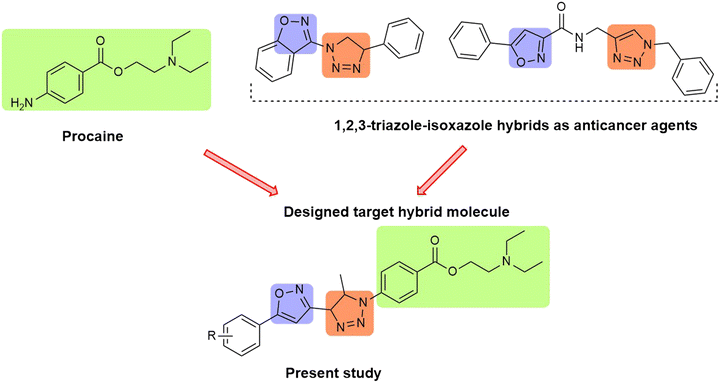
Design, synthesis, and biological evaluation of procaine-based triazole–isoxazoline hybrids as selective PI3K/mTOR inhibitors for esophageal cancer therapy: in vitro and in vivo studies
Yinliang
Sheng
,
Bin
Wu
,
Feng
Li
and
Chunyang
Zhang
 *
*
Department of Thoracic Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China. E-mail: zcy198200@outlook.com
First published on 15th October 2025
Abstract
A novel series of procaine derivatives incorporating 1,2,3-triazole and isoxazoline scaffolds were developed and evaluated for their anticancer potential, particularly against esophageal cancer. Initially, the synthesized compounds were screened for their kinase inhibitory activity against PI3K, mTOR, CDK1, CDK4, EGFR, and VEGFR2, where they exhibited excellent inhibitory potency against PI3K and mTOR. Among the synthesized compounds, 8e, 8f, and 8g emerged as the top-performing kinase inhibitors. These three candidates were subsequently tested against a panel of human cancer cell lines, including breast, cervical, lung, liver, and esophageal cancer cells. Notably, they demonstrated superior cytotoxic activity against esophageal cancer cells. Of these, compound 8e was identified as the most potent and was further evaluated against six esophageal cancer cell lines (Eca109, TE1, TE13, KYSE30, KYSE70, and KYSE150) with diverse genotypic backgrounds. Compound 8e exhibited the highest activity against Eca109 cells. Further investigations revealed that compound 8e significantly inhibited Eca109 cell viability, as confirmed by the MTT assay, and induced apoptosis, as evidenced by annexin V/PI dual staining and DAPI nuclear staining. It also caused G0/G1 cell cycle arrest, decreased mitochondrial membrane potential, and demonstrated marked telomerase inhibitory activity. In addition, wound healing and transwell assays confirmed its ability to suppress the migration and invasion of Eca109 cells. Western blot analysis revealed that compound 8e modulated the expression of key apoptotic regulators (Bcl-2, Bax, and p53) and downregulated the PI3K/Akt/mTOR signaling pathway. In an orthotopic xenograft mouse model, compound 8e significantly reduced tumor volume and increased body weight in a dose-dependent manner, indicating potent in vivo efficacy with favorable tolerability. Biochemical analyses showed that compound 8e mitigated oxidative stress by regulating MDA, SOD, and GSH levels and suppressed pro-inflammatory cytokines including TNF-α, IL-1β, and IL-6. Immunohistochemical staining further confirmed reduced expression of PI3K and p-Akt (Ser473) in tumor tissues. Pharmacokinetic evaluation via both intravenous and oral administration demonstrated that compound 8e possesses excellent bioavailability, highlighting its potential as a promising therapeutic candidate for the treatment of esophageal cancer.
Introduction
In majority of the countries of the world, esophageal squamous cell carcinoma (ESCC) is the leading cause of cancer-related morbidity and mortality. It was classified as a leading cause of death in developing countries as compared to developed nations.1,2 Moreover, due to the lack of early detection programs, as well as challenges in implementing appropriate diagnostic and treatment approaches, the overall survival of ESCC patients is not very encouraging in developing countries.3,4 For instance, in China, the incidence of ESCC has increased in the past decade due to rapid economic development and adoption of western life-style. Studies suggested that ESCC is the fifth most frequently diagnosed cancer and the fourth leading cause of death due to cancer in China, with estimated 286![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 new cases and 211
700 new cases and 211![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2012, according to the National Central Cancer Registry.5 Surgery and chemotherapy are the two distinct modalities used in the premature stage of the disease. However, the resistance has seriously compromised the clinical utility of first-line drugs. Therefore, new and effective agents are worth to be investigated against the BC.
000 deaths in 2012, according to the National Central Cancer Registry.5 Surgery and chemotherapy are the two distinct modalities used in the premature stage of the disease. However, the resistance has seriously compromised the clinical utility of first-line drugs. Therefore, new and effective agents are worth to be investigated against the BC.
Procaine, a well-known local anesthetic of the amino ester group, showed excellent anticancer properties against osteosarcoma6 and colon cancer7,8via the upregulation of microRNA-133b and the inactivation of the ERK/MAPK/FAK pathways by the regulation of RhoA, respectively. Moreover, it showed global genomic DNA hypomethylation and demethylation and reactivation of tumor suppressor genes with hypermethylated CpG islands in cancer cells.9–11 Procaine has also been shown to inhibit growth and to reactivate the expression of RASSF1A mRNA in nasopharyngeal cancer cell lines,12 as well as to inhibit proliferation and DNA methylation in human hepatoma cancer cells.13 Thus, the development of procaine derivatives is advantageous to obtain more potent drugs against esophageal cancer.
A molecular hybridization approach (covalently combining two or more pharmacophores) proved to be an effective tool for the development of novel chemical entities. The hybrid molecules believed to exert improved affinity and efficacy when compared to the parent drugs.14 This technique focuses on the modulation of the pharmacophores giving rise to innovative hybrids that are believed to act by one or more than one mechanism to target pathogenic micro-organisms or diseases. Such an approach has been found more fruitful against devastating diseases such as cancer, bacterial infections, malaria, and HIV.15,16
The PI3K/mTOR signaling pathway plays a pivotal role in the regulation of fundamental cellular processes including cell proliferation, survival, metabolism, and angiogenesis. Aberrant activation of this pathway is frequently observed in esophageal squamous cell carcinoma (ESCC), driven by genetic mutations, gene amplification, and overexpression of key pathway components, thereby contributing to uncontrolled tumor growth and therapeutic resistance.17 Several studies have demonstrated that both PI3K and mTOR are overexpressed or hyperactivated in ESCC patient samples, with such alterations being significantly associated with poor prognosis and enhanced tumor aggressiveness.18–20 Dual inhibition of PI3K and mTOR has emerged as a particularly effective therapeutic strategy, as it circumvents the compensatory feedback mechanisms commonly encountered with single-target inhibitors.21,22 Notably, dual PI3K/mTOR inhibitors, such as BEZ235 (dactolisib), have been reported to markedly suppress tumor growth and induce apoptosis in ESCC models.23 Therefore, the development of selective and potent PI3K/mTOR inhibitors represents a rational and promising approach to improve therapeutic outcomes in ESCC by overcoming resistance mechanisms and enhancing antitumor efficacy.
Recent studies have demonstrated the power of integrated computational and experimental approaches in designing potent PI3K/mTOR dual inhibitors. For example, Cheng et al. employed structure-based drug design (SBDD) on a tricyclic imidazo[1,5]naphthyridine series to optimize both potency and drug-like properties, yielding PF-04979064 – a highly potent, selective dual PI3K/mTOR inhibitor.24 Similarly, docking-guided optimization of the dual PI3K/mTOR inhibitor voxtalisib identified six new analogs with improved binding to PI3Kγ and mTOR.22 In another case, Shen et al. combined medicinal chemistry with modeling simulations: introducing optimized sulfonyl and fluoropyridyl substituents into a 2-morpholino-pyrimidine scaffold produced “compound 26”, which inhibited PI3Kα/β/γ/δ (IC50 = 20–376 nM) and mTOR (IC50 = 189 nM) and potently blocked Akt phosphorylation in cancer cells.25 These examples illustrate how iterative cycles of molecular docking, structure-based design and SAR can pinpoint substituents that balance dual PI3K/mTOR affinity while retaining good ADMET profiles, thereby advancing novel multi-target agents.
Various heterocyclic cores have been leveraged in such dual-inhibitor programs. One strategy is to hybridize known pharmacophores into new scaffolds: for instance, 1,2,4-triazole-linked bis-indole conjugates were designed and found to kill cancer cells by triggering apoptosis and downregulating PI3K pathway markers, in agreement with in silico docking predictions.26 Similarly, fused isoxazolopyrimidines (e.g. morpholino-piperidinyl isoxazolopyrimidine derivatives) have been synthesized as PI3K inhibitors; lead compounds from this series showed sub-micromolar PI3Kδ activity and potent anti-proliferative effects.27 Although dual inhibitors specifically tailored to esophageal squamous cell carcinoma (ESCC) are still emerging, preliminary data underscore the promise of dual targeting in this cancer, for example, the dual PI3K/mTOR inhibitor BEZ235 (dactolisib) synergized with an HDAC inhibitor to induce autophagy and enhance cytotoxicity in ESCC cell models.28 Notably, a recent ESCC review emphasizes that monotherapy often triggers compensatory PI3K/Akt activation, and therefore multi-kinase or combination strategies – including dual PI3K/mTOR blockade – are expected to improve efficacy and overcome resistance in ESCC.17 Together, these studies support an integrated design paradigm: combining computational modeling (docking, pharmacophore/hybrid strategies) with experimental validation can efficiently generate heterocycle-rich dual PI3K/mTOR inhibitors with potential in hard-to-treat cancers like ESCC.
Studies suggest that despite their clinical utility in treating various cancers, existing PI3K/mTOR inhibitors (Fig. 1) suffer from significant limitations, including pronounced toxicity and side effects such as hyperglycemia, diarrhea, rash, increased infection risk, stomatitis, and fatigue, which often lead to high discontinuation rates of up to 50% in trials.29 Resistance mechanisms, such as feedback activation of parallel pathways like MAPK or incomplete mTORC2 inhibition in rapalogs, frequently result in acquired resistance and limited efficacy, particularly in solid tumors where response rates hover around 10–20% due to tumor heterogeneity and stromal interactions.30 Additionally, many lack sufficient selectivity, causing off-target effects and genotoxicity, while pharmacokinetic issues like poor oral bioavailability, short half-lives, and inadequate brain penetration restrict dosing convenience and applicability to CNS malignancies. The current strategies for the discovery of new compounds aim to overcome these challenges through structural modifications that selectivity; provide balanced dual inhibition to prevent feedback loops and target resistant; optimize pharmacokinetics for better oral absorption, extended half-lives, and enhanced tissue penetration; minimize specific side effects like hyperglycemia and immunosuppression to enable safer combination therapies.
Considering the significant role of diverse azaheterocyclic ring systems as versatile scaffolds in the development of small molecule cancer therapeutics, the present study focuses on designing a novel class of dual PI3K and mTOR inhibitors. The 1,2,3-triazole ring is well known for its excellent pharmacological properties including high stability and bioactivity, making it a valuable core structure in drug design.31 Similarly, isoxazole is a privileged heterocyclic motif extensively reported for its anticancer activity against various cancer cell lines.32 For instance, a series of resorcinylic 4,5-diarylisoxazole amides have been developed as potent heat shock protein 90 (HSP90) inhibitors, with compounds such as NVP-AUY922 (luminespib), demonstrating significant activity against multiple tumor xenografts, particularly breast cancer, and advancing to phase II clinical trials.33,34 Furthermore, the immunosuppressant drug leflunomide, an isoxazole derivative, has recently been repurposed and investigated for its potential anticancer effects.35
Prompted by these successful examples, our study aims to develop novel procaine–triazole–isoxazole hybrids as potent anticancer agents targeting the PI3K/mTOR signaling pathway (Fig. 1). This integrated scaffold design leverages the pharmacological advantages of triazole and isoxazole moieties, while incorporating procaine to potentially enhance anticancer activity, drug-likeness, and selectivity toward key oncogenic pathways.
In the present study, the rationale for designing ester derivatives as part of the procaine–triazole–isoxazole hybrid scaffold is based on well-established medicinal chemistry principles that highlight the advantages of ester functionality in drug development, particularly in kinase inhibitor programs.36 Esters are commonly used to improve drug-like properties such as aqueous solubility, membrane permeability, metabolic stability, and bioavailability.37,38 These properties are often critical for ensuring sufficient systemic exposure of small molecule inhibitors, especially when targeting intracellular kinases such as PI3K and mTOR.
Several studies in the literature have demonstrated that esterification of kinase inhibitors can significantly enhance their pharmacokinetic profile without compromising potency. For example, ester prodrugs of the VEGFR/PDGFR dual inhibitor CEP-5214 (CEP-7055) showed markedly improved aqueous solubility and oral bioavailability compared to the parent compound, enabling effective in vivo target engagement and tumor growth inhibition.39 Similarly, C-11 ester derivatives of the natural product wortmannin exhibited enhanced PI3K inhibitory activity in vitro and improved cytotoxicity in cancer cells.40 Furthermore, glycine methyl ester derivatives of EGFR and VEGFR inhibitors maintained strong kinase inhibition while benefiting from better solubility and cell permeability.41
In our design strategy (Fig. 2), the introduction of ester groups into the procaine-triazole-isoxazole hybrid compounds was motivated by these documented advantages. The ester functionality is expected to increase lipophilicity, thereby improving membrane permeability and potentially enhancing oral absorption.
Experimental
Chemicals and other details
The chemicals used in the present study were obtained from Sigma Aldrich, USA, unless otherwise stated. The chemical used in the present study was procured from Sigma Aldrich (USA). The 1H NMR and 13C NMR spectra were recorded using a Bruker Avance 400 and a Bruker Avance 100 Spectrophotometer, respectively. The chemical shifts are expressed in parts per million (ppm), and coupling constants are expressed in hertz (Hz). The low-resolution mass spectrum (MS) was recorded using a Waters ZQ LC/MS single quadrupole system equipped with an electrospray ionization (ESI) source. The elemental analysis of the final derivatives was performed using a Vario Elemental analyser. Thin-layer chromatography was performed using 0.25 mm Merck silica gel plates (60F-254) visualized under UV light.Synthesis of 2-(diethylamino)ethyl 4-aminobenzoate (4)
The synthesis of 2-(diethylamino)ethyl 4-aminobenzoate (4) intermediate was achieved as per the previously reported procedure.42Synthesis of 2-(diethylamino)ethyl 4-azidobenzoate (5)
2-(Diethylamino)ethyl 4-aminobenzoate (4) (1 mol) was dissolved in a few millilitres of distilled water and acidic ionic liquid (4 mol) and ground using a mortar and pestle for 2 to 3 minutes. Sodium nitrite (NaNO2) (2.5 mol) was added to the above mixture and further ground for 20 to 25 minutes. After that, NaN3 (2.5 mol, 0.163 g) was added to the above mixture and further ground for 10 to 15 minutes until nitrogen gas has been completely evolved. The above mixture was diluted with water and the product was extracted with petroleum ether (3 × 15 mL) using a separating funnel. The organic layer was washed with 10% HCl solution. The organic layer was dried to give intermediate compound 5.Yield: 79%; M.p.: 114–115 °C; MW: 262.31; Rf: 0.52; FTIR (νmax; cm−1 KBr): 3017 (aromatic C–H stretching), 2963 (alkyl C–H stretching) 2938 (CH2 stretching), 2868 (CH3 stretching), 2096 (N3 stretching), 1728 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1627 (C
O stretching), 1627 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1469 (CH3 bending), 1408 (COO stretching), 1374 (CH2 bending), 1226 (C–N stretching), 832; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.2 Hz, CH3 × 2), 2.68 (q, 4H, J = 7.3 Hz, CH2 × 2), 2.92 (t 2H, J = 6.9 Hz, aliphatic CH2), 4.43 (t, 2H, J = 6.9 Hz, aliphatic CH2), 8.03 (d, 2H, J = 1.6 Hz, Ar–H), 8.74 (d, 2H, J = 1.9 Hz, Ar–H); 13C NMR (100 MHz, DMSO-d6, TMS) δ ppm: 165.9, 148.2, 130.2, 129.8, 128.6, 63.3, 54.1, 49.8, 13.4; mass: 263.32 (M + H)+; elemental analysis for C13H18N4O2: calculated: C, 59.53; H, 6.92; N, 21.36. Found: C, 59.56; H, 6.91; N, 21.39.
C stretching), 1469 (CH3 bending), 1408 (COO stretching), 1374 (CH2 bending), 1226 (C–N stretching), 832; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.2 Hz, CH3 × 2), 2.68 (q, 4H, J = 7.3 Hz, CH2 × 2), 2.92 (t 2H, J = 6.9 Hz, aliphatic CH2), 4.43 (t, 2H, J = 6.9 Hz, aliphatic CH2), 8.03 (d, 2H, J = 1.6 Hz, Ar–H), 8.74 (d, 2H, J = 1.9 Hz, Ar–H); 13C NMR (100 MHz, DMSO-d6, TMS) δ ppm: 165.9, 148.2, 130.2, 129.8, 128.6, 63.3, 54.1, 49.8, 13.4; mass: 263.32 (M + H)+; elemental analysis for C13H18N4O2: calculated: C, 59.53; H, 6.92; N, 21.36. Found: C, 59.56; H, 6.91; N, 21.39.
Synthesis of 2-(diethylamino)ethyl 4-(4-acetyl-5-methyl-4,5-dihydro-1H-1,2,3-triazol-1-yl)benzoate (6)
Sodium metal (0.01 mol) was dissolved in 20 mL of anhydrous C2H5OH in a 250 mL round-bottom flask. Compound 5 and 3 mL of acetyl acetone (0.01 mol) was added to the above solution and stirred for 1 h. The resulting reaction mixture was refluxed for 13–17 h at 130 °C to 140 °C. The reaction was monitored by TLC. When the reaction was completed, the mixture was poured into ice water. The product was precipitated, washed with water, dried and recrystallized from ethanol.Yield: 73%; M.p.: 127–128 °C; MW: 346.43; Rf: 0.59; FTIR (νmax; cm−1 KBr): 3035 (aromatic C–H stretching), 2967 (alkyl C–H stretching) 2931 (CH2 stretching), 2865 (CH3 stretching), 1718 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1624 (C
O stretching), 1624 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1462 (CH3 bending), 1418 (COO stretching), 1408 (C–N stretching), 1378 (CH2 bending), 832; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.98 (t, 6H, J = 7.4 Hz, CH3 × 2), 1.18 (s, 3H, triazole-CH3), 2.16 (s, 3H, CH3), 2.64 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.9 Hz, CH2), 4.29 (t, 2H, J = 6.9 Hz, CH2), 4.61 (d, 1H, J = 7.0 Hz, triazole-H), 4.79 (d, 1H, J = 4.3 Hz, triazole-H), 7.83 (d, 2H, J = 1.3 Hz, Ar–H), 8.08 (d, 2H, J = 1.7 Hz, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 208.4, 165.7, 148.1, 130.9, 118.5, 113.4, 82.9, 69.8, 63.1, 54.2, 49.8, 25.9, 13.4, 10.5; mass: 347.43 (M + H)+; elemental analysis for C18H26N4O3: calculated: C, 62.41; H, 7.57; N, 16.17. Found: C, 62.46; H, 7.62; N, 16.21.
C stretching), 1462 (CH3 bending), 1418 (COO stretching), 1408 (C–N stretching), 1378 (CH2 bending), 832; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.98 (t, 6H, J = 7.4 Hz, CH3 × 2), 1.18 (s, 3H, triazole-CH3), 2.16 (s, 3H, CH3), 2.64 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.9 Hz, CH2), 4.29 (t, 2H, J = 6.9 Hz, CH2), 4.61 (d, 1H, J = 7.0 Hz, triazole-H), 4.79 (d, 1H, J = 4.3 Hz, triazole-H), 7.83 (d, 2H, J = 1.3 Hz, Ar–H), 8.08 (d, 2H, J = 1.7 Hz, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 208.4, 165.7, 148.1, 130.9, 118.5, 113.4, 82.9, 69.8, 63.1, 54.2, 49.8, 25.9, 13.4, 10.5; mass: 347.43 (M + H)+; elemental analysis for C18H26N4O3: calculated: C, 62.41; H, 7.57; N, 16.17. Found: C, 62.46; H, 7.62; N, 16.21.
Synthesis of substituted 2-(diethylamino)ethyl 4-(4-cinnamoyl-5-methyl-4,5-dihydro-1H-1,2,3-triazol-1-yl)benzoate derivatives 7(a–h)
Corresponding aromatic benzaldehyde (a–h) (0.01 mol) and compound 6 were dissolved in a 250 mL conical flask using 95% ethanol as the solvent. After that, few grams of sodium hydroxide pellets were added and the mixture was stirred for 40 minutes. The product was precipitated, washed with water and recrystallized with ethanol to afford the corresponding target compound.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1628 (C
O stretching), 1628 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1467 (CH3 bending), 1419 (COO stretching), 1409 (C–N stretching), 1374 (CH2 bending), 839; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.07 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.1 Hz, aliphatic CH2 × 2), 2.94 (t, 2H, J = 6.8 Hz, CH2), 4.31 (t, 2H, J = 6.7 Hz, CH2), 4.64 (d, 1H, J = 7.1 Hz, triazole-H), 4.82 (d, 1H, J = 4.5 Hz, triazole-H), 6.83 (d, 1H, J = 15.2 Hz), 7.82 (d, 1H, J = 15.2 Hz), 7.81 (d, 2H, J = 1.37 Hz, Ar–H), 8.05 (d, 2H, J = 1.75 Hz, Ar–H), 7.37–7.52 (m, 5H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.8, 148.2, 142.9, 165.9, 135.2, 130.9, 128.6, 128.5, 127.8, 126.2, 118.5, 113.6, 80.8, 69.9, 63.4, 54.2, 49.8, 13.4, 10.5; mass: 435.57 (M + H)+; elemental analysis for C25H30N4O3: calculated: C, 69.10; H, 6.96; N, 12.89. Found: C, 69.14; H, 6.98; N, 12.93.
C stretching), 1467 (CH3 bending), 1419 (COO stretching), 1409 (C–N stretching), 1374 (CH2 bending), 839; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.07 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.1 Hz, aliphatic CH2 × 2), 2.94 (t, 2H, J = 6.8 Hz, CH2), 4.31 (t, 2H, J = 6.7 Hz, CH2), 4.64 (d, 1H, J = 7.1 Hz, triazole-H), 4.82 (d, 1H, J = 4.5 Hz, triazole-H), 6.83 (d, 1H, J = 15.2 Hz), 7.82 (d, 1H, J = 15.2 Hz), 7.81 (d, 2H, J = 1.37 Hz, Ar–H), 8.05 (d, 2H, J = 1.75 Hz, Ar–H), 7.37–7.52 (m, 5H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.8, 148.2, 142.9, 165.9, 135.2, 130.9, 128.6, 128.5, 127.8, 126.2, 118.5, 113.6, 80.8, 69.9, 63.4, 54.2, 49.8, 13.4, 10.5; mass: 435.57 (M + H)+; elemental analysis for C25H30N4O3: calculated: C, 69.10; H, 6.96; N, 12.89. Found: C, 69.14; H, 6.98; N, 12.93.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1624 (C
O stretching), 1624 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1469 (CH3 bending), 1412 (COO stretching), 1403 (C–N stretching), 1379 (CH2 bending), 849; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.4 Hz, CH3 × 2), 1.09 (s, 3H, triazole-CH3), 2.29 (s, 3H, Ar–CH3), 2.64 (q, 4H, J = 7.5 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.9 Hz, CH2), 4.29 (t, 2H, J = 6.9 Hz, CH2), 4.62 (d, 1H, J = 7.3, Hz, triazole-H), 4.88 (d, 1H, J = 4.3 Hz, triazole-H), 6.85 (d, 1H, J = 15.7, Hz), 7.87 (d, 1H, J = 15.1, Hz), 7.79 (d, 2H, J = 1.31 Hz, Ar–H), 8.07 (d, 2H, J = 1.71 Hz, Ar–H), 7.24–7.74 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 198.7, 165.8, 148.2, 142.8, 137.6, 132.3, 130.8, 128.7, 128.2, 126.2, 118.5, 113.5, 80.7, 69.8, 63.1, 54.2, 49.7, 21.4, 13.3, 10.5; mass: 449.59 (M + H)+; elemental analysis for C26H32N4O3: calculated: C, 69.62; H, 7.19; N, 12.49. Found: C, 69.65; H, 7.14; N, 12.46.
C stretching), 1469 (CH3 bending), 1412 (COO stretching), 1403 (C–N stretching), 1379 (CH2 bending), 849; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.4 Hz, CH3 × 2), 1.09 (s, 3H, triazole-CH3), 2.29 (s, 3H, Ar–CH3), 2.64 (q, 4H, J = 7.5 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.9 Hz, CH2), 4.29 (t, 2H, J = 6.9 Hz, CH2), 4.62 (d, 1H, J = 7.3, Hz, triazole-H), 4.88 (d, 1H, J = 4.3 Hz, triazole-H), 6.85 (d, 1H, J = 15.7, Hz), 7.87 (d, 1H, J = 15.1, Hz), 7.79 (d, 2H, J = 1.31 Hz, Ar–H), 8.07 (d, 2H, J = 1.71 Hz, Ar–H), 7.24–7.74 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 198.7, 165.8, 148.2, 142.8, 137.6, 132.3, 130.8, 128.7, 128.2, 126.2, 118.5, 113.5, 80.7, 69.8, 63.1, 54.2, 49.7, 21.4, 13.3, 10.5; mass: 449.59 (M + H)+; elemental analysis for C26H32N4O3: calculated: C, 69.62; H, 7.19; N, 12.49. Found: C, 69.65; H, 7.14; N, 12.46.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1621 (C
O stretching), 1621 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1473 (CH3 bending), 1418 (COO stretching), 1407 (C–N stretching), 1378 (CH2 bending), 842, 794 (C–Cl stretching), 703; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.08 (s, 3H, triazole-CH3), 2.62 (q, 4H, J = 7.3 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.4 Hz, CH2), 4.27 (t, 2H, J = 6.8 Hz, CH2), 4.64 (d, 1H, J = 6.9 Hz, triazole-H), 4.86 (d, 1H, J = 4.1 Hz, triazole-H), 6.83 (d, 1H, J = 15.4 Hz), 7.83 (d, 1H, J = 15.2 Hz), 7.82 (d, 2H, J = 1.34 Hz, Ar–H), 8.05 (d, 2H, J = 1.75 Hz, Ar–H), 7.56–7.72 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.4, 165.8, 148.2, 142.9, 133.6, 133.1, 130.7, 129.1, 128.7, 126.2, 118.5, 113.4, 80.6, 69.7, 63.2, 54.2, 49.6, 13.4, 10.5; mass: 469.97 (M + H)+; elemental analysis for C26H29ClN4O3: calculated: C, 64.03; H, 6.23; N, 11.95. Found: C, 64.01; H, 6.27; N, 11.97.
C stretching), 1473 (CH3 bending), 1418 (COO stretching), 1407 (C–N stretching), 1378 (CH2 bending), 842, 794 (C–Cl stretching), 703; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.08 (s, 3H, triazole-CH3), 2.62 (q, 4H, J = 7.3 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.4 Hz, CH2), 4.27 (t, 2H, J = 6.8 Hz, CH2), 4.64 (d, 1H, J = 6.9 Hz, triazole-H), 4.86 (d, 1H, J = 4.1 Hz, triazole-H), 6.83 (d, 1H, J = 15.4 Hz), 7.83 (d, 1H, J = 15.2 Hz), 7.82 (d, 2H, J = 1.34 Hz, Ar–H), 8.05 (d, 2H, J = 1.75 Hz, Ar–H), 7.56–7.72 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.4, 165.8, 148.2, 142.9, 133.6, 133.1, 130.7, 129.1, 128.7, 126.2, 118.5, 113.4, 80.6, 69.7, 63.2, 54.2, 49.6, 13.4, 10.5; mass: 469.97 (M + H)+; elemental analysis for C26H29ClN4O3: calculated: C, 64.03; H, 6.23; N, 11.95. Found: C, 64.01; H, 6.27; N, 11.97.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1625 (C
O stretching), 1625 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1495 (C–Br stretching), 1478 (CH3 bending), 1412 (COO stretching), 1402 (C–N stretching), 1373 (CH2 bending), 852; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.97 (t, 6H, J = 7.5 Hz, CH3 × 2), 1.06 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.8 Hz, CH2), 4.29 (t, 2H, J = 6.9 Hz, CH2), 4.62 (d, 1H, J = 6.7 Hz, triazole-H), 4.89 (d, 1H, J = 4.3 Hz, triazole-H), 6.81 (d, 1H, J = 15.6 Hz), 7.78 (d, 1H, J = 15.5 Hz), 7.79 (d, 2H, J = 1.32 Hz, Ar–H), 8.03 (d, 2H, J = 1.79 Hz, Ar–H), 7.48–7.70 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.8, 165.7, 148.2, 142.9, 134.3, 131.5, 130.8, 128.7, 126.2, 122.3, 118.6, 113.5, 80.7, 69.8, 63.2, 54.2, 49.7, 13.4, 10.5; mass: 514.47 (M + H)+; elemental analysis for C26H29BrN4O3: calculated: C, 58.48; H, 5.69; N, 10.91. Found: C, 58.51; H, 5.64; N, 10.89.
C stretching), 1495 (C–Br stretching), 1478 (CH3 bending), 1412 (COO stretching), 1402 (C–N stretching), 1373 (CH2 bending), 852; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.97 (t, 6H, J = 7.5 Hz, CH3 × 2), 1.06 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.8 Hz, CH2), 4.29 (t, 2H, J = 6.9 Hz, CH2), 4.62 (d, 1H, J = 6.7 Hz, triazole-H), 4.89 (d, 1H, J = 4.3 Hz, triazole-H), 6.81 (d, 1H, J = 15.6 Hz), 7.78 (d, 1H, J = 15.5 Hz), 7.79 (d, 2H, J = 1.32 Hz, Ar–H), 8.03 (d, 2H, J = 1.79 Hz, Ar–H), 7.48–7.70 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.8, 165.7, 148.2, 142.9, 134.3, 131.5, 130.8, 128.7, 126.2, 122.3, 118.6, 113.5, 80.7, 69.8, 63.2, 54.2, 49.7, 13.4, 10.5; mass: 514.47 (M + H)+; elemental analysis for C26H29BrN4O3: calculated: C, 58.48; H, 5.69; N, 10.91. Found: C, 58.51; H, 5.64; N, 10.89.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1628 (C
O stretching), 1628 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1482 (CH3 bending), 1414 (COO stretching), 1405 (C–N stretching), 1375 (CH2 bending), 1152 (C–F stretching), 863; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.98 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.09 (s, 3H, triazole-CH3), 2.61 (q, 4H, J = 7.4 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.9 Hz, CH2), 4.27 (t, 2H, J = 6.5 Hz, CH2), 4.64 (d, 1H, J = 6.8 Hz, triazole-H), 4.85 (d, 1H, J = 4.2 Hz, triazole-H), 6.84 (d, 1H, J = 15.2 Hz), 7.76 (d, 1H, J = 15.7 Hz), 7.81 (d, 2H, J = 1.42 Hz, Ar–H), 8.04 (d, 2H, J = 1.74 Hz, Ar–H), 7.18–7.80 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.4, 165.5, 162.2, 148.1, 142.9, 130.9, 130.4, 126.2, 118.4, 115.3, 113.4, 80.6, 69.8, 63.2, 54.2, 49.8, 13.4, 10.5; mass: 453.53 (M + H)+; elemental analysis for C26H29FN4O3: calculated: C, 66.35; H, 6.46; N, 12.38. Found: C, 66.38; H, 6.47; N, 12.35.
C stretching), 1482 (CH3 bending), 1414 (COO stretching), 1405 (C–N stretching), 1375 (CH2 bending), 1152 (C–F stretching), 863; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.98 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.09 (s, 3H, triazole-CH3), 2.61 (q, 4H, J = 7.4 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.9 Hz, CH2), 4.27 (t, 2H, J = 6.5 Hz, CH2), 4.64 (d, 1H, J = 6.8 Hz, triazole-H), 4.85 (d, 1H, J = 4.2 Hz, triazole-H), 6.84 (d, 1H, J = 15.2 Hz), 7.76 (d, 1H, J = 15.7 Hz), 7.81 (d, 2H, J = 1.42 Hz, Ar–H), 8.04 (d, 2H, J = 1.74 Hz, Ar–H), 7.18–7.80 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.4, 165.5, 162.2, 148.1, 142.9, 130.9, 130.4, 126.2, 118.4, 115.3, 113.4, 80.6, 69.8, 63.2, 54.2, 49.8, 13.4, 10.5; mass: 453.53 (M + H)+; elemental analysis for C26H29FN4O3: calculated: C, 66.35; H, 6.46; N, 12.38. Found: C, 66.38; H, 6.47; N, 12.35.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1638 (C
O stretching), 1638 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1481 (CH3 bending), 1415 (COO stretching), 1408 (C–N stretching), 1372 (CH2 bending), 868; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.06 (s, 3H, triazole-CH3), 2.63 (q, 4H, J = 7.3 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.8 Hz, CH2), 4.30 (t, 2H, J = 6.9 Hz, CH2), 4.62 (d, 1H, J = 6.5 Hz, triazole-H), 4.83 (d, 1H, J = 4.1 Hz, triazole-H), 6.82 (d, 1H, J = 15.4 Hz), 7.72 (d, 1H, J = 15.2 Hz), 7.83 (d, 2H, J = 1.48 Hz, Ar–H), 8.06 (d, 2H, J = 1.70, Hz, Ar–H), 6.98–7.56 (m, 4H, Ar–H), 9.42 (s, 1H, Ar–OH); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.8, 165.4, 157.8, 148.2, 142.9, 130.9, 130.6, 130.1, 126.2, 118.5, 115.9, 113.4, 80.6, 69.8, 63.2, 54.3, 49.8, 13.2, 10.3; mass: 451.55 (M + H)+; elemental analysis for C25H30N4O4: calculated: C, 66.65; H, 6.71; N, 12.44. Found: C, 66.63; H, 6.69; N, 12.41.
C stretching), 1481 (CH3 bending), 1415 (COO stretching), 1408 (C–N stretching), 1372 (CH2 bending), 868; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.06 (s, 3H, triazole-CH3), 2.63 (q, 4H, J = 7.3 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.8 Hz, CH2), 4.30 (t, 2H, J = 6.9 Hz, CH2), 4.62 (d, 1H, J = 6.5 Hz, triazole-H), 4.83 (d, 1H, J = 4.1 Hz, triazole-H), 6.82 (d, 1H, J = 15.4 Hz), 7.72 (d, 1H, J = 15.2 Hz), 7.83 (d, 2H, J = 1.48 Hz, Ar–H), 8.06 (d, 2H, J = 1.70, Hz, Ar–H), 6.98–7.56 (m, 4H, Ar–H), 9.42 (s, 1H, Ar–OH); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.8, 165.4, 157.8, 148.2, 142.9, 130.9, 130.6, 130.1, 126.2, 118.5, 115.9, 113.4, 80.6, 69.8, 63.2, 54.3, 49.8, 13.2, 10.3; mass: 451.55 (M + H)+; elemental analysis for C25H30N4O4: calculated: C, 66.65; H, 6.71; N, 12.44. Found: C, 66.63; H, 6.69; N, 12.41.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1632 (C
O stretching), 1632 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1483 (CH3 bending), 1412 (COO stretching), 1401 (C–N stretching), 1378 (CH2 bending), 861; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.97 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.09 (s, 3H, triazole-CH3), 2.61 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.86 (t, 2H, J = 6.9 Hz, CH2), 3.78 (s, 3H, Ar–OCH3), 4.26 (t, 2H, J = 6.5 Hz, CH2), 4.56 (d, 1H, J = 6.7 Hz, triazole-H), 4.81 (d, 1H, J = 4.3 Hz, triazole-H), 6.71 (d, 1H, J = 15.6 Hz), 7.74 (d, 1H, J = 15.4 Hz), 7.80 (d, 2H, J = 1.42 Hz, Ar–H), 8.04 (d, 2H, J = 1.73 Hz, Ar–H), 7.21–7.52 (m, 4H, Ar–H), 9.45 (s, 1H, Ar–OH); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.2, 165.4, 159.8, 148.1, 142.9, 130.8, 130.4, 130.1, 126.2, 118.4, 114.2, 113.4, 80.6, 69.8, 63.2, 55.9, 54.2, 49.6, 13.4, 10.4; mass: 465.58 (M + H)+; elemental analysis for C26H32N4O4: calculated: C, 67.22; H, 6.94; N, 12.06. Found: C, 67.26; H, 6.98; N, 12.04.
C stretching), 1483 (CH3 bending), 1412 (COO stretching), 1401 (C–N stretching), 1378 (CH2 bending), 861; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.97 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.09 (s, 3H, triazole-CH3), 2.61 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.86 (t, 2H, J = 6.9 Hz, CH2), 3.78 (s, 3H, Ar–OCH3), 4.26 (t, 2H, J = 6.5 Hz, CH2), 4.56 (d, 1H, J = 6.7 Hz, triazole-H), 4.81 (d, 1H, J = 4.3 Hz, triazole-H), 6.71 (d, 1H, J = 15.6 Hz), 7.74 (d, 1H, J = 15.4 Hz), 7.80 (d, 2H, J = 1.42 Hz, Ar–H), 8.04 (d, 2H, J = 1.73 Hz, Ar–H), 7.21–7.52 (m, 4H, Ar–H), 9.45 (s, 1H, Ar–OH); 13C NMR (100 MHz, DMSO-d6) δ ppm: 197.2, 165.4, 159.8, 148.1, 142.9, 130.8, 130.4, 130.1, 126.2, 118.4, 114.2, 113.4, 80.6, 69.8, 63.2, 55.9, 54.2, 49.6, 13.4, 10.4; mass: 465.58 (M + H)+; elemental analysis for C26H32N4O4: calculated: C, 67.22; H, 6.94; N, 12.06. Found: C, 67.26; H, 6.98; N, 12.04.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1638 (C
O stretching), 1638 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1532 (NO2 stretching), 1489 (CH3 bending), 1415 (COO stretching), 1403 (C–N stretching), 1374 (CH2 bending), 869; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.07 (s, 3H, triazole-CH3), 2.65 (q, 4H, J = 7.3 Hz, aliphatic CH2 × 2), 2.89 (t, 2H, J = 6.8 Hz, CH2), 4.28 (t, 2H, J = 6.8 Hz, CH2), 4.63 (d, 1H, J = 6.9 Hz, triazole-H), 5.13 (d, 1H, J = 4.2 Hz, triazole-H), 7.08 (d, 1H, J = 15.2 Hz), 8.09 (d, 1H, J = 15.1 Hz), 7.81 (d, 2H, J = 1.47 Hz, Ar–H), 8.06 (d, 2H, J = 1.70 Hz, Ar–H), 7.84–8.03 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 198.4, 165.4, 148.1, 147.1, 141.4, 142.9, 130.8, 129.1, 126.2, 123.8, 118.5, 113.4, 80.8, 69.7, 63.2, 54.2, 49.8, 13.4, 10.4; mass: 480.56 (M + H)+; elemental analysis for C25H29N5O5: calculated: C, 62.62; H, 6.10; N, 14.60. Found: C, 62.67; H, 6.14; N, 14.63.
C stretching), 1532 (NO2 stretching), 1489 (CH3 bending), 1415 (COO stretching), 1403 (C–N stretching), 1374 (CH2 bending), 869; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.07 (s, 3H, triazole-CH3), 2.65 (q, 4H, J = 7.3 Hz, aliphatic CH2 × 2), 2.89 (t, 2H, J = 6.8 Hz, CH2), 4.28 (t, 2H, J = 6.8 Hz, CH2), 4.63 (d, 1H, J = 6.9 Hz, triazole-H), 5.13 (d, 1H, J = 4.2 Hz, triazole-H), 7.08 (d, 1H, J = 15.2 Hz), 8.09 (d, 1H, J = 15.1 Hz), 7.81 (d, 2H, J = 1.47 Hz, Ar–H), 8.06 (d, 2H, J = 1.70 Hz, Ar–H), 7.84–8.03 (m, 4H, Ar–H); 13C NMR (100 MHz, DMSO-d6) δ ppm: 198.4, 165.4, 148.1, 147.1, 141.4, 142.9, 130.8, 129.1, 126.2, 123.8, 118.5, 113.4, 80.8, 69.7, 63.2, 54.2, 49.8, 13.4, 10.4; mass: 480.56 (M + H)+; elemental analysis for C25H29N5O5: calculated: C, 62.62; H, 6.10; N, 14.60. Found: C, 62.67; H, 6.14; N, 14.63.
Synthesis of target derivatives 8(a–h)
The target compounds 8(a–h) were synthesized using corresponding derivatives 7(a–h) (0.01 mol), hydroxyl amine hydrochloride (0.02 mol) and potassium hydroxide (10%) by refluxing for 4–5 h with constant stirring at 110 °C. The reaction was monitored by TLC. When the reaction was completed, the mixture was poured into ice water to obtain the solid product. The product was washed with water, dried and re-crystallized with ethanol.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1632 (C
O stretching), 1632 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1612 (C
C stretching), 1612 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1564 (N
N stretching), 1564 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1492 (CH3 bending), 1419 (COO stretching), 1408 (C–N stretching), 1376 (CH2 bending), 1096 (C–O–C stretching), 862; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.62 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.90 (t, 2H, J = 7.6 Hz, CH2), 4.31 (t, 2H, J = 6.3 Hz, CH2), 4.65 (d, 1H, J = 4.2, Hz, triazole-H), 4.91 (d, 1H, J = 4.5 Hz, triazole-H), 6.63 (s, 1H, isoxazole-H), 7.38–7.42 (m, 5H, Ar–H), 7.79 (d, 2H, J = 1.38, Hz,), 8.05 (d, 2H, J = 1.74, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.9, 164.6, 148.1, 142.4, 130.7, 128.8, 127.6, 127.1, 118.5, 113.4, 83.1, 69.6, 69.2, 63.2, 54.2, 49.5, 44.5, 13.4, 11.8; mass: 448.56 (M + H)+; elemental analysis for C25H29N5O3: calculated: C, 67.09; H, 6.53; N, 15.65. Found: C, 67.11H, 6.54; N, 15.67.
N), 1492 (CH3 bending), 1419 (COO stretching), 1408 (C–N stretching), 1376 (CH2 bending), 1096 (C–O–C stretching), 862; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.62 (q, 4H, J = 7.2 Hz, aliphatic CH2 × 2), 2.90 (t, 2H, J = 7.6 Hz, CH2), 4.31 (t, 2H, J = 6.3 Hz, CH2), 4.65 (d, 1H, J = 4.2, Hz, triazole-H), 4.91 (d, 1H, J = 4.5 Hz, triazole-H), 6.63 (s, 1H, isoxazole-H), 7.38–7.42 (m, 5H, Ar–H), 7.79 (d, 2H, J = 1.38, Hz,), 8.05 (d, 2H, J = 1.74, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.9, 164.6, 148.1, 142.4, 130.7, 128.8, 127.6, 127.1, 118.5, 113.4, 83.1, 69.6, 69.2, 63.2, 54.2, 49.5, 44.5, 13.4, 11.8; mass: 448.56 (M + H)+; elemental analysis for C25H29N5O3: calculated: C, 67.09; H, 6.53; N, 15.65. Found: C, 67.11H, 6.54; N, 15.67.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1639 (C
O stretching), 1639 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1611 (C
C stretching), 1611 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1579 (N
N stretching), 1579 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1497 (CH3 bending), 1419 (COO stretching), 1401 (C–N stretching), 1367 (CH2 bending), 1096 (C–O–C stretching), 859; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.68 (q, 4H, J = 7.23 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.6 Hz, CH2), 4.29 (t, 2H, J = 6.3 Hz, CH2), 4.61 (d, 1H, J = 4.2, Hz, triazole-H), 4.95 (d, 1H, J = 4.7 Hz, triazole-H), 6.61 (s, 1H, isoxazole-H), 6.68–7.39 (m, 4H, Ar–H), 7.82 (d, 2H, J = 1.43, Hz,), 8.02 (d, 2H, J = 1.71, Hz), 9.03 (s, 1H, Ar–OH); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.7, 157.4, 148.1, 135.2, 130.8, 127.4, 118.5, 116.1, 113.4, 83.1, 69.6, 69.5, 63.2, 54.1, 49.6, 44.5, 13.4, 11.7; mass: 464.56 (M + H)+; elemental analysis for C25H29N5O4: calculated: C, 64.78; H, 6.31; N, 15.11. Found: C, 64.80; H, 6.29; N, 15.09.
N), 1497 (CH3 bending), 1419 (COO stretching), 1401 (C–N stretching), 1367 (CH2 bending), 1096 (C–O–C stretching), 859; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.2 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.68 (q, 4H, J = 7.23 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.6 Hz, CH2), 4.29 (t, 2H, J = 6.3 Hz, CH2), 4.61 (d, 1H, J = 4.2, Hz, triazole-H), 4.95 (d, 1H, J = 4.7 Hz, triazole-H), 6.61 (s, 1H, isoxazole-H), 6.68–7.39 (m, 4H, Ar–H), 7.82 (d, 2H, J = 1.43, Hz,), 8.02 (d, 2H, J = 1.71, Hz), 9.03 (s, 1H, Ar–OH); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.7, 157.4, 148.1, 135.2, 130.8, 127.4, 118.5, 116.1, 113.4, 83.1, 69.6, 69.5, 63.2, 54.1, 49.6, 44.5, 13.4, 11.7; mass: 464.56 (M + H)+; elemental analysis for C25H29N5O4: calculated: C, 64.78; H, 6.31; N, 15.11. Found: C, 64.80; H, 6.29; N, 15.09.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1638 (C
O stretching), 1638 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1619 (C
C stretching), 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1562 (N
N stretching), 1562 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1495 (CH3 bending), 1417 (COO stretching), 1403 (C–N stretching), 1378 (CH2 bending), 1093 (C–O–C stretching), 851; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.1 Hz, CH3 × 2), 1.18 (s, 3H, triazole-CH3), 2.25 (s, 3H, CH3), 2.64 (q, 4H, J = 7.16 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.8 Hz, CH2), 4.30 (t, 2H, J = 6.4 Hz, CH2), 4.62 (d, 1H, J = 4.3, Hz, triazole-H), 4.89 (d, 1H, J = 4.3 Hz, triazole-H), 6.65 (s, 1H, isoxazole-H), 7.13–7.16 (m, 4H, Ar–H), 7.76 (d, 2H, J = 1.35, Hz,), 8.02 (d, 2H, J = 1.72, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.4, 148.1, 139.5, 137.4, 130.8, 129.2, 125.8, 118.6, 113.5, 83.1, 69.7, 69.2, 63.2, 54.2, 49.6, 44.5, 21.3, 13.2, 11.7; mass: 462.56 (M + H)+; elemental analysis for C26H31N5O3: calculated: C, 67.66; H, 6.77; N, 15.17. Found: C, 67.68; H, 6.74; N, 15.21.
N), 1495 (CH3 bending), 1417 (COO stretching), 1403 (C–N stretching), 1378 (CH2 bending), 1093 (C–O–C stretching), 851; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.1 Hz, CH3 × 2), 1.18 (s, 3H, triazole-CH3), 2.25 (s, 3H, CH3), 2.64 (q, 4H, J = 7.16 Hz, aliphatic CH2 × 2), 2.92 (t, 2H, J = 6.8 Hz, CH2), 4.30 (t, 2H, J = 6.4 Hz, CH2), 4.62 (d, 1H, J = 4.3, Hz, triazole-H), 4.89 (d, 1H, J = 4.3 Hz, triazole-H), 6.65 (s, 1H, isoxazole-H), 7.13–7.16 (m, 4H, Ar–H), 7.76 (d, 2H, J = 1.35, Hz,), 8.02 (d, 2H, J = 1.72, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.4, 148.1, 139.5, 137.4, 130.8, 129.2, 125.8, 118.6, 113.5, 83.1, 69.7, 69.2, 63.2, 54.2, 49.6, 44.5, 21.3, 13.2, 11.7; mass: 462.56 (M + H)+; elemental analysis for C26H31N5O3: calculated: C, 67.66; H, 6.77; N, 15.17. Found: C, 67.68; H, 6.74; N, 15.21.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1635 (C
O stretching), 1635 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1618 (C
C stretching), 1618 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1572 (N
N stretching), 1572 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1495 (CH3 bending), 1412 (COO stretching), 1405 (C–N stretching), 1369 (CH2 bending), 1099 (C–O–C stretching), 851; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.98 (t, 6H, J = 7.5 Hz, CH3 × 2), 1.17 (s, 3H, triazole-CH3), 2.64 (q, 4H, J = 7.29 Hz, aliphatic CH2 × 2), 2.93 (t, 2H, J = 6.8 Hz, CH2), 3.74 (s, 3H, OCH3), 4.32 (t, 2H, J = 6.7 Hz, CH2), 4.58 (d, 1H, J = 4.1, Hz, triazole-H), 4.93 (d, 1H, J = 4.9 Hz, triazole-H), 6.64 (d, 1H, J = 6.81 Hz, isoxazole-H), 6.89–7.36 (m, 4H, Ar–H), 7.84 (d, 2H, J = 1.42, Hz,), 8.05 (d, 2H, J = 1.75, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.7, 164.2, 159.4, 148.3, 134.8, 130.9, 127.0, 118.7, 114.5, 113.6, 83.3, 69.8, 69.3, 63.4, 55.9, 54.3, 49.7, 44.6, 13.5, 11.8; mass: 478.56 (M + H)+; elemental analysis for C26H31N5O4: calculated: C, 65.39; H, 6.54; N, 14.67. Found: C, 65.41; H, 6.52; N, 14.65.
N), 1495 (CH3 bending), 1412 (COO stretching), 1405 (C–N stretching), 1369 (CH2 bending), 1099 (C–O–C stretching), 851; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.98 (t, 6H, J = 7.5 Hz, CH3 × 2), 1.17 (s, 3H, triazole-CH3), 2.64 (q, 4H, J = 7.29 Hz, aliphatic CH2 × 2), 2.93 (t, 2H, J = 6.8 Hz, CH2), 3.74 (s, 3H, OCH3), 4.32 (t, 2H, J = 6.7 Hz, CH2), 4.58 (d, 1H, J = 4.1, Hz, triazole-H), 4.93 (d, 1H, J = 4.9 Hz, triazole-H), 6.64 (d, 1H, J = 6.81 Hz, isoxazole-H), 6.89–7.36 (m, 4H, Ar–H), 7.84 (d, 2H, J = 1.42, Hz,), 8.05 (d, 2H, J = 1.75, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.7, 164.2, 159.4, 148.3, 134.8, 130.9, 127.0, 118.7, 114.5, 113.6, 83.3, 69.8, 69.3, 63.4, 55.9, 54.3, 49.7, 44.6, 13.5, 11.8; mass: 478.56 (M + H)+; elemental analysis for C26H31N5O4: calculated: C, 65.39; H, 6.54; N, 14.67. Found: C, 65.41; H, 6.52; N, 14.65.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1637 (C
O stretching), 1637 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1612 (C
C stretching), 1612 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1578 (N
N stretching), 1578 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1492 (CH3 bending), 1418 (COO stretching), 1403 (C–N stretching), 1365 (CH2 bending), 1162 (C–F stretching), 1092 (C–O–C stretching), 856; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.97 (t, 6H, J = 7.3 Hz, CH3 × 2), 1.17 (s, 3H, triazole-CH3), 2.65 (q, 4H, J = 7.24 Hz, aliphatic CH2 × 2), 2.93 (t, 2H, J = 6.7 Hz, CH2), 4.31 (t, 2H, J = 6.1 Hz, CH2), 4.63 (d, 1H, J = 4.3, Hz, triazole-H), 4.91 (d, 1H, J = 4.1 Hz, triazole-H), 6.63 (s, 1H, isoxazole-H), 7.07–7.49 (m, 4H, Ar–H), 7.79 (d, 2H, J = 1.39, Hz,), 8.07 (d, 2H, J = 1.76, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.6, 161.8, 148.2, 138.2, 130.7, 126.8, 118.5, 115.9, 113.5, 83.1, 69.7, 69.5, 63.2, 54.2, 49.6, 44.5, 13.5, 11.8; mass: 466.53 (M + H)+; elemental analysis for C25H28FN5O3: calculated: C, 64.50; H, 6.06; N, 15.04. Found: C, 64.52; H, 6.05; N, 15.01.
N), 1492 (CH3 bending), 1418 (COO stretching), 1403 (C–N stretching), 1365 (CH2 bending), 1162 (C–F stretching), 1092 (C–O–C stretching), 856; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.97 (t, 6H, J = 7.3 Hz, CH3 × 2), 1.17 (s, 3H, triazole-CH3), 2.65 (q, 4H, J = 7.24 Hz, aliphatic CH2 × 2), 2.93 (t, 2H, J = 6.7 Hz, CH2), 4.31 (t, 2H, J = 6.1 Hz, CH2), 4.63 (d, 1H, J = 4.3, Hz, triazole-H), 4.91 (d, 1H, J = 4.1 Hz, triazole-H), 6.63 (s, 1H, isoxazole-H), 7.07–7.49 (m, 4H, Ar–H), 7.79 (d, 2H, J = 1.39, Hz,), 8.07 (d, 2H, J = 1.76, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.6, 161.8, 148.2, 138.2, 130.7, 126.8, 118.5, 115.9, 113.5, 83.1, 69.7, 69.5, 63.2, 54.2, 49.6, 44.5, 13.5, 11.8; mass: 466.53 (M + H)+; elemental analysis for C25H28FN5O3: calculated: C, 64.50; H, 6.06; N, 15.04. Found: C, 64.52; H, 6.05; N, 15.01.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1639 (C
O stretching), 1639 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1614 (C
C stretching), 1614 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1568 (N
N stretching), 1568 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1497 (CH3 bending), 1419 (COO stretching), 1406 (C–N stretching), 1374 (CH2 bending), 1098 (C–O–C stretching), 856, 784 (C–Cl stretching); 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.3 Hz, CH3), 1.17 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.23 Hz, aliphatic CH3 × 2), 2.86 (t, 2H, J = 6.7 Hz, CH2), 4.31 (t, 2H, J = 6.2 Hz, CH2), 4.64 (d, 1H, J = 4.1, Hz, triazole-H), 4.91 (d, 1H, J = 4.5 Hz, triazole-H), 6.64 (s, 1H, isoxazole-H), 7.42–7.55 (m, 4H, Ar–H), 7.79 (d, 2H, J = 1.38, Hz,), 8.06 (d, 2H, J = 1.75, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.7, 148.1, 140.5, 133.2, 130.8, 129.1, 126.8, 118.5, 113.5, 83.1, 69.6, 69.2, 63.2, 54.4, 49.8, 44.6, 13.4, 11.8; mass: 482.98 (M + H)+; elemental analysis for C25H28ClN5O3: calculated: C, 62.30; H, 5.86; N, 14.53. Found: C, 62.33; H, 5.85; N, 14.55.
N), 1497 (CH3 bending), 1419 (COO stretching), 1406 (C–N stretching), 1374 (CH2 bending), 1098 (C–O–C stretching), 856, 784 (C–Cl stretching); 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.3 Hz, CH3), 1.17 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.23 Hz, aliphatic CH3 × 2), 2.86 (t, 2H, J = 6.7 Hz, CH2), 4.31 (t, 2H, J = 6.2 Hz, CH2), 4.64 (d, 1H, J = 4.1, Hz, triazole-H), 4.91 (d, 1H, J = 4.5 Hz, triazole-H), 6.64 (s, 1H, isoxazole-H), 7.42–7.55 (m, 4H, Ar–H), 7.79 (d, 2H, J = 1.38, Hz,), 8.06 (d, 2H, J = 1.75, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.8, 164.7, 148.1, 140.5, 133.2, 130.8, 129.1, 126.8, 118.5, 113.5, 83.1, 69.6, 69.2, 63.2, 54.4, 49.8, 44.6, 13.4, 11.8; mass: 482.98 (M + H)+; elemental analysis for C25H28ClN5O3: calculated: C, 62.30; H, 5.86; N, 14.53. Found: C, 62.33; H, 5.85; N, 14.55.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1633 (C
O stretching), 1633 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1619 (C
C stretching), 1619 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1572 (N
N stretching), 1572 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1493 (CH3 bending), 1486 (C–Br stretching), 1415 (COO stretching), 1408 (C–N stretching), 1375 (CH2 bending), 1094 (C–O–C stretching), 858; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.5 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.69 (q, 4H, J = 7.43 Hz, aliphatic CH2 × 2), 2.88 (t, 2H, J = 6.8 Hz, CH2), 4.34 (t, 2H, J = 6.1 Hz, CH2), 4.66 (d, 1H, J = 4.5, Hz, triazole-H), 4.94 (d, 1H, J = 4.2 Hz, triazole-H), 6.66 (s, 1H, isoxazole-H), 7.31–7.36 (m, 4H, Ar–H), 7.76 (d, 2H, J = 1.35, Hz,), 8.02 (d, 2H, J = 1.73, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.4, 164.6, 148.1, 141.5, 131.9, 130.8, 127.3, 122.0, 118.5, 113.4, 83.1, 69.8, 69.2, 63.1, 54.2, 49.8, 44.5, 13.4, 11.7; mass: 527.45 (M + H)+; elemental analysis for C25H28BrN5O3: calculated: C, 57.04; H, 5.36; N, 13.30. Found: C, 57.06; H, 5.34; N, 13.28.
N), 1493 (CH3 bending), 1486 (C–Br stretching), 1415 (COO stretching), 1408 (C–N stretching), 1375 (CH2 bending), 1094 (C–O–C stretching), 858; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.95 (t, 6H, J = 7.5 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.69 (q, 4H, J = 7.43 Hz, aliphatic CH2 × 2), 2.88 (t, 2H, J = 6.8 Hz, CH2), 4.34 (t, 2H, J = 6.1 Hz, CH2), 4.66 (d, 1H, J = 4.5, Hz, triazole-H), 4.94 (d, 1H, J = 4.2 Hz, triazole-H), 6.66 (s, 1H, isoxazole-H), 7.31–7.36 (m, 4H, Ar–H), 7.76 (d, 2H, J = 1.35, Hz,), 8.02 (d, 2H, J = 1.73, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.4, 164.6, 148.1, 141.5, 131.9, 130.8, 127.3, 122.0, 118.5, 113.4, 83.1, 69.8, 69.2, 63.1, 54.2, 49.8, 44.5, 13.4, 11.7; mass: 527.45 (M + H)+; elemental analysis for C25H28BrN5O3: calculated: C, 57.04; H, 5.36; N, 13.30. Found: C, 57.06; H, 5.34; N, 13.28.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), 1637 (C
O stretching), 1637 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), 1613 (C
C stretching), 1613 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching), 1579 (N
N stretching), 1579 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1532 (NO2 stretching), 1497 (CH3 bending), 1418 (COO stretching), 1401 (C–N stretching), 1365 (CH2 bending), 1097 (C–O–C stretching), 858; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.3 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.36 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.5 Hz, CH2), 4.31 (t, 2H, J = 6.2 Hz, CH2), 4.63 (d, 1H, J = 4.2, Hz, triazole-H), 4.90 (d, 1H, J = 4.8 Hz, triazole-H), 6.63 (d, 1H, isoxazole-H), 7.43–8.08 (m, 4H, Ar–H), 7.83 (d, 2H, J = 1.46, Hz,), 8.06 (d, 2H, J = 1.70, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.7, 164.7, 148.6, 148.1, 146.9, 130.8, 128.1, 124.2, 118.7, 113.4, 83.2, 69.7, 69.2, 63.2, 54.2, 49.8, 44.3, 13.4, 11.9; mass: 493.54 (M + H)+; elemental analysis for C25H28N6O5: calculated: C, 60.97; H, 5.73; N, 17.06. Found: C, 60.99; H, 5.75; N, 17.05.
N), 1532 (NO2 stretching), 1497 (CH3 bending), 1418 (COO stretching), 1401 (C–N stretching), 1365 (CH2 bending), 1097 (C–O–C stretching), 858; 1H NMR (400 MHz, DMSO-d6, TMS) δ ppm: 0.96 (t, 6H, J = 7.3 Hz, CH3 × 2), 1.19 (s, 3H, triazole-CH3), 2.67 (q, 4H, J = 7.36 Hz, aliphatic CH2 × 2), 2.87 (t, 2H, J = 6.5 Hz, CH2), 4.31 (t, 2H, J = 6.2 Hz, CH2), 4.63 (d, 1H, J = 4.2, Hz, triazole-H), 4.90 (d, 1H, J = 4.8 Hz, triazole-H), 6.63 (d, 1H, isoxazole-H), 7.43–8.08 (m, 4H, Ar–H), 7.83 (d, 2H, J = 1.46, Hz,), 8.06 (d, 2H, J = 1.70, Hz); 13C NMR (100 MHz, DMSO-d6) δ ppm: 165.7, 164.7, 148.6, 148.1, 146.9, 130.8, 128.1, 124.2, 118.7, 113.4, 83.2, 69.7, 69.2, 63.2, 54.2, 49.8, 44.3, 13.4, 11.9; mass: 493.54 (M + H)+; elemental analysis for C25H28N6O5: calculated: C, 60.97; H, 5.73; N, 17.06. Found: C, 60.99; H, 5.75; N, 17.05.
Kinase inhibitory activity43
CDK1 and CDK4 inhibitory activity
In vitro anti-CDK1/cyclin B1 and CDK4/cyclin D3 assays were performed using commercial kits as per the manufacturer's instruction (BPS Biosciences, UK). Buffer, ATP, CDK substrate peptidase, and distilled water were mixed, and then 25 μL of the mixture was added to a 96-well plate. Five microliters of test compounds were sequentially added. CDK1/cyclin B1 was diluted to 1 ng μL−1, and CDK4/cyclin D3 was diluted to 10 ng μL−1 using a buffer. Twenty microliters of the kinase solutions were incubated with the test compounds at 30 °C for 45 min for CDK1/cyclin B1 and for 60 min for CDK4/cyclin D3. Finally, 50 μL of Kinase-Glo Max reagent was added to each well. The 96-well plate was covered with aluminum foil and incubated at room temperature for 15 min. The luminescence of the test compounds was measured using a microplate reader.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) h. The cells were washed with PBS (pH 7.4), fixed with ice-cold 70% ethanol and resuspended in DAPI, and incubated for 15 min at 37 °C wrapped in aluminium foil. The cells were then washed with PBS and examined using a Nikon Eclipse Ti fluorescence microscope (Nikon Instruments Inc., NY, USA).
h. The cells were washed with PBS (pH 7.4), fixed with ice-cold 70% ethanol and resuspended in DAPI, and incubated for 15 min at 37 °C wrapped in aluminium foil. The cells were then washed with PBS and examined using a Nikon Eclipse Ti fluorescence microscope (Nikon Instruments Inc., NY, USA).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 dilution liquid of the culture medium was added to the Transwell chamber. Next, 500 μL per well of Compound 8e was added to a Transwell chamber containing 200 μL of cell suspension at a concentration of 105 cells per mL in serum-free media. The mixture was then incubated at 37 °C for 24 hours. The upper chamber's A549 cells were cleansed with cotton swabs after the media were withdrawn. Following a 20 minute fixation in formaldehyde, the cells were allowed to dry before staining with a 10 minute solution of PBS containing 0.01% crystal violet. Transmembrane cells were counted in five randomly selected images acquired using an inverted microscope after the removal of crystal violet from the surface.
3 dilution liquid of the culture medium was added to the Transwell chamber. Next, 500 μL per well of Compound 8e was added to a Transwell chamber containing 200 μL of cell suspension at a concentration of 105 cells per mL in serum-free media. The mixture was then incubated at 37 °C for 24 hours. The upper chamber's A549 cells were cleansed with cotton swabs after the media were withdrawn. Following a 20 minute fixation in formaldehyde, the cells were allowed to dry before staining with a 10 minute solution of PBS containing 0.01% crystal violet. Transmembrane cells were counted in five randomly selected images acquired using an inverted microscope after the removal of crystal violet from the surface.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was administered to the right flank region of every single creature. Following a period of five days, the mice were classified into four groups, each consisting of five mice: a saline solution is given to the control group, whereas chemical 8e was administered to the three other treatment groups at different doses. Intraperitoneal injections of compound 8e were administered three times a week for a period of fourteen days. The tumor volume was measured according to the previously described procedure, starting from the beginning of the experiment and continuing at defined intervals until the fourteenth day. In the following step, the mice were put under anesthesia and then slaughtered, and the samples were collected for further examination.50–52
1 was administered to the right flank region of every single creature. Following a period of five days, the mice were classified into four groups, each consisting of five mice: a saline solution is given to the control group, whereas chemical 8e was administered to the three other treatment groups at different doses. Intraperitoneal injections of compound 8e were administered three times a week for a period of fourteen days. The tumor volume was measured according to the previously described procedure, starting from the beginning of the experiment and continuing at defined intervals until the fourteenth day. In the following step, the mice were put under anesthesia and then slaughtered, and the samples were collected for further examination.50–52
Group 2: implanted Eca109 cells + compound 8e (2 mg kg−1).
Group 3: implanted Eca109 cells + compound 8e (5 mg kg−1).
Group 4: implanted Eca109 cells + compound 8e (10 mg kg−1).
Treatment started when all tumors reached 100 mm3. The tumor length, width, and mouse body weight were measured, whereas the tumor volume was calculated using the following formula:
| Tumor volume (mm3) = length (mm) × width (mm) × height (mm)/2. Data are presented as mean ± standard error of the mean (SEM). |
Results and discussion
Chemistry
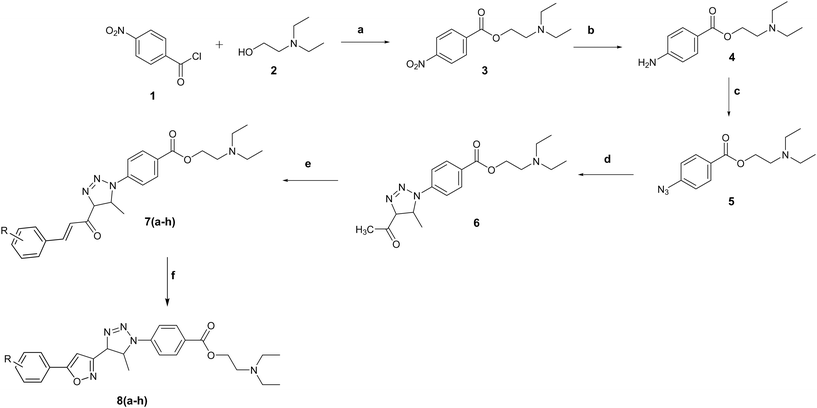
Intermediate compounds 3, 4, 5, 6, and 7(a–h) and title compounds 8(a–h) were synthesized in a facile manner, as shown in Scheme 1. In the first step, compound 3 was obtained by the reaction of p-nitro benzoyl chloride (1) with 2-(diethylamino)ethanol (2). The compound 4 was obtained after reducing para-nitrobenzoyl diethylethanolamine (3) with tin metal in the presence of HCl.53 Furthermore, compound 5 was obtained by the diazotization of amino group of 1-(4-aminophenyl)-3-(diethylamino)-2-hydroxypropan-1-one (4), which further allowed refluxing with acetyl acetone for 12 to 15 h to furnish intermediate compound (6). The compounds 7(a–h) were obtained via the Claisen–Schmidt condensation reaction of compound (6) with different aromatic aldehydes (a–h) in the presence of KOH. The title compounds 8(a–h) were synthesized in good yields by reacting corresponding 7(a–h) with hydroxyl amine hydrochloride in an alkaline medium using KOH as the base.All the synthesized compounds were ascertained by different spectroscopic methods such as FT-IR, 1H-NMR, 13C-NMR, mass and elemental analyses. The FT-IR spectra of title compounds showed an absorption band at 3428 cm−1 due to the presence of OH groups linked with the phenyl ring. The absorption bands at 3047–3024 cm−1 and 2956–2941 cm−1 correspond to the C–H proton of the aromatic ring and the stretching vibration of the aliphatic group, respectively. Furthermore, the stretching vibration of a CH3 group linked with the phenyl ring appeared at 2958–2938 cm−1. Another peak at 1729–1713 cm−1 was attributed to the ester carbonyl group. The aromatic nitro group appeared at 1538 cm−1. The aromatic fluoro group showed an absorption band at 1165 cm−1 and an aromatic bromo group observed at 768 cm−1. The 1H NMR spectra of title compounds showed singlet peaks at 1.16–1.19 ppm due to the triazole methyl group. Furthermore, the isoxazoline and triazole rings showed doublet peaks at 4.92–4.65 and 2.86–3.05 due to the CH2 and CH groups. The aromatic proton appeared as a doublet at 8.03–8.07 ppm due to the C–H groups. The 13C-NMR resonance peaks of title compounds 8(a–h) appeared at 148.2 to 113.4 and 146.2 to 124.4 due to the aromatic ring carbon. The isoxazole carbon atoms correspond to 83.2 to 44.6 ppm. The resonance peaks at 69.7 to 69.3 ppm are assigned to the CH2 group of the triazole ring. Furthermore, the resonance appeared at 63.2 to 54.4 ppm attributed to the aliphatic CH2 group linked with the phenyl ring.
All the synthesized compounds 8(a–h) were ascertained by elemental analysis and mass spectroscopy.
Kinase inhibitory activity
The aberrant activation of kinases has been considered as a major underlying cause for various cancers.54 Many molecules have been pursued to target a variety of kinases such as CDK,55 EGFR,45 VEGFR,56 and Hsp90.57 PI3K and mTOR inhibitors have found a significant place in anticancer therapeutics and many of the molecules are now under various stages of drug development.58Initially, the designed compounds were tested against PI3K and mTOR, and the comparative inhibitory activity is provided in Table 1. It has been found that the entire set of derivatives showed significant to moderate inhibition of both kinases in the nano-molar range. Compound 8a with no substitution showed the lowest inhibitory activity against PI3K (IC50 = 67.78 ± 2.60 nM) with moderate inhibitory activity against mTOR kinase (IC50 = 59.27 ± 2.73 nM). However, upon introduction of substitution, the activity was significantly improved against PI3K with slight reduction in mTOR kinase (compound 8b having p-hydroxy). The inhibitory activity was found to be significantly improved against both the tested kinases upon replacing p-hydroxy with p-methyl in the case of compound 8c. The presence of p-methoxy group in 8d was found to be detrimental for the activity, rendering the compound moderately active against both PI3K and mTOR. Interestingly, the introduction of electron-withdrawing groups as substituents in the subsequent compounds, i.e. compounds 8e, 8f, 8g and 8h, showed drastic improvement in inhibitory activity against the tested kinase. However, all these molecules displayed a range of inhibitory activities. The presence of p-nitro group in 8h showed the least inhibitory activity among the above-mentioned group. Upon replacing the nitro group with bromo in the case of compound 8g, the activity was found to be drastically improved, which was further found to be increased in the case of 8f (p-chloro) and 8e (p-fluoro). The comparative inhibitory activity suggests that none of the synthesized compounds showed better activity than gedatolisib, a standard inhibitor of tested kinases. Moreover, compound 8e was identified as the most potent inhibitor of PI3K and mTOR among the tested series, while a non-substituted derivative was found to be least active. The structure–activity relationship study of compounds suggests that compounds containing an electron-withdrawing group showed a better inhibitory profile than that of their electron-donating derivatives, since apparently the size of the electron-withdrawing group in p-position plays an important role in the activity against the tested kinases.
| Compound | Substitution | Kinase inhibitiona (IC50, in nM) | |||||
|---|---|---|---|---|---|---|---|
| PI3K | mTOR | CDK1 | CDK4 | EGFR | VGEFR2 | ||
| a Mean ± SEM of three replicates. | |||||||
| 8a | H | 67.78 ± 2.60 | 59.27 ± 2.73 | 562.41 ± 18.53 | 2802.34 ± 102.05 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8b | 4-OH | 56.82 ± 2.31 | 61.43 ± 2.45 | 630.24 ± 22.15 | 2236.26 ± 97.45 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8c | 4-CH3 | 39.30 ± 1.32 | 27.74 ± 1.42 | 726.72 ± 39.25 | 1894.42 ± 85.67 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8d | 4-OCH3 | 45.67 ± 1.34 | 40.31 ± 1.05 | 1034.45 ± 43.21 | 1556.75 ± 84.52 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8e | 4-F | 4.23 ± 0.32 | 2.3 ± 0.15 | 945.22 ± 37.12 | 1264.50 ± 93.42 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8f | 4-Cl | 10.04 ± 0.56 | 13.63 ± 0.62 | 846.51 ± 32.71 | 1045.35 ± 95.34 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8g | 4-Br | 19.34 ± 0.27 | 27.04 ± 0.45 | 902.45 ± 42.62 | 1326.30 ± 99.42 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8h | 4-NO2 | 32.26 ± 0.43 | 30.40 ± 0.56 | 1251.34 ± 64.14 | 1444.22 ± 90.32 | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Gedatolisib (standard) | 6.12 ± 0.32 | 2.8 ± 0.31 | — | — | — | — | |
| Palbociclib (standard) | — | — | — | 13.41 ± 0.18 | — | — | |
| Dinaciclib (standard) | — | — | 18.42 ± 0.24 | — | — | — | |
| Erlotinib (standard) | — | — | — | — | 0.12 ± 0.01 | 0.008 ± 0.11 | |
| Vehicle control | >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
|
Moreover, the target compounds were evaluated for their inhibitory activity against key off-target kinases, namely CDK1, CDK4, EGFR, and VEGFR2, to assess their target selectivity profile. The results of these assays are summarized in Table 1. It was observed that none of the synthesized compounds exhibited significant inhibitory activity against EGFR and VEGFR2, with IC50 values exceeding 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM, indicating a lack of meaningful interaction with these kinases. In the case of CDK1 and CDK4, the compounds demonstrated only weak inhibition, showing considerably higher IC50 values compared to the reference standards—dinaciclib for CDK1 and palbociclib for CDK4.
000 nM, indicating a lack of meaningful interaction with these kinases. In the case of CDK1 and CDK4, the compounds demonstrated only weak inhibition, showing considerably higher IC50 values compared to the reference standards—dinaciclib for CDK1 and palbociclib for CDK4.
These findings suggest that the synthesized procaine–triazole–isoxazole derivatives possess high selectivity toward the intended targets, namely PI3K and mTOR, while sparing other important kinases involved in cell cycle regulation and angiogenesis. The data support the conclusion that these compounds are promising dual inhibitors of the PI3K/mTOR pathway with reduced likelihood of off-target kinase inhibition.
Anticancer activity of compound 8e, 8f and 8g on various cancer cells
Under the influence of good results in kinase inhibitory assays, the top-three ranked compounds were subjected for further examinations of anticancer activity against diverse cancer cell lines, such as cervical cancer (HeLa), human breast cancer cell line (MCF-7), liver cancer cell line (HepG2), lung cancer cell line (A549), esophageal cancer (Eca109) and non-tumorigenic epithelial cell (MCF12A), and the results are presented in Table 2. It has been found that the tested compound showed potent to moderate activity against the tested cell lines. Compound 8g showed the lowest activity among all the tested derivatives against entire cell lines. It showed least activity against A549 with significant activity against the MCF-7 cells. The activity was found significantly increased in the case of compound 8f except against HepG2 cells, where it displayed the lowest activity. The most significant inhibitory activity was reported by compound 8e against all the tested cell lines with the highest potency against Eca109 cells, where it showed more potent activity than standard cisplatin. Moreover, none of the compounds were found to be toxic to the normal epithelial cells at the highest tested concentration of 250 μM. The above inhibitory results suggested that the most active compound is with the smallest and the highest electron withdrawing substituent.| Compound | IC50a (in μM) | |||||
|---|---|---|---|---|---|---|
| HeLa | MCF-7 | HepG2 | A549 | Eca109 | MCF12A | |
| a Mean ± SEM of three replicates. | ||||||
| 8e | 23.12 ± 0.67 | 22.03 ± 0.23 | 16.72 ± 1.67 | 17.78 ± 2.43 | 0.28 ± 0.15 | >250 |
| 8f | 12.34 ± 1.05 | 5.62 ± 0.94 | 45.53 ± 2.10 | 38.42 ± 2.71 | 34.15 ± 2.62 | >250 |
| 8g | 34.56 ± 1.39 | 12.34 ± 1.43 | 37.75 ± 2.83 | 54.62 ± 3.53 | 47.24 ± 3.55 | >250 |
| Cisplatin (standard) | 4.22 ± 0.65 | 2.22 ± 0.29 | 4.74 ± 0.85 | 3.54 ± 0.54 | 4.22 ± 0.43 | Not determined |
| Vehicle control | >250 | >250 | >250 | >250 | >250 | >250 |
Moreover, the negative control consisted of cells treated with the vehicle alone (0.1% DMSO in a culture medium) without any test compound. This control group served to account for any non-specific effects of the vehicle on cell viability. The results showed no significant reduction in cell viability, with IC50 values exceeding 250 μM for all tested cell lines, indicating that the observed inhibitory effects in experimental groups were due to the test compounds and not the vehicle or assay conditions.
Anticancer activity of compound 8e on various esophageal cancer cells
Studies have shown that human esophageal cancer is quite complex, and the process of carcinogenesis involves the actions of genes at multiple levels. These inherited genotypes led to the identification of genetically defined ‘intrinsic’ subtypes associated with different clinical outcomes, responses to therapy, and preferential sites of relapse.59–61 Thus, inspired by the excellent inhibitory activity of compound 8e against the human esophageal cancer cell line Eca109, in the next instance, we decided to further quantify the inhibitory potential of compound 8e against the battery of different esophageal squamous cell carcinoma (ESCC) cell lines having different genotypes, reflecting the heterogeneity of ESCC. Eca109 cells, originating from a Chinese ESCC patient, exhibit frequent mutations in TP53 and alterations in cell cycle regulators such as CDKN2A, rendering it a significant model for investigating drug resistance and tumor progression.62,63 TE1 and TE13, originating from poorly differentiated ESCC, are frequently utilized in research concerning cell proliferation and apoptosis. TE1 is characterized by mutations in TP53 and modifications in EGFR signaling pathways,64,65 whereas TE13 frequently displays mutations in KRAS and additional oncogenic pathways.66The KYSE series, comprising KYSE150, KYSE30, and KYSE70, consists of well-characterized cell lines obtained from Japanese ESCC patients, each exhibiting unique genetic profiles.67 KYSE150 exhibits a high frequency of TP53 mutations and amplification of oncogenes such as EGFR and MET, positioning it as a preferred model for molecular and xenograft research. KYSE30 is distinguished by its specific mutations in cell adhesion molecules, particularly CDH1 (E-cadherin), which are pertinent to metastasis studies. KYSE70 shares several molecular characteristics with KYSE150; however, it demonstrates a reduced level of genomic instability, rendering it a valuable model for investigating early-stage ESCC.68
As shown in Table 3, compound 8e showed significant inhibition of numerous ESCC cell lines with potent inhibition of Eca109 cells among all the tested ESCC cell lines with IC50 of 0.28 μM.
Effect of compound 8e on the morphological characteristics of Eca109 cells
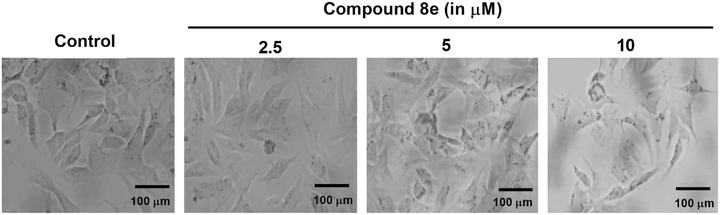
To assess the impact of compound 8e on the morphology of Eca109 esophageal cancer cells, the cells were treated with increasing concentrations of 2.5, 5, and 10 μM. As shown in Fig. 3, a concentration-dependent alteration in cellular morphology was observed. Compared to untreated control cells, which maintained a typical elongated and adherent epithelial morphology, treatment with compound 8e led to noticeable morphological changes, including cell shrinkage, membrane blebbing, and loss of cellular integrity.These morphological disruptions became more pronounced at higher concentrations, indicating a dose-responsive effect. At 10 μM, a significant number of cells appeared rounded, detached, and displayed the condensed cytoplasm—hallmarks commonly associated with apoptotic or necrotic cell death. The observed deterioration in the cellular architecture suggests that compound 8e exerts cytotoxic effects on Eca109 cells, potentially contributing to its anticancer activity by compromising cell viability and triggering cell death pathways.
Effect of compound 8e on the nuclear morphology and apoptosis of Eca109 cells
Apoptosis is the process of programmed cell death by which the body will try to eliminate the dysfunctional cells. In ESCC, the cancerous cells evade this process and make it dysfunctional, which leads to their survival. The process of apoptosis is highly governed with pro-apoptotic and apoptotic genes, and oncogenic mutation of these genes disrupt apoptosis, leading to tumour survival and metastasis.69 A lot of studies have confirmed the role of many cytotoxic agents in promoting apoptosis.70 To explore whether the attenuation of Eca109 cellular proliferation by compound 8e is attributed to apoptosis, the cells were treated with indicated doses of 8e, and the results are presented in Fig. 3. On analysing DAPI-stained cells via fluorescence microscopy (Fig. 4A), the rate of apoptosis was found to be increased in the compound 8e-treated group, as evidenced by the reduction in the number of cancer cells. These results suggest that compound 8e kills Eca109 cancer cells possibly via inducing apoptosis. These results further confirmed that compound 8e increased the rate of apoptosis, as evidenced by flow cytometric analysis through Annexin V-FITC and propidium iodide dual staining of compound 8e-treated Eca109 cells (Fig. 4B and C).Effect of compound 8e on the migration and invasion of Ec109 cells
The cancer treatment has faced serious obstacles due to the metastasis of tumours, which was initiated by the migration and invasion processes.71 Thus, current approaches to control cancer cell metastasis are concentrated on reducing the migration and invasion processes. Thus, the next part of the study was aimed to explore the effect of compound 8e on the migration of Eca109 cells by the wound-healing assay and transwell-Matrigel invasion assay. The results are presented in Fig. 5. It has been found that compound 8e causes a reduction in the migration of cells around the wound corner, and the rate of sclerosis on both the sides decreased with the increase in sclerosis width. Moreover, the scratch-healing rate of the 8e-treated cell group was found to be slowly decreasing in a dose-dependent manner (Fig. 5A and B). The compound 8e-treated group also showed a reduction in the invasion ability of Eca109 cells (Fig. 5C and D). These results indicated that compound 8e possesses the ability to attenuate the tumor metastasis.| Compound | % inhibition |
|---|---|
| Compound 8e | 71.5 |
| BIBR1532 | 51.2 |
| Control | 0 |
Effect of compound 8e on the mitochondria-associated apoptotic protein
Western blot analysis was performed to determine the effect of compound 8e on the expression of mitochondria-associated intrinsic apoptosis protein (p53, Bax and Bcl2), which governs the cell death or apoptosis.84 As shown in Fig. 8, compound 8e-treated group showed increased expression of p-53 and Bax, while the level of Bcl2 was found to be decreased significantly. The highest tested dose of compound 8e (10 μM) causes more significant effects than other tested concentrations, suggesting that 8e causes a dose-dependent reduction of mitochondria-associated apoptotic proteins.Effect on the PI3K/Akt/mTOR signalling cascade
PI3K/Akt/mTOR is a signal transduction pathway, which is directly linked to the growth and development of diverse cancers.21 It has been found that the activation of the PI3K/Akt/mTOR signaling pathway plays an important role in cellular proliferation, apoptosis and metastasis and found aberrantly activated in many cancers including ESCC.85 Therefore, many of the newer anticancer drugs such as duvelisib, copanlisib and idelalisib inhibit this signalling pathway. Recently, alpelisib, an alpha-specific PI3K inhibitor, was approved by the FDA in May 2019 for use in combination with endocrine therapy fulvestrant for the treatment of HR-positive and HER2/neu-negative breast cancer.86 Therefore, inspired by the excellent inhibitory activity of compound 8e against PI3K and mTOR, it was worthwhile to scrutinize its effect on the PI3K/Akt/mTOR downstream signalling proteins in Eca109 cells using western blot analysis. The results are presented in Fig. 9. It has been found that the levels of p-PI3K, p-Akt and p-mTOR decreased significantly, while no significant changes were observed in their non-phosphorylated counterparts in the 8e-treated group in a dose-dependent manner. Thus, it could be understood that compound 8e might exert anticancer effects by attenuating the PI3K/Akt/mTOR downstream signalling cascade.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060. It also showed excellent bioavailability of 89.5%. From the above-mentioned pharmacokinetic parameters, it was suggested that compound 8e has excellent bioavailability in both IV and oral routes.
060. It also showed excellent bioavailability of 89.5%. From the above-mentioned pharmacokinetic parameters, it was suggested that compound 8e has excellent bioavailability in both IV and oral routes.
| Parameters | Intravenous | Oral solution |
|---|---|---|
| t 1/2 (h) | 3.42 ± 0.11 | 9.22 ± 0.05 |
| C max (μg mL−1) | 1.27 ± 0.001 | 2.11 ± 0.001 |
| t max | 0 | 2.00 ± 0.04 |
| K e (h−1) | 0.0752 | |
| K a (h−1) | — | 1.03 |
| AUC (ng h mL−1) | 6270 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060 060 |
| F (%) | — | 89.5 |
Conclusion
In summary, we have successfully synthesized a series of novel procaine derivatives incorporating 1,2,3-triazole and substituted isoxazole moieties via a straightforward and efficient synthetic route. Among the designed compounds, several demonstrated significant inhibitory activities against PI3K and mTOR, with notable anticancer efficacy against esophageal cancer cells, surpassing their effects on other cancer cell lines. Notably, compound 8e emerged as the most potent candidate, exhibiting strong pro-apoptotic effects through the modulation of key mitochondrial apoptotic proteins including p53, Bcl-2, and Bax. Furthermore, it effectively inhibited the migration and invasion of esophageal cancer cells by attenuating the phosphorylation of PI3K, Akt, and mTOR, as confirmed by western blot analysis. In vivo evaluation using an Eca109 cell-derived xenograft mouse model further substantiated the therapeutic potential of compound 8e. Treatment led to a marked reduction in tumor volume, accompanied by an increase in body weight, suggesting favorable systemic tolerance. Additionally, compound 8e significantly reduced oxidative stress and suppressed the production of pro-inflammatory cytokines in tumor tissues. Immunohistochemical staining also revealed a substantial decrease in the expression of PI3K and p-Akt in treated tumors. Pharmacokinetic profiling of compound 8e indicated optimal parameters including excellent bioavailability, supporting its potential for further preclinical development. Collectively, these findings position compound 8e as a promising lead molecule for the development of next-generation procaine–triazole–isoxazole-based anticancer agents targeting the PI3K/Akt/mTOR signaling pathway in esophageal cancer.Conflicts of interest
There are no conflict of interest to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: the spectral data of compounds 8a–g are provided in the supplementary information. See DOI: https://doi.org/10.1039/d5md00554j.References
- J. Ferlay, H. R. Shin, F. Bray, D. Forman, C. Mathers and D. M. Parkin, Int. J. Cancer, 2010, 127, 2893–2917 CrossRef CAS PubMed.
- L. A. Torre, F. Bray, R. L. Siegel, J. Ferlay, J. Lortet-Tieulent and A. Jemal, Ca-Cancer J. Clin., 2015, 65, 87–108 CrossRef PubMed.
- N. Shukla, M. Hagenbuchner, K. T. Win and J. Yang, Comput. Methods Programs Biomed., 2018, 155, 199–208 CrossRef PubMed.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- H. Liang, J. H. Fan and Y. L. Qiao, Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China, Cancer Biol. Med., 2017, 14, 33–41 Search PubMed.
- B. Ying, H. Huang, H. Li, M. Song, S. Wu and H. Ying, Oncol. Res., 2017, 25, 1463–1470 CrossRef PubMed.
- H. Sabit, M. B. Samy, O. A. M. Said and M. M. El-Zawahri, Procaine Induces Epigenetic Changes in HCT116 Colon Cancer Cells, Genet. Res. Int., 2016, 2016, 8348450, DOI:10.1155/2016/8348450.
- C. Li, S. Gao, X. Li, C. Li and L. Ma, Oncol. Res., 2018, 26, 209–217 CrossRef.
- A. Villar-Garea, M. F. Fraga, J. Espada and M. Esteller, Cancer Res., 2003, 63, 4984–4989 CAS.
- Z. Gao, Z. Xu, M. S. Hung, Y. C. Lin, T. Wang, M. Gong, X. Zhi, D. M. Jablons and L. You, Oncol. Rep., 2009, 22, 1479–1484 CAS.
- Y. C. Li, Y. Wang, D. D. Li, Y. Zhang, T. C. Zhao and C. F. Li, J. Cell. Biochem., 2018, 119, 2440–2449 CrossRef CAS PubMed.
- H. Zhou, M. Xu, G. Luo and Y. Zhang, Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi, 2007, 21, 1118–1121 Search PubMed.
- M. Tada, F. Imazeki, K. Fukai, A. Sakamoto, M. Arai, R. Mikata, T. Tokuhisa and O. Yokosuka, Hepatol. Int., 2007, 1, 355–364 CrossRef.
- E. Kucuksayan and T. Ozben, Curr. Top. Med. Chem., 2016, 17, 907–918 CrossRef.
- V. Dubey, M. Pathak, H. R. H. R. Bhat and U. P. U. P. Singh, Chem. Biol. Drug Des., 2012, 80, 598–604 CrossRef.
- U. P. Singh, M. Pathak, V. Dubey, H. R. Bhat, P. Gahtori and R. K. Singh, Chem. Biol. Drug Des., 2012, 80, 572–583 CrossRef CAS PubMed.
- Q. Luo, R. Du, W. Liu, G. Huang, Z. Dong and X. Li, PI3K/Akt/mTOR Signaling Pathway: Role in Esophageal Squamous Cell Carcinoma, Regulatory Mechanisms and Opportunities for Targeted Therapy, Front. Oncol., 2022, 12, 852383 CrossRef CAS.
- X. Zhang, Y. Wang, X. Zhang, Y. Shen, K. Yang, Q. Ma, Y. Qiao, J. Shi, Y. Wang, L. Xu, B. Yang, G. Ge, L. Hu, X. Kong, C. Yang, Y. Chen, J. Ding and L. Meng, Intact regulation of G1/S transition renders esophageal squamous cell carcinoma sensitive to PI3Kα inhibitors, Signal Transduction Targeted Ther., 2023, 8, 153 CrossRef CAS.
- R. Huang, Q. Dai, R. Yang, Y. Duan, Q. Zhao, J. Haybaeck and Z. Yang, A Review: PI3K/AKT/mTOR Signaling Pathway and Its Regulated Eukaryotic Translation Initiation Factors May Be a Potential Therapeutic Target in Esophageal Squamous Cell Carcinoma, Front. Oncol., 2022, 12, 817916 CrossRef CAS PubMed.
- J. j. Shi, H. Xing, Y. x. Wang, X. Zhang, Q. m. Zhan, M. y. Geng, J. Ding and L. h. Meng, PI3Kα inhibitors sensitize esophageal squamous cell carcinoma to radiation by abrogating survival signals in tumor cells and tumor microenvironment, Cancer Lett., 2019, 459, 145–155 CrossRef CAS PubMed.
- O. Ekshyyan, A. Anandharaj and C. A. O. Nathan, Clin. Cancer Res., 2013, 19, 3719–3721 CrossRef CAS.
- X. Wu, Y. Xu, Q. Liang, X. Yang, J. Huang, J. Wang, H. Zhang and J. Shi, Advances in Dual PI3K/mTOR Inhibitors for Tumour Treatment, Front. Pharmacol., 2022, 13, 875372 CrossRef CAS PubMed.
- T. M. Wise-Draper, G. Moorthy, M. A. Salkeni, N. A. Karim, H. E. Thomas, C. A. Mercer, M. S. Beg, S. O'Gara, O. Olowokure, H. Fathallah, S. C. Kozma, G. Thomas, O. Rixe, P. Desai and J. C. Morris, A Phase Ib Study of the Dual PI3K/mTOR Inhibitor Dactolisib (BEZ235) Combined with Everolimus in Patients with Advanced Solid Malignancies, Target. Oncol., 2017, 12, 323–332 CrossRef PubMed.
- H. Cheng, C. Li, S. Bailey, S. M. Baxi, L. Goulet, L. Guo, J. Hoffman, Y. Jiang, T. O. Johnson, T. W. Johnson, D. R. Knighton, J. Li, K. K. C. Liu, Z. Liu, M. A. Marx, M. Walls, P. A. Wells, M. J. Yin, J. Zhu and M. Zientek, Discovery of the highly potent PI3K/mTOR dual inhibitor PF-04979064 through structure-based drug design, ACS Med. Chem. Lett., 2013, 4, 91–97 CrossRef CAS PubMed.
- S. Shen, X. He, Z. Yang, L. Zhang, Y. Liu, Z. Zhang, W. Wang, W. Liu, Y. Li, D. Huang, K. Sun, X. Ni, X. Yang, X. Chu, Y. Cui, Q. Lv, J. Lan and F. Zhou, Discovery of an Orally Bioavailable Dual PI3K/mTOR Inhibitor Based on Sulfonyl-Substituted Morpholinopyrimidines, ACS Med. Chem. Lett., 2018, 9, 719–724 CrossRef CAS.
- P. A. Yakkala, S. R. Panda, S. Shafi, V. G. M. Naidu, M. S. Yar, P. N. Ubanako, S. A. Adeyemi, P. Kumar, Y. E. Choonara, E. V. Radchenko, V. A. Palyulin and A. Kamal, Synthesis and Cytotoxic Activity of 1,2,4-Triazolo-Linked Bis-Indolyl Conjugates as Dual Inhibitors of Tankyrase and PI3K, Molecules, 2022, 27, 7642 CrossRef CAS PubMed.
- J. L. Guo, Y. Y. Liu and Y. Z. Pei, Synthesis and biological evaluation of 3-(piperidin-4-yl)isoxazolo[4,5-d]pyrimidine derivatives as novel PI3Kδ inhibitors, Chin. Chem. Lett., 2015, 26, 1283–1288 CrossRef CAS.
- N. Wu, Y. Zhu, X. Xu, Y. Zhu, Y. Song, L. Pang and Z. Chen, The anti-tumor effects of dual PI3K/mTOR inhibitor BEZ235 and histone deacetylase inhibitor Trichostatin A on inducing autophagy in esophageal squamous cell carcinoma, J. Cancer, 2018, 9, 987–997 CrossRef.
- R. Mishra, H. Patel, S. Alanazi, M. K. Kilroy and J. T. Garrett, PI3K inhibitors in cancer: Clinical implications and adverse effects, Int. J. Mol. Sci., 2021, 22, 3464 CrossRef CAS PubMed.
- J. Yang, R. Butti, S. Cohn, V. Toffessi-Tcheuyap, A. Mal, M. Nguyen, C. Stevens, A. Christie, A. Mishra, Y. Ma, J. Kim, R. Abraham, P. Kapur, R. E. Hammer and J. Brugarolas, Unconventional mechanism of action and resistance to rapalogs in renal cancer, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2310793121 CrossRef CAS PubMed.
- K. Bozorov, J. Zhao and H. A. Aisa, 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: A recent overview, Bioorg. Med. Chem., 2019, 27, 3511–3531 CrossRef CAS PubMed.
- J. Zhu, J. Mo, H. z. Lin, Y. Chen and H. p. Sun, The recent progress of isoxazole in medicinal chemistry, Bioorg. Med. Chem., 2018, 26, 3065–3075 CrossRef CAS PubMed.
- M. R. Jensen, J. Schoepfer, T. Radimerski, A. Massey, C. T. Guy, J. Brueggen, C. Quadt, A. Buckler, R. Cozens, M. J. Drysdale, C. Garcia-Echeverria and P. Chène, NVP-AUY922: a small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models, Breast Cancer Res., 2008, 10, R33 CrossRef.
- J. Jung, J. Kwon, S. Hong, A. N. Moon, J. Jeong, S. Kwon, J. a. Kim, M. Lee, H. Lee, J. H. Lee and J. Lee, Discovery of novel heat shock protein (Hsp90) inhibitors based on luminespib with potent antitumor activity, Bioorg. Med. Chem. Lett., 2020, 30, 127165 CrossRef CAS PubMed.
- F. Behrens, M. Koehm and H. Burkhardt, Update 2011: leflunomide in rheumatoid arthritis – strengths and weaknesses, Curr. Opin. Rheumatol., 2011, 23, 282–287 CrossRef CAS PubMed.
- H. Y. Lin, H. W. Han, W. X. Sun, Y. S. Yang, C. Y. Tang, G. H. Lu, J. L. Qi, X. M. Wang and Y. H. Yang, Design and characterization of α-lipoic acyl shikonin ester twin drugs as tubulin and PDK1 dual inhibitors, Eur. J. Med. Chem., 2018, 144, 137–150 CrossRef CAS PubMed.
- O. H. Chan and B. H. Stewart, Physicochemical and drug-delivery considerations for oral drug bioavailability, Drug Discovery Today, 1996, 1, 461–473 CrossRef CAS.
- H. M. R. de Souza, J. S. Guedes, R. H. C. N. Freitas, L. G. V. Gelves, H. H. Fokoue, C. M. R. Sant'Anna, E. J. Barreiro and L. M. Lima, Comparative chemical and biological hydrolytic stability of homologous esters and isosteres, J. Enzyme Inhib. Med. Chem., 2022, 37, 718–727, DOI:10.1080/14756366.2022.2027933.
- B. Ruggeri, J. Singh, D. Gingrich, T. Angeles, M. Albom, H. Chang, C. Robinson, K. Hunter, P. Dobrzanski, S. Jones-Bolin, L. Aimone, A. Klein-Szanto, J. M. Herbert, F. Bono, P. Schaeffer, P. Casellas, B. Bourie, R. Pili, J. Isaacs, M. Ator, R. Hudkins, J. Vaught, J. Mallamo and C. Dionne, CEP-7055: A novel, orally active pan inhibitor of vascular endothelial growth factor receptor tyrosine kinases with potent antiangiogenic activity and antitumor efficacy in preclinical models, Cancer Res., 2003, 63, 5978–5991 CAS.
- L. C. Creemer, H. A. Kirst, C. J. Vlahos and R. M. Schultz, Synthesis and in vitro evaluation of new wortmannin esters: Potent inhibitors of phosphatidylinositol 3-kinase, J. Med. Chem., 1996, 39, 5021–5024 CrossRef CAS.
- L. Xi, J. Q. Zhang, Z. C. Liu, J. H. Zhang, J. F. Yan, Y. Jin and J. Lin, Novel 5-anilinoquinazoline-8-nitro derivatives as inhibitors of VEGFR-2 tyrosine kinase: Synthesis, biological evaluation and molecular docking, Org. Biomol. Chem., 2013, 11, 4367–4378 RSC.
- Z. Qiang, W. Yu and Y. Yu, Pharmacology, 2019, 104, 126–138 CrossRef CAS PubMed.
- J. Hu, Y. Zhang, N. Tang, Y. Lu, P. Guo and Z. Huang, Bioorg. Med. Chem., 2021, 32, 115997 CrossRef CAS PubMed.
- U. Singh and H. Bhat, J. Thorac. Oncol., 2017, 12, S1494–S1495 CrossRef.
- J. K. Srivastava, G. G. Pillai, H. R. Bhat, A. Verma and U. P. Singh, Sci. Rep., 2017, 7, 5851 CrossRef PubMed.
- H. R. Bhat, A. Masih, A. Shakya, S. K. Ghosh and U. P. Singh, J. Heterocyclic Chem., 2020, 57, 390–399 CrossRef CAS.
- W. Geng, X. Feng, C. Li and S. Ni, Bull. Chem. Soc. Ethiop., 2024, 38, 1091–1101 CrossRef CAS.
- L. Zhang and C. Li, In Vitro Cell. Dev. Biol.: Anim., 2024, 60, 365–373 CrossRef CAS PubMed.
- X. I. Cheng, Z. Zuo, Q. Zhou, P. Ma and X. Liu, Synthesis Of Curcumin-Ferulic Acid Ester Via Steglich Esterification And Anti-Lung Cancer Activity Against Human Non-Small Lung Cancer Cells, J. Chil. Chem. Soc., 2024, 69, 6086–6091 CrossRef CAS.
- J. Jung, Toxicol. Res., 2014, 30, 1–5 CrossRef PubMed.
- W. Xing, D. T. Chen, J. H. Pan, Y. H. Chen, Y. Yan, Q. Li, R. F. Xue, Y. F. Yuan and W. A. Zeng, Lidocaine Induces Apoptosis and Suppresses Tumor Growth in Human Hepatocellular Carcinoma Cells in Vitro and in a Xenograft Model in Vivo, Anesthesiology, 2017, 126, 868–881 CrossRef CAS.
- Q. Su, B. Xu, Z. Tian and Z. Gong, Chem. Biol. Drug Des., 2022, 99, 320–330 CrossRef CAS.
- Z. Qiang, W. Yu and Y. Yu, Pharmacology, 2019, 104, 126–138 CrossRef CAS PubMed.
- J. Zhang, P. L. Yang and N. S. Gray, Targeting cancer with small molecule kinase inhibitors, Nat. Rev. Cancer, 2009, 9, 28–39 CrossRef CAS PubMed.
- T. G. Davies, J. Bentley, C. E. Arris, F. T. Boyle, N. J. Curtin, J. A. Endicott, A. E. Gibson, B. T. Golding, R. J. Griffin, I. R. Hardcastle, P. Jewsbury, L. N. Johnson, V. Mesguiche, D. R. Newell, M. E. M. Noble, J. A. Tucker, L. Wang and H. J. Whitfield, Nat. Struct. Biol., 2002, 9, 745–749 CrossRef CAS PubMed.
- L. Jakobsson, J. Kreuger, K. Holmborn, L. Lundin, I. Eriksson, L. Kjellén and L. Claesson-Welsh, Dev. Cell, 2006, 10, 625–634 CrossRef CAS PubMed.
- G. Garg, A. Khandelwal and B. S. J. Blagg, in Advances in Cancer Research, 2016, vol. 129, pp. 51–88 Search PubMed.
- H. Pópulo, J. M. Lopes and P. Soares, The mTOR signalling pathway in human cancer, Int. J. Mol. Sci., 2012, 13, 1886–1918 CrossRef.
- H. Simba, H. Kuivaniemi, V. Lutje, G. Tromp and V. Sewram, Systematic review of genetic factors in the etiology of esophageal squamous cell carcinoma in african populations, Front. Genet., 2019, 10, 642 CrossRef CAS PubMed.
- X. Liu, M. Zhang, S. Ying, C. Zhang, R. Lin, J. Zheng, G. Zhang, D. Tian, Y. Guo, C. Du, Y. Chen, S. Chen, X. Su, J. Ji, W. Deng, X. Li, S. Qiu, R. Yan, Z. Xu, Y. Wang, Y. Guo, J. Cui, S. Zhuang, H. Yu, Q. Zheng, M. Marom, S. Sheng, G. Zhang, S. Hu, R. Li and M. Su, Genetic Alterations in Esophageal Tissues From Squamous Dysplasia to Carcinoma, Gastroenterology, 2017, 53, 166–177 CrossRef PubMed.
- U. Testa, G. Castelli and E. Pelosi, The Molecular Characterization of Genetic Abnormalities in Esophageal Squamous Cell Carcinoma May Foster the Development of Targeted Therapies, Curr. Oncol., 2023, 30, 610–640 CrossRef.
- N. Zhang, J. Shi, X. Shi, W. Chen and J. Liu, Mutational Characterization and Potential Prognostic Biomarkers of Chinese Patients with Esophageal Squamous Cell Carcinoma, Onco Targets Ther, 2020, 13, 12797–12809, DOI:10.2147/OTT.S275688.
- N. Jiang, W. Q. Zhang, Y. T. Hao, L. M. Zhang, L. Shan, X. D. Yang, C. L. Peng and H. Dong, SMAC exhibits anti-tumor effects in ECA109 cells by regulating expression of inhibitor of apoptosis protein family, World J. Clin. Cases, 2021, 9, 5019–5027 CrossRef.
- M. K. Kashyap and O. Abdel-Rahman, Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma, Mol. Cancer, 2018, 17, 54 CrossRef.
- K. Yoshida, E. Kyo, T. Tsuda, T. Tsujino, M. Ito, M. Niimoto and E. Tahara, EGF and TGF-α, the ligands of hyperproduced EGFR in human esophageal carcinoma cells, act as autocrine growth factors, Int. J. Cancer, 1990, 45, 131–135, DOI:10.1002/ijc.2910450124.
- C. Galiana, A. Fusco, N. Martel, T. Nishihira, S. Hirohashi and H. Yamasaki, Possible role of activated ras genes in human esophageal carcinogenesis, Int. J. Cancer, 1993, 54, 978–982 CrossRef CAS PubMed.
- H. Wang, X. Yang, Y. Guo, L. Shui, S. Li, Y. Bai, Y. Liu, M. Zeng and J. Xia, HERG1 promotes esophageal squamous cell carcinoma growth and metastasis through TXNDC5 by activating the PI3K/AKT pathway, J. Exp. Clin. Cancer Res., 2019, 38, 324 CrossRef.
- C. Zhang, C. Li, J. Z. Su, K. Zhao, L. Shao and J. Deng, The genomic landscape of esophageal squamous cell carcinoma cell lines, Cancer Cell Int., 2025, 25, 174 CrossRef CAS.
- Y. Tsujimoto and S. Shimizu, Role of the mitochondrial membrane permeability transition in cell death, Apoptosis, 2007, 12, 835–840 CrossRef CAS PubMed.
- B. A. Carneiro and W. S. El-Deiry, Targeting apoptosis in cancer therapy, Nat. Rev. Clin. Oncol., 2020, 17, 395–417 CrossRef PubMed.
- P. S. Steeg, Targeting metastasis, Nat. Rev. Cancer, 2016, 16, 201–218 CrossRef CAS PubMed.
- S. Figel and R. A. Fenstermaker, in Handbook of Brain Tumor Chemotherapy, Molecular Therapeutics, and Immunotherapy, 2nd edn, 2018, pp. 257–269 Search PubMed.
- M. B. Kastan and J. Bartek, Cell-cycle checkpoints and cancer, Nature, 2004, 432, 316–323 CrossRef CAS PubMed.
- B. O’Leary, R. S. Finn and N. C. Turner, Treating cancer with selective CDK4/6 inhibitors, Nat. Rev. Clin. Oncol., 2016, 13, 417–430 CrossRef.
- M. H. Rahaman, F. Lam, L. Zhong, T. Teo, J. Adams, M. Yu, R. W. Milne, C. Pepper, N. A. Lokman, C. Ricciardelli, M. K. Oehler and S. Wang, Mol. Oncol., 2019, 13, 2178–2193 CrossRef CAS.
- L. Vadlakonda, M. Pasupuleti and R. Pallu, Role of PI3K-AKT-mTOR and Wnt signaling pathways in transition of G1-S phase of cell cycle in cancer cells, Front. Oncol., 2013, 3, 85 Search PubMed.
- P. M. Lansdorp, Telomerase and Cancer, Arch. Med. Res., 2022, 53, 741–746 CrossRef CAS PubMed.
- M. M. Ouellette, W. E. Wright and J. W. Shay, Targeting telomerase-expressing cancer cells, J. Cell Mol. Med., 2011, 15, 1433–1442 CrossRef CAS.
- H. C. Fan, F. W. Chang, J. D. Tsai, K. M. Lin, C. M. Chen, S. Z. Lin, C. A. Liu and H. J. Harn, Telomeres and Cancer, Life, 2021, 11, 1405 CrossRef CAS PubMed.
- X. Tian, B. Chen and X. Liu, Telomere and Telomerase as Targets for Cancer Therapy, Appl. Biochem. Biotechnol., 2010, 160, 1460–1472 CrossRef CAS PubMed.
- W. H. Talib, A. R. Alsayed, M. Barakat, M. I. Abu-Taha and A. I. Mahmod, argeting Drug Chemo-Resistance in Cancer Using Natural Products, Biomedicines, 2021, 9, 1353 CrossRef CAS PubMed.
- L. Xi, J. C. Schmidt, A. J. Zaug, D. R. Ascarrunz and T. R. Cech, A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression, Genome Biol., 2015, 16, 231 CrossRef.
- J. D. Ly, D. R. Grubb and A. Lawen, The mitochondrial membrane potential (Δψm) in apoptosis; an update, Apoptosis, 2003, 8, 115–128 CrossRef CAS PubMed.
- A. Basu and S. Haldar, Mol. Hum. Reprod., 1998, 4, 1099–1109 CrossRef CAS.
- J. J. X. Lee, K. Loh and Y. S. Yap, PI3K/Akt/mTOR inhibitors in breast cancer, Cancer Biol. Med., 2015, 12, 342–354 CAS.
- F. André, E. Ciruelos, G. Rubovszky, M. Campone, S. Loibl, H. S. Rugo, H. Iwata, P. Conte, I. A. Mayer, B. Kaufman, T. Yamashita, Y. S. Lu, K. Inoue, M. Takahashi, Z. Pápai, A. S. Longin, D. Mills, C. Wilke, S. Hirawat and D. Juric, N. Engl. J. Med., 2019, 380, 1929–1940 CrossRef.
- OECD Guidelines, OECD.
- O. Bedi and P. Krishan, Naunyn-Schmiedeberg's Arch. Pharmacol., 2020, 393, 565–571 CrossRef CAS PubMed.
- S. Navaneethakrishnan, J. L. Rosales and K. Y. Lee, in Handbook of Oxidative Stress in Cancer: Mechanistic Aspects, 2022, vol. 1 Search PubMed.
- M. Benhar, D. Engelberg and A. Levitzki, ROS, stress-activated kinases and stress signaling in cancer, EMBO Rep., 2002, 3, 420–425 CrossRef CAS PubMed.
- V. Sosa, T. Moliné, R. Somoza, R. Paciucci, H. Kondoh and M. E. LLeonart, Oxidative stress and cancer: an overview, Ageing Res. Rev., 2013, 12, 376–390 CrossRef CAS PubMed.
- S. Reuter, S. C. Gupta, M. M. Chaturvedi and B. B. Aggarwal, Oxidative stress, inflammation, and cancer: How are they linked?, Free Radical Biol. Med., 2010, 49, 1603–1616 CrossRef CAS PubMed.
- C. I. Diakos, K. A. Charles, D. C. McMillan and S. J. Clarke, Cancer-related inflammation and treatment effectiveness, Lancet Oncol., 2014, 15, e493–e503 CrossRef.
- P. Viatour, M. P. Merville, V. Bours and A. Chariot, Phosphorylation of NF-kB and IkB proteins: implications in cancer and inflammation, Trends Biochem. Sci., 2005, 30, 43–52 CrossRef CAS PubMed.
- K. H. Khan, T. A. Yap, L. Yan and D. Cunningham, Targeting the PI3K-AKT-mTOR signaling network in cancer, Chin. J. Cancer, 2013, 32, 253–265 CrossRef CAS PubMed.
- N. Jiang, Q. Dai, X. Su, J. Fu, X. Feng and J. Peng, Role of PI3K/AKT pathway in cancer: the framework of malignant behavior, Mol. Biol. Rep., 2020, 47, 4587–4629 CrossRef CAS PubMed.
- K. Okkenhaug, M. Graupera and B. Vanhaesebroeck, Targeting PI3K in Cancer: Impact on Tumor Cells, Their Protective Stroma, Angiogenesis, and Immunotherapy, Cancer Discovery, 2016, 6, 1090–1105 CrossRef CAS.
- Y. Lai, X. Chu, L. Di, W. Gao, Y. Guo, X. Liu, C. Lu, J. Mao, H. Shen, H. Tang, C. Q. Xia, L. Zhang and X. Ding, Recent advances in the translation of drug metabolism and pharmacokinetics science for drug discovery and development, Acta Pharm. Sin. B, 2022, 12, 2751–2777 CrossRef CAS PubMed.
- J. Tibbitts, D. Canter, R. Graff, A. Smith and L. A. Khawli, Key factors influencing ADME properties of therapeutic proteins: A need for ADME characterization in drug discovery and development, mAbs, 2016, 8, 229–245 CrossRef CAS PubMed.
- G. T. Tucker, Pharmacokinetic considerations and challenges in oral anticancer drug therapy, Pharm. J., 2019, 11, 6 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |