 8.57 (s, 2H), 8.45 (s, 2H), 8.04 (dd, J = 8.7, 5.7 Hz, 4H), 7.35 (t, J = 8.8 Hz, 4H). 13C NMR (101 MHz, DMSO-d6) δ 165.2, 162.7, 153.3, 130.7, 130.6, 130.4, 130.4, 116.4, 116.1. HRMS: m/z [M + H]+ calculated for C15H14N5F2, 302.1212; found 302.1212.
Synthesis of N,N′-di(4-chlorophenylmethanimino)guanidine hydrochloride (1c). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and 4-chlorobenzaldehyde (2 eq.) yielded compound 1c as a white powder with a 76% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.43 (s, 2H), 8.62 (s, 2H), 8.45 (s, 2H), 8.00 (d, J = 8.6 Hz, 4H), 7.57 (d, J = 8.5 Hz, 4H). 13C NMR (101 MHz, DMSO-d6) δ 153.3, 148.1, 135.7, 132.7, 130.0, 129.3. HRMS: m/z [M + H]+ calculated for C15H14N5Cl2, 334.0621; found 334.0628.
Synthesis of N,N′-di((3,5-dimethyl)phenylmethanimino)guanidine hydrochloride (1d). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and 3,5-dimethylbenzaldehyde (2 eq.) yielded compound 1d as a white powder with a 93% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.30 (s, 2H), 8.49 (s, 2H), 8.39 (s, 2H), 7.55 (s, 4H), 7.13 (s, 2H), 2.34 (s, 12H). 13C NMR (101 MHz, DMSO-d6) δ 153.1, 149.5, 138.3, 133.6, 132.7, 126.0, 21.2. HRMS: m/z [M + H]+ calculated for C19H24N5, 322.2026; found 322.2031.
Synthesis of N,N′-di((3,5-bis(trifluoromethyl))phenylmethanimino)guanidine hydrochloride (1e). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and 3,5-bis(trifluoromethyl)benzaldehyde (2 eq.) yielded compound 1e as a white powder with a 93% yield. 1H NMR (700 MHz, DMSO-d6) δ 12.64 (s, 2H), 8.90 (s, 2H), 8.70 (s, 4H), 8.58 (s, 2H), 8.21 (s, 2H). 13C NMR (176 MHz, DMSO-d6) δ 154.0, 146.6, 136.7, 131.6, 131.4, 131.2, 131.0, 128.8, 126.0, 124.5, 123.9, 122.9, 121.4. HRMS: m/z [M + H]+ calculated for C19H12F12N5, 538.0896; found 538.0903.
Synthesis of N,N′-di(2-naphtylmethanimino)guanidine hydrochloride (2a). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and 2-naphtaldehyde (2 eq.) yielded compound 2a as a white powder with a 78% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.49 (s, 2H), 8.65 (s, 4H), 8.33 (dd, J = 13.6, 4.9 Hz, 4H), 8.10–7.92 (m, 6H), 7.61 (p, J = 5.4 Hz, 4H). 13C NMR (101 MHz, DMSO-d6) δ 153.2, 149.5, 134.5, 133.1, 131.5, 130.3, 128.9, 128.8, 128.3, 127.9, 127.3, 123.7. HRMS: m/z [M + H]+ calculated for C23H20N5, 366.1713; found 366.1722.
Synthesis of N,N′-di(phenantrylmethanimino)guanidine hydrochloride (2b). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and phenantrene-9-carboxaldehyde (2 eq.) yielded compound 2b as an off-white powder with a 18% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.51 (s, 2H), 9.45 (s, 2H), 9.05–8.94 (m, 2H), 8.92 (d, J = 8.1 Hz, 2H), 8.75 (s, 2H), 8.72 (s, 2H), 8.59 (dd, J = 6.4 Hz, 3.2 Hz, 2H), 8.16 (d, J = 6.9 Hz, 2H), 7.80 (dddd, J = 15.8 Hz, 14.7 Hz, 6.2 Hz, 1.6 Hz, 8H). 13C NMR (101 MHz, DMSO-d6) δ 153.0, 131.2, 131.0, 130.5, 129.9, 129.4, 128.6, 128.1, 127.9, 127.7, 124.5, 124.2, 123.5. HRMS: m/z [M + H]+ calculated for C31H24N5, 466.2026; found 466.2026.
Synthesis of N,N′-di(pyrenylmethanimino)guanidine hydrochloride (2c). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and pyrene-1-carbaldehyde (2 eq.) yielded compound 2c as a green powder with a 52% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.60 (s, 2H), 9.79 (s, 2H), 9.07 (d, J = 8.2 Hz, 2H), 8.80 (s, 2H), 8.63 (d, J = 9.5 Hz, 2H), 8.50–8.34 (m, 8H), 8.30 (q, J = 9.0 Hz, 4H), 8.15 (t, J = 7.6 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) δ 132.9, 131.2, 130.5, 129.7, 129.4, 129.2, 127.8, 127.1, 126.7, 126.4, 126.4, 125.6, 124.4, 124.2, 122.1. HRMS: m/z [M + H]+ calculated for C35H24N5, 514.2026; found 514.2021.
Synthesis of N,N′-di(thiophenylmethanimino)guanidine hydrochloride (3a). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and 2-thiophenecarboxaldehyde (2 eq.) yielded compound 3a as an orange powder with a 34% yield. 1H NMR (500 MHz, DMSO-d6) δ 12.29 (s, 2H), 8.65 (s, 2H), 8.24 (s, 2H), 7.79 (d, J = 5.0 Hz, 2H), 7.62 (dd, J = 3.6, 0.9 Hz, 2H), 7.19 (dd, J = 5.0, 3.6 Hz, 2H), 3.40 (s, 2H). 13C NMR (126 MHz, DMSO-d6) δ 152.6, 144.5, 137.8, 132.7, 130.8, 128.4. HRMS: m/z [M + H]+ calculated for C11H12N5S2, 278.0529; found 278.0530.
Synthesis of N,N′-di(imidazolemethan-2-imino)guanidine hydrochloride (3b). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and imidazole-2-carboxaldehyde (2 eq.) yielded compound 3b as a beige powder with a 15% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.53 (s, 4H), 7.97 (s, 2H), 7.38 (s, 2H), 7.26 (s, 4H). 13C NMR (176 MHz, DMSO-d6) δ 144.3, 141.8, 133.8, 123.8. HRMS: m/z [M + H]+ calculated for C9H12N9, 246.1210; found 246.1220.
Synthesis of N,N′-di(pyridinylmethan-2-imino)guanidine hydrochloride (3c). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and pyridine-2-carboxaldehyde (2 eq.) yielded compound 3c as a yellow powder with a 36% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.73 (s, 2H), 8.75 (s, 2H), 8.67–8.66 (d, J = 4 Hz, 2H), 8.52 (s, 2H), 8.48–8.40 (d, J = 8 Hz, 2H), 8.00–7.96 (t, J = 7.2 Hz, 2H), 7.52–7.49 (t, J = 4.8 Hz, 2H). 13C NMR (126 MHz, DMSO-d6) δ 152.5, 149.7, 137.8, 125.5, 121.9. HRMS: m/z [M + H]+ calculated for C13H14N7, 268.1305; found 268.1315.
Synthesis of N,N′-di(pyrole-2-methanimino)guanidine hydrochloride (3d). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and pyrole-2-carboxaldehyde (2 eq.) yielded compound 3d as a khaki powder with a 71% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.69 (s, 2H), 11.80 (s, 2H), 8.24 (s, 4H), 7.05 (s, 2H), 6.52 (s, 2H), 6.15–6.14 (m, 2H). 13C NMR (126 MHz, DMSO-d6) δ 152.8, 139.7, 127.5, 123.0, 114.2, 109.8. HRMS: m/z [M + H]+ calculated for C11H14N7, 244.1305; found 244.1315.
Synthesis of N,N′-di(indole-5-methanimino)guanidine hydrochloride (3e). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and indole-5-carboxaldehyde (2 eq.) yielded compound 3e as a pink powder with a 68% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.04 (s, 2H), 11.49 (s, 2H), 8.48 (s, 2H), 8.32 (s, 2H), 8.03 (s, 2H), 7.82–7.80 (d, J = 8.5 Hz, 2H), 7.50 (s, 2H), 7.48 (s, 2H), 7.44 (s, 2H), 6.54 (s, 2H). 13C NMR (126 MHz, DMSO-d6) δ 131.0, 127.1, 122.3, 120.7, 112.6, 102.5. HRMS: m/z [M + H]+ calculated for C19H18N7, 344.1618; found 344.1629.
Synthesis of N,N′-di(pyrimidine-5-methanimino)guanidine hydrochloride (3f). Following the general procedure, N,N′-diaminoguanidine hydrochloride (1 eq.) and 5-formylpyrimidine (2 eq.) yielded compound 3f as a yellow powder with a 53% yield. 1H NMR (400 MHz, DMSO-d6) δ 12.70 (s, 2H), 9.37 (s, 4H), 9.26 (s, 2H), 8.77 (s, 2H), 8.50 (s, 2H). 13C NMR (126 MHz, DMSO-d6) δ 159.4, 156.3, 143.6, 128.3. HRMS: m/z [M + H]+ calculated for C11H12N9, 270.121; found 270.1220.
Biological assays
Author contributions
A. R. S. and S. L. for conceptualization of the project. S. L. for investigation and synthesis of the compounds and biological studies. S. L. for the original draft preparation. S. L. and A. R. S. for writing, review and editing. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI).Supplementary information: 1H and 13C NMR spectra, mass spectrometry, purity, biological tests and IC50 determination, selectivity indexes on PANC1 cancer cells, excitation and emission spectra of diiminoguanidines, stability assays, confocal microscopy images. See DOI: https://doi.org/10.1039/d5md00808e.
Acknowledgements
This research was funded by Natural Sciences and Engineering Research Council, grant number RGPIN-2021-03128. We thank G. Ferbeyre from the Département de Biochimie et de Médecine moléculaire, S. Cecioni from the Département de Chimie and the Plateforme de microcopie de l'Université de Montréal for access to their laboratories and instruments. Access to the NMR via the Regional Centre for Magnetic Resonance (UdeM – Chemistry) was possible due to funding from the Canada Foundation for Innovation and the Institute Courtois.Notes and references
- C. J. Cabasag, J. Ferlay, M. Laversanne, J. Vignat, A. Weber, I. Soerjomataram and F. Bray, Gut, 2022, 71(8), 1686–1687 Search PubMed.
- C. J. Halbrook, C. A. Lyssiotis, M. Pasca di Magliano and A. Maitra, Cell, 2023, 186(8), 1729–1754 CrossRef.
- L. Rahib, M. R. Wehner, L. M. Matrisian and K. T. Nead, JAMA Netw. Open, 2021, 4(4), e214708–e214708 CrossRef PubMed.
- L. Rahib, B. D. Smith, R. Aizenberg, A. B. Rosenzweig, J. M. Fleshman and L. M. Matrisian, Cancer Res., 2014, 74(11), 2913–2921 CrossRef CAS PubMed.
- A. P. Klein, Nat. Rev. Gastroenterol. Hepatol., 2021, 18(7), 493–502 CrossRef.
- E. E. Montalvo-Javé, N. Nuño-Lámbarri, G. N. López-Sánchez, E. A. Ayala-Moreno, G. Gutierrez-Reyes, J. Beane and T. M. Pawlik, J. Gastrointest. Surg., 2023, 27(5), 1001–1010 CrossRef PubMed.
- R. Pothuraju, S. Rachagani, W. M. Junker, S. Chaudhary, V. Saraswathi, S. Kaur and S. K. Batra, J. Exp. Clin. Cancer Res., 2018, 37(1), 319 CrossRef.
- J. Zheng, M. A. Guinter, A. T. Merchant, M. D. Wirth, J. Zhang, R. Z. Stolzenberg-Solomon and S. E. Steck, Nutr. Rev., 2017, 75(11), 883–908 CrossRef PubMed.
- T. J. Grant, K. Hua and A. Singh, Prog. Mol. Biol. Transl. Sci., 2016, 144, 241–275 Search PubMed.
- S. E. Kern, R. H. Hruban, M. Hidalgo and C. J. Yeo, Cancer Biol. Ther., 2002, 1(6), 607–613 CrossRef PubMed.
- G. Z. Murimwa, J. D. Karalis, J. Meier, M. Nehrubabu, M. Thornton, M. Porembka, S. Wang, H. J. Zeh, A. C. Yopp and P. M. Polanco, J. Surg. Oncol., 2023, 128(4), 540–548 CrossRef PubMed.
- C. W. Choi, I. K. Choi, J. H. Seo, B. S. Kim, J. S. Kim, C. D. Kim, S. H. Um, J. S. Kim and Y. H. Kim, Am. J. Clin. Oncol., 2000, 23(4), 425–428 CrossRef PubMed.
- D. B. Longley, D. P. Harkin and P. G. Johnston, Nat. Rev. Cancer, 2003, 3(5), 330–338 CrossRef PubMed.
- W. B. Wang, Y. Yang, Y. P. Zhao, T. P. Zhang, Q. Liao and H. Shu, World J. Gastroenterol., 2014, 20(42), 15682–15690 CrossRef PubMed.
- N. S. Siddiqui, A. Godara, M. M. Byrne and M. W. Saif, Expert Opin. Pharmacother., 2019, 20(4), 399–409 CrossRef PubMed.
- D. B. Smith and J. P. Neoptolemos, Core Evidence, 2007, 2(2), 111–119 Search PubMed.
- T. H. Cartwright, A. Cohn, J. A. Varkey, Y.-M. Chen, T. P. Szatrowski, J. V. Cox and J. J. Schulz, J. Clin. Oncol., 2002, 20(1), 160–164 CrossRef PubMed.
- M. K. Chllamma, N. Cook, N. C. Dhani, K. Giby, A. Dodd, L. Wang, D. W. Hedley, M. J. Moore and J. J. Knox, Br. J. Cancer, 2016, 115(6), 649–654 Search PubMed.
- C. Nitipir, R. Vrabie, A. I. Parosanu, R. Tulin, B. Cretu, A. Cursaru, I. Slavu, A. Miron, V. Calu and M. C. Orlov Slavu, Cureus, 2021, 13(11), e19361 Search PubMed.
- H. L. Kindler, American Society of Clinical Oncology Educational Book, 2012, vol. 32, pp. 232–237 Search PubMed.
- D.-C. Chi, F. Brogan, I. Turenne, S. Zelonis, L. Schwartz and M. W. Saif, Anticancer Res., 2012, 32(9), 4147 Search PubMed.
- H. A. Burris, M. J. Moore, J. Andersen, M. R. Green, M. L. Rothenberg, M. R. Modiano, M. C. Cripps, R. K. Portenoy, A. M. Storniolo, P. Tarassoff, R. Nelson, F. A. Dorr, C. D. Stephens and D. D. V. Hoff, J. Clin. Oncol., 2023, 41(36), 5482–5492 Search PubMed.
- Y. J. Min, K. R. Joo, N. H. Park, T. K. Yun, Y. W. Nah, C. W. Nam and J. H. Park, Korean J. Intern. Med., 2002, 17(4), 259–262 Search PubMed.
- W. Plunkett, P. Huang, Y. Z. Xu, V. Heinemann, R. Grunewald and V. Gandhi, Semin. Oncol., 1995, 22(4), 3–10 Search PubMed.
- B. Hryciuk, B. Szymanowski, A. Romanowska, E. Salt, B. Wasąg, B. Grala, J. Jassem and R. Duchnowska, Oncol. Lett., 2018, 15(2), 1912–1916 Search PubMed.
- J. S. Armstrong, Br. J. Pharmacol., 2007, 151(8), 1154–1165 CrossRef PubMed.
- C. Wang and R. J. Youle, Annu. Rev. Genet., 2009, 43, 95–118 CrossRef PubMed.
- F. Fontanesi, Mitochondria: Structure and Role in Respiration, eLS, pp. 1–13 Search PubMed.
- H. Wang, B. Fang, B. Peng, L. Wang, Y. Xue, H. Bai, S. Lu, N. H. Voelcker, L. Li, L. Fu and W. Huang, Front. Chem., 2021, 9, 683220 CrossRef PubMed.
- F. J. Bock and S. W. G. Tait, Nat. Rev. Mol. Cell Biol., 2020, 21(2), 85–100 Search PubMed.
- T. T. Nguyen, S. Wei, T. H. Nguyen, Y. Jo, Y. Zhang, W. Park, K. Gariani, C.-M. Oh, H. H. Kim, K.-T. Ha, K. S. Park, R. Park, I.-K. Lee, M. Shong, R. H. Houtkooper and D. Ryu, Exp. Mol. Med., 2023, 55(8), 1595–1619 CrossRef PubMed.
- D. R. Green and J. C. Reed, Science, 1998, 281(5381), 1309–1312 CrossRef PubMed.
- A. Hébert, M. Parisotto, G. Ferbeyre and A. R. Schmitzer, Supramol. Chem., 2019, 31(3), 127–139 CrossRef.
- A. Hébert, M. Parisotto, M.-C. Rowell, A. Doré, A. Fernandez Ruiz, G. Lefrançois, P. Kalegari, G. Ferbeyre and A. R. Schmitzer, Sci. Rep., 2021, 11(1), 9854 CrossRef PubMed.
- J. Vatté, V. Bourdeau, G. Ferbeyre and A. R. Schmitzer, Molecules, 2024, 29(16), 3773–3786 CrossRef PubMed.
- J. Vatté, V. Bourdeau, G. Ferbeyre and A. R. Schmitzer, Molecules, 2024, 29(22), 5329–5355 CrossRef PubMed.
- B. Juliana de Oliveira Carneiro, F. Tanos Celmar Costa, R. L. Steven and V. José Daniel Figueroa, Mini-Rev. Med. Chem., 2020, 20(5), 342–368 CrossRef.
| This journal is © The Royal Society of Chemistry 2026 |