Design and synthesis of sorafenib-inspired benzofuran hybrids as VEGFR-2 inhibitors: antiproliferative evaluation, mechanistic insights, and docking studies in hepatocellular carcinoma
Mohamed S.
Shehda
 *a,
Aya M.
Almatary
a,
Mohamed S. H.
Salem
*a,
Aya M.
Almatary
a,
Mohamed S. H.
Salem
 *bc,
Mohamed H.
Aboutaleb
a,
Shinobu
Takizawa
*bc,
Mohamed H.
Aboutaleb
a,
Shinobu
Takizawa
 c,
Yasmine M.
Abdel Aziz
c,
Yasmine M.
Abdel Aziz
 b,
Magda A. A.
El-Sayed
b,
Magda A. A.
El-Sayed
 *de and
Ranza
Elrayess
*de and
Ranza
Elrayess
 bf
bf
aPharmaceutical Chemistry Department, Faculty of Pharmacy, Horus University-Egypt, New Damietta 34518, Egypt. E-mail: mshehda@horus.edu.eg
bPharmaceutical Organic Chemistry Department, Faculty of Pharmacy, Suez Canal University, Ismailia 41522, Egypt
cSANKEN, The University of Osaka, Mihogaoka, Ibaraki, Osaka 567-0047, Japan. E-mail: mohamedsalem43@sanken.osaka-u.ac.jp
dPharmaceutical Organic Chemistry Department, Faculty of Pharmacy, Mansoura University, Mansoura 35516, Egypt. E-mail: magdaaziz1@yahoo.com
eDepartment of pharmaceutic organic chemistry, Faculty of Pharmacy, Mansoura National University, Gamasa 7731168, Egypt
fPharmaceutical Organic Chemistry Department, College of Pharmacy, Al-Ayen Iraqi University, AUIQ, An Nasiriyah, 6400, Iraq
First published on 12th December 2025
Abstract
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related mortality worldwide owing to its high metastatic potential. Vascular endothelial growth factor receptor-2 (VEGFR-2), a key regulator of angiogenesis and cell proliferation, is a critical therapeutic target in cancer, particularly in HCC. However, the clinical use of various VEGFR-2 inhibitors is limited by adverse effects, poor selectivity and drug resistance. In this study, two novel series of sorafenib-inspired benzofuran hybrids, azines (6a–k) and thiosemicarbazones (7a–g), were synthesized and evaluated for their cytotoxicity against HepG-2 cell lines. Compounds 6b, 6c, 7a, 7b, 7e, 7f, and 7g exhibited potent activity with IC50 values of 7.21–18.01 μM. Among them, 7a, 7b, 7f, and 7g demonstrated high selectivity (SI = 5.7–11.2) toward HepG-2 cells over normal WI-38 cells. Enzyme assays confirmed significant VEGFR-2 inhibition, led by 7g (IC50 = 0.072 μM), comparable to sorafenib (IC50 = 0.069 μM). In HepG-2 cells, 7g induced G0/G1 arrest and apoptosis, upregulating Bax, caspase-8 and -9, and downregulating Bcl-2. Molecular docking confirmed the strong binding affinity of 7g within the VEGFR-2 active site through key interactions with Glu885, Asp1046, and Cys1045, mirroring sorafenib. A 100 ns molecular dynamics simulation further demonstrated that compound 7g retains a highly stable binding mode within VEGFR-2, supported by low RMSD fluctuations and persistent key hydrogen-bond and hydrophobic interactions. These findings nominate 7g as a promising VEGFR-2-targeted lead for HCC upon further optimization.
1. Introduction
Cancer remains one of the most significant global public health challenges, as it ranks among the top causes of death in the 21st century.1 Primary liver cancer, mainly hepatocellular carcinoma (HCC), is considered the 6th most commonly diagnosed cancer and the third leading cause of cancer-related deaths worldwide. Globally, in 2020, HCC was estimated to have caused around 906![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases and approximately 830
000 new cases and approximately 830![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths, with an incidence-to-mortality ratio close to 1
000 deaths, with an incidence-to-mortality ratio close to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The high mortality rate of HCC and the very low five-year survival rate is believed to be owing to its high tendency for metastasis.2 On the other hand, colorectal cancer (CRC) represents the third most commonly diagnosed malignancy, with an estimated 1.9 million new cases and 0.9 million deaths in 2020.3 Liver metastasis is a frequent complication in CRC patients and remains a major cause of cancer-related mortality.4,5
1. The high mortality rate of HCC and the very low five-year survival rate is believed to be owing to its high tendency for metastasis.2 On the other hand, colorectal cancer (CRC) represents the third most commonly diagnosed malignancy, with an estimated 1.9 million new cases and 0.9 million deaths in 2020.3 Liver metastasis is a frequent complication in CRC patients and remains a major cause of cancer-related mortality.4,5
One of the most attractive targets in the development of modern anticancer agents, is tyrosine kinase. These enzymes play a critical role in regulating key physiological and biochemical processes, including cell growth, differentiation, and apoptosis, as well as maintaining normal cellular function.6,7 Aberrant activation of their receptors (RTKs) has been strongly linked to the initiation and progression of numerous types of cancer.8 Among these, vascular endothelial growth factor receptor (VEGFR), especially the VEGFR-2 isoform, is the most overexpressed RTKs in hepatocellular carcinoma (HCC) and notably expressed in colon cancer.9 Accordingly, HepG-2 (HCC) and HCT-116 (colorectal) are widely used in vitro models to assess cytotoxicity and VEGFR-2-directed inhibition,10,11 providing translationally relevant readouts that align with clinical use of VEGFR-2 inhibitors. The VEGF–VEGFR signaling pathway regulates both cell proliferation and angiogenesis,12 with the latter playing a key role in tumor growth and metastasis.13 Therefore, the inhibition of VEGFR-2 is regarded as a highly effective strategy for suppressing tumor angiogenesis and metastasis. VEGFR-2 inhibitors, such as sorafenib (I) and fruquintinib (II) are among the most widely used therapeutics for hepatocellular carcinoma and colorectal cancer, respectively.14,15
Sorafenib (I) is globally considered the most accepted first-line systemic treatment for advanced HCC.16 Moreover, some studies have demonstrated its significant cytotoxic effects against colon cancer cells.17 It acts primarily as a tyrosine kinase inhibitor that targets growth factor receptors, mainly VEGFR-2.18 It is classified as a type II tyrosine kinase inhibitor, which binds to the inactive (DFG-out) conformation of the kinase and occupies both the ATP-binding site and the adjacent allosteric pocket, resulting in more selectivity and inhibition.19–21
Despite its therapeutic effectiveness, the clinical use of sorafenib is often limited by significant adverse effects, its poor kinase selectivity and the frequent development of drug resistance.22 Therefore, the design and synthesis of new derivatives through structural modification of sorafenib have attracted significant attention.23,24
Several privileged pharmacophoric moieties have been recognized for their role in VEGFR-2 inhibition. Among them, benzofuran represents a privileged heterocyclic scaffold of high value in medicinal chemistry due to its well-documented anticancer potential.25 Numerous benzofuran-based agents have demonstrated antiproliferative activity through VEGFR-2 inhibition.26 For instance, fruquintinib (II) is a benzofuran-based selective and potent VEGFR-2 inhibitor approved for the treatment of metastatic colorectal cancer,27 while compound III has also shown remarkable VEGFR-2 inhibitory activity15,28 (Fig. 1).
2. Rationale and design
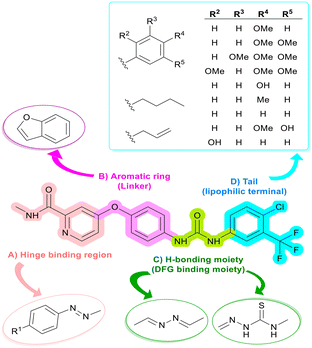
Based on the well-established role of VEGFR-2 in tumor angiogenesis and progression, along with the clinical success of sorafenib as a first-line treatment for HCC via VEGFR-2 inhibition, and guided by our interest in the development of novel tyrosine kinase inhibitors with improved selectivity and efficacy,16,29 we designed a series of novel hybrid molecules as potential therapeutics for treatment of HCC through structural modification of sorafenib scaffold by incorporating privileged pharmacophoric chemical moieties known for their ability to inhibit VEGFR-2 and cytotoxic activity.As outlined in Fig. 2, the molecular design was guided by the pharmacophoric model of most VEGFR-2 type II inhibitors, such as sorafenib (I), which includes four essential features:30,31 A) a peripheral substituted aromatic ring to occupy the hinge region (ATP binding pocket), usually incorporating a heteroaromatic moiety that accepts a hydrogen bond from the hinge backbone NH (Cys919), anchoring the ligand in the ATP pocket, B) an aromatic ring as a linker to bridge the hinge binding region and the core pharmacophore, C) a hydrogen bond donor/acceptor group (DFG-binding moiety) as the core pharmacophore to interact with Glu885 and/or Asp1046 within the activation loop, and D) a hydrophobic tail to fill the hydrophobic allosteric pocket.
 | ||
| Fig. 1 Structure of sorafenib and representative benzofuran-based compounds previously reported as VEGFR-2 inhibitors. | ||
In the current work, as seen in Fig. 2, the hinge-binding region is represented by a diazenyl-substituted phenyl ring, which has proven to be an effective moiety for anticancer activity, as observed in compounds IV and V, which show potent cytotoxicity against multiple cancer cell lines, including HepG-2 cells.32,33 Although this nonclassical hinge-binding moiety cannot form a direct hydrogen bond with Cys919, the extended conjugated system of the diazenyl-phenyl group provides strong hydrophobic interactions that can offset this loss; notably, potent VEGFR-2 inhibitors lacking this interaction, such as compounds VI and VII, have been reported.34,35 On the other hand, the aromatic linker was represented by a benzofuran ring, which, as previously mentioned, is a privileged scaffold reported in several VEGFR-2 inhibitors, as demonstrated by the previously reported compounds VIII and IX.36,37 This moiety also offers favorable π–π interactions that enhance binding affinity.
The DFG-binding region was designed by incorporating azine and thiosemicarbazone moieties, both recognized as core pharmacophores in various anticancer agents, as exemplified by compounds X and XI.35,38 Their hydrogen-bonding capabilities facilitate key interactions with the catalytic residues Glu885 and/or Asp1046, which are crucial for effective kinase inhibition. The hydrophobic tail was optimized using phenyl, substituted phenyl, or aliphatic groups in selected analogs to enhance hydrophobic interactions within the allosteric pocket and to evaluate the impact of molecular flexibility and electronic properties on biological activity35 (Fig. 3).
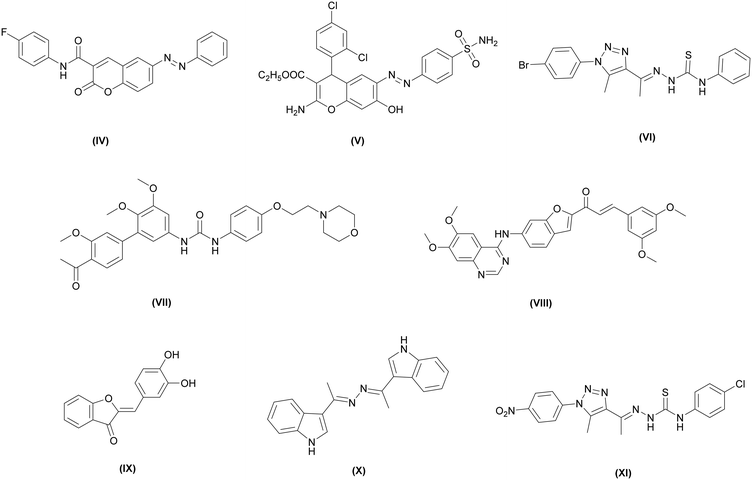 | ||
| Fig. 3 Reported anticancer and VEGFR-2 inhibitory compounds featuring alternative pharmacophoric incorporations. | ||
This rational design approach led to the synthesis of two novel series of compounds (6a–k and 7a–g) through structural modification of the sorafenib scaffold via molecular hybridization, incorporating alternative privileged pharmacophores, as illustrated in Fig. 4. The synthesized compounds were evaluated for their in vitro cytotoxicity against HepG-2 cell line and for VEGFR-2 kinase inhibitory activity. The most potent compound was further subjected to detailed biological investigations, including assessment of its effects on cell cycle distribution, induction of apoptosis and necrosis, and modulation of apoptotic and anti-apoptotic gene expression in HepG-2 cells. Furthermore, molecular docking studies were performed to rationalize the observed inhibitory activities and to elucidate the binding modes of these compounds within the VEGFR-2 active site.
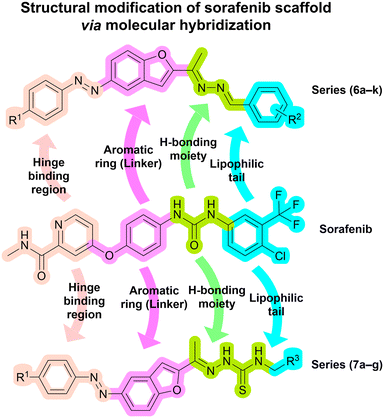 | ||
| Fig. 4 Newly designed series 6a–k and 7a–g obtained through structural modification of the sorafenib scaffold via molecular hybridization, illustrating alterations in each key pharmacophoric region. | ||
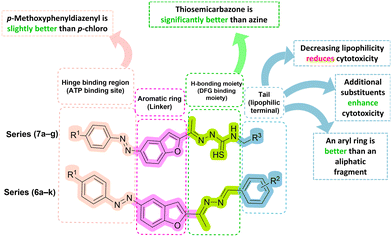 | ||
| Fig. 5 Visual summarization of SAR study for the anti-cancer activity of the novel compounds (6a–k and 7a–g). | ||
3. Results and discussion
3.1. Chemistry
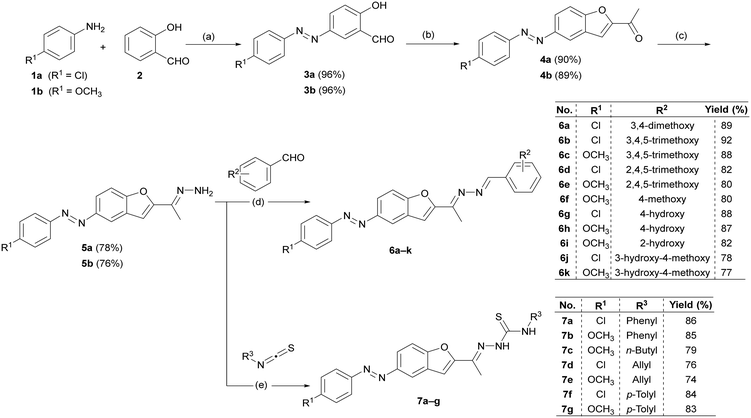
The target compounds 6a–k and 7a–g were prepared as presented in Scheme 1. The starting hydrazone derivatives 5a, b were obtained through three steps, as shown in Scheme 1. First, 2-hydroxy-5-(phenyldiazenyl)benzaldehyde derivatives 3a, b were prepared by the coupling reaction of salicylaldehyde (2) with the diazonium salts of aniline derivatives 1a, b.39,40 Then 1-(benzofuran-2-yl)ethan-1-one derivatives 4a, b were afforded via a Rap–Stoermer reaction, in which a cyclocondensation reaction between compounds 3a, b and chloroacetone (as an α-haloketone) was conducted under basic conditions using potassium carbonate while refluxing in acetone.41 The chemical structures of the 1-(benzofuran-2-yl)ethan-1-one derivatives 4a, b were confirmed by the full spectral analysis (IR, 1H NMR, 13C NMR, HRMS). The 1H NMR spectra of both compounds displayed characteristic aromatic signals integrating for eight protons, corresponding to the benzofuran core and the para-substituted phenyldiazenyl moiety. Additionally, a singlet at δ 2.5–2.6 ppm was assigned to the methyl group of the ethanone fragment. The IR spectra showed strong absorption bands in the range of 1673–1682 cm−1, characteristic of the conjugated carbonyl group and sharp absorption bands in the range of 1600–1577 cm−1, corresponding to the azo group.The third step involved the condensation of compounds 4a, b with hydrazine hydrate to give the desired hydrazone derivatives 5a, b according to the previously reported procedure.42 Their chemical structures were assigned by the full spectral analysis where 1H NMR spectroscopy revealed a downfield shift of the methyl singlet from δ ∼2.6 ppm to δ 2.0 ppm. Moreover, the spectra showed a new singlet signal around δ 6.9 ppm corresponding to the protons of the NH2 group, supporting the successful transformation of the carbonyl group into a hydrazone moiety.
As demonstrated in Scheme 1, towards the synthesis of the targeted azines (bis-Schiff base) 6a–k, the hydrazone derivatives 5a, b were refluxed with various substituted aromatic aldehydes in absolute ethanol through nucleophilic condensation, in the presence of a catalytic amount of glacial acetic acid.43 The formation of the azines 6a–k was confirmed by the full spectral analysis (IR, 1H NMR, 13C NMR, HRMS). 1H NMR spectra revealed the appearance of a singlet signal around 8.50 ppm corresponding to azomethine proton N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH, along with the complete disappearance of the NH2 signal present in the starting hydrazone derivatives. They also showed the characteristic signals corresponding to the protons of the substituted benzylidene rings. For example, compounds 6a–f, 6j, and 6k each displayed one or more singlet signals in the range of δ 3.8–3.7 ppm, with each singlet integrating for three protons, corresponding to the number of methoxy groups in each compound. Moreover, compounds 6g–k showed singlet a signal in the range of δ 3.8–3.7 ppm, with each singlet integrating for one proton, corresponding to the hydroxy group in each compound.
CH, along with the complete disappearance of the NH2 signal present in the starting hydrazone derivatives. They also showed the characteristic signals corresponding to the protons of the substituted benzylidene rings. For example, compounds 6a–f, 6j, and 6k each displayed one or more singlet signals in the range of δ 3.8–3.7 ppm, with each singlet integrating for three protons, corresponding to the number of methoxy groups in each compound. Moreover, compounds 6g–k showed singlet a signal in the range of δ 3.8–3.7 ppm, with each singlet integrating for one proton, corresponding to the hydroxy group in each compound.
On the other hand, the target thiosemicarbazones 7a–g were synthesized by refluxing the corresponding hydrazone intermediates 5a, b with different isothiocyanates in ethanol.44 Their chemical structures were assigned by the full spectral analysis (IR, 1H NMR, 13C NMR, HRMS). 1H NMR analysis revealed the appearance of two new singlet signals corresponding to the newly introduced NH protons. Additionally, new signals were observed, attributed to the substituents introduced via the isothiocyanate moieties, confirming the successful formation of the thiosemicarbazone derivatives 7a–g. For example, compound 7c showed the four characteristic signals of the butyl group in the range of δ 3.57–0.89 ppm, integrating for seven protons. On the other hand, compounds 7d and 7e exhibited four characteristic signals in the range of δ 5.90–4.24 ppm, integrating for five protons, which are consistent with the presence of an allyl group.
3.2. Biological evaluation
| In vitro cytotoxicity IC50a (μM) | |||||
|---|---|---|---|---|---|
| HepG-2 | HCT-116 | HepG-2 | HCT-116 | ||
| a IC50 (μM): 1–10 (highly potent), 11–20 (potent), 21–50 (moderate), 51–100 (weak), and >100 (not active). b DOX (doxorubicin), SOR (sorafenib). | |||||
| 6a | 37.50 ± 2.2 | 41.17 ± 2.5 | 6j | 53.27 ± 3.1 | 59.77 ± 3.2 |
| 6b | 14.03 ± 1.2 | 23.19 ± 1.6 | 6k | 47.55 ± 2.8 | 35.10 ± 2.2 |
| 6c | 18.01 ± 1.4 | 16.14 ± 1.3 | 7a | 13.44 ± 1.1 | 11.62 ± 0.9 |
| 6d | 34.93 ± 2.1 | 38.46 ± 2.4 | 7b | 9.81 ± 0.7 | 7.21 ± 0.5 |
| 6e | 27.63 ± 1.8 | 31.57 ± 2.0 | 7c | 32.65 ± 2.0 | 29.81 ± 1.9 |
| 6f | 39.62 ± 2.4 | 45.65 ± 2.6 | 7d | 21.81 ± 1.6 | 28.74 ± 1.8 |
| 6g | 58.35 ± 3.3 | 64.11 ± 3.5 | 7e | 19.59 ± 1.4 | 12.90 ± 1.0 |
| 6h | 54.06 ± 3.1 | 48.61 ± 2.8 | 7f | 10.29 ± 0.9 | 6.94 ± 0.5 |
| 6i | 72.63 ± 3.6 | 63.59 ± 3.4 | 7g | 7.71 ± 0.5 | 8.11 ± 0.7 |
| DOXb | 4.50 ± 0.2 | 5.23 ± 0.3 | |||
| SORb | 9.18 ± 0.6 | 5.47 ± 0.3 | |||
As demonstrated in Table 2. Compounds 7b and 7g exhibited high selectivity toward HepG-2 cells, with selectivity index (SI) values of 8.5 and 11.2, respectively. Similarly, compounds 7a and 7f demonstrated favorable selectivity, with SI values of 5.7 and 7.6, respectively. On the other hand, compounds 6b, 6c, and 7e showed lower selectivity with SI values of 3, 2.4, and 2.7, respectively, in comparison to doxorubicin, which displayed the lowest SI value of 1.5 under the same conditions.
| In vitro cytotoxicity IC50 (μM) | |||
|---|---|---|---|
| Comp. | HepG-2 | WI-38 | SI |
| 6b | 14.03 ± 1.2 | 42.83 ± 2.5 | 3 |
| 6c | 18.01 ± 1.4 | 43.67 ± 2.9 | 2.4 |
| 7a | 13.44 ± 1.1 | 76.68 ± 3.9 | 5.7 |
| 7b | 9.81 ± 0.7 | 83.18 ± 2.3 | 8.5 |
| 7e | 19.59 ± 1.4 | 52.76 ± 3.1 | 2.7 |
| 7f | 10.29 ± 0.9 | 78.51 ± 3.6 | 7.6 |
| 7g | 7.71 ± 0.5 | 86.19 ± 2.7 | 11.2 |
| DOX | 4.50 ± 0.2 | 6.72 ± 0.5 | 1.5 |
| Comp. | MW (g mol−1) | VEGFR-2 |
|---|---|---|
| IC50 (μM) ± SD | ||
| 7a | 447.94 | 0.823 ± 0.023 |
| 7b | 443.53 | 0.209 ± 0.006 |
| 7f | 461.91 | 0.378 ± 0.01 |
| 7g | 457.55 | 0.072 ± 0.002 |
| Sorafenib | 464.83 | 0.069 ± 0.002 |
These results indicate that the anticancer activity of these compounds may be mediated through interaction with the active site of the VEGFR-2 target protein. This assumption will be validated by a molecular docking study.
The SAR study for VEGFR-2 inhibitory activity of the compounds demonstrated that the presence of a hydrogen bond-acceptor group (OCH3) in the hinge-binding region significantly enhanced the inhibitory activity. Moreover, increasing the hydrophobic substitution (p-tolyl instead of phenyl) in the hydrophobic tail led to a remarkable improvement in inhibitory potency.
Additionally, for the most potent compound 7g, EGFR inhibitory activity was also evaluated to investigate its dual kinase inhibition. The results, showing notable EGFR inhibition, are provided in the SI.
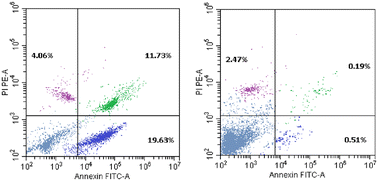
| Sample | Apoptosis | Necrosis | ||
|---|---|---|---|---|
| Total | Early | Late | ||
| 7g/HepG-2 | 35.42 | 19.63 | 11.73 | 4.06 |
| Control/HepG-2 | 3.17 | 0.51 | 0.19 | 2.47 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
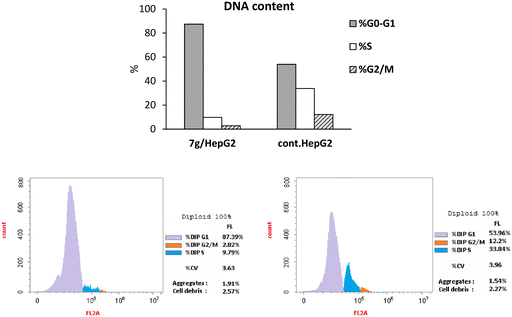
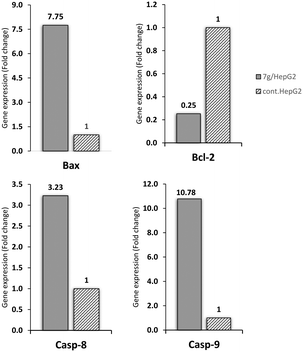
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Moreover, the apoptotic genes, caspase-8 and caspase-9, showed elevation in their expression levels by 3.23-fold and 10.776-fold, respectively, compared to the control cells. Based on these results, compound 7g is suggested to exert a significant apoptotic effect through both the intrinsic and extrinsic apoptotic pathways.
1). Moreover, the apoptotic genes, caspase-8 and caspase-9, showed elevation in their expression levels by 3.23-fold and 10.776-fold, respectively, compared to the control cells. Based on these results, compound 7g is suggested to exert a significant apoptotic effect through both the intrinsic and extrinsic apoptotic pathways.
3.3. Molecular docking study
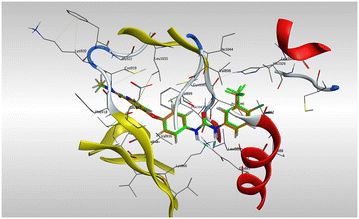
In light of the significant in vitro VEGFR-2 inhibitory activity exhibited by compounds 7a, 7b, 7f, and 7g, molecular docking studies were conducted to elucidate their potential binding modes within the VEGFR-2 active site. The docking simulations were performed using the Molecular Operating Environment (MOE, version 2024.06), employing a rigid ligand docking protocol and utilizing the crystal structure of VEGFR-2 co-crystallized with sorafenib (PDB ID: 3WZE), which was retrieved from the Protein Data Bank and used as the receptor model throughout the study. Prior to the docking studies, a validation step was performed by re-docking the co-crystallized sorafenib ligand into the VEGFR-2 active site. The validity of the docking protocol was confirmed by the superimposition of the redocked ligand onto the original co-crystallized ligand (Fig. 9), yielding a low RMSD value of 1.47 Å and successfully reproducing the experimentally observed binding mode.53The experimental binding mode of sorafenib revealed the formation of multiple key interactions within the VEGFR-2 active site, as illustrated in Fig. 10. Importantly, Glu885 formed two hydrogen bonds with the two nitrogen atoms of the semicarbazone group located in the DFG-binding moiety. Additionally, Asp1046 was involved in two types of interactions: a hydrogen bond with the carbonyl oxygen of the semicarbazone group, and a π–H interaction with the aromatic ring in the tail moiety. These two residues are well recognized as essential for VEGFR-2 inhibition. Furthermore, Cys919 formed two hydrogen bonds with the hinge-binding moiety of sorafenib, while Phe1047 engaged in a π–π stacking interaction with the central aromatic linker. The overall binding interaction energy was calculated to be −8.79 kcal mol−1, reflecting strong and stable binding within the active site of 3WZE.
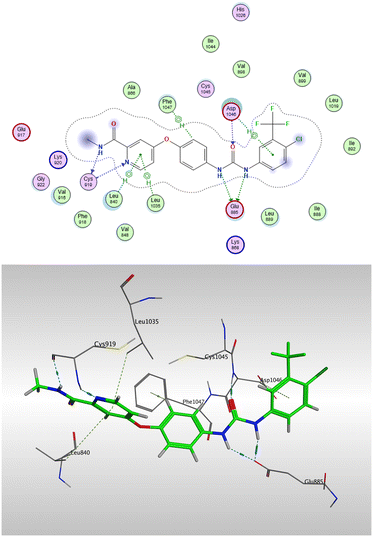 | ||
| Fig. 10 2D interaction diagram of sorafenib with VEGFR-2 active site (upper) and its 3D binding mode (lower). | ||
The docking results of compounds 7a, 7b, 7f, and 7g were analyzed and compared to sorafenib, the co-crystallized ligand, as demonstrated in Fig. 11. Among them, compound 7g (which exhibited the highest VEGFR-2 inhibitory activity) showed the most favorable binding energy (−8.90 kcal mol−1), while maintaining the key interactions with essential amino acid residues. Notably, Glu885 formed two hydrogen bonds with the thiosemicarbazone group located in the DFG-binding moiety.
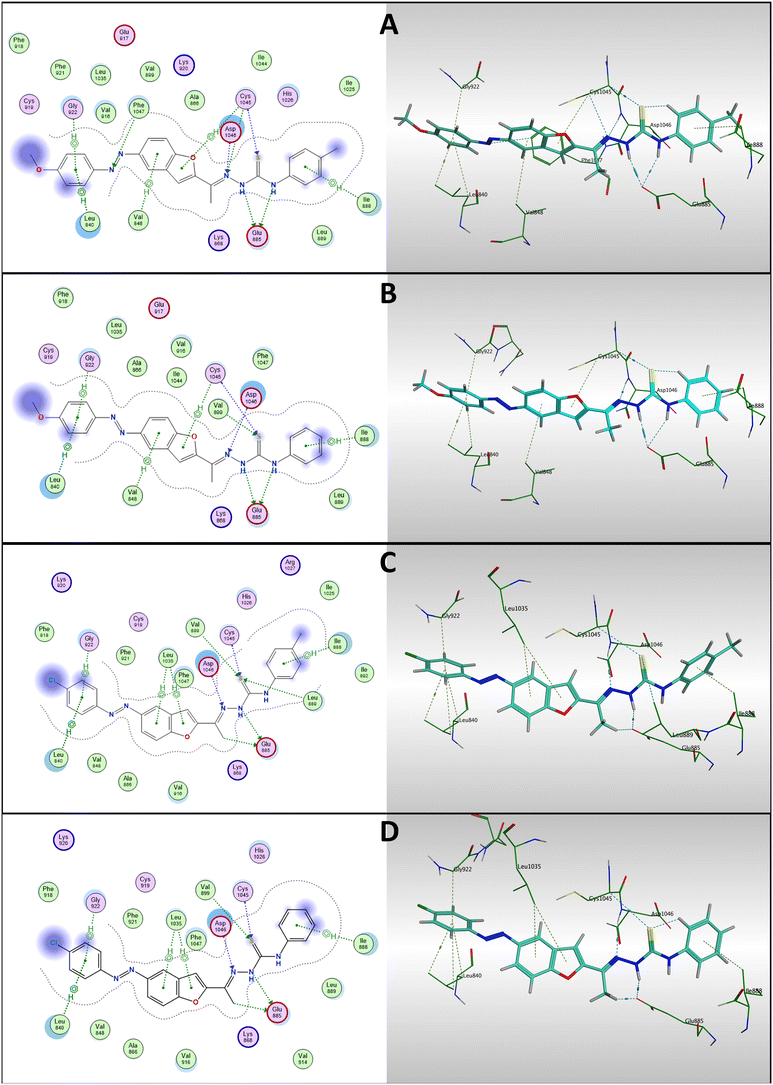 | ||
| Fig. 11 2D interaction diagrams of compounds (A) 7g, (B) 7b, (C) 7f, and (D) 7a with VEGFR-2 active site (left), and their corresponding 3D binding modes (right). | ||
Additionally, Asp1046 and Cys1045 each formed hydrogen bonds with the iminic nitrogen and sulfur atom, respectively, within the same moiety. Compound 7g also exhibited multiple π–H interactions with hydrophobic residues, including Leu840, Gly922, Val848, Cys1045, and Ile888, which interacted with the hinge-binding moiety, the aromatic linker, and the hydrophobic tail. These interactions support a well-oriented and stable binding conformation within the hydrophobic pocket, closely resembling the binding pose of sorafenib. Furthermore, the involvement of Phe1047 through an H-acceptor interaction (a residue also engaged by sorafenib) likely contributes to the enhanced binding affinity of compound 7g.
Compound 7b displayed a high binding affinity (−8.86 kcal mol−1), with almost the same binding mode that of compound 7g. It formed the same hydrogen bonds with Glu885, Asp1046 and Cys1045, contributing to stabilization within the DFG-binding region. Also, π–H interactions were observed with several hydrophobic residues similar to compound 7g. These interactions mirror those observed in compound 7g and sorafenib, highlighting compound 7b as another strong VEGFR-2 inhibitor candidate.
For compounds 7a and 7f, they exhibited a binding energy of −8.16 kcal mol−1 and −8.20 kcal mol−1 respectively and retained the essential binding features required for VEGFR-2 inhibition. Both formed only one hydrogen bond with Glu885 and another with Asp1046, reinforcing its anchoring in the DFG region. Additionally, they formed H–acceptor interactions via their sulfur atom with Cys1045. Hydrophobic π–H interactions were established with Leu840, Ile888, Gly922, and Leu1035, stabilizing the aromatic components of them both inside the hydrophobic pocket. Although slightly weaker in total binding energy of compounds 7a and 7f compared to compounds 7a and 7g, both still demonstrate a relevant binding orientation, which may contribute to moderate VEGFR-2 inhibition.
In summary, all tested compounds preserved critical pharmacophoric interactions, particularly with Glu885 and Asp1046, which are essential for VEGFR-2 inhibition. The superior performance of compounds 7a and 7g in terms of binding energy and interaction profiles, especially their engagement with the DFG-motif and the hydrophobic back pocket, highlights their potential as promising VEGFR-2 inhibitors. The additional interactions observed with Phe1047 in compound 7g may further enhance its binding stability, aligning it closely with the behavior of sorafenib.
3.4. Molecular dynamic simulation
While molecular docking is a useful method for studying enzyme–ligand interactions, it provides only a static representation and does not account for the dynamic behavior of molecules under physiological conditions. To address this limitation, a molecular dynamic simulation was conducted for the most potent compounds 7g in relation to the VEGR-2 enzyme (PDB: 3WZE). The simulation began with the best-docked poses of 7g from the docking study using this pose as the initial frame (frame-0). A 100 nanosecond molecular dynamics simulation was performed in an explicit hydration environment for all structures. System stability and molecular trajectories were evaluated based on the root-mean-square deviation (RMSD) of protein Cα-atoms and ligand heavy atoms.Analysis of the RMSD profiles of the VEGFR-2–compound 7g complex over the 100 ns MD simulation demonstrates that the system achieves and maintains a high degree of stability throughout the simulation time Fig. 12. The RMSD of the protein Cα fluctuates within a narrow range of 1–2 Å after an initial equilibration period, without exhibiting any substantial drift, which reflects good structural integrity of the kinase domain with no major conformational rearrangements. The ligand RMSD of compound 7g remains consistently close to the protein RMSD, fluctuating mainly between 1.2 and 1.8 Å. This overlap suggests that compound 7g is well-anchored within the ATP-binding site and does not undergo significant positional changes relative to the receptor. The absence of large spikes or long-duration deviations further supports a stable ligand–receptor association with no evidence of dissociation or conformational rearrangements.
 | ||
| Fig. 12 The 100 ns simulation RMSD values for the proteins VEGFR-2 Cα-atoms (blue) and compound 7g heavy atoms (red). | ||
In summary, both the protein and the ligand exhibited a narrow range of RMSD fluctuation, which highlights the high stability of VEGFR-2–compound 7g complex. The results confirm that compound 7g maintains a stable binding mode during the entire simulation, supporting its potential as a potent VEGFR-2 inhibitor.
Analysis of the simulation interaction diagram of compound 7g within the ATP-binding site of VEGFR-2 revealed that the ligand's stability within the binding pocket is predominantly supported by a network of direct hydrogen bonds, water-bridged hydrogen bonds, and hydrophobic interactions with multiple key residues (Fig. 13). The most dominant interaction is a strong, persistent hydrogen bond with Glu-885, which exhibits the highest occupancy among all contacts for 160% of the simulation time. This interaction is a hallmark of potent VEGFR-2 inhibitors, as Glu-885 is a key catalytic residue that anchors ligands within DFG motif area. A second major stabilizing interaction is the hydrogen bonding and water-bridged contact with Asp-1046, where the ligand maintains both direct and water-mediated polar interactions for more than 80% of the simulation time. Asp-1046 lies in the DFG motif, and its high interaction persistence indicates that compound 7g effectively stabilizes the DFG-out conformation, which is characteristic of type-II kinase inhibitors. In addition to these polar anchors, the ligand benefits from extensive hydrophobic interactions with residues such as Leu-840, Ala-866, Val-916, His-1026, Leu-1035 and Phe-1047. These residues form a deep hydrophobic pocket surrounding the ligand's aromatic scaffold, with interaction fractions in the range of 30% to 60% of the simulation time. His-1026 and Phe-1047 provides both hydrophobic contact and Pi–Pi stacking support for 31% and 22% of the trajectory, respectively, contributing to the alignment of the ligand aromatic core.
This combination of the stable DFG motif hydrogen bonds, robust water-mediated bridging, and dense hydrophobic interactions explains the stable RMSD behavior and supports the type-II inhibitor binding mode. Overall, the interaction profile highlights compound 7g as a ligand capable of establishing a balanced network of polar and hydrophobic contacts essential for potent and sustained VEGFR-2 inhibition.
3.5. Pharmacokinetic and physicochemical profiling
The pharmacokinetic properties and key physicochemical descriptors of the most active thiosemicarbazone derivatives (7a, 7b, 7f, and 7g) were evaluated using the SwissADME platform to explore their potential drug-likeness and absorption characteristics. The calculated parameters, including molecular weight, topological polar surface area (TPSA), hydrogen-bonding features, and lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P), are summarized in Table 5.
P), are summarized in Table 5.
| Parameter | 7a | 7b | 7f | 7g |
|---|---|---|---|---|
| Molecular weight | 447.94 | 443.53 | 461.97 | 457.55 |
| Heavy atoms | 31 | 32 | 32 | 33 |
| Aromatic heavy atoms | 21 | 21 | 21 | 21 |
| Fraction Csp3 | 0.04 | 0.08 | 0.08 | 0.12 |
| Rotatable bonds | 7 | 8 | 7 | 8 |
| H-bond acceptors | 4 | 5 | 4 | 5 |
| H-bond donors | 2 | 2 | 2 | 2 |
| Molar refractivity | 129.53 | 131.01 | 134.49 | 135.97 |
| TPSA (Å2) | 106.37 | 115.60 | 106.37 | 115.60 |
Consensus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
5.73 | 5.16 | 6.07 | 5.49 |
| GI absorption | Low | Low | Low | Low |
| BBB permeant | No | No | No | No |
| Lipinski violations | 0 | 0 | 0 | 0 |
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.55 |
All compounds showed molecular weights within the range favorable for oral candidates (<500 g mol−1) and displayed moderate polarity, with TPSA values between 106–116 Å2. Each molecule possessed two hydrogen-bond donors and four to five acceptors, consistent with good balance between permeability and solubility. The consensus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values (5.16–6.07) suggest sufficient lipophilicity to support membrane diffusion. None of the compounds were predicted to cross the blood–brain barrier, indicating limited central nervous system exposure. Gastrointestinal absorption was predicted as low across the series, which may reflect their extended conjugation and limited flexibility. All derivatives satisfied Lipinski's rule of five and had identical bioavailability scores (0.55), denoting acceptable oral-drug profiles.
P values (5.16–6.07) suggest sufficient lipophilicity to support membrane diffusion. None of the compounds were predicted to cross the blood–brain barrier, indicating limited central nervous system exposure. Gastrointestinal absorption was predicted as low across the series, which may reflect their extended conjugation and limited flexibility. All derivatives satisfied Lipinski's rule of five and had identical bioavailability scores (0.55), denoting acceptable oral-drug profiles.
In addition, we characterized the electronic properties of compound 7g by DFT calculations.54,55 The optimized geometry of compound 7g is shown along with its HOMO and LUMO electron density distributions in Fig. 14. The frontier orbital maps indicate uniform electron delocalization over the molecular scaffold, consistent with electronic stability. The DFT-calculated parameters for the 6- and 7-series are summarized in SI.
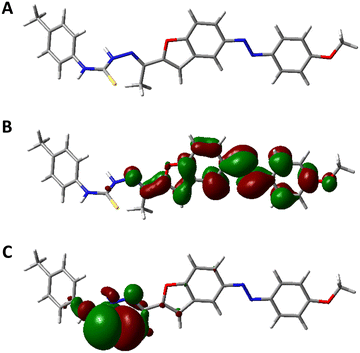 | ||
| Fig. 14 Frontier Kohn–Sham molecular orbitals of compounds 7g; (A) optimized structure, (B) LUMO, (C) HOMO. | ||
Across both series, the compounds show comparable electronic features with only slight variations in their energy levels. Within the 7-series, 7g exhibits a HOMO–LUMO energy gap (ΔE) of 3.284 eV, which corresponds well with its high VEGFR-2 inhibitory potency (IC50 = 0.072 μM) and strong cytotoxic activity against HepG-2 and HCT-116 cells (IC50 = 7.71 μM and 8.11 μM, respectively). Compounds 7b (ΔE = 3.351 eV) and 7f (ΔE = 3.472 eV) also display relatively narrow energy gaps, aligning with their moderate activities. In contrast, members of the 6-series such as 6d (ΔE = 2.941 eV) show similar electronic behaviour but lower biological efficacy, indicating that factors beyond the energy gap contribute to activity differences.
Additional descriptors such as nucleophilicity (Nu), hardness (η), softness (σ), and chemical potential (μ) provide further context. Compound 7g has Nu = 0.201 eV, η = 1.642 eV, σ = 0.304 eV, and μ = −4.043 eV, reflecting a balanced electronic profile supportive of stable yet reactive character. Other derivatives with slightly higher Nu values, including 6e and 7e, did not show improved activity, suggesting that these parameters alone do not determine biological outcomes.
4. Experimental
4.1. Chemistry
All reagents utilized in the synthesis were commercially obtained from Sigma-Aldrich, Merck, and Alfa Aesar, and were used as received without further purification. The progress of the reactions was monitored using thin-layer chromatography (TLC), employing various solvent systems as mobile phases and silica gel 60 F254 (Merck) as the stationary phase. Spots were visualized under ultraviolet light at wavelengths of 254 and 366 nm. Melting points were determined using a Stuart SMP10 melting point apparatus and are reported without correction. NMRs were recorded with Bruker AVANCE II (1H NMR 600 or 400, 13C NMR 150 or 100) at Osaka University, Japan. Chemical shifts (δ) are reported in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal reference. FT-IR spectra were recorded on a JASCO FT-IR system (FT/IR4100), and the absorption bands are expressed in wavenumbers (cm−1) at Osaka University, Japan. ESI mass spectra were obtained by using JMS-T100LC (JEOL) at Osaka University, Japan. Compounds 3a and 3b were synthesized following previously reported procedures.39,40 The synthetic routes for the final target compounds are presented in Scheme 1.(E)-1-(5-((4-Chlorophenyl)diazenyl)benzofuran-2-yl)ethan-1-one (4a). Yellowish brown powder; yield 90%, m.p. 182–183 °C. IR (KBr, cm−1) 3113, 3094 (CH sp2), 2925 (CH sp3), 1682 (conjugated CO). 1H NMR (DMSO-d6, 400 MHz) δ 8.37 (s, 1H), 8.07 (d, J = 8.8, 1H), 8.03 (s, 1H), 7.91–7.88 (m, 3H), 7.65 (d, J = 8.5 Hz, 2H), 2.57 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 188.42, 157.03, 154.09, 150.91, 149.20, 136.52, 130.18, 128.27, 124.77, 122.90, 120.10, 115.49, 113.89, 27.08. HRMS (ESI): m/z calcd for C16H11ClN2O2 [M + Na]+: 321.0401; found: 321.0406.
(E)-1-(5-((4-Methoxyphenyl)diazenyl)benzofuran-2-yl)ethan-1-one (4b). Orange-brown powder; yield 89%, m.p. 140–142 °C. IR (KBr, cm−1) 3116, 3068 (CH sp2), 2974, 2847, 2838, (CH sp3), 1673 (conjugated CO). 1H NMR (DMSO-d6, 400 MHz) δ 8.26 (s, 1H), 8.01 (dd, J = 8.8, 1.7 Hz, 1H), 7.97 (s, 1H), 7.87 (d, J = 8.7 Hz, 2H), 7.83 (d, J = 8.8 Hz, 1H), 7.10 (d, J = 8.7 Hz, 2H), 3.83 (s, 3H), 2.55 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 188.40, 162.54, 156.54, 153.94, 149.43, 146.56, 128.17, 125.11, 122.79, 119.08, 115.49, 115.17, 113.67, 56.19, 27.06. HRMS (ESI): m/z calcd for C17H14N2O3 [M + Na]+: 317.0897; found: 317.0895.
(E)-1-(4-Chlorophenyl)-2-(2-((E)-1-hydrazineylideneethyl)benzofuran-5-yl)diazene (5a). Yellowish brown powder; yield 78%, m.p. 176–178 °C. IR (KBr, cm−1) 3355, 3216 (NH2), 3055 (CH sp2), 2952, 2923 (CH sp3). 1H NMR (DMSO-d6, 600 MHz) δ 8.07 (d, J = 1.9 Hz, 1H), 7.85 (d, J = 8.5 Hz, 2H), 7.81 (dd, J = 8.5, 2.0 Hz, 1H), 7.65 (d, J = 8.7 Hz, 1H), 7.60 (d, J = 8.8 Hz, 2H), 7.02 (s, 1H), 6.94 (s, 2H), 2.02 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 158.28, 156.40, 151.00, 148.75, 136.07, 133.12, 130.13, 130.07, 124.58, 119.88, 116.26, 112.11, 102.66, 11.68. HRMS (ESI): m/z calcd for C16H13ClN4O [M + Na]+: 335.0670; found: 335.0670.
(E)-1-(2-((E)-1-Hydrazineylideneethyl)benzofuran-5-yl)-2-(4-methoxyphenyl)diazene (5b). Yellowish brown powder; yield 76%, m.p. 148–149 °C. IR (KBr, cm−1) 3375, 3281 (NH2), 3216 (CH sp2), 2969, 2841 (CH sp3). 1H NMR (DMSO-d6, 600 MHz) δ 8.02 (d, J = 1.8 Hz, 1H), 7.85 (d, J = 8.8 Hz, 2H), 7.77 (dd, J = 8.7, 1.9 Hz, 1H), 7.62 (d, J = 8.7 Hz, 1H), 7.09 (d, J = 8.8 Hz, 2H), 7.01 (s, 1H), 6.89 (s, 2H), 3.82 (s, 3H), 2.02 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 162.22, 158.08, 155.85, 148.96, 146.65, 133.28, 130.03, 124.88, 119.55, 115.52, 115.10, 111.95, 102.71, 56.15, 11.69. HRMS (ESI): m/z calcd for C17H16N4O2 [M + Na]+: 331.1165; found: 331.1161.
(E)-1-(4-Chlorophenyl)-2-(2-((E)-1-(((E)-3,4-dimethoxybenzylidene)hydrazineylidene)ethyl)benzofuran-5-yl)diazene (6a). Yellow powder; yield 89%, m.p. 172–174 °C. IR (KBr, cm−1) 3006, 3963 (CH sp2) 2936, 2843 (CH sp3) 1618 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1600 (C
C), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 8.57 (s, 1H), 8.31 (s, 1H), 8.03 (d, J = 8.9 Hz, 1H), 7.96 (d, J = 8.5 Hz, 2H), 7.89 (d, J = 8.8 Hz, 1H), 7.73 (s, 1H), 7.70 (d, J = 8.5 Hz, 2H), 7.55 (s, 1H), 7.45 (d, J = 8.2 Hz, 1H), 7.11 (d, J = 8.3 Hz, 1H), 3.86 (s, 6H), 2.55 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 160.21, 157.01, 155.82, 152.35, 149.53, 149.00, 141.17, 138.98, 136.34, 130.17, 129.20, 127.38, 124.72, 124.21, 121.61, 117.94, 113.05, 112.02, 110.66, 110.07, 56.19, 55.99, 14.89. HRMS (ESI): m/z calcd for C25H21ClN4O3 [M + Na]+: 483.1194; found: 483.1198.
C). 1H NMR (DMSO-d6, 400 MHz) δ 8.57 (s, 1H), 8.31 (s, 1H), 8.03 (d, J = 8.9 Hz, 1H), 7.96 (d, J = 8.5 Hz, 2H), 7.89 (d, J = 8.8 Hz, 1H), 7.73 (s, 1H), 7.70 (d, J = 8.5 Hz, 2H), 7.55 (s, 1H), 7.45 (d, J = 8.2 Hz, 1H), 7.11 (d, J = 8.3 Hz, 1H), 3.86 (s, 6H), 2.55 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 160.21, 157.01, 155.82, 152.35, 149.53, 149.00, 141.17, 138.98, 136.34, 130.17, 129.20, 127.38, 124.72, 124.21, 121.61, 117.94, 113.05, 112.02, 110.66, 110.07, 56.19, 55.99, 14.89. HRMS (ESI): m/z calcd for C25H21ClN4O3 [M + Na]+: 483.1194; found: 483.1198.
(E)-1-(4-Chlorophenyl)-2-(2-((E)-1-(((E)-3,4,5-trimethoxybenzylidene)hydrazineylidene)ethyl)benzofuran-5-yl)diazene (6b). Yellow powder; yield 92%, m.p. 178–180 °C. IR (KBr, cm−1) 3085 (CH sp2), 2995, 2952, 2936, 2835 (CH sp3), 1619 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 600 MHz) δ 8.48 (s, 1H), 8.27 (s, 1H), 7.99 (dd, J = 9.0, 1.2 Hz, 1H), 7.91 (d, J = 8.7 Hz, 2H), 7.86 (d, J = 9.1 Hz, 1H), 7.73 (s, 1H), 7.65 (d, J = 8.8 Hz, 2H), 7.22 (s, 2H), 3.82 (s, 6H), 3.70 (s, 3H), 2.50 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 159.60, 157.02, 156.18, 155.64, 153.71, 150.98, 149.01, 140.79, 136.35, 130.17, 130.09, 129.16, 124.72, 121.66, 118.01, 113.08, 110.90, 106.23, 60.72, 56.50, 14.97. HRMS (ESI): m/z calcd for C26H23ClN4O4 [M + Na]+: 513.1300; found: 513.1290.
C). 1H NMR (DMSO-d6, 600 MHz) δ 8.48 (s, 1H), 8.27 (s, 1H), 7.99 (dd, J = 9.0, 1.2 Hz, 1H), 7.91 (d, J = 8.7 Hz, 2H), 7.86 (d, J = 9.1 Hz, 1H), 7.73 (s, 1H), 7.65 (d, J = 8.8 Hz, 2H), 7.22 (s, 2H), 3.82 (s, 6H), 3.70 (s, 3H), 2.50 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 159.60, 157.02, 156.18, 155.64, 153.71, 150.98, 149.01, 140.79, 136.35, 130.17, 130.09, 129.16, 124.72, 121.66, 118.01, 113.08, 110.90, 106.23, 60.72, 56.50, 14.97. HRMS (ESI): m/z calcd for C26H23ClN4O4 [M + Na]+: 513.1300; found: 513.1290.
(E)-1-(4-Methoxyphenyl)-2-(2-((E)-1-(((E)-3,4,5-trimethoxybenzylidene)hydrazineylidene)ethyl)benzofuran-5-yl)diazene (6c). Yellow powder; yield 88%, m.p. 170–172 °C. IR (KBr, cm−1) 2952, 2840 (CH sp3), 1616 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 8.47 (s, 1H), 8.18 (d, J = 1.8 Hz, 1H), 7.93 (dd, J = 8.8, 1.8 Hz, 1H), 7.88 (d, J = 8.9 Hz, 2H), 7.80 (d, J = 8.8 Hz, 1H), 7.68 (s, 1H), 7.21 (s, 2H), 7.11 (d, J = 8.9 Hz, 2H), 3.84 (s, 3H), 3.80 (s, 6H), 3.70 (s, 3H), 2.48 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.41, 159.56, 156.50, 156.24, 155.44, 153.71, 149.21, 146.62, 140.77, 130.12, 129.05, 125.02, 121.42, 117.13, 115.15, 112.86, 110.88, 106.21, 60.72, 56.49, 56.18, 14.96. HRMS (ESI): m/z calcd for C27H26N4O5 [M + Na]+: 509.1795; found: 509.1785.
C). 1H NMR (DMSO-d6, 400 MHz) δ 8.47 (s, 1H), 8.18 (d, J = 1.8 Hz, 1H), 7.93 (dd, J = 8.8, 1.8 Hz, 1H), 7.88 (d, J = 8.9 Hz, 2H), 7.80 (d, J = 8.8 Hz, 1H), 7.68 (s, 1H), 7.21 (s, 2H), 7.11 (d, J = 8.9 Hz, 2H), 3.84 (s, 3H), 3.80 (s, 6H), 3.70 (s, 3H), 2.48 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.41, 159.56, 156.50, 156.24, 155.44, 153.71, 149.21, 146.62, 140.77, 130.12, 129.05, 125.02, 121.42, 117.13, 115.15, 112.86, 110.88, 106.21, 60.72, 56.49, 56.18, 14.96. HRMS (ESI): m/z calcd for C27H26N4O5 [M + Na]+: 509.1795; found: 509.1785.
(E)-1-(4-Chlorophenyl)-2-(2-((E)-1-(((E)-2,4,5-trimethoxybenzylidene)hydrazineylidene)ethyl)benzofuran-5-yl)diazene (6d). Yellow powder; yield 82%, m.p. 181–183 °C. IR (KBr, cm−1) 3001 (CH sp2), 2964, 2936, 2838 (CH sp3), 1619, 1605 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 600 MHz) δ 8.75 (s, 1H), 8.25 (d, J = 1.6 Hz, 1H), 7.98 (dd, J = 8.7, 1.6 Hz, 1H), 7.91 (d, J = 8.4 Hz, 2H), 7.84 (d, J = 8.7 Hz, 1H), 7.68 (s, 1H), 7.65 (d, J = 8.5 Hz, 2H), 7.50 (s, 1H), 6.76 (s, 1H), 3.87 (s, 3H), 3.86 (s, 3H), 3.75 (s, 3H), 2.50 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 156.99, 155.96, 155.93, 155.84, 155.57, 155.52, 154.06, 150.99, 148.98, 143.71, 136.32, 130.16, 129.24, 124.71, 121.58, 117.83, 113.01, 110.34, 109.15, 98.27, 57.19, 56.42, 14.87. HRMS (ESI): m/z calcd for C26H23ClN4O4 [M + Na]+: 513.1300; found: 513.1297.
C). 1H NMR (DMSO-d6, 600 MHz) δ 8.75 (s, 1H), 8.25 (d, J = 1.6 Hz, 1H), 7.98 (dd, J = 8.7, 1.6 Hz, 1H), 7.91 (d, J = 8.4 Hz, 2H), 7.84 (d, J = 8.7 Hz, 1H), 7.68 (s, 1H), 7.65 (d, J = 8.5 Hz, 2H), 7.50 (s, 1H), 6.76 (s, 1H), 3.87 (s, 3H), 3.86 (s, 3H), 3.75 (s, 3H), 2.50 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 156.99, 155.96, 155.93, 155.84, 155.57, 155.52, 154.06, 150.99, 148.98, 143.71, 136.32, 130.16, 129.24, 124.71, 121.58, 117.83, 113.01, 110.34, 109.15, 98.27, 57.19, 56.42, 14.87. HRMS (ESI): m/z calcd for C26H23ClN4O4 [M + Na]+: 513.1300; found: 513.1297.
(E)-1-(4-Methoxyphenyl)-2-(2-((E)-1-(((E)-2,4,5-trimethoxybenzylidene)hydrazineylidene)ethyl)benzofuran-5-yl)diazene (6e). Yellow powder; yield 80%, m.p. 174–175 °C. IR (KBr, cm−1) 2997, 2937, 2837 (CH sp3), 1603 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 8.75 (s, 1H), 8.17 (s, 1H), 7.92–7.88 (m, 3H), 7.79 (d, J = 8.5 Hz, 1H), 7.64 (s, 1H), 7.49 (s, 1H), 7.11 (d, J = 8.2 Hz, 2H), 6.75 (s, 1H). 3.84 (s, 9H), 3.74 (s, 3H), 2.49 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.39, 156.47, 155.89, 155.75, 155.50, 154.03, 149.18, 146.63, 143.70, 129.14, 125.01, 123.54, 121.33, 116.95, 115.15, 113.73, 112.79, 110.31, 109.14, 98.25, 57.18, 56.41, 56.18, 14.86. HRMS (ESI): m/z calcd for C27H26N4O5 [M + Na]+: 509.1795; found: 509.1788.
C). 1H NMR (DMSO-d6, 400 MHz) δ 8.75 (s, 1H), 8.17 (s, 1H), 7.92–7.88 (m, 3H), 7.79 (d, J = 8.5 Hz, 1H), 7.64 (s, 1H), 7.49 (s, 1H), 7.11 (d, J = 8.2 Hz, 2H), 6.75 (s, 1H). 3.84 (s, 9H), 3.74 (s, 3H), 2.49 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.39, 156.47, 155.89, 155.75, 155.50, 154.03, 149.18, 146.63, 143.70, 129.14, 125.01, 123.54, 121.33, 116.95, 115.15, 113.73, 112.79, 110.31, 109.14, 98.25, 57.18, 56.41, 56.18, 14.86. HRMS (ESI): m/z calcd for C27H26N4O5 [M + Na]+: 509.1795; found: 509.1788.
(E)-1-(2-((E)-1-(((E)-4-Methoxybenzylidene)hydrazineylidene)ethyl)benzofuran-5-yl)-2-(4-methoxyphenyl)diazene (6f). Yellow powder; yield 80%, m.p. 182–183 °C. IR (KBr, cm−1) 3065, 3008 (CH sp2), 2933, 2840 (CH sp3), 1614 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 8.54 (s, 1H), 8.18 (d, J = 1.8 Hz, 1H), 7.93 (dd, J = 8.8, 2.0 Hz, 1H), 7.88 (d, J = 9.0 Hz, 2H), 7.83 (d, J = 8.7 Hz, 2H), 7.80 (d, J = 8.9 Hz, 1H), 7.67 (s, 1H), 7.11 (d, J = 8.9 Hz, 2H), 7.04 (d, J = 8.7 Hz, 2H), 3.84 (s, 3H), 3.80 (s, 3H), 2.48 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.57, 160.25, 156.65, 156.29, 155.75, 149.34, 146.76, 131.02, 130.67, 129.25, 127.44, 125.18, 121.52, 117.24, 115.32, 115.13, 113.00, 110.89, 56.35, 56.12, 15.00. HRMS (ESI): m/z calcd for C25H22N4O3 [M + Na]+: 449.1584; found: 449.1570.
C). 1H NMR (DMSO-d6, 400 MHz) δ 8.54 (s, 1H), 8.18 (d, J = 1.8 Hz, 1H), 7.93 (dd, J = 8.8, 2.0 Hz, 1H), 7.88 (d, J = 9.0 Hz, 2H), 7.83 (d, J = 8.7 Hz, 2H), 7.80 (d, J = 8.9 Hz, 1H), 7.67 (s, 1H), 7.11 (d, J = 8.9 Hz, 2H), 7.04 (d, J = 8.7 Hz, 2H), 3.84 (s, 3H), 3.80 (s, 3H), 2.48 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.57, 160.25, 156.65, 156.29, 155.75, 149.34, 146.76, 131.02, 130.67, 129.25, 127.44, 125.18, 121.52, 117.24, 115.32, 115.13, 113.00, 110.89, 56.35, 56.12, 15.00. HRMS (ESI): m/z calcd for C25H22N4O3 [M + Na]+: 449.1584; found: 449.1570.
4-((E)-(((E)-1-(5-((E)-(4-Chlorophenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazineylidene)methyl)phenol (6g). Yellow powder; yield 88%, m.p. 238–240 °C. IR (KBr, cm−1) 3309 (OH), 3127, 3029 (CH sp2), 2930 (CH sp3), 1608 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 600 MHz) δ 10.30 (s, 1H), 8.50 (s, 1H), 8.25 (d, J = 1.2 Hz, 1H), 7.98 (dd, J = 8.7, 1.6 Hz, 1H), 7.90 (d, J = 8.7 Hz, 2H), 7.84 (d, J = 8.9 Hz, 1H), 7.72 (d, J = 8.6 Hz, 2H), 7.67 (s, 1H), 7.65 (d, J = 8.6 Hz, 2H), 6.85 (d, J = 8.6 Hz, 2H), 2.47 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 161.51, 160.56, 156.96, 155.88, 155.68, 150.94, 148.94, 136.29, 131.11, 130.10, 129.17, 124.68, 121.45, 117.91, 112.94, 110.37, 14.76. HRMS (ESI): m/z calcd for C23H17ClN4O2 [M + H]+: 417.1118; found: 417.1107.
C). 1H NMR (DMSO-d6, 600 MHz) δ 10.30 (s, 1H), 8.50 (s, 1H), 8.25 (d, J = 1.2 Hz, 1H), 7.98 (dd, J = 8.7, 1.6 Hz, 1H), 7.90 (d, J = 8.7 Hz, 2H), 7.84 (d, J = 8.9 Hz, 1H), 7.72 (d, J = 8.6 Hz, 2H), 7.67 (s, 1H), 7.65 (d, J = 8.6 Hz, 2H), 6.85 (d, J = 8.6 Hz, 2H), 2.47 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 161.51, 160.56, 156.96, 155.88, 155.68, 150.94, 148.94, 136.29, 131.11, 130.10, 129.17, 124.68, 121.45, 117.91, 112.94, 110.37, 14.76. HRMS (ESI): m/z calcd for C23H17ClN4O2 [M + H]+: 417.1118; found: 417.1107.
4-((E)-(((E)-1-(5-((E)-(4-Methoxyphenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazineylidene)methyl)phenol (6h). Yellow powder; yield 87%, m.p. 223–224 °C. IR (KBr, cm−1) 3079 (OH), 3006 (CH sp2), 2966, 2835 (CH sp3), 1606 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 10.12 (s, 1H), 8.49 (s, 1H), 8.15 (d, J = 1.6 Hz, 1H), 7.90 (dd, J = 8.9, 1.8 Hz, 1H), 7.87 (d, J = 8.8 Hz, 2H), 7.77 (d, J = 8.8 Hz, 1H), 7.72 (d, J = 8.5 Hz, 2H), 7.61 (s, 1H), 7.09 (d, J = 8.9 Hz, 2H), 6.85 (d, J = 8.5 Hz, 2H), 3.82 (s, 3H), 2.45 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.37, 161.18, 160.42, 156.46, 155.79, 155.69, 149.18, 146.62, 131.07, 129.09, 124.99, 121.25, 117.03, 116.35, 115.12, 112.75, 110.40, 56.15, 14.76. HRMS (ESI): m/z calcd for C24H20N4O3 [M + Na]+: 435.1428; found: 435.1421.
C). 1H NMR (DMSO-d6, 400 MHz) δ 10.12 (s, 1H), 8.49 (s, 1H), 8.15 (d, J = 1.6 Hz, 1H), 7.90 (dd, J = 8.9, 1.8 Hz, 1H), 7.87 (d, J = 8.8 Hz, 2H), 7.77 (d, J = 8.8 Hz, 1H), 7.72 (d, J = 8.5 Hz, 2H), 7.61 (s, 1H), 7.09 (d, J = 8.9 Hz, 2H), 6.85 (d, J = 8.5 Hz, 2H), 3.82 (s, 3H), 2.45 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.37, 161.18, 160.42, 156.46, 155.79, 155.69, 149.18, 146.62, 131.07, 129.09, 124.99, 121.25, 117.03, 116.35, 115.12, 112.75, 110.40, 56.15, 14.76. HRMS (ESI): m/z calcd for C24H20N4O3 [M + Na]+: 435.1428; found: 435.1421.
2-((E)-(((E)-1-(5-((E)-(4-Methoxyphenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazineylidene)methyl)phenol (6i). Yellow powder; yield 82%, m.p. 171–173 °C. IR (KBr, cm−1) 3121, 3069, 3007 (CH sp2), 2934 (OH), 2840 (CH sp3), 1615 (N
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1602 (C
N), 1602 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 11.28 (s, 1H), 8.91 (s, 1H), 8.20 (s, 1H), 7.95 (d, J = 8.8 Hz, 1H), 7.89 (d, J = 7.9 Hz, 2H), 7.81 (d, J = 8.3 Hz, 1H), 7.78 (s, 1H), 7.70 (d, J = 6.4 Hz, 1H), 7.41–7.35 (m, 1H), 7.11 (d, J = 7.3 Hz, 2H), 6.99–6.91 (m, 2H), 3.84 (s, 3H), 2.46 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.43, 159.37, 157.14, 156.65, 155.06, 149.24, 146.60, 133.84, 131.75, 129.01, 125.05, 123.56, 121.67, 120.16, 119.08, 117.28, 117.01, 115.17, 112.94, 111.92, 56.19, 15.13. HRMS (ESI): m/z calcd for C24H20N4O3 [M + Na]+: 435.1428; found: 435.1417.
C). 1H NMR (DMSO-d6, 400 MHz) δ 11.28 (s, 1H), 8.91 (s, 1H), 8.20 (s, 1H), 7.95 (d, J = 8.8 Hz, 1H), 7.89 (d, J = 7.9 Hz, 2H), 7.81 (d, J = 8.3 Hz, 1H), 7.78 (s, 1H), 7.70 (d, J = 6.4 Hz, 1H), 7.41–7.35 (m, 1H), 7.11 (d, J = 7.3 Hz, 2H), 6.99–6.91 (m, 2H), 3.84 (s, 3H), 2.46 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.43, 159.37, 157.14, 156.65, 155.06, 149.24, 146.60, 133.84, 131.75, 129.01, 125.05, 123.56, 121.67, 120.16, 119.08, 117.28, 117.01, 115.17, 112.94, 111.92, 56.19, 15.13. HRMS (ESI): m/z calcd for C24H20N4O3 [M + Na]+: 435.1428; found: 435.1417.
5-((E)-(((E)-1-(5-((E)-(4-Chlorophenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazineylidene)methyl)-2-methoxyphenol (6j). Yellow powder; yield 78%, m.p. 198–200 °C. IR (KBr, cm−1) 3518 (OH), 3028 (CH sp2), 2948, 2847 (CH sp3), 1616 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 9.29 (s, 1H), 8.44 (s, 1H), 8.24 (d, J = 1.6 Hz, 1H), 7.97 (dd, J = 8.8, 1.8 Hz, 1H), 7.89 (d, J = 8.6 Hz, 2H), 7.82 (d, J = 8.8 Hz, 1H), 7.82 (d, J = 8.8 Hz, 1H), 7.66 (s, 1H), 7.63 (d, J = 8.6 Hz, 2H), 7.39 (d, J = 1.8 Hz, 1H), 7.24 (dd, J = 8.1, 1.7 Hz, 1H), 7.00 (d, J = 8.3 Hz, 1H), 3.80 (s, 1H), 2.46 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 160.46, 157.00, 155.84, 151.47, 150.98, 148.98, 147.33, 136.32, 131.76, 130.15, 129.56, 129.19, 127.50, 124.71, 122.82, 121.55, 117.94, 114.02, 113.01, 112.32, 110.59, 56.17, 14.80. HRMS (ESI): m/z calcd for C24H19ClN4O3 [M + Na]+: 469.1038; found: 469.1031.
C). 1H NMR (DMSO-d6, 400 MHz) δ 9.29 (s, 1H), 8.44 (s, 1H), 8.24 (d, J = 1.6 Hz, 1H), 7.97 (dd, J = 8.8, 1.8 Hz, 1H), 7.89 (d, J = 8.6 Hz, 2H), 7.82 (d, J = 8.8 Hz, 1H), 7.82 (d, J = 8.8 Hz, 1H), 7.66 (s, 1H), 7.63 (d, J = 8.6 Hz, 2H), 7.39 (d, J = 1.8 Hz, 1H), 7.24 (dd, J = 8.1, 1.7 Hz, 1H), 7.00 (d, J = 8.3 Hz, 1H), 3.80 (s, 1H), 2.46 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 160.46, 157.00, 155.84, 151.47, 150.98, 148.98, 147.33, 136.32, 131.76, 130.15, 129.56, 129.19, 127.50, 124.71, 122.82, 121.55, 117.94, 114.02, 113.01, 112.32, 110.59, 56.17, 14.80. HRMS (ESI): m/z calcd for C24H19ClN4O3 [M + Na]+: 469.1038; found: 469.1031.
2-Methoxy-5-((E)-(((E)-1-(5-((E)-(4-methoxyphenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazineylidene)methyl)phenol (6k). Yellow powder; yield 77%, m.p. 185–187 °C. IR (KBr, cm−1) 3195 (OH), 3071. 3002 (CH sp2), 2941, 2943 (CH sp3), 1615 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 9.32 (s, 1H), 8.44 (s, 1H), 8.14 (d, J = 1.5 Hz, 1H), 7.90 (dd, J = 8.8, 1.8 Hz, 1H), 7.86 (d, J = 8.8 Hz, 2H), 7.76 (d, J = 8.8 Hz, 1H), 7.61 (s, 1H), 7.39 (d, J = 1.6 Hz, 1H), 7.23 (d, J = 8.2 Hz, 1H), 7.09 (d, J = 8.8 Hz, 2H), 6.99 (d, J = 8.3 Hz, 1H), 3.82 (s, 3H), 3.80 (s, 3H), 2.45 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.39, 160.39, 156.48, 155.92, 155.63, 151.45, 149.19, 147.33, 146.62, 129.09, 127.53, 125.01, 122.79, 121.32, 117.07, 115.15, 114.01, 112.80, 112.32, 110.58, 56.17, 14.79. HRMS (ESI): m/z calcd for C25H22N4O4 [M + Na]+: 465.1533; found: 465.1525.
C). 1H NMR (DMSO-d6, 400 MHz) δ 9.32 (s, 1H), 8.44 (s, 1H), 8.14 (d, J = 1.5 Hz, 1H), 7.90 (dd, J = 8.8, 1.8 Hz, 1H), 7.86 (d, J = 8.8 Hz, 2H), 7.76 (d, J = 8.8 Hz, 1H), 7.61 (s, 1H), 7.39 (d, J = 1.6 Hz, 1H), 7.23 (d, J = 8.2 Hz, 1H), 7.09 (d, J = 8.8 Hz, 2H), 6.99 (d, J = 8.3 Hz, 1H), 3.82 (s, 3H), 3.80 (s, 3H), 2.45 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 162.39, 160.39, 156.48, 155.92, 155.63, 151.45, 149.19, 147.33, 146.62, 129.09, 127.53, 125.01, 122.79, 121.32, 117.07, 115.15, 114.01, 112.80, 112.32, 110.58, 56.17, 14.79. HRMS (ESI): m/z calcd for C25H22N4O4 [M + Na]+: 465.1533; found: 465.1525.
(E)-2-(1-(5-((E)-(4-Chlorophenyl)diazenyl)benzofuran-2-yl)ethylidene)-N-phenylhydrazine-1-carbothioamide (7a). Yellow powder; yield 86%, m.p. 222–225 °C. IR (KBr, cm−1) 3318 (NH), 3176 (NH), 2973 (CH sp3), 1600 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 600 MHz) δ 10.95 (s, 1H), 10.09 (s, 1H), 8.21 (s, 1H), 7.91 (m, 3H), 7.85–7.76 (m, 2H), 7.64 (d, J = 8.6 Hz, 2H), 7.57 (d, J = 7.8 Hz, 2H), 7.37 (t, J = 7.7 Hz, 2H), 7.20 (t, J = 7.3 Hz, 1H), 2.41 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 177.47, 156.79, 155.87, 150.99, 149.02, 140.35, 139.45, 136.30, 130.15, 129.51, 128.77, 126.15, 124.70, 122.34, 120.42, 118.04, 112.94, 108.23, 14.28. HRMS (ESI): m/z calcd for C23H18ClN5OS [M + Na]+: 470.0818; found: 470.0807.
C). 1H NMR (DMSO-d6, 600 MHz) δ 10.95 (s, 1H), 10.09 (s, 1H), 8.21 (s, 1H), 7.91 (m, 3H), 7.85–7.76 (m, 2H), 7.64 (d, J = 8.6 Hz, 2H), 7.57 (d, J = 7.8 Hz, 2H), 7.37 (t, J = 7.7 Hz, 2H), 7.20 (t, J = 7.3 Hz, 1H), 2.41 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 177.47, 156.79, 155.87, 150.99, 149.02, 140.35, 139.45, 136.30, 130.15, 129.51, 128.77, 126.15, 124.70, 122.34, 120.42, 118.04, 112.94, 108.23, 14.28. HRMS (ESI): m/z calcd for C23H18ClN5OS [M + Na]+: 470.0818; found: 470.0807.
(E)-2-(1-(5-((E)-(4-Methoxyphenyl)diazenyl)benzofuran-2-yl)ethylidene)-N-phenylhydrazine-1-carbothioamide (7b). Yellow powder; yield 85%, m.p. 189–190 °C. IR (KBr, cm−1) 3312 (NH), 3173 (NH), 2833 (CH sp3), 1601 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 10.92 (s, 1H), 10.07 (s, 1H), 8.12 (d, J = 1.9 Hz, 1H), 7.91–7.84 (m, 3H), 7.77 (d, J = 6.7 Hz, 2H), 7.74 (s, 1H), 7.57 (d, J = 7.4 Hz, 2H), 7.36 (t, J = 7.8 Hz, 2H), 7.20 (t, J = 7.3 Hz, 1H), 7.10 (d, J = 9.0 Hz, 2H), 3.83 (s, 3H), 2.41 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 177.4, 162.38, 156.27, 155.65, 149.22, 146.62, 140.46, 139.47, 129.45, 128.76, 126.14, 125.00, 123.41, 120.17, 117.18, 115.14, 112.74, 108.22, 56.18, 14.29. HRMS (ESI): m/z calc for C24H21N5O2S [M + Na]+: 466.1308; found: 466.1309.
C). 1H NMR (DMSO-d6, 400 MHz) δ 10.92 (s, 1H), 10.07 (s, 1H), 8.12 (d, J = 1.9 Hz, 1H), 7.91–7.84 (m, 3H), 7.77 (d, J = 6.7 Hz, 2H), 7.74 (s, 1H), 7.57 (d, J = 7.4 Hz, 2H), 7.36 (t, J = 7.8 Hz, 2H), 7.20 (t, J = 7.3 Hz, 1H), 7.10 (d, J = 9.0 Hz, 2H), 3.83 (s, 3H), 2.41 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 177.4, 162.38, 156.27, 155.65, 149.22, 146.62, 140.46, 139.47, 129.45, 128.76, 126.14, 125.00, 123.41, 120.17, 117.18, 115.14, 112.74, 108.22, 56.18, 14.29. HRMS (ESI): m/z calc for C24H21N5O2S [M + Na]+: 466.1308; found: 466.1309.
(E)-N-Butyl-2-(1-(5-((E)-(4-methoxyphenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazine-1-carbothioamide (7c). Yellow powder; yield 79%, m.p. 173–175 °C. IR (KBr, cm−1) 3356 (NH), 3217 (NH), 2911, 2883 (CH sp3), 1601 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 10.47 (s, 1H), 8.46 (s, 1H), 8.11 (s, 1H), 7.86 (d, J = 8.1 Hz, 3H), 7.72 (d, J = 8.7 Hz, 2H), 7.63 (s, 1H), 7.10 (d, J = 8.2 Hz, 2H), 3.83 (s, 3H), 3.57 (q, J = 7.7 Hz, 2H), 2.32 (s, 3H), 1.62–1.50 (m, 2H), 1.37–1.23 (m, 2H), 0.89 (t, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 178.23, 162.36, 156.19, 155.80, 149.19, 146.61, 139.35, 129.44, 124.98, 120.06, 117.11, 115.13, 112.63, 107.79, 56.17, 43.93, 31.40, 20.16, 14.35, 14.06. HRMS (ESI): m/z calc for C22H25N5O2S [M + Na]+: 446.1621; found: 446.1612.
C). 1H NMR (DMSO-d6, 400 MHz) δ 10.47 (s, 1H), 8.46 (s, 1H), 8.11 (s, 1H), 7.86 (d, J = 8.1 Hz, 3H), 7.72 (d, J = 8.7 Hz, 2H), 7.63 (s, 1H), 7.10 (d, J = 8.2 Hz, 2H), 3.83 (s, 3H), 3.57 (q, J = 7.7 Hz, 2H), 2.32 (s, 3H), 1.62–1.50 (m, 2H), 1.37–1.23 (m, 2H), 0.89 (t, J = 6.9 Hz, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 178.23, 162.36, 156.19, 155.80, 149.19, 146.61, 139.35, 129.44, 124.98, 120.06, 117.11, 115.13, 112.63, 107.79, 56.17, 43.93, 31.40, 20.16, 14.35, 14.06. HRMS (ESI): m/z calc for C22H25N5O2S [M + Na]+: 446.1621; found: 446.1612.
(E)-N-Allyl-2-(1-(5-((E)-(4-chlorophenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazine-1-carbothioamide (7d). Yellow powder; yield 76%, m.p. 227–229 °C. IR (KBr, cm−1) 3343 (NH), 3224 (NH), 2974, 2924 (CH sp3), 1577 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 600 MHz) δ 10.64 (s, 1H), 8.62 (s, 1H), 8.21 (d, J = 1.4 Hz, 1H), 7.92 (dd, J = 8.9, 1.9 Hz, 2H), 7.90 (d, J = 8.7 Hz, 2H), 7.78 (d, J = 8.8 Hz, 1H), 7.69 (s, 1H), 7.65 (d, J = 8.5 Hz, 2H), 5.93–5.88 (m, 1H), 5.16 (dd, J = 17.2, 1.5 Hz, 1H), 5.12–5.07 (m, 1H), 4.24 (t, J = 5.3 Hz, 2H), 2.35 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 178.61, 156.73, 156.00, 150.99, 148.99, 139.59, 136.29, 135.32, 130.15, 129.53, 124.69, 120.34, 118.00, 116.32, 112.86, 107.93, 46.46, 14.12. HRMS (ESI): m/z calcd for C20H18ClN5OS [M + Na]+: 434.0813; found: 434.0810.
C). 1H NMR (DMSO-d6, 600 MHz) δ 10.64 (s, 1H), 8.62 (s, 1H), 8.21 (d, J = 1.4 Hz, 1H), 7.92 (dd, J = 8.9, 1.9 Hz, 2H), 7.90 (d, J = 8.7 Hz, 2H), 7.78 (d, J = 8.8 Hz, 1H), 7.69 (s, 1H), 7.65 (d, J = 8.5 Hz, 2H), 5.93–5.88 (m, 1H), 5.16 (dd, J = 17.2, 1.5 Hz, 1H), 5.12–5.07 (m, 1H), 4.24 (t, J = 5.3 Hz, 2H), 2.35 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 178.61, 156.73, 156.00, 150.99, 148.99, 139.59, 136.29, 135.32, 130.15, 129.53, 124.69, 120.34, 118.00, 116.32, 112.86, 107.93, 46.46, 14.12. HRMS (ESI): m/z calcd for C20H18ClN5OS [M + Na]+: 434.0813; found: 434.0810.
(E)-N-Allyl-2-(1-(5-((E)-(4-methoxyphenyl)diazenyl)benzofuran-2-yl)ethylidene)hydrazine-1-carbothioamide (7e). Yellow powder; yield 74%, m.p. 188–190 °C. IR (KBr, cm−1) 3350 (NH), 3205 (NH), 2953, 2939 (CH sp3), 1602 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C). 1H NMR (DMSO-d6, 400 MHz) δ 10.60 (s, 1H), 8.59 (s, 1H), 8.11 (d, J = 1.8 Hz, 1H), 7.87–7.84 (m, 3H), 7.72 (d, J = 8.8 Hz, 1H), 7.65 (s, 1H), 7.10 (d, J = 9.0 Hz, 2H), 5.95–5.86 (m, 1H), 5.16 (dd, J = 17.3, 1.6 Hz, 1H), 5.09 (dd, J = 10.2, 1.4 Hz, 1H), 4.24 (t, J = 5.7 Hz, 2H), 3.83 (s, 3H), 2.34 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 178.59, 162.37, 156.21, 155.76, 149.20, 146.61, 139.70, 135.33, 129.43, 124.99, 120.09, 117.14, 116.32, 115.13, 112.65, 107.96, 56.18, 46.46, 14.13. HRMS (ESI): m/z calc for C21H21N5O2S [M + Na]+: 430.1308; found: 430.1316.
C). 1H NMR (DMSO-d6, 400 MHz) δ 10.60 (s, 1H), 8.59 (s, 1H), 8.11 (d, J = 1.8 Hz, 1H), 7.87–7.84 (m, 3H), 7.72 (d, J = 8.8 Hz, 1H), 7.65 (s, 1H), 7.10 (d, J = 9.0 Hz, 2H), 5.95–5.86 (m, 1H), 5.16 (dd, J = 17.3, 1.6 Hz, 1H), 5.09 (dd, J = 10.2, 1.4 Hz, 1H), 4.24 (t, J = 5.7 Hz, 2H), 3.83 (s, 3H), 2.34 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 178.59, 162.37, 156.21, 155.76, 149.20, 146.61, 139.70, 135.33, 129.43, 124.99, 120.09, 117.14, 116.32, 115.13, 112.65, 107.96, 56.18, 46.46, 14.13. HRMS (ESI): m/z calc for C21H21N5O2S [M + Na]+: 430.1308; found: 430.1316.
(E)-2-(1-(5-((E)-(4-Chlorophenyl)diazenyl)benzofuran-2-yl)ethylidene)-N-(p-tolyl)hydrazine-1-carbothioamide (7f). Yellow powder; yield 84%, m.p. 233–235 °C. IR (KBr, cm−1) 3326 (NH), 3224 (NH), 3058 (CH sp2), 2974, 2919 (CH sp3). 1H NMR (DMSO-d6, 600 MHz) δ 10.89 (s, 1H), 10.01 (s, 1H), 8.21 (d, J = 1.8 Hz, 1H), 7.92 (dd, J = 8.8, 1.9 Hz, 1H), 7.89 (d, J = 8.6 Hz, 2H), 7.81 (s, 1H), 7.80 (d, J = 8.9 Hz, 1H), 7.64 (d, J = 8.6 Hz, 2H), 7.42 (d, J = 8.3 Hz, 2H), 7.16 (d, J = 8.2 Hz, 2H). 10.01 (s, 1H), 2.40 (s, 3H), 2.29 (s, 3H). 13C NMR (DMSO-d6, 150 MHz) δ 177.51, 156.78, 155.91, 150.99, 149.02, 140.19, 136.89, 136.30, 135.25, 130.15, 129.56, 129.51, 129.22, 126.12, 124.70, 122.33, 120.39, 118.04, 112.93, 108.14, 21.15, 14.26. HRMS (ESI): m/z calcd for C24H20ClN5OS [M + Na]+: 484.0969; found: 484.0963.
(E)-2-(1-(5-((E)-(4-Methoxyphenyl)diazenyl)benzofuran-2-yl)ethylidene)-N-(p-tolyl)hydrazine-1-carbothioamide (7g). Yellow powder; yield 83%, m.p. 228–229 °C. IR (KBr, cm−1) 3314 (NH), 3242 (NH), 2964, 2942, 2916, 2836, 1601 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), (CH sp3). 1H NMR (DMSO-d6, 400 MHz) δ 10.92 (s, 1H), 10.06 (s, 1H), 8.18 (d, J = 1.9 Hz, 1H), 7.99–7.85 (m, 3H), 7.84 (s, 1H), 7.81 (d, J = 8.9 Hz, 1H), 7.48 (d, J = 8.3 Hz, 2H), 7.22 (d, J = 8.2 Hz, 2H), 7.16 (d, J = 9.0 Hz, 2H), 3.89 (s, 3H), 2.45 (s, 3H), 2.34 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 177.47, 162.36, 156.25, 155.66, 149.21, 146.61, 140.29, 136.89, 135.23, 129.45, 129.21, 126.09, 124.99, 120.13, 117.17, 115.12, 112.71, 108.16, 56.17, 21.15, 14.26. HRMS (ESI): m/z calc for C25H23N5O2S [M + Na]+: 480.1465; found: 480.1459.
C), (CH sp3). 1H NMR (DMSO-d6, 400 MHz) δ 10.92 (s, 1H), 10.06 (s, 1H), 8.18 (d, J = 1.9 Hz, 1H), 7.99–7.85 (m, 3H), 7.84 (s, 1H), 7.81 (d, J = 8.9 Hz, 1H), 7.48 (d, J = 8.3 Hz, 2H), 7.22 (d, J = 8.2 Hz, 2H), 7.16 (d, J = 9.0 Hz, 2H), 3.89 (s, 3H), 2.45 (s, 3H), 2.34 (s, 3H). 13C NMR (DMSO-d6, 100 MHz) δ 177.47, 162.36, 156.25, 155.66, 149.21, 146.61, 140.29, 136.89, 135.23, 129.45, 129.21, 126.09, 124.99, 120.13, 117.17, 115.12, 112.71, 108.16, 56.17, 21.15, 14.26. HRMS (ESI): m/z calc for C25H23N5O2S [M + Na]+: 480.1465; found: 480.1459.
4.2. Biology
4.3. Molecular modelling simulation methodology
Molecular docking studies were carried out using the Molecular Operating Environment (MOE) software, version 2024.06. The VEGFR-2 crystal structure complexed with sorafenib (PDB ID: 3WZE) was obtained from the Protein Data Bank and employed as the docking target. Compounds 7a, 7b, 7f, and 7g were energy-minimized and their partial charges optimized, preparing them for docking. The protein was prepared through structure correction, 3D hydrogen addition, and energy minimization, with removal of water molecules, and the protocol was validated by re-docking the co-crystallized sorafenib. Docking was conducted using the triangle matcher placement method, London dG scoring, and GBVI/WSA dG refinement. Thirty poses were initially generated for each ligand, then refined to ten poses using the co-crystalline ligand (sorafenib) as a reference, and the best pose for each compound was selected for comparison with it.35,614.4. Molecular dynamic simulation methodology
For the most active compound 7g, molecular dynamic simulation was conducted using the VEGFR-2 protein structure (PDB: 3WZE). The simulation was initiated from the docked best pose. The non-commercial academic version of Desmond software (2022.4) integrated within the Maestro interface (https://www.deshawresearch.com/resources.html) was employed for all-atom molecular dynamics simulations. All calculations utilized OPLS_2005 force field parameters. Each complex underwent identical simulation protocols: the protein–ligand systems were placed in orthorhombic periodic boundary boxes extending 10 Å3 beyond the complex and filled with TIP3P explicit water molecules. Systems were neutralized with counter-ions and supplemented with 0.15 M salt to mimic physiological conditions. Prior to the production simulations, each system underwent preparatory relaxation through brief energy minimization followed by 12 ps equilibration simulations under NVT and NPT ensembles. The 100 ns production runs employed the NPT ensemble with 2 ps integration time steps at 300 K and 1 atm pressure. Van der Waals interactions were calculated with a 9 Å cutoff radius, while long-range electrostatic interactions were computed using the particle mesh Ewald (PME) method. Trajectory visualization was performed within the Maestro environment, and interaction analyses were conducted using Desmond's interaction diagram tools.4.5. Pharmacokinetic and physicochemical profiling
Key compounds were further assessed for ADMET and drug-likeness using SwissADME and PreADMET, while geometry optimizations and DFT calculations were carried out at the B3LYP/6-311G level in Gaussian 16 to evaluate their frontier molecular orbitals and derived reactivity descriptors (ΔE, χ, η, σ, ω, Nu).5. Conclusions
Two novel series of sorafenib-inspired benzofuran hybrids, azine derivatives (6a–k) and thiosemicarbazone derivatives (7a–g), were successfully designed, synthesized, and evaluated for their cytotoxicity against HepG-2 and HCT-116 cell lines. Among them, azines 6b and 6c, together with thiosemicarbazones 7a, 7b, 7e, 7f, and 7g, exhibited superior cytotoxicity against both cancer cell lines. Notably, compounds 7a, 7b, 7f, and 7g demonstrated high selectivity toward cancer cells over normal cells, prompting further VEGFR-2 inhibitory evaluation. Compound 7g emerged as the most potent candidate, showing strong VEGFR-2 inhibition comparable to sorafenib along with additional EGFR inhibition. Mechanistic studies revealed that 7g induced G0–G1 cell cycle arrest, triggered apoptosis via intrinsic and extrinsic pathways, and favorably modulated apoptotic gene expression. Furthermore, molecular docking studies validated the strong binding affinity of 7g within the VEGFR-2 active site, closely resembling sorafenib interactions. Overall, these findings highlight thiosemicarbazone–benzofuran hybrids, particularly compound 7g, as promising multitargeted kinase inhibitors warranting further optimization and preclinical investigation for the treatment of hepatocellular carcinoma.Author contributions
Conceptualization and methodology: M. S. S., A. M. A., M. S. H. S., M. A. A. E.-S.; investigation and software: M. S. S., M. H. A., M. A. A. E.-S.; supervision: S. T., Y. M. A. A., M. A. A. E.-S., R. E.; writing – original draft: M. S. S., A. M. A., R. E.; writing – review & editing: M. S. H. S., S. T., Y. M. A. A., M. A. A. E.-S.; visualization: M. S. S., M. S. H. S.; validation: M. S. S., A. M. A., M. A. A. E.-S.; funding acquisition: M. S. H. S., S. T.Conflicts of interest
The authors declare that they have no conflicts of interest related to the content of this article.Data availability
Data for this article, including spectral analysis (NMR), HRMS, and biological data are available in the supplementary information (SI).Supplementary information is available. See DOI: https://doi.org/10.1039/d5md00837a.
Acknowledgements
The authors would like to acknowledge the Computational Chemistry and Molecular Modelling Laboratory, Department of Pharmaceutical Organic Chemistry, Faculty of Pharmacy, Mansoura University, in which the molecular docking studies carried out. The DFT calculations were performed using Research Center for Computational Science, Okazaki, Japan (Project: 25-IMS-C280). We acknowledge the technical staff of the Comprehensive Analysis Center of SANKEN, The University of Osaka. This work was supported by JSPS KAKENHI Grant Numbers 24k17681, JP21H05207, JP21H05217, JP22KK0073, and JP22K06502 from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), and the Japan Society for the Promotion of Science (JSPS), JST CREST (No. JPMJCR20R1).References
- J. Ferlay, M. Colombet, I. Soerjomataram, D. M. Parkin, M. Piñeros, A. Znaor and F. Bray, Int. J. Cancer, 2021, 149, 778–789 CrossRef.
- F. Bray, M. Laversanne, H. Sung, J. Ferlay, R. L. Siegel, I. Soerjomataram and A. Jemal, Ca-Cancer J. Clin., 2024, 74, 229–263 CrossRef.
- Y. Xi and P. Xu, Transl. Oncol., 2021, 14, 101174 CrossRef.
- A. W. C. Kow, J. Gastrointest. Oncol., 2019, 10, 1274–1298 CrossRef.
- N. C. Bird, D. Mangnall and A. W. Majeed, J. Surg. Oncol., 2006, 94, 68–80 CrossRef PubMed.
- M. A. Zeidan, A. S. Mostafa, R. M. Gomaa, L. A. Abou-zeid, M. El-Mesery, M. A. A. El-Sayed and K. B. Selim, Eur. J. Med. Chem., 2019, 168, 315–329 CrossRef.
- A. K. El-Damasy, M. S. H. Salem, M. M. Sebaiy and M. S. Elgawish, p38 mitogen-activated protein kinase inhibitors, in Current Molecular Targets of Heterocyclic Compounds for Cancer Therapy, Elsevier, 2024, pp. 219–254 Search PubMed.
- W. A. Ewes, S. S. Tawfik, A. M. Almatary, M. Ahmad Bhat, H. W. El-Shafey, A. A. B. Mohamed, A. Haikal, M. A. El-Magd, A. A. Elgazar, M. Balaha and A. Hamdi, Molecules, 2024, 29, 3186 CrossRef PubMed.
- M. M. Kandeel, M. K. AbdElhameid, M. Adel, M. Y. Al-Shorbagy and A. T. Negmeldin, Molecules, 2025, 30, 1105 CrossRef.
- A.-G. A. El-Helby, H. Sakr, I. H. Eissa, H. Abulkhair, A. A. Al-Karmalawy and K. El-Adl, Arch. Pharm., 2019, 352, 1900113 CrossRef.
- A. Elwan, A. E. Abdallah, H. A. Mahdy, M. A. Dahab, M. S. Taghour, E. B. Elkaeed, A. B. M. Mehany, A. Nabeeh, M. Adel, A. A. Alsfouk, H. Elkady and I. H. Eissa, Molecules, 2022, 27, 5047 CrossRef.
- C. Jayasinghe, N. Simiantonaki, S. Habedank and C. J. Kirkpatrick, J. Exp. Clin. Cancer Res., 2015, 34, 42 CrossRef PubMed.
- A. Altharawi, M. M. Alanazi, M. A. Alossaimi, A. S. Alanazi, S. M. Alqahtani, M. H. Geesi and Y. Riadi, Molecules, 2023, 28, 5548 CrossRef PubMed.
- M. Li, X. Yu, W. Li, T. Liu, G. Deng, W. Liu, H. Liu and F. Gao, Oncotarget, 2018, 9, 152–166 CrossRef.
- O. M. Abdelhafez, K. M. Amin, H. I. Ali, M. M. Abdalla and E. Y. Ahmed, RSC Adv., 2014, 4, 11569–11579 RSC.
- L. K. D. Cabral, C. Tiribelli and C. H. C. Sukowati, Cancers, 2020, 12, 1576 CrossRef PubMed.
- T. Kacan, E. Nayir, A. Altun, S. Kilickap, N. A. Babacan, H. Ataseven and T. Kaya, J. Oncol. Sci., 2016, 2, 53–57 CrossRef.
- Z. K. Otrock, J. A. Makarem and A. I. Shamseddine, Blood Cells, Mol., Dis., 2007, 38, 258–268 CrossRef.
- Z. Zhao, H. Wu, L. Wang, Y. Liu, S. Knapp, Q. Liu and N. S. Gray, ACS Chem. Biol., 2014, 9, 1230–1241 CrossRef.
- M. S. H. Salem, Y. M. A. Aziz, M. S. Elgawish, M. M. Said and K. A. Abouzid, Bioorg. Chem., 2020, 94, 103472 CrossRef PubMed.
- M. S. Elgawish, A. M. Almatary, S. A. Zaitone and M. S. H. Salem, RSC Med. Chem., 2025, 16, 4698–4720 RSC.
- S. Wilhelm, C. Carter, M. Lynch, T. Lowinger, J. Dumas, R. A. Smith, B. Schwartz, R. Simantov and S. Kelley, Nat. Rev. Drug Discovery, 2006, 5, 835–844 CrossRef PubMed.
- M. H. Potashman, J. Bready, A. Coxon, T. M. DeMelfi, L. DiPietro, N. Doerr, D. Elbaum, J. Estrada, P. Gallant, J. Germain, Y. Gu, J.-C. Harmange, S. A. Kaufman, R. Kendall, J. L. Kim, G. N. Kumar, A. M. Long, S. Neervannan, V. F. Patel, A. Polverino, P. Rose, S. van der Plas, D. Whittington, R. Zanon and H. Zhao, J. Med. Chem., 2007, 50, 4351–4373 CrossRef.
- M. Kim, J. Lee, K. Jung, H. Kim, W. Aman, J.-S. Ryu and J.-M. Hah, Bioorg. Med. Chem. Lett., 2014, 24, 3600–3604 CrossRef.
- A. J. Ayoub, G. A. El-Achkar, S. E. Ghayad, L. Hariss, R. H. Haidar, L. M. Antar, Z. I. Mallah, B. Badran, R. Grée, A. Hachem, E. Hamade and A. Habib, Int. J. Mol. Sci., 2023, 24, 10399 CrossRef PubMed.
- M. M. Al-Sanea, G. H. Al-Ansary, Z. M. Elsayed, R. M. Maklad, E. B. Elkaeed, M. A. Abdelgawad, S. N. A. Bukhari, M. M. Abdel-Aziz, H. Suliman and W. M. Eldehna, J. Enzyme Inhib. Med. Chem., 2021, 36, 987–999 CrossRef.
- W. M. Eldehna, H. O. Tawfik, M. S. Nafie, O. Al Kamaly, A. A. El-Hamaky, M. A. El Hassab, Z. M. Elsayed, Y. S. R. Elnaggar, A. A. Al-Karmalawy, V. di Giacomo and M. Balaha, Bioorg. Chem., 2025, 160, 108494 CrossRef PubMed.
- M. Shirley, Drugs, 2018, 78, 1757–1761 CrossRef PubMed.
- A. M. Omar, S. Ihmaid, E. L. S. E. Habib, S. S. Althagfan, S. Ahmed, H. S. Abulkhair and H. E. A. Ahmed, Bioorg. Chem., 2020, 99, 103781 CrossRef.
- S. J. Modi and V. M. Kulkarni, J. Biomol. Struct. Dyn., 2022, 40, 5712–5727 CrossRef PubMed.
- M. S. Nafie, M. I. Youssef, A. A. El-Hamaky, Z. M. Elsayed, E. F. Khaleel, E. Roshdy, M. M. Al-Sanea, M. M. Elbadawi, M. Abe, W. M. Eldehna, H. A. Abdel-Aziz and H. O. Tawfik, Bioorg. Chem., 2025, 163, 108702 CrossRef PubMed.
- B. De Filippis, A. Della Valle, A. Ammazzalorso, C. Maccallini, G. Tesse, R. Amoroso, A. Mollica and L. Giampietro, Molecules, 2024, 29, 5872 CrossRef.
- F. F. Alblewi, M. H. Alsehli, Z. M. Hritani, A. Eskandrani, W. H. Alsaedi, M. O. Alawad, A. A. Elhenawy, H. Y. Ahmed, M. S. A. El-Gaby, T. H. Afifi and R. M. Okasha, Int. J. Mol. Sci., 2023, 24, 16716 CrossRef.
- C. Wang, H. Gao, J. Dong, Y. Zhang, P. Su, Y. Shi and J. Zhang, Bioorg. Med. Chem., 2014, 22, 277–284 CrossRef.
- E. M. Othman, E. A. Fayed, E. M. Husseiny and H. S. Abulkhair, Bioorg. Chem., 2022, 127, 105968 CrossRef.
- C. Zhang, Y. Liu, X. Zhang, C. Wan and Z. Mao, RSC Med. Chem., 2024, 16, 392–399 RSC.
- F. Xie, H. Zhu, H. Zhang, Q. Lang, L. Tang, Q. Huang and L. Yu, Eur. J. Med. Chem., 2015, 89, 310–319 CrossRef PubMed.
- M. S. Ibrahim, B. Farag, J. Y. Al-Humaidi, M. E. A. Zaki, M. Fathalla and S. M. Gomha, Molecules, 2023, 28, 3869 CrossRef.
- H. N. Chopde, C. P. Pandhurnekar, J. S. Meshram and R. Pagadala, J. Heterocyclic Chem., 2017, 54, 758–763 CrossRef.
- S. Menati, A. Azadbakht, R. Azadbakht, A. Taeb and A. Kakanejadifard, Dyes Pigm., 2013, 98, 499–506 CrossRef.
- M. Koca, A. S. Ertürk and O. Bozca, ChemistrySelect, 2022, 7, e202202243 CrossRef.
- H. S. Abulkhair, S. Elmeligie, A. Ghiaty, A. El-Morsy, A. H. Bayoumi, H. E. A. Ahmed, K. El-Adl, M. F. Zayed, M. H. Hassan, E. N. Akl and M. S. El-Zoghbi, Arch. Pharm., 2021, 354, 2000449 CrossRef PubMed.
- M. S. Alam, J. U. Ahmed and D.-U. Lee, Chem. Pharm. Bull., 2014, 62, 1259–1268 CrossRef PubMed.
- M. Anary-Abbasinejad, N. Karimi, H. Mehrabi and R. Ranjbar-Karimi, J. Sulfur Chem., 2012, 33, 653–659 CrossRef.
- J. Huang, X. Zhang, Q. Tang, F. Zhang, Y. Li, Z. Feng and J. Zhu, J. Clin. Pathol., 2011, 64, 343–348 Search PubMed.
- Y. Wang, P. Ji, J. Liu, R. R. Broaddus, F. Xue and W. Zhang, Mol. Cancer, 2009, 8, 8 CrossRef PubMed.
- K. H. Kim and J. M. Sederstrom, Curr. Protoc. Mol. Biol., 2015, 111, 28 Search PubMed.
- S. Elmore, Toxicol. Pathol., 2007, 35, 495–516 CrossRef PubMed.
- H. W. El-Shafey, R. M. Gomaa, S. M. El-Messery and F. E. Goda, Bioorg. Chem., 2020, 101, 103987 CrossRef PubMed.
- R. A. Hassan, Z. Mahmoud, A. A. Elmaaty, M. R. Elnagar, A. M. Abdou, A. H. A. El-Haleem and M. I. A. Hamed, Bioorg. Chem., 2025, 164, 108921 CrossRef PubMed.
- Z. Jia, H. H. Yang, Y.-J. Liu and X.-Z. Wang, Appl. Biochem. Biotechnol., 2018, 186, 145–160 CrossRef PubMed.
- N. A. Nawareg, A. S. Mostafa, S. M. El-Messery and M. N. A. Nasr, Bioorg. Chem., 2022, 127, 106038 Search PubMed.
- Y. A. Mostafa, J. A. Assoud, A. Y. Desoky, S. Mohamady, N. M. Mohamed, O. I. A. Salem, Z. M. Almarhoon, S. Bräse and B. G. M. Youssif, Front. Chem., 2024, 12, 1498104 CrossRef PubMed.
- A. M. Almatary, M. S. H. Salem, M. R. Elnagar, M. H. Aboutaleb, T. S. Ibrahim, A. Hamdi and M. A.-A. El-Sayed, Bioorg. Chem., 2025, 161, 108511 CrossRef PubMed.
- M. S. H. Salem, R. Sharma, S. Suzuki, Y. Imai, M. Arisawa and S. Takizawa, Chirality, 2024, 36, e23673 CrossRef PubMed.
- O. A. El-Khouly, M. A. Henen, M. A. A. El-Sayed and S. M. El-Messery, Sci. Rep., 2022, 12, 17104 CrossRef PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef PubMed.
- K. Sharma, P. S. Suresh, R. Mullangi and N. R. Srinivas, Biomed. Chromatogr., 2015, 29, 803–834 CrossRef PubMed.
- I. Vermes, C. Haanen, H. Steffens-Nakken and C. Reutellingsperger, J. Immunol. Methods, 1995, 184, 39–51 CrossRef PubMed.
- M. S. Mohamed, A. O. Abdelhamid, F. M. Almutairi, A. G. Ali and M. K. Bishr, Bioorg. Med. Chem., 2018, 26, 623–629 CrossRef PubMed.
- A. M. Moussa, H. Abdelrasheed Allam, M. K. El-Ashrey, M. A. Fouad and A. A. Al-Karmalawy, Bioorg. Chem., 2024, 153, 107911 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |