An integrated microfluidic system with shear force control for an automatically modified mRNA display technique for screening high-specificity peptide probes†
Hao-Yen
Wang
a,
Shih-Yu
Shen
b,
Lily Hui-Ching
Wang
*b and
Gwo-Bin
Lee
 *ac
*ac
aInstitute of NanoEngineering and MicroSystems, National Tsing Hua University, Hsinchu, Taiwan
bInstitute of Molecular and Cellular Biology, National Tsing Hua University, Hsinchu, 30013, Taiwan. E-mail: lilywang@life.nthu.edu.tw; Fax: +886 3 5715934; Tel: +886 3 5715131 Ext. 33493
cDepartment of Power Mechanical Engineering, National Tsing Hua University, Hsinchu, 30013, Taiwan. E-mail: gwobin@pme.nthu.edu.tw; Fax: +886 3 5742495; Tel: +886 3 5715131 Ext. 33765
First published on 12th November 2025
Abstract
We developed a low-cost, integrated microfluidic system (IMS) featuring micropumps, microvalves, micromixers, and thermoelectric modules that fully automated a modified-mRNA display technique for the screening of high-specificity peptide probes in under a day. Peptides targeting KIF2C, a protein overexpressed in various cancers, which has been extensively explored for the development of biomolecular therapeutics, were screened on this new platform. The IMS enabled precise control of shear force (ranging from 0.96 to 9.6 nN) during washing, eliminating off-target peptides under physiologically relevant conditions. As a result, the total screening time was reduced from several days to less than one day. The overall process of the mRNA display could be divided into two stages: (i) mRNA preparation and (ii) mRNA display selection. To the best of our knowledge, it is the first work of integrating mRNA display selection into a single microfluidic chip. Among the screened candidates, one peptide, OCTRD 14, demonstrated both high binding affinity and high target specificity in three independent in vitro assays. In the dissociation constant (Kd) measurements, OCTRD14 exhibited a Kd of 2.4 μM. Although this Kd value was not particularly low, the binding affinity observed in the in vitro assay was comparable to that of the positive control (i.e. antibody). Since the compared signal was detected by using an antibody, this suggests that OCTRD14 may behave similarly to antibody-like candidates. Overall, this high-efficiency peptide screening IMS shows promise for future biomedical applications.
Introduction
When identifying suitable drugs for established and emerging diseases, it is critical that they not only demonstrate high efficacy but also low toxicity.1 Although monoclonal antibody-based drugs are already on the market (e.g. trastuzumab),2 they tend to suffer from issues related to their large sizes, low bio-availability, immunogenic side effects, and production difficulties. Peptides, in contrast, are smaller, demonstrate high specificity as molecular probes, and can be more readily screened and synthesized, making them promising candidates for therapeutic applications.3There are three major technologies for peptide screening: phage display,4 ribosome display,5–8 and mRNA display.9,10 The former requires bacterial culture, while ribosome and mRNA displays are in vitro techniques.4–10 Culture-based methods are relatively time-consuming and may be limited in library size (∼108) due to bacterial transformation efficiency.4 Alternatively, the ribosome display faces issues related to the large sizes of ribosomes (which interferes with peptide-target binding) and instability of peptide–ribosome–mRNA complexes.9,10 In contrast, the mRNA display features a covalent mRNA–peptide linkage via puromycin, offering higher stability, larger library sizes (∼10 (ref. 14)), and ease of accommodating high peptide diversity.9,10 However, the mRNA display is time-consuming, labour-intensive, and costly. To overcome these challenges, “transcription–translation coupled with the association of puromycin linker” (TRAP) display technology was developed11 to rapidly screen peptides. While the TRAP display might accelerate peptide screening, it requires manual intervention and inefficient screening derived from low-stringency steps. Furthermore, traditional selection methods do not precisely control the washing shear force after the binding processes, risking the loss of high-affinity peptides and removal of weakly-bound and/or off-target candidates.
The kinesin family member 2C (KIF2C), also known as mitotic centromere-associated kinesin (MCAK), belongs to the kinesin-13 family. Unlike most kinesins that transport cargo by moving along microtubules, KIF2C acts primarily as a microtubule-depolymerizing motor protein, regulating microtubule dynamics.12,13 It is overexpressed in various cancers, such as breast invasive carcinoma, lung adenocarcinoma, and hepatocellular carcinoma.14 It has been linked to several cancer-associated processes, including evasion of apoptosis, genomic instability, metastasis, and immune escape.15,16 KIF2C is also involved in 1) DNA double-strand break repair17,18 and 2) the TBX15/miR-152/KIF2C signalling pathway for mediating doxorubicin resistance in breast cancer through modulation of PKM2.19 Furthermore, KIF2C serves as a prognostic biomarker and a regulator of the tumor microenvironment in breast and several other cancers.20,21 A recently identified small-molecule inhibitor, 7S9, was shown to restore paclitaxel sensitivity in chemo-resistant triple-negative breast cancer cells and exert synergistic effects with other microtubule-targeting agents,22 highlighting KIF2C as a promising therapeutic target. However, conventional small-molecule inhibitors face limitations, including off-target activity, limited specificity, and poor intracellular delivery. Designing a peptide inhibitor that selectively binds and blocks KIF2C may overcome these challenges, providing higher specificity and efficacy with reduced side effects.
Microfluidics has recently emerged as a promising technique for various biomedical applications,23,24 offering advantages of reduced sample/reagent consumption, precise fluid control, rapid, high-throughput analysis, and improved reproducibility. For instance, our group has reported integrated microfluidic systems (IMSs) with a phage display for cancer-specific peptide selection.25–27 These IMSs enable efficient screening of peptides with high affinity and specificity, incorporating both positive and negative selection on-chip. Using IMSs, peptides targeting colon cancer cells were successfully identified, demonstrating high capture specificity and highlighting the potential of microfluidic-assisted peptide screening for early cancer diagnosis and targeted therapy. Several previous works have reported mRNA display methods whereby biotinylated antigens were immobilized on SA sensor chips (Biacore) with a Biacore 3000 instrument (Biacore) for selection; however, other steps were still performed off-chip.28
To the best of our knowledge, no prior work has integrated the full mRNA display workflow onto a single automated chip. Herein we implemented a modified mRNA (M-mRNA) display merged with the puromycin annealing step from the TRAP display approach (Fig. 1) on-chip to simplify the generation of mRNA–PuL complexes. The chip featured an S-shaped micropump29 to precisely control washing shear force based on theoretical peptide–protein binding forces calculated from a Kd value of ∼10 nM and hydrogen bond lengths of 1.5–3 Å (ref. 3 and 30) (0.25–1.23 nN for one hydrogen bond). Considering that 3–10 hydrogen bonds were expected to contribute to the binding interactions in the benchtop M-mRNA display assay, the binding force ranged from 0.75 to 12.3 nN, and the desired selection shear forces were set within this range. Moreover, the unique screening process consisted of six positive selection rounds with KIF2C-coated beads (with increasing shear force), two negative selection rounds with bare beads, as well as bovine serum albumin (BSA), Von Hippel–Lindau (VHL), and KIF2A-coated beads under low shear force, and one competitive selection round with KIF2C-coated beads incubated in HeLa cell lysate to simulate microenvironments of cancer cells. Table S1 summarizes the comparison between this study and previous reports, demonstrating the significant improvements achieved in the present work. We hypothesized that high-affinity, high-specificity peptides could be screened automatically with this approach.
 | ||
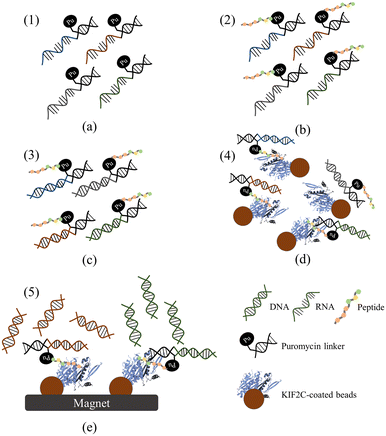
| Fig. 1 Overview of drug development via peptide screening on a fully automated IMS capable of M-mRNA display. (a) The DNA library was first constructed (115 bp). (b) The new system enabled precise control of shear force during waste removal and washing to optimize selection. (c and d) After NGS sequencing and scoring, the predicted candidates were synthesized for subsequent validation. (e) In vitro validation included binding assays, specificity tests, and Kd measurements. (f) Future applications in Trim-TAC (Tripartite motif-containing protein 21 (TRIM21)-based PROTACs) technology,31 which consists of a target recognition domain (TRD), the selected peptide candidates, and a degron signal that triggers selective degradation of the target protein via the proteasome or lysosomal pathways. | ||
Materials and methods
Overview of M-mRNA display
The M-mRNA display process included (1) benchtop mRNA preparation and (2) peptide selection on-chip (Fig. 2). A random DNA library was first constructed (∼2010) with a random region (NNK) that would be translated into 10 amino acids, i.e., the TRD sequence:11 5′-ATACTAATACGACTCACTATAGGATTAAGGAGGTGATATTTATGNNKNNKNNKNNKNNKNNKNNKNNKNNKNNKGGTGGAGGAGGAGGTAGCTAGGACGGGGGGCGGGAGGCGGG-3′. The DNA library (1 μg) was transcribed to mRNA with an EzRNA™ T7 High Yield RNA synthesis kit (IT1100, SMOBIO, Taiwan) at 37 °C for 1 h with 2 μL of 10X T7 buffer, 2 μL of T7 RNA polymerase mix, 8 μL of NTP (Ψ) mix (25 mM each), and diethylpyrocarbonate (DEPC)-treated water to 20 μL. Template DNA was then degraded by RQ1 RNase-Free DNase (1 U μL−1) provided with the kit at 37 °C for 15 min, and mRNA was extracted via precipitation with 7.5 M 10.5 μL LiCl at −20 °C for 30 min, followed by centrifugation at 20600 RPM for 40 min. The pellet was washed with 200 μl of 70% ethanol, air-dried for 20 min, dissolved in 30 μl of DEPC-treated water, and quantified by an EzDrop1000 (Blue-Ray, Taiwan).For peptide selection on-chip, 5 μg of mRNA were mixed with 1.4 μL of 100 μM puromycin linker9 at 25 °C for 30 min (Fig. 2a). Peptide synthesis was carried out using an in vitro translation (IVT) system (E6850S, PURExpress® Δ RF123 Kit, NEB, USA) in a 25 μL reaction volume incubated at 37 °C for 2 h (Fig. 2b). The samples were then cooled down. To prevent degradation and dissociation of the mRNA-PuL-peptide complexes, reverse transcription (RT) was performed using an ExcelR™ RT Kit II (RB1400, SMOBIO, Taiwan) with 20 μL mRNA-PuL-peptide complex, 1.3 μL of 100 μM G5S-4.R20 primer (3′-CCACCTCCTCCTCCATCGAT-5′, Integrated DNA Technologies [IDT], USA), 12 μL of 5X RT buffer, 3 μL of RTase/RI enzyme mix, and 18.7 μL of DEPC-treated water (Fig. 2c).
Positive selection (six rounds; Table 1) was performed with KIF2C-coated beads (Dynabeads™ His-Tag Isolation and Pulldown, Thermo Scientific™, USA). First, 2 μg of KIF2C were mixed with 1 μg of magnetic beads at room temperature for 30 min at 35 RPM (mode: UU; Elmi Intelli-Mixer RM-2; Latvia). Two negative selection rounds were carried out by mixing bare beads and beads coated with kinesin family member 2A (KIF2A), VHL, and BSA (proteins were bound to beads as described above). KIF2A was chosen because of its structural similarity to KIF2C, BSA was used as a blocking protein, and VHL was chosen to function as a target for the degron signal region in subsequent Trim-TAC applications.
| NS | CS | PS | Washing | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Round | T | D | T | D | T | D | POI | T | D | I | V | Temp. |
| 1st | — | — | — | — | 1 | 60 min | 2 μg | 3 | 1 min | 0.96 nN | 100 μL | 25 °C |
| 2nd | — | — | — | — | 1 | 60 min | 2 μg | 3 | 1 min | 0.96 nN | 100 μL | 25 °C |
| 3rd | — | — | — | — | 1 | 60 min | 2 μg | 3 | 1 min | 4.8 nN | 100 μL | 25 °C |
| 4th | — | — | — | — | 1 | 60 min | 2 μg | 3 | 1 min | 4.8 nN | 100 μL | 25 °C |
| 5th | — | — | — | — | 1 | 60 min | 2 μg | 3 | 1 min | 9.6 nN | 100 μL | 25 °C |
| 6th | — | — | — | — | 1 | 60 min | 2 μg | 3 | 1 min | 9.6 nN | 100 μL | 25 °C |
| 7th | 1 | 30 min | — | — | 1 | 30 min | 2 μg | 3 | 1 min | 0.96 nN | 100 μL | 25 °C |
| 8th | 1 | 30 min | — | — | 1 | 30 min | 2 μg | 3 | 1 min | 0.96 nN | 100 μL | 25 °C |
| 9th | — | — | 1 | 60 min | — | — | — | 3 | 1 min | 0.96 nN | 100 μL | 25 °C |
Competitive selection was next conducted using KIF2C-coated beads in a solution containing 3 μg μL−1 of HeLa cell lysate, 100 μL of RNase inhibitor (1 U μL−1) (IT1100, SMOBIO, Taiwan), 1 μL of EDTA (50 mM) (ACID-EDT01, Jhih shuo chemical, Taiwan), 60 μL of protease inhibitor (0.1 g mL−1) (A32963, Thermo Scientific™, USA), and 60 μL of phosphatase inhibitor (0.1 g mL−1) (A32957, Thermo Scientific™, USA). We used cell lysate in this to mimic the in vivo microenvironment, in which DNAs/proteins would fold properly. This also ensured that the target protein could bind the probes even amidst a plethora of off-target proteins. Wash steps were conducted with a binding/washing buffer (BWB; 100 mM Na2HPO4, 600 mM NaCl, 0.02% Tween-20, & DEPC treated H2O) under specifically controlled shear forces modulated by an S-shaped micropump (Table 1). The shear force during washing was increased with each round of positive selection, starting at 0.96 nN and ending at 9.6 nN (Fig. 2d). By increasing the stringency in this way, weakly-bound and/or off-target candidates were removed mechanically. In rounds 7 and 8, a negative selection step was introduced to eliminate peptides that could also bind to BSA, KIF2A, and VHL, thereby increasing specificity. Low shear forces were used in rounds 7–8 to prevent loss of previously enriched KIF2C-specific candidates. In round 9, competitive selection was conducted under low shear force to minimize the risk of washing out promising candidates. After selection, a second RT step was conducted to confirm the presence of cDNA. PCR was performed with forward (T7SD8M2.F44, 5′-ATACTAATACGACTCACTATAGGATTAAGGAGGTGATATTTATG-3′; IDT) and reverse primers (G5S-4an21, 5′-CCACCTCCTCCTCCATCGATCCTGCCCCCCGCCCTCCGCCC-3′; IDT.nR41 primers, stock = 100 μM for each), using the SMO-HiFi™ DNA polymerase system (TF1000, SMOBIO) under the following conditions: 95 °C for 5 min and 30 cycles of 94 °C for 20 s, 60 °C for 20 s, and 68 °C for 15 s (Fig. 2e). The PCR products were analysed by electrophoresis on a 2% agarose gel, and DNA was purified using a Zymoclean™ gel DNA recovery kit (D4008, Zymo Research, USA) for the subsequent round of M-mRNA display.
Chip design
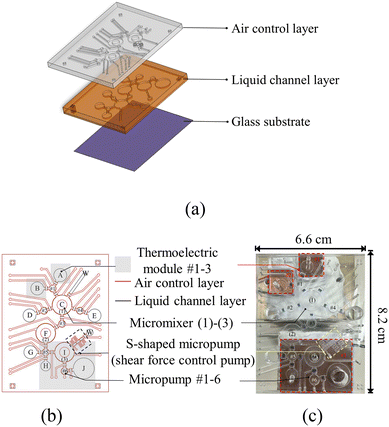
The 6.6 × 8.2 cm chip consisted of three layers (Fig. 3). The first and second layers were made from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polydimethylsiloxane (PDMS, Sylgard 184A/184B, Dow Corning, US) as in our prior works29 and as shown in Fig. S2. The thickness of the air control and liquid channel layers was 1.5 and 0.5 mm, respectively; the thickness of the PDMS membrane between these two layers for micropumps and micromixers was 250 μm. The third layer was a hydrophilic glass substrate (thickness = 700 μm) that facilitated liquid flow (Fig. 3a). The chip was equipped with 17 electromagnetic microvalves (EMVs; 1.0 × 1.2 × 1.5 mm), three donut-shaped micromixers (radius = 6 mm & thickness = 1.5 mm), six circular micropumps (radius = 2.2 mm & thickness = 1.5 mm), and one S-shaped micropump29 that could control the shear force (Fig. 3b; length of channels = 1 mm & cross-sectional area = 0.5 × 1.4 mm2). The S-shaped micropump generated a peristaltic-like flow that could establish stable, uniform fluid movement through sequential compressions, enabling precise flow control. For the bonding of the air control and liquid channel layers onto the glass, surfaces were treated with O2 plasma (CUTE MP/R, Femto Science, Korea) at 30 W for 90 s.
1 polydimethylsiloxane (PDMS, Sylgard 184A/184B, Dow Corning, US) as in our prior works29 and as shown in Fig. S2. The thickness of the air control and liquid channel layers was 1.5 and 0.5 mm, respectively; the thickness of the PDMS membrane between these two layers for micropumps and micromixers was 250 μm. The third layer was a hydrophilic glass substrate (thickness = 700 μm) that facilitated liquid flow (Fig. 3a). The chip was equipped with 17 electromagnetic microvalves (EMVs; 1.0 × 1.2 × 1.5 mm), three donut-shaped micromixers (radius = 6 mm & thickness = 1.5 mm), six circular micropumps (radius = 2.2 mm & thickness = 1.5 mm), and one S-shaped micropump29 that could control the shear force (Fig. 3b; length of channels = 1 mm & cross-sectional area = 0.5 × 1.4 mm2). The S-shaped micropump generated a peristaltic-like flow that could establish stable, uniform fluid movement through sequential compressions, enabling precise flow control. For the bonding of the air control and liquid channel layers onto the glass, surfaces were treated with O2 plasma (CUTE MP/R, Femto Science, Korea) at 30 W for 90 s.
After reagent loading, four pieces of 20 μm single-sided tape (68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546, Tesa®, USA) were used to encapsulate four open chambers (two for RT, one for IVT, & one for PCR; Fig. 3b) where three thermoelectric modules were placed underneath to prevent reagent evaporation. The temperature control modules included three thermoelectric modules (one each for IVT, RT, & PCS), paired with the corresponding MAX6675 temperature sensors (Centenary Materials, Taiwan), TK-39 K-type thermocouples (He-Ying Electronic Materials, Taiwan), and a feedback control system using a 5-V relay (Centenary Materials; Fig. 3c). One mixer each was used for the positive, negative, and competitive selection steps. Chambers A–J were designed for processes ranging from IVT with PuL annealing to PCR (Fig. S1a). To test the feasibility of on-chip M-mRNA, version-1 of the M-mRNA display chip was first designed and tested, as schematically shown in Fig. S1 (see Fig. S1b for a top-down view of the device). As discussed below, this “version 1” design was later modified (“version 2”) such that the entire 9 selection rounds could be performed on a single chip.
546, Tesa®, USA) were used to encapsulate four open chambers (two for RT, one for IVT, & one for PCR; Fig. 3b) where three thermoelectric modules were placed underneath to prevent reagent evaporation. The temperature control modules included three thermoelectric modules (one each for IVT, RT, & PCS), paired with the corresponding MAX6675 temperature sensors (Centenary Materials, Taiwan), TK-39 K-type thermocouples (He-Ying Electronic Materials, Taiwan), and a feedback control system using a 5-V relay (Centenary Materials; Fig. 3c). One mixer each was used for the positive, negative, and competitive selection steps. Chambers A–J were designed for processes ranging from IVT with PuL annealing to PCR (Fig. S1a). To test the feasibility of on-chip M-mRNA, version-1 of the M-mRNA display chip was first designed and tested, as schematically shown in Fig. S1 (see Fig. S1b for a top-down view of the device). As discussed below, this “version 1” design was later modified (“version 2”) such that the entire 9 selection rounds could be performed on a single chip.
On-chip procedures and system integration
The selection process on the version-2 chip (Table 1) began with six rounds of positive selection; incubations were performed in chamber I for 60 min at room temperature, and the supernatant was removed using the S-shaped micropump. The following two rounds were negative selection in chamber C for 30 min at room temperature, after which the supernatant was transferred to chamber I for additional positive selection for 30 min at room temperature; this was followed by removal of the supernatant using the S-shaped micropump. The final round, competitive selection, was conducted in chamber F for 60 min at room temperature, with the supernatant removal performed once again using the S-shaped micropump (Table 1).
Sequencing and scoring
After selection, DNAs from the 7th, 9th, and 10th rounds of version-1 screening, and from the 4th, 6th, 8th, and 9th rounds of version-2 screening, were sequenced by next-generation sequence techniques (NGS, Illumina NovaSeq X Plus, Illumina, USA) at Genomics (Taiwan). After sequencing, the peptide sequences were simulated using CABS-dock32,33 and PyMOL (the PyMOL Molecular Graphics System, Version 3.0 Schrödinger, LLC), and the binding affinities were predicted using Area-Affinity34–36 and the Prodigy Webserver.37–39In version-1 screening, a large number of candidates were generated; therefore, a series of criteria were implemented to rank them.
1. Identify those sequences that emerged from both the on-bench and on-chip M-mRNA display methods.
2. Remove the sequences that were also present in the NGS results of M-mRNA display using VHL as the target protein.
3. Use CABS-dock and PyMOL to remove those candidates that were predicted to bind the C-terminal of KIF2C because selected peptides were designed to be fused with a flexible tail (N′-GGGGGS-C′) and a degron signal for later studies.
4. Use Area-Affinity and Prodigy to calculate the Kd of the remaining candidates and rank them accordingly.
5. When two candidates had a similar Kd, the one with more hydrogen bonds was prioritized.
6. When both Kd and number of hydrogen bonds were similar, the peptide with the higher hydrophobicity was deemed superior (Fig. S3).
There was more variability in the binding properties of the candidates selected by the version 2 chip and so the following criteria were further used (Fig. S3).
1. The most common sequence was prioritized.
2. Candidates predicted by CABS-dock to bind the C-terminal of KIF2C were removed.
3. Peptide candidates that were also sequenced from the negative selection round were removed.
Validation of candidates
To validate the selected peptide candidates, a Strep-tag (N′-candidate sequence-SAWSHPQFEK-C′) was appended to the C-terminus of each selected peptide sequence for detection. An in vitro binding assay, Kd measurements, and a specificity assay featuring “far-western” blotting and enzyme-linked immunosorbent assay (ELISA) were used to verify the most promising candidates. For the former, 100 nM of KIF2C protein in 50 μL of 1× PBS were immobilized on a high-binding 96-well microplate (655![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 075, Greiner Bio-One, Austria) overnight at 4 °C. The wells were then blocked with 50 μL of 1% BSA in 1× PBS overnight at 4 °C, followed by three washes with PBS with 0.05% Tween 20 (PBST). During Kd measurement, a peptide concentration of 15 μM was often sufficient to achieve high or full binding. Based on this observation, we selected 15 μM for our comparative assay to evaluate the peptide's binding capacity relative to the antibody. Subsequently, 15 μM of each peptide candidate in 50 μL of 1× PBS were added to the wells and incubated overnight at 4 °C. After incubation, the wells were washed thrice with PBST. For detection, a 1
075, Greiner Bio-One, Austria) overnight at 4 °C. The wells were then blocked with 50 μL of 1% BSA in 1× PBS overnight at 4 °C, followed by three washes with PBS with 0.05% Tween 20 (PBST). During Kd measurement, a peptide concentration of 15 μM was often sufficient to achieve high or full binding. Based on this observation, we selected 15 μM for our comparative assay to evaluate the peptide's binding capacity relative to the antibody. Subsequently, 15 μM of each peptide candidate in 50 μL of 1× PBS were added to the wells and incubated overnight at 4 °C. After incubation, the wells were washed thrice with PBST. For detection, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution of streptavidin–Alexa Fluor 488 (Invitrogen) in PBST was added and incubated for h at room temperature, followed by three washes with PBST. As the positive control, wells were treated with a 1
2000 dilution of streptavidin–Alexa Fluor 488 (Invitrogen) in PBST was added and incubated for h at room temperature, followed by three washes with PBST. As the positive control, wells were treated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of 1 mg mL−1 anti-MCAK primary antibody (ab70536, Abcam, Singapore). After washing three times with PBST (1× PBS, 0.05% Tween-20), a 1
1000 dilution of 1 mg mL−1 anti-MCAK primary antibody (ab70536, Abcam, Singapore). After washing three times with PBST (1× PBS, 0.05% Tween-20), a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution 2 mg mL−1 Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488, A-11008, Invitrogen, USA) was used to confirm KIF2C coating. Finally, 50 μL of 1× PBS were added to each well, and the fluorescence signal was measured using a microplate reader (BMG LabTech, Germany) with excitation and emission at 488 and 520 nm, respectively.
2000 dilution 2 mg mL−1 Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488, A-11008, Invitrogen, USA) was used to confirm KIF2C coating. Finally, 50 μL of 1× PBS were added to each well, and the fluorescence signal was measured using a microplate reader (BMG LabTech, Germany) with excitation and emission at 488 and 520 nm, respectively.
K d was determined in a similar fashion, though with peptide concentrations ranging from 380 pM to 100 μM in 4-fold serial dilutions. KIF2C was immobilized onto Pierce™ nickel-coated plates (Thermo Scientific™, USA). The fluorescence intensity was curve-fitted using GraphPad Prism software (USA), and the Kd values were calculated using the following equation:
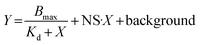 | (1) |
To assess specificity, far-western blotting was performed. KIF2C and VHL proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (0.45 μm, HVLP04700, Millipore, USA) alongside a protein ladder (PM2510, SMOBIO, 0.6 mg ml−1, 5 μL). Membranes were blocked with 3% BSA in PBS for 1.5 h at room temperature, incubated overnight at 4 °C with 1 mg mL−1 of each peptide in 1× PBS, washed thrice with PBST for 5 min, incubated overnight in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of mouse anti-strep II-tag monoclonal antibody (mAb, ABclonal, Taiwan), washed thrice with PBST, incubated in a 1
1000 dilution of mouse anti-strep II-tag monoclonal antibody (mAb, ABclonal, Taiwan), washed thrice with PBST, incubated in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution of goat anti-mouse IgG antibody conjugated with horseradish peroxidase (HRP; GeneTex) for 1 h, washed thrice with PBST as above, and luminescence signals were quantified using an ImageQuant™ LAS 4000 (GE Healthcare, USA).
5000 dilution of goat anti-mouse IgG antibody conjugated with horseradish peroxidase (HRP; GeneTex) for 1 h, washed thrice with PBST as above, and luminescence signals were quantified using an ImageQuant™ LAS 4000 (GE Healthcare, USA).
Alternatively, ELISA was performed whereby 100 nM of BSA, VHL, and KIF2C were added to a high-binding 96-well microplate, incubated for 24 h, and blocked with 1% BSA in PBS for 24 h. Each peptide was added (10 μM) and incubated with the coated proteins for 24 h, followed by three PBST washes. To detect strep-tag peptides, the anti-strep tag Alexa Fluor 488 antibody was incubated with the peptide–protein complexes for 1 h, followed by three PBST washes. Finally, 50 μL of 1× PBS were added to each well, and fluorescence signals were measured on the microplate reader.
Results and discussion
Characterization of the device
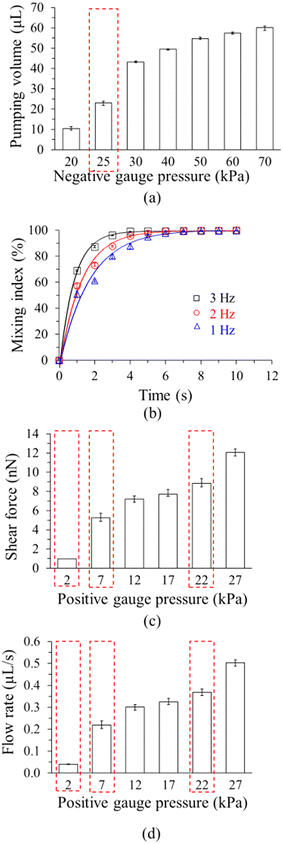
After chip microfabrication, its operating conditions were optimized (Fig. 4), starting with the pumping rates of the circular micropumps (Fig. 4a). The pumping rate increased with applied pressures and driving frequency (1, 2, & 3 Hz, respectively), and 24.0 ± 1.1 μL could be transported in 5 s at +30- and −25 kPa gauge pressures; since this was close to the desired volume of 20 μL, these conditions were used subsequently.The donut-shaped, pneumatically-driven micromixers were actuated by alternating positive and negative gauge pressures controlled by the EMVs. To evaluate mixing efficiency, a mixing index was quantified by ImageJ (https://github.com/imagej/ImageJ) under driving frequencies of 1, 2, and 3 Hz at constant positive and negative gauge pressures of +20 and −20 kPa, respectively (Fig. 4b).
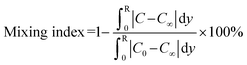 | (2) |
A driving frequency of 2 Hz was selected since it was relatively gentle, yet stringent enough to minimize binding of off-target candidates.
The pumping rates and shear forces of the S-shaped micropump were also measured (Fig. 4c and d), and the pumping rate increased with applied pressure and driving frequency. The shear force was calculated by the following formula.
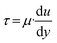 | (3) |
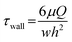 | (4) |
| Fshear = τ·A | (5) |
Characterization of the temperature control module
Temperature control for IVT, RT, and PCR was achieved using an Arduino-based thermoelectric control module, and temperatures of 37 and 42 °C for the former two, respectively, showed only ±0.5 and ±0.4 °C variation over 120 and 50 min, respectively (Fig. S3a). PCR temperatures were also satisfactory (Fig. S3b).IVT and RT-PCR on the microfluidic chip
After realizing that protein samples were evaporating, 50 μL of mineral oil (330760-1 L; Sigma-Aldrich) were added; note that a single-sided tape was also used to prevent evaporation, and peptides were successfully synthesized thereafter (Fig. S4a). DNA content was maintained after IVT (Fig. S4b).To assess the chip's capacity to conduct RT-PCR, the amplicons were electrophoresed (Fig. S5), and a band was evident, indicating that on-chip RT-PCR was successfully performed.
Frequency of the top 10 candidates
The frequencies of the top 10 candidates were monitored to assess the degree of enrichment (Fig. 5). An increase in the frequency of a specific sequence during the selection process, as observed through NGS, typically indicates that the sequence was preferentially enriched due to its advantageous binding characteristics. However, in some cases, the increased frequency could be influenced by secondary factors such as lower nonspecific binding, improved solubility, or even PCR amplification efficiency.40 Based on this analysis and additional selection criteria (Fig. S6), OCTRD5, OCTRD11, OCTRD14, OCTRD15, and OCTRD17 were selected for peptide synthesis. We assessed their performance by analysing the Shannon entropy, Pielou's evenness, and sequencing frequency (Fig. 5 and S7). In the version-1 chip, the Shannon entropy increased over successive rounds, suggesting a lack of enrichment for specific sequences. Pielou's evenness remained high, indicating a uniform distribution of candidate frequencies without dominant binders.The top 10 candidates in the final round had frequencies ranging from 0.04 to 0.4%. This weak signal may have been influenced by the shallow sequencing depth, potentially missing suitable candidates and amplifying low-affinity ones. The Shannon entropy decreased from 18.9 in round four to 15.6 in the final round, while Pielou's evenness slightly decreased from 0.97 to 0.91. This trend indicates that although the sequence diversity remained relatively stable, certain candidates began to dominate the pool, pointing to the effectiveness of gradually increasing the shear force. The frequencies of the top 10 candidates increased from 0.04–0.8% in round four to 0.3–1.7% in the final round. Importantly, OCTRD14 did not appear in round four or six but emerged post-negative selection, suggesting that this step effectively removed nonspecific binders and enabled the enrichment of more specific ones. OCTRD14 reached a frequency of 0.3% from round eight onwards.
K d measurements
All candidates exhibited Kd values in the micromolar to sub-micromolar range (Fig. S8). Despite being relatively high when compared to antibodies, each peptide demonstrated consistent binding and specificity to KIF2C. Importantly, TRD13, OCTRD 11, and OCTRD14 exhibited particularly high binding affinities and specificities. The native structure of KIF2C may have been compromised when immobilized in the ELISA plate; the Kd values would likely be superior when using methods like surface plasmon resonance41 or microscale thermophoresis.42Specificity tests by far-western blotting and ELISA
To confirm the KIF2C selectivity of the peptide candidates, two validation steps were performed (Fig. 6). Although peptides did not bind to the membrane space regions blocked by BSA and VHL (28 kDa), binding was observed with KIF2A, despite the use of KIF2A-coated beads during the negative selection steps. This residual binding was likely due to the high structural similarity between the motor domains of KIF2C and KIF2A, which can result in cross-reactivity.22 Most peptide candidates demonstrated preferential binding to KIF2C over KIF2A,Next, ELISA was performed to compare the specificity of each peptide candidate (Fig. 6k), and OCTRD14 demonstrated the highest binding signal and specificity. In the future work, we plan to perform another round of selection using the full-length KIF2C as the target, followed by a comparative analysis of the enriched peptide candidates. With this approach, the specificity of these peptides against both KIF2A and KIF2C may be enhanced accordingly. This improvement may reduce cross-reactivity and further validate the effectiveness of our shear-force–controlled selection strategy.
In vitro binding assay
Only OCTRD14 exhibited a fluorescence signal comparable to the positive control (Fig. 7), suggesting that it demonstrates antibody-like binding affinity towards KIF2C. It also demonstrated the strongest binding affinity at 15 μM and highest specificity in both the ELISA and the far-western blot, yet its Kd was relatively high: ∼2.4 μM. However, a high Kd does not necessarily indicate weak biological activity, as it might result from a combination of slow association/dissociation kinetics rather than weak binding. For example, BRD-5110, a hetero-bifunctional degrader, was reported to exhibit a high Kd (∼3 μM) when binding to truncated CRBN, but it still successfully induced ternary complex formation and degradation.43 Significantly, high specificity and favourable binding behaviour could be more meaningful indicators of probe potential than Kd alone. Therefore, OCTRD14 may still be a promising probe.Potential cell-based validation
We evaluated the degradation efficiency of TRDs selected from our screening by introducing TRD-linker-TDS combinations into HEK 293 T, HeLa, and MDA-MB-231 cell lines. We observed that degradation efficiency varied among different cell types and recruitment systems (VHL, CMA, NPGY, and IκBα). Notably, some TRD candidates (e.g., TRD4-5G1S-CMA and TRD15-5G1S-NPGY) exhibited partial degradation of endogenous KIF2C. Although the degradation effect was not consistent and significant, these findings indicated the potential of extending our microfluidic mRNA display platform toward cell-based functional validation.Conclusions
This study has developed an integrated microfluidic system to automate an M-mRNA display to accelerate and simplify the conventional mRNA display. The advantages of the on-chip M-mRNA display were not just to speed up the selection process (<1 day) nor to integrate the whole process into a single microfluidic chip. The most important benefit was to precisely control the shear force precisely in the range from 0.96 to 9.6 nN as the protein–protein interaction in the physiological environment in each washing step, which was crucial for selecting the high-affinity candidates. Among the candidates selected from the on-bench M-mRNA display and on-chip M-mRNA display (on version-1 and version-2 chips, respectively), OCTRD14 exhibited the best potential as an anti-KIF2C antibody, ranking third in Kd measurement (∼2.4 μM) and demonstrating the highest specificity, as validated by far-western blotting and ELISA.Author contributions
Hao-Yen Wang: conceptualization, methodology, investigation, data curation, microfluidic system design, formal analysis, validation and writing – original draft; Shih-Yu Shen: conceptualization, methodology, investigation, data curation, formal analysis and validation; Lily Hui-Ching Wang: methodology, validation, provided resources and writing – review & editing; Gwo-Bin Lee: conceptualization, methodology, provided resources, validation, funding acquisition, supervision and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
Data will be made available on request.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5lc00914f.
Acknowledgements
The authors would like to thank the National Science and Technology Council (NSTC) of Taiwan (NSTC 113-2218-E-007-027-MY3; NSTC 113-2221-E-007-144-MY3; NSTC 114-2221-E-007-009-MY3; 114-2311-B-007-006).References
- M. D. Segall and C. Barber, Addressing toxicity risk when designing and selecting compounds in early drug discovery, Drug Discovery Today, 2014, 19(5), 688–693 CrossRef CAS PubMed.
- M. Harries and I. Smith, The development and clinical use of trastuzumab (Herceptin), Endocr.-Relat. Cancer, 2002, 9(2), 75–85 CAS.
- M. Crawford, R. Woodman and P. K. Ferrigno, Peptide aptamers: tools for biology and drug discovery, Briefings Funct. Genomics Proteomics, 2023, 2(1), 72–79 CrossRef.
- J. Bazan, I. Całkosiński and A. Gamian, Phage display--a powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications, Hum. Vaccines Immunother., 2012, 8(12), 1817–1828 CrossRef CAS.
- C. Zahnd, P. Amstutz and A. Plückthun, Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target, Nat. Methods, 2007, 4(3), 269–279 CrossRef CAS PubMed.
- T. Lamla and V. A. Erdmann, Searching sequence space for high-affinity binding peptides using ribosome display, J. Mol. Biol., 2003, 329(2), 381–388 CrossRef CAS.
- L. C. Mattheakis, R. R. Bhatt and W. J. Dower, An in vitro polysome display system for identifying ligands from very large peptide libraries, Proc. Natl. Acad. Sci. U. S. A., 1994, 91(19), 9022–9026 CrossRef CAS.
- G. M. Gersuk, M. J. Corey, E. Corey, J. E. Stray, G. H. Kawasaki and R. L. Vessella, High-affinity peptide ligands to prostate-specific antigen identified by polysome Selection, Biochem. Biophys. Res. Commun., 1997, 232(2), 578–582 CrossRef CAS PubMed.
- N. Nemoto, E. Miyamoto-Sato, Y. Husimi and H. Yanagawa, In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro, FEBS Lett., 1997, 414(2), 405–408 CrossRef CAS PubMed.
- R. W. Roberts and J. W. Szostak, RNA-peptide fusions for the in vitro selection of peptides and proteins, Proc. Natl. Acad. Sci. U. S. A., 1997, 94(23), 12297–12302 CrossRef CAS PubMed.
- T. Ishizawa, T. Kawakami, P. C. Reid and H. Murakami, TRAP display: a high-speed selection method for the generation of functional polypeptides, J. Am. Chem. Soc., 2013, 135(14), 5433–5440 CrossRef CAS PubMed.
- S. Montenegro Gouveia, K. Leslie, L. C. Kapitein, R. M. Buey, I. Grigoriev, M. Wagenbach, I. Smal, E. Meijering, C. C. Hoogenraad, L. Wordeman, M. O. Steinmetz and A. Akhmanova, In Vitro Reconstitution of the Functional Interplay between MCAK and EB3 at Microtubule Plus Ends, Curr. Biol., 2010, 20(19), 1717–1722 CrossRef CAS PubMed.
- S. Honnappa, S. M. Gouveia, A. Weisbrich, F. F. Damberger, N. S. Bhavesh, H. Jawhari, I. Grigoriev, F. J. A. van Rijssel, R. M. Buey, A. Lawera, I. Jelesarov, F. K. Winkler, K. Wüthrich, A. Akhmanova and M. O. Steinmetz, An EB1-Binding Motif Acts as a Microtubule Tip Localization Signal, Cell, 2009, 138(2), 366–376 CrossRef CAS PubMed.
- X. Zhang, Y. Li, P. Hu, L. Xu and H. Qiu, KIF2C is a Biomarker Correlated with Prognosis and Immunosuppressive Microenvironment in Human Tumors, Front. Genet., 2022, 13, 891408 CrossRef CAS.
- D. Hanahan and R. A. Weinberg, Hallmarks of Cancer: The Next Generation, Cell, 2011, 144(5), 646–674 CrossRef CAS.
- N. N. Kreis, H. H. Moon, L. Wordeman, F. Louwen, C. Solbach, J. Yuan and A. Ritter, KIF2C/MCAK a prognostic biomarker and its oncogenic potential in malignant progression, and prognosis of cancer patients: a systematic review and meta-analysis as biomarker, Crit. Rev. Clin. Lab. Sci., 2024, 61(6), 404–434 CrossRef CAS.
- S. Zhu, M. Paydar, F. Wang, Y. Li, L. Wang, B. Barrette, T. Bessho, B. H. Kwok and A. Peng, Kinesin Kif2C in regulation of DNA double strand break dynamics and repair, eLife, 2020, 9, e53402 CrossRef PubMed.
- M. Shokrollahi, M. Stanic, A. Hundal, J. N. Y. Chan, D. Urman, C. A. Jordan, A. Hakem, R. Espin, J. Hao, R. Krishnan, P. G. Maass, B. C. Dickson, M. P. Hande, M. A. Pujana, R. Hakem and K. Mekhail, DNA double-strand break–capturing nuclear envelope tubules drive DNA repair, Nat. Struct. Mol. Biol., 2024, 31(9), 1319–1330 CrossRef CAS.
- C. F. Jiang, Y. X. Xie, Y.-C. Qian, M. Wang, L.-Z. Liu, Y.-Q. Shu, X.-M. Bai and B.-H. Jiang, TBX15/miR-152/KIF2C pathway regulates breast cancer doxorubicin resistance via promoting PKM2 ubiquitination, Cancer Cell Int., 2021, 21(1), 542 CrossRef CAS.
- S. Liu, Z. Ye, V. W. Xue, Q. Sun, H. Li and D. Lu, KIF2C is a prognostic biomarker associated with immune cell infiltration in breast cancer, BMC Cancer, 2023, 23(1), 307 CrossRef CAS PubMed.
- B. Zhang, P. Liu, Y. Li, Q. Hu, H. Li, X. Pang and H. Wu, Multi-omics analysis of kinesin family member 2C in human tumors: novel prognostic biomarker and tumor microenvironment regulator, Am. J. Cancer Res., 2022, 12(11), 4954–4976 CAS.
- Y.-S. Pao, K.-J. Liao, Y.-C. Shiau, M.-H. Chao, M.-C. Li, L.-M. Lin, H.-H. Chang, H.-W. Yeh, Y.-J. Chen, Y.-T. Chiu, M. Y.-C. Pan, Y.-H. Chang, S.-Y. Shen, S.-Y. Lin, H.-C. Cheng, Y.-C. Lin, Y.-J. Sun, C.-C. Kuo, H.-P. Hsieh and L. H.-C. Wang, KIF2C promotes paclitaxel resistance by depolymerizing polyglutamylated microtubules, Dev. Cell, 2025, 60(15), 2097–2113 CrossRef CAS PubMed.
- C. Y. Lee, G. B. Lee, J. L. Lin, F. C. Huang and C. S. Liao, Integrated microfluidic systems for cell lysis, mixing/pumping and DNA amplification, J. Micromech. Microeng., 2005, 15(6), 1215 CrossRef CAS.
- M. A. Eddings and B. K. Gale, A PDMS-based gas permeation pump for on-chip fluid handling in microfluidic devices, J. Micromech. Microeng., 2006, 16(11), 2396 CrossRef.
- C.-W. Yu, C.-Y. Fu, L.-Y. Hung, C.-H. Wang, N.-J. Chiang, Y.-C. Wang, Y.-S. Shan and G.-B. Lee, Screening of peptide specific to cholangiocarcinoma cancer cells using an integrated microfluidic system and phage display technology, Microfluid. Nanofluid., 2017, 21(9), 145 CrossRef.
- Y. J. Che, H. W. Wu, L. Y. Hung, C. A. Liu, H. Y. Chang, K. Wang and G. B. Lee, An integrated microfluidic system for screening of phage-displayed peptides specific to colon cancer cells and colon cancer stem cells, Biomicrofluidics, 2015, 9(5), 054121 CrossRef.
- C. H. Wang, C. H. Weng, Y. J. Che, K. Wang and G. B. Lee, Cancer cell-specific oligopeptides selected by an integrated microfluidic system from a phage display library for ovarian cancer diagnosis, Theranostics, 2015, 5(4), 431–442 CrossRef CAS PubMed.
- N. Tabata, Y. Sakuma, Y. Honda, N. Doi, H. Takashima, E. Miyamoto-Sato and H. Yanagawa, Rapid antibody selection by mRNA display on a microfluidic chip, Nucleic Acids Res., 2009, 37(8), e64–e64 CrossRef PubMed.
- C.-H. Wang and G.-B. Lee, Pneumatically driven peristaltic micropumps utilizing serpentine-shaped channels, J. Micromech. Microeng., 2006, 16(2), 341 CrossRef CAS.
- J. S. Jiménez and M. J. Benítez, Gibbs free energy and enthalpy–entropy compensation in protein–ligand interactions, Biophysica, 2024, 4(2), 298–309 CrossRef.
- P. Lu, Y. Cheng, L. Xue, X. Ren, X. Xu, C. Chen, L. Cao, J. Li, Q. Wu, S. Sun, J. Hou, W. Jia, W. Wang, Y. Ma, Z. Jiang, C. Li, X. Qi, N. Huang and T. Han, Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders, Cell, 2024, 187(25), 7126–7142 CrossRef CAS PubMed.
- M. Blaszczyk, M. Kurcinski, M. Kouza, L. Wieteska, A. Debinski, A. Kolinski and S. Kmiecik, Modeling of protein–peptide interactions using the CABS-dock web server for binding site search and flexible docking, Methods, 2016, 93, 72–83 CrossRef CAS PubMed.
- M. Kurcinski, M. Jamroz, M. Blaszczyk, A. Kolinski and S. Kmiecik, CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site, Nucleic Acids Res., 2015, 43(W1), W419–W424 CrossRef CAS PubMed.
- Y. X. Yang, P. Wang and B. T. Zhu, Importance of interface and surface areas in protein-protein binding affinity prediction: A machine learning analysis based on linear regression and artificial neural network, Biophys. Chem., 2022, 283, 106762 CrossRef CAS PubMed.
- Y. X. Yang, P. Wang and B. T. Zhu, Binding affinity prediction for antibody–protein antigen complexes: a machine learning analysis based on interface and surface areas, J. Mol. Graphics Modell., 2023, 118, 108364 CrossRef CAS PubMed.
- Y. X. Yang, J. Y. Huang, P. Wang and B. T. Zhu, AREA-AFFINITY: An academic free web server for machine learning-based prediction of protein–protein and antibody–protein antigen binding affinities, J. Chem. Inf. Model., 2023, 63, 3230–3237 CrossRef CAS PubMed.
- A. Vangone and A. M. J. J. Bonvin, Contacts-based prediction of binding affinity in protein–protein complexes, eLife, 2015, 4, e07454 CrossRef PubMed.
- L. C. Xue, J. P. Rodrigues, P. L. Kastritis, A. M. Bonvin and A. Vangone, PRODIGY: a web server for predicting the binding affinity of protein–protein complexes, Bioinformatics, 2016, 32(23), 3676–3678 CrossRef CAS PubMed.
- F. Jalali-Yazdi, L. H. Lai, T. T. Takahashi and R. W. Roberts, High-throughput measurement of binding kinetics by mRNA display and next-generation sequencing, Angew. Chem., Int. Ed., 2016, 55(12), 4007–4010 CrossRef CAS.
- L. Heinrich, N. Tissot, D. J. Hartmann and R. Cohen, Comparison of the results obtained by ELISA and surface plasmon resonance for the determination of antibody affinity, J. Immunol. Methods, 2010, 352(1), 13–22 CrossRef CAS.
- L. Khavrutskii, J. Yeh, O. Timofeeva, S. G. Tarasov, S. Pritt, K. Stefanisko and N. Tarasova, Protein purification-free method of binding affinity determination by microscale thermophoresis, J. Visualized Exp., 2013, 78, 50541 Search PubMed.
- A. J. Ameri and Z. A. Lewis, Shannon entropy as a metric for conditional gene expression in Neurospora crassa, G3:Genes, Genomes, Genet., 2021, 11(4), jkab055 CrossRef CAS.
- W. Jiang and H. Soutter, The development and application of biophysical assays for evaluating ternary complex formation induced by proteolysis targeting chimeras (PROTACS), J. Visualized Exp., 2024, 203, 9 Search PubMed.
Footnote |
| † Preliminary results in this work were presented at MicroTAS 2024. |
| This journal is © The Royal Society of Chemistry 2026 |