Novel pyrido[2,3-b][1,4]oxazine-based EGFR-TK inhibitors: rational design and synthesis of potent and selective agents targeting resistance mutations in non-small cell lung cancer
Vaibhav B.
Yadav
a,
Shailee V.
Tiwari
b,
S. Hemant
Kumar
cd and
Santosh R.
Deshmukh
 *a
*a
aDepartment of Chemistry, Ahmednagar College, Ahilyanagar (Affiliated to S.P. Pune University), Maharashtra 414001, India. E-mail: sant.deshmukh@gmail.com
bDepartment of Pharmaceutical Chemistry, Shri Ramkrishna Paramhans College of Pharmacy, Hasnapur, Parbhani, Maharashtra 431401, India
cResearch Scholar, Manipal Academy of Higher Education, Manipal, India
dInstitute of Bioinformatics and Applied Biotechnology, Biotech Park, Electronic City, Phase I, Bengaluru-560100, India
First published on 14th November 2025
Abstract
Several first-, second-, and third-generation EGFR-TKIs have proven effective as anti-cancer therapeutics. However, the rapid development of drug resistance and mutations continues to be a major challenge in EGFR-TKI therapy. Addressing both intrinsic and acquired resistance resulting from EGFR mutations requires further exploration and the identification of novel inhibitors. In this study, we identified a new class of pyrido[2,3-b][1,4]oxazine-based inhibitors that exhibited potent EGFR kinase inhibitory activity. These compounds demonstrated significant anti-proliferative effects against EGFR-mutated non-small cell lung cancer (NSCLC) cell lines, including HCC827 (EGFR exon 19 deletion), H1975 (EGFR L858R/T790M double mutation), and A549 (wild-type EGFR overexpression). These novel pyrido[2,3-b][1,4]oxazine analogues were rationally designed and synthesized using the Suzuki cross-coupling reaction in a multi-step synthetic route. Anticancer evaluation of these derivatives using the MTT assay showed promising therapeutic potential. The most promising compounds were 7f, 7g, and 7h, with 7f showing potency (IC50 values: 0.09, 0.89, and 1.10 μM, in the HCC827, NCI-H1975 and A-549 cell lines, respectively) equivalent to clinically approved osimertinib. Interestingly, these compounds are selectively cytotoxic against cancer cells while not harming normal BEAS-2B cells at doses over 61 μM. Mechanistic studies demonstrated that compound 7f acts as an EGFR-TK autophosphorylation inhibitor, causing significant apoptosis (33.7% early and 9.1% late) compared to control conditions (2.4% early and 1.8% late). Molecular docking showed that the compounds scored similar to osimertinib, with the di-fluorophenyl group engaging the glycine-rich loop, pyridine substituents forming front pocket interactions, and essential hinge region interactions maintained, suggesting effective EGFR target engagement. These findings identify pyrido[2,3-b][1,4]oxazine derivatives as potential anticancer candidates worth further exploration for the development of targeted therapies against non-small cell lung cancer.
1. Introduction
Cancer causes 16.8% of global deaths.1 In 2022, there were over 20 million new cancer cases and 9.7 million deaths, with a lifetime incidence rate of 20% and mortality rates of 11.1% for men and 8.3% for women. Lung carcinoma had the highest incidence (2.5 million cases, 12.4% of total diagnoses) and mortality (1.8 million deaths, 18.7% of cancer-related deaths), followed by breast, colorectal, prostate, and stomach cancers. Females are most likely to have breast cancer, while males are more likely to develop lung cancer. Demographical predictions predict a 77% increase in annual cancer incidence by 2050, with 35 million more cases.2 The rising number of cancer diagnoses and deaths globally has prompted the development of efficient cancer treatments.The epidermal growth factor receptor (EGFR), a member of the ErbB family of tyrosine kinases, is a well-known therapeutic target in non-small cell lung cancer (NSCLC). EGFR inhibition has become a key strategy in NSCLC treatment, leading to the development of first-generation tyrosine kinase inhibitors (TKIs) such as gefitinib and erlotinib.3,4 These agents showed high initial efficacy in patients with EGFR-mutant NSCLC. However, resistance typically emerges within 6–12 months in approximately 60% of cases, significantly limiting long-term clinical benefits.5,6 To address this limitation, second-generation EGFR-TKIs such as afatinib, neratinib, and dacomitinib were developed, offering improved potency and broader antitumor activity. However, their effectiveness is frequently diminished by development of resistance when targeting the T790M mutation, and inadequate selectivity for WT-EGFR continues to be a major problem during treatment, resulting in serious side effects from the second-generation EGFR-TKIs.7 Third-generation EGFR-TKIs were subsequently designed to overcome this resistance. Among all third-generation EGFR-TKIs, including nazartinib, almonertinib, avitinib, rociletinib, maverlertinib, naquotinib, lazertinib and olmutinib, only osimertinib selectively and irreversibly targets EGFR-activating mutations, including T790M, while exhibiting reduced affinity for wild-type EGFR.7 It demonstrates greater efficacy than first-generation TKIs and chemotherapy in patients with both treatment-naive and T790M-positive NSCLC. Nevertheless, resistance to osimertinib also arises, most commonly through the C797S mutation, which disrupts covalent binding at the C797 residue, thereby significantly reducing drug efficacy.8–10
Given these challenges, there is an urgent need to develop novel, potent, and selective EGFR inhibitors capable of targeting resistant mutations such as T790M and L858R, including C797S. Additionally, dual-effective inhibitors that can target both wild-type and mutant EGFR (T790M/L858R) forms may offer promising therapeutic alternatives. The goal of this study is to discover novel EGFR-TK inhibitors by synthesizing 7-(4-pyrimidyl)-2,3-dihydro-1-phenylsulphonyl-1H-pyrido[2,3-b][1,4]oxazine hybrids (7) using a molecular hybridization approach.
Heterocyclic moieties are fundamental pharmacophores in many clinically approved pharmaceuticals, demonstrating their value for developing pharmacologically active molecules.11–13 About 70% of drugs are heterocyclic. Many bicyclic heterocycles discovered in recent decades have clinical approval and strong pharmacological efficacy.14,15
The pyrimidine pharmacophore, usually combined with functional groups like sulfonamide, indole, furan, and tetrahydrofuran, provides the structural basis for several small-molecule EGFR inhibitors, including lapatinib, osimertinib, mobocertinib, and afatinib.16,17 Sulfonamide derivatives have antifungal, antibacterial, anti-inflammatory, anti-oxidant, antidiabetic, and anticancer properties, making them a valuable medicinal chemistry scaffold. Belinostat, the third FDA-approved medication for T-cell lymphoma after vorinostat and romidepsin, is a sulfonamide-based HDAC inhibitor.18 Furthermore, sulfonamide-containing compounds inhibit EGFR, reducing cancer cell proliferation and viability.19–21 Hybrid drug candidates containing pyrimidine and sulfonamide moieties have improved therapeutic effectiveness due to their antibacterial, anticancer, and antiviral characteristics.22 As shown in Fig. 1, enantioenriched 3,4-dihydro-2H-1,4-benzoxazines represent important N,O-heterocyclic scaffolds found in natural products, pharmaceuticals, and bioactive compounds.23–25
 | ||
| Fig. 1 Previously reported bioactive compounds containing the 3,4-dihydro-2H-1,4-benzoxazine moiety. | ||
PI3K inhibitors thieno[2,3-d]pyrimidine and thiazolo[5,4-d]pyrimidine derivatives have nanomolar potency, notable antiproliferative activity against cancer cell lines, favourable drug-like properties, and significant in vivo anticancer efficacy in preclinical models.26 To build on these previous findings and optimize lead compound attributes, we designed a number of novel compounds with systematic structural alterations to the existing pharmacophore with the goal of understanding structure–activity relationships (SARs). Fig. 2 depicts our rational design strategy, which included four major structural changes. To minimize rotational flexibility and increase binding affinity, conformational restriction was accomplished by intramolecular cyclisation between the sulfonamide nitrogen and the methoxy carbon of the pyridine moiety. The heterocyclic core was simplified by substituting the thieno[2,3-d]pyrimidine scaffold with a pyrimidine moiety. Fluorine substituents were strategically repositioned from the 2,4-disubstitution pattern to 2,5-positions on the phenyl ring to optimize electronic and steric interactions, and a trifluoromethyl group was added to the pyrimidine ring as an additional pharmacophoric element to improve metabolic stability, lipophilicity, and target selectivity. The C-2 position of the pyrimidine ring is an important position for the SAR study, encouraging systematic substitution with various aliphatic, heterocyclic, and cyclic amine moieties to optimize inhibitory activity.
 | ||
| Fig. 2 Design of novel compounds with systematic structural modifications on the thieno[2,3-d]pyrimidine scaffold. | ||
To improve therapeutic efficacy, three unique bioactive pharmacophores were strategically incorporated into a single molecular scaffold: pyrido[2,3-b][1,4]oxazine, phenyl sulfonamide, and pyrimidine. This molecular hybridization strategy sought to increase anticancer potency by combining the beneficial characteristics of various pharmacophores into a single molecular scaffold.
Using the standard MTT assay, the synthesized hybrid compounds were tested for anticancer activity against HCC827 (EGFR exon 19 deletion mutation), H1975 (EGFR L858R/T790M double mutation), and A549 (WT-EGFR overexpression) cell lines. The three most potent derivatives were then tested for cytotoxicity against BEAS-2B normal lung cells to assess selectivity. Western blot analysis was carried out to determine the lead compound's potential to inhibit EGFR-TK autophosphorylation and downstream signaling pathways in HCC827 cells. In addition, molecular docking experiments were performed on all synthesized hybrids within the EGFR active site to understand their binding interactions and confirm their potential as EGFR inhibitors.
To our knowledge, this present work is the first complete effort in the rational design, synthesis, and anticancer evaluation of novel pyrido[2,3-b][1,4]oxazine derivatives, therefore introducing a new structural class for potential anticancer agents.
2. Results and discussion
2.1 Chemistry
Scheme 1 describes the five-step synthesis of target compounds 4b, 7, and 8 with pyrido[2,3-b][1,4]oxazine, pyrimidine, and sulfonamide pharmacophores. Three bioactive motifs were systematically incorporated into a molecular scaffold using a multi-component synthesis approach to evaluate their synergistic anticancer effects. The reaction of 7-bromo-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazine (1) with 2,5-difluorobenzenesulfonyl chloride (1a) in the presence of pyridine and DCM allowed for the planned incorporation of pyrido-oxazine and sulfonamide pharmacophores, producing the corresponding sulfonamide derivative (2) in 54.9% yield.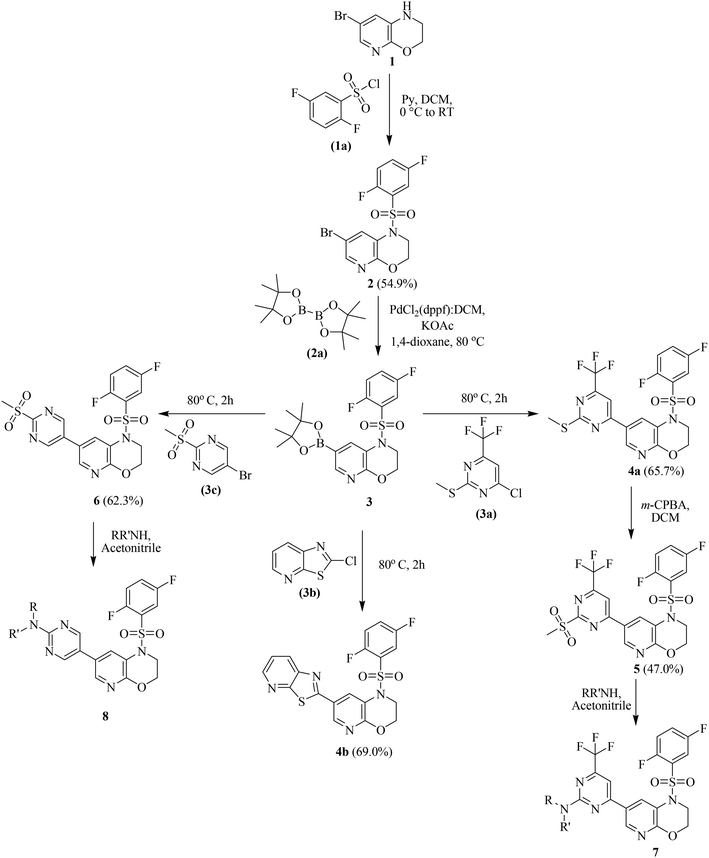 | ||
| Scheme 1 Synthetic route for the preparation of compounds 4b, 7, and 8 from 1 (detailed experimental procedures and characterization data are provided in section 4.1). | ||
The Miyaura borylation of derivative 2 was accomplished through cross-coupling with bis(pinacolato)diboron (2a) using Pd(dppf)Cl2 in DCM as the catalyst, potassium acetate as the base, and 1,4-dioxane as the solvent under an inert atmosphere at 80 °C for 2 hours, yielding the intermediate boronate (3). Thereafter, the in situ addition of heterocyclic halides, followed by continuous heating at 80 °C for 2 hours, facilitates Suzuki cross-coupling with the boronate intermediate (3). Compound 4a was obtained in 65.7% yield by Suzuki cross-coupling of boronate (3) with 4-chloro-2-(methylthio)-6-(trifluoromethyl)pyrimidine (3a). In order to evaluate the impact of the position of pyrimidine nitrogen atoms and the lack of a trifluoromethyl group on anticancer activity in the SAR study, boronate (3) was coupled with 5-bromo-2-(methylsulfonyl)pyrimidine (3c) under the same conditions to give derivative 6 in 52.6% yield. Based on our recent findings revealing the anticancer potential of thiazolo[5,4-b]pyridine-containing derivatives,24 compound 4b was designed to incorporate this heterocyclic scaffold for anticancer evaluation. Compound 4b was obtained in 69% yield by Suzuki cross-coupling of boronate (3) with 2-chlorothiazolo[5,4-b]pyridine (3b).
Furthermore, the S-methyl group in derivative 4a was oxidized to the corresponding methylsulfone 5 (47%) using m-chloroperbenzoic acid in DCM for 18 hours with stirring from 0 °C to room temperature.
Previous research has demonstrated that cyclic amines directly linked to the C-4 position of thieno[2,3-d]pyrimidine scaffolds have powerful anticancer properties.28 Based on these findings, we hypothesized that incorporating cyclic amines to the C-2 position of pyrimidine in our scaffold would increase conformational flexibility and potentially alter anticancer efficacy, encouraging the synthesis of substituted-C-2 analogues for the investigation of the structure–activity connection. In the final synthetic step, target compounds 7a–k and 8a–b were synthesized in good to excellent yields via nucleophilic aromatic substitution reactions of sulfones 5 and 6 with different cyclic amines under reflux conditions in acetonitrile (Scheme 1, Fig. 3).
The 7k compound was synthesized from sulfone 5 in two steps (Scheme 2). In the first stage, N-Boc-protected amine 9a was combined with compound 5 to produce the N-Boc-protected intermediate 9 in 64% yield. The subsequent de-protection of 9 under acidic conditions resulted in compound 7k in 58.8% yield.
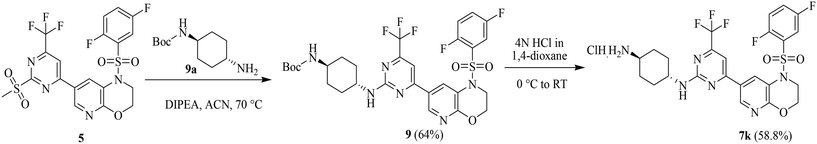 | ||
| Scheme 2 Synthetic route for the preparation of 7k from 5 (detailed experimental procedures and characterization data are provided in section 4.1). | ||
These hybrid compounds 4a–b, 5, 6, 7a–k, and 8a–b were investigated for anticancer efficacy against HCC827, NCI-H1975, and A549 cancer cell lines using the MTT assay. The three most effective compounds (7f, 7g, and 7h) were subsequently evaluated for cytotoxicity against BEAS-2B normal lung cells to assess selectivity. The lead compound 7f was tested for its inhibitory effects on EGFR-TK autophosphorylation and downstream signalling pathways in HCC827 cells by the western blot assay, as well as its early or late apoptotic pathway induction. The EGFR active site binding interactions and inhibitory characteristics of all synthesised compounds were examined using molecular docking.
2.2 Biological evaluations
| Entry | Compounds | IC50 (μM) | |||
|---|---|---|---|---|---|
| HCC827 | NCI-H1975 | A-549 | BEAS-2B | ||
| 1 | 4a | 4.11 ± 0.09 | 5.11 ± 0.26 | 5.43 ± 0.08 | |
| 2 | 4b | 12.17 ± 0.11 | 10.46 ± 0.18 | 12.66 ± 0.41 | |
| 3 | 5 | 4.18 ± 0.08 | 5.03 ± 0.25 | 5.35 ± 0.26 | |
| 4 | 6 | 6.33 ± 0.22 | 8.27 ± 0.30 | 9.14 ± 0.14 | |
| 5 | 8a | 10.21 ± 0.20 | 8.64 ± 0.36 | 10.36 ± 0.42 | |
| 6 | 8b | 10.63 ± 0.14 | 8.90 ± 0.24 | 10.44 ± 0.39 | |
| 7 | 7a | 2.90 ± 0.07 | 2.81 ± 0.12 | 3.80 ± 0.35 | |
| 8 | 7b | 2.99 ± 0.18 | 3.07 ± 0.37 | 3.93 ± 0.10 | |
| 9 | 7c | 2.18 ± 0.25 | 2.40 ± 0.31 | 3.79 ± 0.07 | |
| 10 | 7d | 1.78 ± 0.33 | 2.32 ± 0.14 | 3.44 ± 0.16 | |
| 11 | 7e | 0.36 ± 0.12 | 1.46 ± 0.20 | 2.85 ± 0.09 | |
| 12 | 7f | 0.09 ± 0.16 | 0.89 ± 0.08 | 1.10 ± 0.28 | 61 ± 0.24 |
| 13 | 7g | 0.32 ± 0.24 | 1.41 ± 0.10 | 2.28 ± 0.34 | 52 ± 0.10 |
| 14 | 7h | 0.12 ± 0.26 | 1.04 ± 0.17 | 2.01 ± 0.25 | 58 ± 0.19 |
| 15 | 7i | 1.59 ± 0.19 | 2.18 ± 0.09 | 3.25 ± 0.11 | |
| 16 | 7j | 2.28 ± 0.10 | 2.55 ± 0.07 | 3.87 ± 0.12 | |
| 17 | 7k | 1.11 ± 0.06 | 2.01 ± 0.18 | 3.19 ± 0.20 | |
| 18 | Osimertinib | 0.007 ± 0.07 | 0.088 | 0.76 ± 0.11 | |
Three compounds (7f, 7g, and 7h) exhibited the most effective cytotoxic effects against the chosen cancer cell lines. Compound 7f showed superior anticancer activity against HCC827, H1975, and A549 cell lines, with IC50 values of 0.09 μM, 0.89 μM, and 1.10 μM, respectively, when compared to all other synthesized derivatives.
The in vitro anticancer activity data enabled the clarification of the structure–activity relationship for the pyrido[2,3-b][1,4]oxazine derivatives. Thorough SAR analysis indicated that optimal cytotoxic activity depended on several important structural characteristics: the pyrido-oxazine ring system as the primary scaffold, a pyrimidine moiety, an amine functional group, a trifluoromethyl substituent on the pyrimidine ring, and a difluorophenyl sulfonyl group, all of which collectively enhanced the anticancer efficacy of the most potent compounds.
SAR analysis gave deeper insights, demonstrating that derivatives containing amino groups have higher anticancer activity. Compounds 7a–k had an amino group in their molecular structures and had much better anticancer efficacy than the other synthesized derivatives that lacked this functional group (4a, 4b, 5, and 6). The absence of a pyrimidine ring in compound 4b, as well as its low activity against the tested cancer cell lines, demonstrates that the presence of a pyrimidine ring is essential to increase anticancer activity. Compounds 4a, 5, and 7a–k were synthesized with a trifluoromethyl (–CF3) group connected to the pyrimidine ring and showed higher anticancer activity than compounds without this –CF3 substitution (6, 8a, and 8b). This SAR demonstrates that the –CF3 group is required for maximum anticancer effects.
The presence of a free “NH” group in the structure may promote interactions with the target enzyme. Compounds with an unsubstituted “NH” group (7c, 7d, 7e, 7f, 7g, 7h, 7i, 7j, and 7k) have considerably higher anticancer activity than compounds without this free “NH” functionality (4a, 4b, 5, 6, 7a, 7b, 8a, and 8b). This finding indicates that the free “NH” group is significant in the binding affinity and subsequent biological activity of these molecules.
Compounds with six-membered cyclic rings (7a–d and 7i–k) in amines have a negative impact on anticancer activity. In contrast, compounds with a five-membered cyclic ring (7e, 7g and 7h) or an aliphatic group substitution (7f) on the “NH” group showed promising anticancer potential. This suggests that smaller ring systems with aliphatic substituents have the best steric and electronic features for increased anticancer activity.
To assess the cytotoxic effects of the synthesized compounds on non-malignant cells, the BEAS-2B normal lung epithelial cell line was used. The findings in Table 1 show that the most effective compounds have superior selectivity for cancer cell lines over normal cells. The highly potent anticancer compounds 7f, 7g, and 7h demonstrated exceptional selectivity for cancer cells, with no cytotoxicity seen against the BEAS-2B cell line at dosages up to 61 μM. Compound 7f exhibited notable selectivity, being approximately 680-fold, 70-fold, and 55-fold more selective towards HCC827, H1975, and A549 cancer cell lines, respectively, compared to the BEAS-2B normal lung cell line (Fig. 4). This selective anticancer profile is a very desirable therapeutic feature, indicating that these compounds can preferentially target cancer cells while protecting the vitality of normal cells, reducing possible off-target effects.
The p-EGFR and p-Akt bands (top and third panels) show a dose-dependent decrease in phosphorylation intensity compared to the DMSO-treated control, indicating inhibition of EGFR activation and suppression of the downstream Akt pathway. Reduction in p-EGFR and p-Akt levels with increasing concentrations of compound 7f confirms the effective blockade of EGFR signaling in a concentration-dependent manner.
Compound 7f treatment caused a significant increase in apoptotic cell death, with 33.7% of cells suffering early apoptosis and 9.1% displaying late apoptosis, compared to control conditions, which exhibited negligible apoptotic activity (2.4% early apoptosis and 1.8% late apoptosis). As shown in Fig. 6, the significant increase in both early and late apoptotic populations demonstrates that compound 7f effectively activates programmed cell death pathways, exhibiting strong pro-apoptotic activity against HCC827 cancer cells.
In the current series, the hinge-binding motif is composed of a fused pyrido[2,3-b][1,4]oxazine scaffold with dual hydrogen bond acceptor functionalities. As a result, EGFR complexes with structurally suitable hinge binders were chosen to generate the docking grid. The co-crystallized ligands in EGFR structures (PDB IDs: 8sc7,29 4jq7,30 4jq8,30 4jr3,30 and 4jrv30) have no hydrogen bond donor groups in their core scaffolds for hinge interactions, making them appropriate for ensemble docking grid creation with the present compound series. This docking grid was used to understand the interactions of our ligands with the WT-EGFR. Additionally, an ensemble docking grid was generated for the double mutant variant of EGFR (T790M/L858R).
On the EGFR-WT docking grid, osimertinib had a docking score of −24 and effectively reproduced the binding mode shown in its XRD structure with EGFR (PDB ID: 4zau31). Table 2 shows that the fused pyrido[2,3-b][1,4]oxazine derivatives had a greater docking score, with an average docking score of −25. As shown in Fig. 7, the pyridine nitrogen of the fused core made a hinge connection with Met793, while Lys745 made a cation–π interaction with the difluorophenyl moiety and was within hydrogen bonding distance with the sulfone group in some cases. In the majority of compounds, the pyrido[2,3-b][1,4]oxazine oxygen was close enough to the gatekeeper residue Thr790 to form a weak hydrogen bond (3.1 Å to 3.5 Å). Three compounds (4b, 8a, and 8b) did not attain favourable docking conformations, most likely due to steric conflicts between their substituents and a lack of appropriate receptor conformations in the ensemble grid.
| Entry | Comp. | Docking score | Entry | Comp. | Docking score | ||
|---|---|---|---|---|---|---|---|
| WT | T790M/L858R | WT | T790M/L858R | ||||
| 1 | 4a | −30.6 | −26.2 | 10 | 7f | −27.3 | −25.9 |
| 2 | 4b | — | — | 11 | 7g | −20.1 | −24.4 |
| 3 | 5 | −29.4 | −26.2 | 12 | 7h | −27.4 | −25.1 |
| 4 | 6 | −22.8 | — | 13 | 7i | −25.3 | — |
| 5 | 7a | −28.9 | −24.5 | 14 | 7j | −24.4 | −17.5 |
| 6 | 7b | −30.1 | −25.9 | 15 | 7k | −21.3 | −18.7 |
| 7 | 7c | −27.6 | −21.6 | 16 | 8a | — | — |
| 8 | 7d | −26.7 | — | 17 | 8b | — | — |
| 9 | 7e | −28.3 | −24.3 | 18 | Osimertinib | −24 | −24 |
 | ||
| Fig. 7 Docking modes and 2D interaction diagrams of osimertinib, 4a, 7a, 7f, 7g and 7h on the EGFR-WT docking grid. | ||
Osimertinib had a docking score of −24 on the EGFR-double mutant (T790M/L858R) grid as well. Its binding mode was reproduced with a slight deviation from the X-ray structure (PDB ID: 6z4b32) due to the difference in the conformations of the active site residues and the presence of another ligand in the active site of the experimental structure (Fig. 8A). Eleven among the seventeen compounds docked well on this docking grid (Fig. 8). Their docking scores have been captured in Table 2. The pyridine N makes a hinge interaction with the backbone –NH of Met793. The mutation of the gatekeeper residue from threonine to methionine causes the loss of the possibility of a hydrogen bond between the oxazine –O and that of Thr790. However, Met790 along with Ala793 and Leu844 makes strong hydrophobic interactions with the difluoro phenyl moiety.
 | ||
| Fig. 8 Docking modes and 2D interaction diagrams of osimertinib, 4a, 7a, 7f, 7g and 7h on the EGFR-double mutant (T790M/L858R) grid. | ||
The docking modes of the majority of the compounds in both the docking grids are similar to each other with the pyrido[2,3-b][1,4]oxazine core making a hinge interaction and the difluoro phenyl making cation–π or hydrophobic interactions. While the highly inactive compounds did not have favourable docking modes, the trend in docking scores did not reflect the same in that of the cellular IC50.
3. Conclusions
In conclusion, we effectively designed and synthesized novel substituted pyrido[2,3-b][1,4]oxazine analogues using a multi-step synthetic strategy that included the Suzuki cross-coupling reaction. A comprehensive anticancer study using HCC827, H1975, and A549 cancer cell lines demonstrated great therapeutic potential, with compounds 7f, 7g, and 7h emerging as the most promising candidates. The lead compound 7f demonstrated high potency, with IC50 values of 0.09 μM, 0.89 μM, and 1.10 μM against HCC827, H1975, and A549 cancer cell lines, comparable to clinically approved osimertinib. These potent compounds showed excellent selectivity for malignant cells, with negligible cytotoxicity against normal BEAS-2B cells at dosages up to 61 μM, indicating a key therapeutic advantage. Mechanistic investigations revealed that compound 7f functions as an EGFR-TK autophosphorylation inhibitor in HCC827 cells, elucidating its primary mode of action. The apoptosis study of 7f revealed a significant rise in programmed cell death (33.7% early and 9.1% late apoptosis) compared to control conditions (2.4% early and 1.8% late apoptosis). The molecular docking study suggests the binding mode of the novel pyrido[2,3-b][1,4]oxazine derivatives expounded in this work. The osimertinib-alternative compounds have similar docking scores to osimertinib while the cellular IC50 is poor compared to the latter. Since the cellular IC50 is a function of various other factors such as cell permeability, further DMPK studies are needed to understand the activity of these compounds.These findings show that pyrido[2,3-b][1,4]oxazine derivatives are promising candidates for further development as targeted therapies for non-small cell lung cancer and other cancers.
4. Experimental
4.1 Chemistry
All reactions were carried out in an inert nitrogen atmosphere with oven-dried glassware. Before use, solvents were dried and degassed in accordance with standard procedures. Thin-layer chromatography (TLC) was performed on Merck silica gel 60 F254 (0.25 mm thick) plates, and the results were visualised using short (254 nm) and long (365 nm) UV light and KMnO4 stain. Column chromatography was carried out using 230–400 mesh silica gel. The 1H and 13C NMR spectra were taken using 400 and 100 MHz NMR spectrometers, respectively, with CDCl3 or DMSO-d6 as solvents. Chemical shifts (δ) are expressed in ppm relative to a tetramethylsilane (Me4Si) internal reference. Signal multiplicities are denoted as follows: s (singlet), d (doublet), t (triplet), q (quartet), and m (multiplet). High-resolution mass spectra (HRMS) were obtained using a micromass-QTOF spectrometer in electrospray ionisation (ESI) mode and a Thermo Scientific Velos Pro Orbitrap mass spectrometer. The purity of the compounds was determined using ultra-performance liquid chromatography-MS (UPLC–MS) Nexera series-Modular UHPLC/HPLC Systems and the ACQUITY UPLC H-Class PLUS System.Cream solid (2.20 g, 69.0% yield), LC-MS m/z, [M + H]+ = 446.95, 1H NMR (400 MHz, DMSO-d6) δ 8.73 (s, 1H), 8.63 (d, J = 6.8 Hz, 2H), 8.48 (d, J = 8.0 Hz, 1H), 7.89 (m, 1H), 7.74 (m, 1H), 7.65–7.57 (m, 2H), 4.18 (d, J = 4.8 Hz, 2H), 4.04 (d. J = 4 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) δ 163.7, 158.7 (dd, JC–F = 250.5 Hz), 157.3, 154.7, 153.1, 147.6, 146.3, 143.1, 130.3, 128.8, 126.3, 124.2 (dd, JC–F = 34.3, 10.1 Hz), 123.4, 122.3, 120.3 (dd, JC–F = 24.2, 8.1 Hz), 120.1, 117.8 (d, JC–F = 27.3 Hz), 64.8, 43.1. HRMS (ESI): m/z [M + H]+ calcd. for C19H13F2N4O3S2, 447.0397, found 447.0380.
Cream solid, (yield: 62.3%). LC-MS m/z, [M + H]+ = 469.00, 1H NMR (400 MHz, DMSO-d6) δ 9.37 (s, 2H), 8.56 (d, J = 1.6 Hz, 1H), 8.37 (d, J = 2.0 Hz, 1H), 7.99 (d, J = 8.0 Hz, 1H), 7.72 (dd, JH–F = 16.8, 8.8 Hz, 1H), 7.61–7.55 (tt, JH–F = 9.2, 4 Hz, 1H), 4.14–4.02 (dd, J = 4.4 Hz, 4H), 3.45 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 164.8, 156.8, 154.1, 144.0, 132.9, 130.7, 127.1(d, JC–F = 17.4 Hz), 124.3, 123.4, 120.6, 120.4, 120.3, 118.5 (d, JC–F = 28.0 Hz), 104.3, 64.9, 43.7, 34.9. HRMS (ESI): m/z [M + H]+ calcd. for C18H15F2N4O5S2, 469.0452, found 469.0430.
1-((2,5-Difluorophenyl)sulfonyl)-7-(2-(4-methylpiperazin-1-yl)-6-(trifluoromethyl)pyrimidin-4-yl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazine (7a). Cream solid, 42% yield, LC-MS m/z, [M + H]+ = 557.0, 1H NMR (400 MHz, DMSO-d6) δ 8.80 (d, J = 2.0 Hz, 1H), 8.62 (d, J = 2.0 Hz, 1H), 7.81–7.77 (ddd, JH–F = 10.8, 5.2, 3.2 Hz, 1H), 7.70–7.64 (ddd, JH–F = 12, 9.2, 3.6 Hz, 1H), 7.58–7.52 (tt, JH–F = 9.6, 4 Hz, 1H), 7.50 (s, 1H), 4.31 (t, J = 4.4 Hz, 2H), 4.05 (t, J = 4.8 Hz, 2H), 3.87 (s, 4H), 2.53 (s, 4H), 2.32 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 163.7, 160.9, 158.7 (d, JC–F = 281.6 Hz), 156.2, 155.6 (d, JC–F = 252 Hz), 154.6, 143.8, 128.6, 127.0 (dd, JC–F = 17.5, 7.2 Hz), 126.0, 124.1 (dd, JC–F = 24, 8.8 Hz), 122.1 (d, JC–F = 276.3 Hz), 120.3 (dd, JC–F = 24.6, 8.5 Hz), 119.8, 117.3 (d, JC–F = 27.6 Hz), 100.4, 65.0, 53.9, 45.4, 43.3, 43.1. HRMS (ESI): m/z [M + H]+ calcd. for C23H22F5N6O3S, 557.1394, found 558.1434.
1-(4-(4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-6-(trifluoromethyl)pyrimidin-2-yl)piperazin-1-yl)ethan-1-one (7b). Cream solid, 57.4% yield, LC-MS m/z, 25 [M + H]+ = 584.95, 1H NMR (400 MHz, DMSO-d6) δ 8.89 (d, J = 2.4 Hz, 1H), 8.65 (s, 1H), 7.89 (s, 1H), 7.73 (s, 2H), 7.67 (s, 1H), 4.24 (s, 2H), 4.05 (s, 2H), 3.89 (s, 2H), 3.81 (s, 2H), 3.60 (s, 4H), 2.07 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 168.5, 163.8, 161.0, 158.7 (d, JC–F = 284.4 Hz), 156.2, 155.6 (d, JC–F = 254.2 Hz), 155.0, 143.8, 128.7, 126.9 (dd, JC–F = 17.4, 7.4 Hz), 126.0, 124.1 (dd, JC–F = 24.1, 9.2 Hz),122.1 (d, JC–F = 276.2 Hz), 120.3 (dd, JC–F = 24.4, 8.3 Hz), 119.8, 117.5 (d, JC–F = 27.6 Hz), 100.6, 64.9, 45.1, 43.6, 43.3, 21.3. HRMS (ESI): m/z [M + Na]+ calcd. for C24H21F5N6O4SNa, 607.1163, found 607.1168.
4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-N-(tetrahydro-2H-pyran-4-yl)-6-(trifluoromethyl)pyrimidin-2-amine (7c). Cream solid, 84.3% yield, LC-MS m/z, [M + H]+ = 557.90, 1H NMR (400 MHz, DMSO-d6) δ 8.77 (d, J = 4.8 Hz, 2H), 7.81 (bs, 1H), 7.71–7.62 (m, 2H), 7.55–7.50 (tt, JH–F = 9.2, 4 Hz, 1H), 7.45 (s, 1H), 4.26 (t, J = 4.4 Hz, 2H), 4.02 (t, J = 4.4 Hz, 3H), 3.91 (d, J = 11.6 Hz, 2H), 3.47–3.42 (m, 2H), 1.93 (d, J = 11.6 Hz, 2H), 1.67–1.57 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 163.5, 161.4, 158.6, 156.2, 155.6 (d, JC–F = 252.1 Hz), 154.6, 143.6, 129.2, 126.2, 124.1 (d, JC–F = 23.8 Hz), 122.2 (d, JC–F = 276.7 Hz), 120.3 (m), 119.7, 117.5 (d, JC–F = 27.7 Hz), 99.9, 66.0, 64.6, 47.5, 43.3, 32.0. HRMS (ESI): m/z [M + Na]+ calcd. for C23H20F5N5O4SNa, 580.1054, found 580.1053.
4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-N-(1-methylpiperidin-4-yl)-6-(trifluoromethyl)pyrimidin-2-amine (7d). Cream solid, 58.8% yield, LC-MS m/z, [M + H]+ = 571.00, 1H NMR (400 MHz, DMSO-d6) δ 8.79–8.68 (d, 2H), 7.81–7.76 (m, 2H), 7.65 (bs, 1H), 7.53 (bs, 1H), 7.46 (s, 1H), 4.25 (m, 2H), 4.02–3.97 (m, 3H), 3.10 (d, J = 10.0 Hz, 2H), 2.61 (t, 2H), 2.17 (d, J = 7.6 Hz, 2H), 1.81 (d, J = 10.0 Hz, 2H), 1.25 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 163.6, 162.3, 161.5, 158.6 (d, JC–F = 247.6 Hz), 155.5 (d, JC–F = 252.3 Hz), 154.6, 144.0, 129.3, 126.2, 124.1 (d, JC–F = 24.6, 9.1 Hz), 122.2 (d, JC–F = 276.2 Hz), 120.0 (m), 119.6, 117.5 (d, JC–F = 27.3 Hz), 100.0, 64.6, 52.9, 46.6, 43.2, 44.1, 29.4. HRMS (ESI): m/z [M + H]+ calcd. for C24H24F5N6O3S, 571.1551, found 572.1607.
4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-N-(thiazol-2-ylmethyl)-6-(trifluoromethyl)pyrimidin-2-amine (7e). Cream solid, 47% yield, LC-MS m/z, [M + H]+ = 570.90, 1H NMR (400 MHz, DMSO-d6) δ 8.79 (dd, J = 2.0 Hz, 12.4 Hz, 1H), 8.65 (d, J = 1.6 Hz, 1H), 8.50 (t, JH–F = 6.0 Hz, 1H), 7.84 (t, J = 3.2 Hz, 1H), 7.72 (d, J = 3.2 Hz, 1H), 7.65–7.59 (ddd, JH–F = 16.4, 9.2, 3.6, Hz, 1H), 7.54–7.42 (m, 3H), 4.90 (d, J = 6.0 Hz, 2H), 4.26 (t, J = 4.4 Hz, 2H), 4.02 (t, J = 4.4 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) δ 169.2, 163.8, 161.9, 158.7, 156.2, 155.5 (d, JC–F = 252.4 Hz), 154.7, 143.9, 142.2, 129.1, 125.8, 124.0 (m), 122.1 (d, JC–F = 276.3 Hz), 120.1 (m), 120.0, 119.8, 119.7, 117.5 (d, JC–F = 25 Hz), 101.2, 64.9, 43.2, 42.5. HRMS (ESI): m/z [M + H]+ calcd. for C22H16F5N6O3S2, 571.0645, found 571.0646.
4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-N-(2,2-difluoropropyl)-6-(trifluoromethyl)pyrimidin-2-amine (7f). Cream solid, 30.4% yield, LC-MS m/z, [M + H]+ = 551.90, 1H NMR (400 MHz, DMSO-d6) 8.81 (d, J = 2.0 Hz,1H), 8.74 (d, J = 2.0 Hz, 1H), 8.05 (t, J = 6.0 Hz, 1H), 7.82 (s, 1H), 7.67–7.62 (ddd, JH–F = 16.4, 8.8, 3.6 Hz, 1H), 7.56–7.49 (tt, JH–F = 9.2, 5.2 Hz, 2H), 4.27 (t, J = 4.4 Hz, 2H), 4.03 (t, J = 4.4 Hz, 2H), 3.94–3.86 (tt, JH–F = 13.6, 6.8 Hz, 2H), 1.66 (t, JH–F = 19.2 Hz, 3H). 13C NMR (101 MHz, DMSO-d6) δ 163.5, 162.4, 158.7, 156.2, 155.5 (d, JC–F = 252.2 Hz), 154.7, 143.8, 129.1, 126.0, 124.0 (d, JC–F = 23.7 Hz), 123.3, 122.1 (d, JC–F = 276.3 Hz), 120.9, 120.0 (m), 119.7, 117.1 (d), 114.6, 101.1, 64.7, 43.2, 29.0. HRMS (ESI): m/z [M + H]+ calcd. for C21H17F7N5O3S, 552.0940, found 552.0943.
4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-N-(oxazol-4-ylmethyl)-6-(trifluoromethyl)pyrimidin-2-amine (7g). Cream solid, 58.5% yield, LC-MS m/z, [M + H]+ = 554.90, 1H NMR (400 MHz, DMSO-d6) δ 8.78 (d, J = 2.0 Hz, 1H), 8.68 (d, J = 2.0 Hz, 1H), 8.21 (s, 1H), 8.06 (t, JH–F = 5.2 Hz, 1H), 7.89 (s, 1H), 7.82 (bs, 1H), 7.66–7.61 (ddd, JH–F = 16.8, 8.8, 5.6 Hz, 1H), 7.54 (tt, JH–F = 9.6, 4.4 Hz, 1H), 7.49 (s, 1H), 4.53 (d, J = 6.0 Hz, 2H), 4.27 (t, J = 4.4 Hz, 2H), 4.02 (t, J = 4.4 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) δ 163.6, 162.0, 158.7, 156.2, 155.5 (d, JC–F = 252.1 Hz), 154.6, 152.1, 143.8, 137.5, 135.9, 129.0, 126.1, 124.1 (d, JC–F = 23.7, 8.4 Hz), 122.1 (d, JC–F = 276.9 Hz), 119.9 (m), 119.7, 117.1 (d, JC–F = 29.31 Hz), 100.5, 64.8, 43.2, 36.8. HRMS (ESI): m/z [M + H]+ calcd. for C22H16F5N6O4S, 555.0874, found 555.0875.
N-Cyclopentyl-4-(1-((2,5-difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-6-(trifluoromethyl)pyrimidin-2-amine (7h). Cream solid, 81% yield, LC-MS m/z, [M + H]+ = 542.05, 1H NMR (400 MHz, DMSO-d6) δ 8.77 (d, J = 2.0 Hz, 1H), 8.72 (s, 1H), 7.82 (bs, 1H), 7.71–7.62 (m, 2H), 7.55–7.50 (tt, JH–F = 9.6, 4.0, Hz, 1H), 7.41 (s, 1H), 4.29–4.23 (m, 3H), 4.03 (t, J = 4.8 Hz, 2H), 2.04–1.98 (m, 2H), 1.74–1.68 (m, 2H), 1.61–1.58 (m, 4H). 13C NMR (101 MHz, DMSO-d6) δ 163.4, 161.8, 158.7, 156.2, 155.6 (d, JC–F = 252.1 Hz), 154.5, 143.5, 128.8, 126.8 (d, JC–F = 17.6 Hz), 126.3, 124.0 (dd, JC–F = 23.4, 8.5 Hz), 122.2 (d, JC–F = 276.4 Hz), 120.3 (d, JC–F = 31.7 Hz), 119.8, 117.2 (d, d, JC–F = 17.6 Hz = 28.1 Hz), 99.5, 64.9, 52.7, 43.3, 31.9, 23.5. HRMS (ESI): m/z [M + H]+ calcd. for C23H20F5N5O3S, 542.1285, found 542.1288.
4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-N-(piperidin-4-yl)-6-(trifluoromethyl)pyrimidin-2-amine (7i). Purified by Prep-HPLC, cream solid, 63.4% yield, LC-MS m/z, [M + H]+ = 557.27, 1H NMR (400 MHz, DMSO-d6) δ 8.82 (d, J = 2.0 Hz, 1H), 8.66 (d, J = 2.4 Hz, 1H), 7.91 (s, 3H), 7.80–7.76 (ddd, JH–F = 10.8, 5.2, 3.2 Hz, 1H), 7.70–7.64 (ddd, JH–F = 14.8, 7.2, 3.2 Hz, 1H), 7.57–7.51 (tt, JH–F = 9.6, 4.0 Hz, 1H), 4.71 (d, J = 13.6 Hz, 2H), 4.29 (t, J = 4.0 Hz, 2H), 4.04 (t, J = 4.0 Hz, 2H), 3.45–3.40 (m, 1H), 3.22–3.15 (m, 2H), 2.09 (d, J = 2.4 Hz, 2H) 1.61–1.51 (m, 2H). 13C NMR (101 MHz, DMSO-d6) δ 163.9, 160.9, 158.7, 156.2, 155.6 (d, JC–F = 253.9 Hz), 154.7, 143.9, 129.0, 126.9 (dd, JC–F = 17.4, 7.1 Hz), 126.0, 124.1 (dd, JC–F = 23.8, 9.0 Hz), 122.1 (d, JC–F = 276.1 Hz), 120.3 (dd, JC–F = 24.4,8.6 Hz), 119.8, 117.4 (d, JC–F = 27.3 Hz), 100.5, 64.9, 47.6, 43.2, 41.7, 29.1. HRMS (ESI): m/z [M + H]+ calcd. for C23H22F5N6O3S, 557.1394, found 557.1468.
1-(4-((4-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-6-(trifluoromethyl)pyrimidin-2-yl)amino)piperidin-1-yl)ethan-1-one (7j). Cream solid, 30% yield, LC-MS m/z, [M + H]+ = 599.05, 1H NMR (400 MHz, DMSO-d6) δ 8.79 (s, 2H), 7.81–7.63 (m, 3H), 7.55–7.49 (tt, JH–F = 9.2, 3.6 Hz, 1H), 7.46 (s, 1H), 4.26 (s, 3H), 4.02 (d, J = 4.0 Hz, 3H), 2.90 (s, 1H), 2.74 (s, 1H), 2.01 (s, 5H), 1.25 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 168.0, 163.6, 161.7, 158.6, 156.2, 155.6 (d, JC–F = 252.3 Hz), 154.6, 143.7, 129.2, 126.2 (m), 126.0, 124.1 (dd, JC–F = 24.3, 8.9 Hz), 122.2 (d, JC–F = 276.5 Hz), 120.2 (m), 119.6, 117.5 (d, JC–F = 27.1 Hz), 99.9, 64.6, 48.4, 43.3, 40.1, 31.3, 21.3. HRMS (ESI): m/z [M + H]+ calcd. for C25H24F5N6O4S, 599.1500, found 599.1501.
Tert-butyl (4-((4-(1-((2,5-difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-6-(trifluoromethyl)pyrimidin-2-yl)amino)cyclohexyl)carbamate (9). Cream solid, 64% yield, LC-MS m/z, [M + H]+ = 671.00, 1H NMR (400 MHz, DMSO-d6) δ 8.83 (s, 2H), 8.02 (d, J = 5.2 Hz, 1H), 7.92 (d, J = 7.2 Hz, 1H), 7.70 (bs, 1H), 7.58 (t, J = 8.8 Hz, 2H), 6.81 (s, 1H), 4.23 (s, 2H), 4.04 (s, 2H), 3.23 (d, J = 6.4 Hz, 2H), 2.65 (d, J = 11.6 Hz, 2H), 2.03–1.98 (s, 2H), 1.93 (s, 1H), 1.82 (s, 3H), 1.37 (s, 9H).
1-((2,5-Difluorophenyl)sulfonyl)-7-(2-morpholinopyrimidin-5-yl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazine (8a). Cream solid, yield: 0.04 g, 47%, LC-MS m/z, [M + H]+ = 476.00, 1H NMR (400 MHz, DMSO-d6) δ 8.65 (s, 2H), 8.30 (s, 1H), 8.10 (s, 1H), 7.94 (m, 1H), 7.70 (m, 1H), 7.57–7.56 (tt, JH–F = 4.0 Hz, 1H), 4.10 (d, 2H), 3.98 (d, 2H), 3.75 (d, J = 4.8 Hz, 4H), 3.68 (d, J = 4.4 Hz, 4H). 13C NMR (101 MHz, DMSO-d6) δ 160.6, 158.7, 156.3, 155.6, 155.5 (d, JC–F = 253.3 Hz) 151.8, 141.3 (2C), 128.1, 126.6 (dd, JC–F = 17.5, 10.1 Hz), 125.2, 124.0 (dd, JC–F = 24.0, 8.8 Hz), 120.1 (dd, JC–F = 24.3, 8.3 Hz), 119.9, 117.9 (d, JC–F = 27.6 Hz), 65.9, 64.1, 44.0, 43.4. HRMS (ESI): m/z [M + H]+ calcd. for C21H20F2N5O4S, 476.1204, found 476.1190.
1-(4-(5-(1-((2,5-Difluorophenyl)sulfonyl)-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)pyrimidin-2-yl)piperazin-1-yl)ethan-1-one (8b). Cream solid, yield: 0.025 g, 49%. LC-MS m/z, [M + H]+ = 517.05, 1H NMR (400 MHz, DMSO-d6) 8.66 (s, 2H), 8.31 (s, 1H), 8.10–7.95 (s, 2H), 7.71–7.59 (s, 2H), 4.05 (d, 4H), 3.80 (d, 4H), 3.54 (s, 4H), 2.06 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 168.5, 160.5, 158.8 (d, JC–F = 247.4 Hz), 155.7, 153.0, 151.9, 141.3 (2C), 128.2, 126.6 (dd, JC–F = 17.2, 11.1 Hz), 125.2, 124.0 (m), 120.2 (m), 119.5, 118.9, 117.9 (d, JC–F = 28.3 Hz), 64.1, 59.8, 45.3, 43.7, 43.3, 40.6, 21.3. HRMS (ESI): m/z [M + H]+ calcd. for C23H22F2N6O4S, 517.1470, found 517.1445.
| Sr. no. | Comp. | HPLC % purity | Sr. no. | Comp. | HPLC % purity | Sr. no. | Comp. | HPLC % purity |
|---|---|---|---|---|---|---|---|---|
| 1 | 4a | 95.72 | 7 | 7c | 96.58 | 13 | 7i | 96.44 |
| 2 | 4b | 96.72 | 8 | 7d | 91.97 | 14 | 7j | 97.70 |
| 3 | 5 | 96.71 | 9 | 7e | 96.63 | 15 | 7k | 97.65 |
| 4 | 6 | 92.04 | 10 | 7f | 92.66 | 16 | 8a | 93.97 |
| 5 | 7a | 97.99 | 11 | 7g | 95.57 | 17 | 8b | 96.97 |
| 6 | 7b | 95.09 | 12 | 7h | 99.09 |
4.2 Biology
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm; the insoluble material was then removed. Equal quantities of proteins were loaded, separated using 8% SDS-PAGE, and subsequently transferred to polyvinylidene fluoride membranes (Millipore). The anti-EGFR, anti-pEGFR (Tyr1068), and anti-AKT antibodies were diluted to a ratio of 1
000 rpm; the insoluble material was then removed. Equal quantities of proteins were loaded, separated using 8% SDS-PAGE, and subsequently transferred to polyvinylidene fluoride membranes (Millipore). The anti-EGFR, anti-pEGFR (Tyr1068), and anti-AKT antibodies were diluted to a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, whereas the anti-pAKT antibody was diluted to a ratio of 1
1000, whereas the anti-pAKT antibody was diluted to a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000. All antibodies mentioned have been purchased from Sigma Aldrich. Anti-β-actin was diluted to a ratio of 1
2000. All antibodies mentioned have been purchased from Sigma Aldrich. Anti-β-actin was diluted to a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, and the secondary antibodies were utilized at a concentration of 1
5000, and the secondary antibodies were utilized at a concentration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000. The results were identified using an enhanced chemiluminescence system (Millipore).34
5000. The results were identified using an enhanced chemiluminescence system (Millipore).34
4.3 Docking study
EGFR-WT grid. The EGFR complexes with PDB IDs 8sc7, 4jq7, 4jq8, 4jr3, and 4jrv were obtained from the PDB (https://www.rcsb.org). These structures were uploaded to ICM-Pro35 and converted into ICM objects. Water molecules were removed during this procedure, hydrogen atoms were optimally added, and histidine protonation states and asparagine and glutamine orientations were optimized using the ICM force field.36 The protein structures from the five ICM objects were extracted, overlaid with the EGFR structure stored under PDB ID 8sc7, and stacked. The residues within 5 Å from the ligand in the complex with PDB ID 8sc7 were identified as active site residues. The box dimensions for generating the maps were auto calculated based on the chosen residues. An ensemble docking grid (4D docking grid) was created from a stack of five structures. The co-crystallized ligands in the five chosen EGFR complexes were redocked onto this grid with an effort (thoroughness) of 10 and their experimental binding modes were reproduced.
EGFR-double mutant (T790M/L858R) grid. The T790M/L858R EGFR-double mutant complexes with PDB IDs 3w2o,37 4rj5, 4rj6, 4rj7, 4rj8,38 5edq and 5edr39 were used in the aforementioned manner to generate an ensemble docking grid. The co-crystallized ligands in the seven chosen EGFR complexes were redocked onto this grid with an effort of 10 and their experimental binding modes were reproduced.
EGFR-WT grid. The PDB (https://www.rcsb.org) was used to download the osimertinib-bound EGFR complex and it was uploaded into ICM Pro. A two-dimensional structure of the ligand osimertinib was extracted and stored as an SDF file. The fused pyrido[2,3-b][1,4]oxazine compounds were sketched and saved to an SDF file. These were then docked onto the two 4D grids with a 10-point effort (thoroughness). The charge group determining function was set to auto to ensure that the pH-appropriate tautomer of the ligand is docked. The top five docking modes were saved based on their docking scores.
Author contributions
VBY performed the chemical synthesis and analytical characterization under the supervision of SRD. SVT conducted the MTT assay, western blot, and apoptosis studies, and contributed to the investigation and writing of the biological sections. SHK carried out the molecular docking studies and related investigations. SRD contributed to the supervision, conceptualization, study design, investigation, original draft writing, and manuscript editing. All authors reviewed and approved the final version of the manuscript prior to submission.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI).Supplementary information: 1H NMR, 13C NMR, LCMS, HRMS and HPLC purity for synthesized compounds as well as the docking modes of the co-crystal ligands of the structures used to generate the docking grids. See DOI: https://doi.org/10.1039/d5md00861a.
Acknowledgements
We dedicate this work to the memory of the late Dr. Aniket P. Sarkate, whose guidance, support, and inspiration were pivotal to this research and left a lasting impact on our work and the scientific community. The authors also would like to convey their sincere thanks to Dr. R. J. Barnabas, Chairman of the B. P. H. E. Society Ahilyanagar, for his important guidance, ongoing support, and encouragement throughout the research process.References
- F. Bray, M. Laversanne, E. Weiderpass and I. Soerjomataram, The ever-increasing importance of cancer as a leading cause of premature death worldwide, Cancer, 2021, 127, 3029, DOI:10.1002/cncr.33587.
- F. Bray, M. Laversanne, H. Sung, J. Ferlay, R. L. Siegel, A. Jemal and I. Soerjomataram, Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2024, 74, 229, DOI:10.3322/caac.21834.
- X. Du, B. Yang, Q. An, Y. G. Assaraf, X. Cao and J. Xia, Acquired resistance to third-generation EGFR-TKIs and emerging next-generation EGFR inhibitors, Innovation, 2021, 2, 100103, DOI:10.1016/j.xinn.2021.100103.
- K. Kashima, H. Kawauchi, H. Tanimura, Y. Tachibana, T. Chiba, T. Torizawa and H. Sakamoto, CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation, Mol. Cancer Ther., 2020, 19, 2288, DOI:10.1158/1535-7163.MCT-20-0229.
- A. Gazdar, Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors, Oncogene, 2009, 28, S24–S31, DOI:10.1038/onc.2009.198.
- Y. Sun, R. Fu, S. Lin, J. Zhang and M. Ji, et al. Discovery of new thieno[2,3-d]pyrimidine and thiazolo[5,4-d]pyrimidine derivatives as orally active phosphoinositide 3-kinase inhibitors, Bioorg. Med. Chem., 2021, 29, 115890, DOI:10.1016/j.bmc.2020.115890.
- S. Guardiola, M. Varese, M. Sánchez-Navarro and E. Giralt, A Third Shot at EGFR: New Opportunities in Cancer Therapy, Trends Pharmacol. Sci., 2019, 40, 941, DOI:10.1016/j.tips.2019.10.004.
- C. Mehlman, J. Cadranel, G. Rousseau-Bussac, R. Lacave and A. Pujals, et al. Resistance mechanisms to osimertinib in EGFR-mutated advanced non-small-cell lung cancer: A multicentric retrospective French study, Lung Cancer, 2019, 137, 149, DOI:10.1016/j.lungcan.2019.09.019.
- D. Rangachari, C. To, J. E. Shpilsky, P. A. VanderLaan and S. S. Kobayashi, et al. EGFR-Mutated Lung Cancers Resistant to Osimertinib through EGFR C797S Respond to First-Generation Reversible EGFR Inhibitors but Eventually Acquire EGFR T790M/C797S in Preclinical Models and Clinical Samples, J. Thorac. Oncol., 2019, 14, 1995, DOI:10.1016/j.jtho.2019.07.016.
- S. Singh, S. Sadhukhan and A. Sonawane, 20 years since the approval of first EGFR-TKI, gefitinib: Insight and foresight, Biochim. Biophys. Acta, Rev. Cancer, 2023, 1878, 188967, DOI:10.1016/j.bbcan.2023.188967.
- B. F. Abdel-Wahab, S. Shaaban and G. A. El-Hiti, Synthesis of sulfur-containing heterocycles via ring enlargement, Mol. Diversity, 2018, 22, 517, DOI:10.1007/s11030-017-9810-3.
- S. R. Deshmukh and S. R. Thopate, C2/C3 alkynylation of L-ascorbic acid by Sonogashira coupling and efficient access to some potent and highly selective novel anticancer agents, New J. Chem., 2019, 43, 208, 10.1039/C8NJ04477E.
- S. R. Deshmukh, A. S. Nalkar, A. P. Sarkate, S. V. Tiwari, D. K. Lokwani and S. R. Thopate, Design, synthesis, and biological evaluation of novel 2,3-Di-O-Aryl/Alkyl sulfonate derivatives of l-ascorbic acid: Efficient access to novel anticancer agents via in vitro screening, tubulin polymerization inhibition, molecular docking study and ADME predictions, Bioorg. Chem., 2024, 147, 107402, DOI:10.1016/j.bioorg.2024.107402.
- E. Vitaku, D. T. Smith and J. T. Njardarson, Analysis of the Structural Diversity, Substitution Patterns, and Frequency of Nitrogen Heterocycles among U.S. FDA Approved Pharmaceuticals, J. Med. Chem., 2014, 57, 10257, DOI:10.1021/jm501100b.
- A. P. Taylor, R. P. Robinson, Y. M. Fobian, D. C. Blakemore, L. H. Jones and O. Fadeyi, Modern Advances in Heterocyclic Chemistry in Drug Discovery, Org. Biomol. Chem., 2016, 14, 6611, 10.1039/C6OB00936K.
- T. T. Yadav, G. M. Shaikh, M. S. Kumar, M. Chintamaneni and M. YC, A Review on Fused Pyrimidine Systems as EGFR Inhibitors and Their Structure-Activity Relationship, Front. Chem., 2022, 10, 861288, DOI:10.3389/fchem.2022.861288.
- H. Yao, K. Yang, L. Cao, Y. Ren, P. Hou, M. Yan and X. Li, Synthesis and Evaluation of Novel Amino Pyrimidine Derivatives Containing Sulfonamide and Their Application as EGFR Inhibitors, Bioorg. Chem., 2025, 160, 108467, DOI:10.1016/j.bioorg.2025.108467.
- Y. Wan, G. Fang, H. Chen, X. Deng and Z. Tang, Sulfonamide derivatives as potential anti-cancer agents and their SARs elucidation, Eur. J. Med. Chem., 2021, 226, 113837, DOI:10.1016/j.ejmech.2021.113837.
- M. S. Alsaid, A. A. Al-Mishari, A. M. Soliman, F. A. Ragab and M. M. Ghorab, Discovery of benzo[g]quinazolin benzenesulfonamide derivatives as dual EGFR/HER2 inhibitors, Eur. J. Med. Chem., 2017, 141, 84, DOI:10.1016/j.ejmech.2017.09.061.
- M. A. Bhat, B. Tüzün, N. A. Alsaif, A. Ali Khan and A. M. Naglah, Synthesis, characterization, molecular modeling against EGFR target and ADME/T analysis of novel purine derivatives of sulfonamides, J. Mol. Struct., 2022, 1257, 132600, DOI:10.1016/j.molstruc.2022.132600.
- M. M. Ghorab, A. M. Soliman, K. El-Adl and N. S. Hanafy, New quinazoline sulfonamide derivatives as potential anticancer agents: Identifying a promising hit with dual EGFR/VEGFR-2 inhibitory and radiosensitizing activity, Bioorg. Chem., 2023, 140, 106791, DOI:10.1016/j.bioorg.2023.106791.
- A. S. Borude, S. R. Deshmukh, S. V. Tiwari, S. H. Kumar and S. R. Thopate, Design and synthesis of novel thiazolo[5,4-b]pyridine derivatives as potent and selective EGFR-TK inhibitors targeting resistance mutations in non-small cell lung cancer, Eur. J. Med. Chem., 2024, 276, 116727, DOI:10.1016/j.ejmech.2024.116727.
- V. R. Anderson and C. M. Perry, Levofloxacin: A Review of Its Use as a High-Dose, Short-Course Treatment for Bacterial Infection, Drugs, 2008, 68, 535, DOI:10.2165/00003495-200868040-00011.
- S. Itadani, S. Takahashi, M. Ima, T. Sekiguchi, M. Fujita, Y. Nakayama and J. Takeuchi, Discovery of Highly Potent Dual CysLT1 and CysLT2 Antagonist, ACS Med. Chem. Lett., 2014, 5, 1230, DOI:10.1021/ml500298y.
- J. M. Rook, J. L. Bertron, H. P. Cho, P. M. Garcia-Barrantes, S. P. Moran, J. T. Maksymetz, K. D. Nance, J. W. Dickerson, D. H. Remke, S. Chang, J. M. Harp, A. L. Blobaum, C. M. Niswender, C. K. Jones, S. R. Stauffer, P. J. Conn and C. W. Lindsley, A Novel M1 PAM VU0486846 Exerts Efficacy in Cognition Models without Displaying Agonist Activity or Cholinergic Toxicity, ACS Chem. Neurosci., 2018, 9, 2274, DOI:10.1021/acschemneuro.8b00131.
- Y. Sun, R. Fu, S. Lin, J. Zhang, M. Ji, Y. Zhang, D. Wu, K. Zhang, H. Tian, M. Zhang, L. Sheng, Y. Li, J. Jin, X. Chen and H. Xu, Discovery of New Thieno[2,3-d]pyrimidine and Thiazolo[5,4-d]pyrimidine Derivatives as Orally Active Phosphoinositide 3-Kinase Inhibitors, Bioorg. Med. Chem., 2021, 29, 115890, DOI:10.1016/j.bmc.2020.115890.
- K. S. Karnik, A. P. Sarkate, S. V. Tiwari, R. Azad and P. S. Wakte, Free energy perturbation guided Synthesis with Biological Evaluation of Substituted Quinoline derivatives as small molecule L858R/T790M/C797S mutant EGFR inhibitors targeting resistance in Non-Small Cell Lung Cancer (NSCLC), Bioorg. Chem., 2021, 115, 105226, DOI:10.1016/j.bioorg.2021.105226.
- K. S. Karnik, A. P. Sarkate, S. V. Tiwari, R. Azad, P. V. L. S. Burra and P. S. Wakte, Computational and Synthetic approach with Biological Evaluation of Substituted Quinoline derivatives as small molecule L858R/T790M/C797S triple mutant EGFR inhibitors targeting resistance in Non-Small Cell Lung Cancer (NSCLC), Bioorg. Chem., 2021, 107, 104612, DOI:10.1016/j.bioorg.2020.104612.
- C. E. Whitehead, E. K. Ziemke, C. L. Frankowski-McGregor, R. A. Mumby, J. Chung, J. Li, N. Osher, O. Coker, V. Baladandayuthapani, S. Kopetz and J. S. Sebolt-Leopold, A first-in-class selective inhibitor of EGFR and PI3K offers a single-molecule approach to targeting adaptive resistance, Nat. Cancer, 2024, 5, 1250, DOI:10.1038/s43018-024-00781-6.
- Y. H. Peng, H. Y. Shiao, C. H. Tu, P. M. Liu, J. T. A. Hsu and P. K. Amancha, et al. Protein kinase inhibitor design by targeting the Asp-Phe-Gly (DFG) motif: The role of the DFG motif in the design of epidermal growth factor receptor inhibitors, J. Med. Chem., 2013, 56, 3889, DOI:10.1021/jm400072p.
- Y. Yosaatmadja, S. Silva, J. M. Dickson, A. V. Patterson, J. B. Smaill, J. U. Flanagan, M. J. McKeage and C. J. Squire, Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed, J. Struct. Biol., 2015, 192, 539, DOI:10.1016/j.jsb.2015.10.018.
- S. V. Tiwari, A. P. Sarkate, D. K. Lokwani, D. N. Pansare, S. G. Gattani, S. S. Sheaikh, S. P. Jain and S. V. Bhandari, Explorations of novel pyridine-pyrimidine hybrid phosphonate derivatives as aurora kinase inhibitors, Bioorg. Med. Chem., 2022, 67, 128747, DOI:10.1016/j.bmcl.2022.128747.
- M. A. Kashem, R. M. Nelson, J. D. Yingling, S. S. Pullen, A. S. Prokopowicz III, J. W. Jones, J. P. Wolak, G. R. Rogers, M. M. Morelock, R. J. Snow, C. A. Homon and S. Jakes, Three mechanistically distinct kinase assays compared: measurement of intrinsic ATPase activity identified the most comprehensive set of ITK inhibitors, J. Biomol. Screening, 2007, 12, 70, DOI:10.1177/1087057106296047.
- M. A. C. Neves, M. Totrov and R. Abagyan, Docking and scoring with ICM: The benchmarking results and strategies for improvement, J. Comput.-Aided Mol. Des., 2012, 26, 675, DOI:10.1007/s10822-012-9547-0.
- J. Niggenaber, L. Heyden, T. Grabe, M. P. Müller, J. Lategahn and D. Rauh, Complex Crystal Structures of EGFR with Third-Generation Kinase Inhibitors and Simultaneously Bound Allosteric Ligands, ACS Med. Chem. Lett., 2020, 12, 2484, DOI:10.1021/acsmedchemlett.0c00472.
- Y. A. Arnautova, R. A. Abagyan and M. Totrov, Development of a new physics-based internal coordinate mechanics force field and its application to protein loop modeling, Proteins: Struct., Funct., Bioinf., 2011, 79, 477, DOI:10.1002/prot.22896.
- S. Sogabe, Y. Kawakita, S. Igaki, H. Iwata, H. Miki and D. R. Cary, et al. Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors, ACS Med. Chem. Lett., 2012, 4, 201, DOI:10.1021/ml300327z.
- E. J. Hanan, C. Eigenbrot, M. C. Bryan, D. J. Burdick, B. K. Chan and Y. Chen, et al. Discovery of Selective and Noncovalent Diaminopyrimidine-Based Inhibitors of Epidermal Growth Factor Receptor Containing the T790M Resistance Mutation, J. Med. Chem., 2014, 57, 10176, DOI:10.1021/jm501578n.
- E. J. Hanan, M. Baumgardner, M. C. Bryan, Y. Chen, C. Eigenbrot and P. Fan, et al. 4-Aminoindazolyl-dihydrofuro[3,4-d]pyrimidines as non-covalent inhibitors of mutant epidermal growth factor receptor tyrosine kinase, Bioorg. Med. Chem. Lett., 2016, 26, 534, DOI:10.1016/j.bmcl.2015.11.078.
| This journal is © The Royal Society of Chemistry 2026 |




