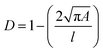Open Access Article
Open Access ArticleMechanobiological regulation of T cells via transient viscoelastic microfluidic confinement
Mohammad
Asghari
 a,
Morteza
Aramesh
b,
Prerit
Mathur
a,
Morteza
Aramesh
b,
Prerit
Mathur
 a,
Yingchao
Meng
a,
Yingchao
Meng
 a,
Margherita
Bernero
a,
Margherita
Bernero
 c,
Viola
Vogel
c,
Stavros
Stavrakis
c,
Viola
Vogel
c,
Stavros
Stavrakis
 *a and
Andrew J.
deMello
*a and
Andrew J.
deMello
 *a
*a
aInstitute for Chemical and Bioengineering, ETH Zürich, Vladimir Prelog Weg 1, Zürich, 8093, Switzerland. E-mail: stavros.stavrakis@chem.ethz.ch; andrew.demello@chem.ethz.ch
bInstitute for Biomedical Engineering, Department of Information Technology and Electrical Engineering, ETH Zürich, Gloriastrasse 37/39, Zürich, 8092, Switzerland
cInstitute of Translational Medicine, Department for Health Sciences and Technology, ETH Zürich, Gloriastrasse 39, Zürich, 8092, Switzerland
First published on 17th November 2025
Abstract
T cell migration through confined spaces, such as venular blood vessel walls or tumor tissues, is crucial for delivering immune effectors to sites of infection or disease. While mechanical confinement has been shown to trigger signaling pathways in T cells, the effects of transient (millisecond) mechanical confinement remain largely unknown. Here we demonstrate that even rapid, sub-millisecond flow-induced deformations can significantly impact T cell function. Using a high-throughput microfluidic system to confine large populations of activated and non-activated T cells in narrow channels of varying sizes, we show that transient microscale confinement regulates protein and gene expression in a channel size-dependent manner, impacting both immediate and long-term phenotypic changes (≥4 h after confinement) in gene regulation and T cell function. Notably, we demonstrate that confinement in narrow channels (5 μm high and 15 μm wide) primarily disrupts chromatin integrity and leads to nuclear envelope rupture. In contrast, post-confinement effects induced by wider channels (10 μm high and 15 μm wide) enhance T cell activation. These results establish a new and promising approach for promoting population growth in immunotherapy applications, such as CAR T cell therapy.
Introduction
Transmigration of T cells across confined spaces is a critical process occurring at various locations in the body, such as when T cells enter the thymus for maturation, when they migrate to lymph nodes for activation, or when they infiltrate cancerous tissues to mount an immune response.1,2 Decades of studies have investigated T cell trafficking through various tissues and synthetic hydrogels,3 revealing the underlying molecular and cellular mechanisms.4–6 However, while much attention has been directed towards understanding the biochemical cues that regulate T cell transmigration, less emphasis has been given to assessing the role of mechanical forces during this process.7Mechanobiological studies, either active (propelled by cell migration) or passive (pushed by fluid flow) have shown that as cells traverse confined spaces, their nuclei undergo significant deformation due to mechanical constraints.7–9 The extent of the forces induced by mechanical confinement can lead to nuclear envelope rupture, lamin disruption, and nucleo–cytoplasmic exchange, raising concerns about cell recovery and function under such stress.10,11 For instance, it has been shown that migration through densely packed tissues can compromise the integrity of the nuclear envelope, resulting in the depletion or rupture of lamin proteins12 and uncontrolled exchange of nucleo-cytoplasmic content.13,14 Others have shown that cell deformations have both rapid and long-term effects on nuclear and cellular functions, including muscle contraction and human disease pathogenesis.10,15,16 In addition, other studies on timescales ranging from minutes to hours have revealed significant reorganization of cytoskeletal components due to mechanical stresses, leading to changes in cellular metabolism17 and modifications in gene regulation.18 Accordingly, to better understand the impact of spatial constraints on T cell function on short timescales, it is important to develop new tools able to generate mechanical forces and image confined cell migration with high spatial and temporal resolution.
Several approaches have been developed to study active migration in confined spaces in vitro. One such approach involves Transwell invasion assays, where “Boyden chambers” are used to simulate cellular escape through narrow spaces using hollow plastic chambers.19 Whilst such approaches provide useful information, they are unable to directly monitor cells during migration, since migration occurs perpendicular to the imaging plane. An alternative method to study confined migration involves placing cells within a scaffold that resembles the extracellular matrix.20 Such scaffolds are generated through random self-assembly of matrix fibers, creating an environment that mimics biological tissue but with only limited control over pore size. Recently, microfluidic systems have been used to generate and mimic physical confinement of cells within tissue-based environments.12,21–23 These devices typically incorporate fluidic channels of controllable geometry through which the cells must actively migrate, either spontaneously or in response to a chemotactic gradient. While such platforms are versatile and provide the user with flexibility in both design and degree of constriction, they do not capture events induced by transient, sub-millisecond-scale confinement imposed by fluid flow.
Herein, we investigate the effects of passive confinement on T cell function by using flow to push T cells through microcapillaries. Specifically, we use viscoelastic deformability cytometry24 to rapidly confine and capture the transient translocation cells through micron-sized channels. Our approach can be described as transient mechanical confinement using a viscoelastic carrier fluid in a microfluidic device. The viscoelastic fluid (0.1% (v/w), 1 MDa PEO) serves to position cells for uniform deformation while passing through the confinement region. In brief, cells are driven through a network of parallel microchannels containing height constrictions between 5 μm and 15 μm. The high-throughput nature of the approach enables statistically rigorous evaluation of how transient confinement regulates protein and gene expression. In the context of our study, transient or immediate refers to sub-millisecond timescales of the confinement (i.e., while the cell traverses the constriction). In contrast, long-term effects refer to cellular responses observed hours after confinement (4 hours or more post-exposure). To elucidate how T cells adaptively respond to transient confinement, we combine in situ deformability measurements with conventional flow cytometry, reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and confocal microscopy to reveal the significance of transient stress in regulating T cell responses at both early and late times. Finally, we present potential mechanisms by which transient deformability translates into altered cell function.
Results and discussion
The microfluidic platform
To determine how confinement and fluidic pressure affect cell deformability, we used our recently developed viscoelastic deformability cytometer to focus and deform cells in a sheathless and high-throughput manner.24 The microfluidic device is simple in construction, having two primary components: a cell focusing zone and a downstream cell deformation zone (Fig. 1a). In the former, cells are focused both laterally and vertically by balancing elastic forces (originating from the inherent viscoelasticity of the carrier medium) with viscous forces as they move along the channel.25,26 In the latter, flow through a narrow constriction induces high shear27 and normal stresses24 on cells, causing them to deform. It is important to note that the viscoelastic properties of the medium obviate any need for sheath fluids, greatly simplifying microfluidic device design. Moreover, multiple channels having a single inlet and outlet can be arranged in parallel, leading to the in situ measurement and calculation of cell size and deformability at rates in excess of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per second;24 a value two orders of magnitude higher than the previous state of the art.28,29
000 cells per second;24 a value two orders of magnitude higher than the previous state of the art.28,29
To investigate the effect of transient spatial confinement on the deformability of both non-activated and activated T cells, we conducted experiments using constriction microchannels of varying cross section. Specifically, T cells were confined in channels with heights of 5, 10, and 15 μm, a constriction channel length of 300 μm, and a width of 15 μm. Initially, non-activated cells were driven through constrictions using a constant pressure of 1.5 bar, with their deformability and size being measured in a region close to the end of the constriction section. Single-cell images are used to measure cell size (A) and deformability (D) using,
Channel height regulates protein/gene expression in non-activated T cells
To evaluate whether sub-millisecond confinement affects early (4 hours) and/or late (24 hours) gene/protein expression in primary human T cells, we analyzed specific genes and proteins that are relevant for T cell activation. Early gene expression analysis was conducted 4 hours after passing T cells through 5, 10, and 15 μm high microchannels using a constant pressure of 1.5 bar and a constriction channel length of 300 μm. RT-qPCR analysis revealed that multiple genes, notably ITGAL, PTPRC, TCRA, TAZ, and MTOR, were differentially regulated when compared to cells collected from the inlet that had not been subjected to mechanical confinement (Fig. 2a). ITGAL, PTPRC, and TCRA encode for membrane proteins LFA-1 (integrin), CD45 (phosphatase), and TCR (T cell receptor), respectively, and play a vital role in T cell activation and antigen recognition.30,31TAZ and MTOR, on the other hand, encode for intracellular proteins that are sensitive to environmental stress factors. As part of the Hippo pathway, a signaling pathway involved in the control of organ size and development, TAZ is activated under the influence of mechanical forces such as stress, strain and physiological distortion, impacting cell growth and differentiation,32 while MTOR encodes for kinases that respond to stresses such as DNA damage and nutrient deprivation, which in turn controls both cell growth and proliferation.33Results confirm that sub-millisecond confinement of T cells can modulate the expression of specific genes involved in T cell activation and environmental stress response. The expression of ITGAL was found to be lower in T cells after their mechanical confinement (Fig. 2a). Specifically, T cells that have passed through 5 μm channels exhibited significantly lower expression of ITGAL compared to cells driven through 10 and 15 μm channels, which exhibited equivalent gene expression levels. Additionally, PTPRC expression was significantly higher in cells driven through 5 μm channels compared to those passed through 10 μm and 15 μm channels. Finally, the expression levels of MTOR, TAZ and TCRA did not significantly differ when T cells were passed through channels of different heights. No additional genes were assayed beyond the five shown, which were pre-selected for their relevance to T cell activation.
To confirm that the observed changes in gene expression are subsequently translated to the protein level, we performed flow cytometry analysis for LFA-1 (CD11a stained), TCR, and CD45 proteins, at 4 hours (Fig. 2b–d) and 24 hours (Fig. 2e–g) after exposure to sub-millisecond spatial confinement. Significant changes in protein expression were observed at both time points, compared to cells not passed through the device, with expression also depending on the channel size. Confinement in 5 μm channels resulted in significantly lower expression of both TCR (Fig. 2b) and LFA-1 (CD11a) (Fig. 2d), but higher expression of CD45 (Fig. 2c). Notably, the reductions in surface TCR and LFA-1 at 4 h post-confinement likely reflect rapid receptor internalization rather than complete protein degradation. T cells can downregulate surface TCR within minutes of stimulation,34 for instance by endocytosis of TCR–CD3 complexes, and LFA-1 surface levels can also be modulated through internalization or shedding. Thus, a mechanically induced signaling event could prompt removal of these receptors from the cell surface, explaining the 4 h flow cytometry results. Expression of TCR also decreased after confinement through 10 and 15 μm channels with respect to the inlet (Fig. 2b), whilst CD45 expression levels were increased significantly in the case of the 10 μm channel (Fig. 2c). Furthermore, relative to cells that did not pass through the device, these confinement-induced changes in protein expression persisted for at least 24 hours post-treatment.
These results are in excellent agreement with previous studies suggesting that mechanical confinement of cells is transduced to the molecular machinery, with cells adapting both their structure and molecular content in response to confinement.35,36 The study by King et al.36 reports that T cells exposed to shear stresses for extended durations (on the order of minutes) exhibit activation of mechanosensitive ion channels. Our findings complement this work by showing that even extremely brief mechanical cues (rather than prolonged shear) can modulate T cell behavior. In addition, our observations are in line with a recent mechanobiology report which highlights that even subtle mechanical cues can alter T cell signaling.35 Specifically, the current data indicate that the transduction of fluidic forces alters the expression of genes and proteins that are associated with T cell activation. Additionally, our results confirm that accommodation of external forces occurs extremely rapidly, since the transit time through the constriction zone is approximately 300 microseconds. This indicates that despite the brief exposure to confinement, the effects on protein expression are long-lasting.
We next evaluated cellular viability 4 hours after mechanical confinement using a Zombie NIR live/dead cell viability assay. For all tested channel heights, the viability of “squeezed” cells was more than 85%, a value equivalent to that of cells that had not been squeezed (Fig. S2). Similarly, apoptosis analysis using FITC-annexin V indicated a viability of more than 87% for all tested channel heights (Fig. S3). Finally, to investigate any possible “crowding effects” induced by PEO on cellular function, a control experiment was performed using 400 kDa Ficoll, which replicates the degree of crowding present in blood plasma. Flow cytometry analysis (Fig. S4) demonstrated no significant difference in protein expression between the PEO and the Ficoll sample sets, both at the inlet (i.e. without squeezing) and at the 15 μm channel outlet (i.e. after squeezing). This confirms a negligible effect of macromolecular crowding on protein expression.
Non-activated T cell activation is largely influenced by mechanical confinement
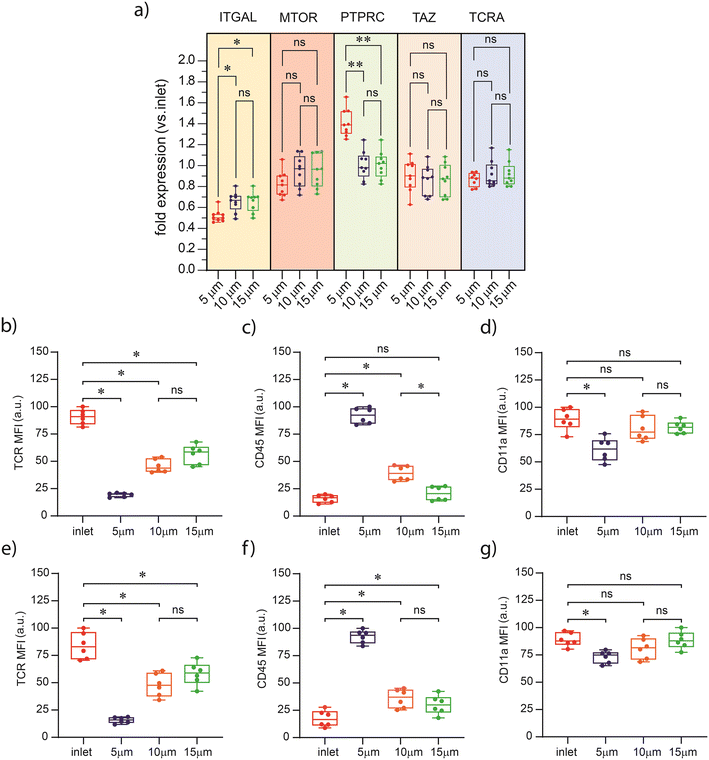
To study the influence of rapid confinement on protein expression of activation markers, we used our microfluidic deformability platform to activate T cells. To induce polyclonal activation and expansion, cells were exposed to culture plates coated with activating antibodies against CD3 (αCD3; TCR stimulus) and CD28 (αCD28; costimulatory cue). The term “costimulatory cue” refers to the role of CD28 signaling in providing costimulation to T cells during activation. Protein expression analysis revealed differences in cell activation between the sample groups, as measured by activation markers, i.e. CD69 and IL-2 expression (24 hours after activation) and CD25 expression (4 days after activation). CD25 and CD69 are activation markers for lymphocytes. CD25, part of the IL-2 receptor, reaches its highest expression level after at least 4 days, while CD69, expressed after 24 hours, serves as an early activation marker.37 CD69 expression was highest for cells that had passed through the 10 μm high channels, while those exposed to 5 μm confinement showed the lowest amount of expression (Fig. 3a). Similarly, cells exposed to 10 μm and 5 μm confinements exhibited the highest and lowest secretion of IL-2 protein, respectively (Fig. 3b). An identical trend was observed for CD25 expression measured 4 days after activation (Fig. 3c). Additionally, in all cases no significant changes in protein expression were observed after passage through 15 μm channels relative to the control (inlet). All assays were performed using 5000 viable cells per condition. Accordingly, the observed phenotypic differences are attributable to mechanical deformation and not to variation in culture density or cell recovery. These data indicate that enhanced activation of T cells in response to spatial constraints has significant potential utility in a range of immunotherapy applications, particularly in cases where there is a need to expand large numbers of T cells in a short time.Cytoskeletal remodeling post confinement
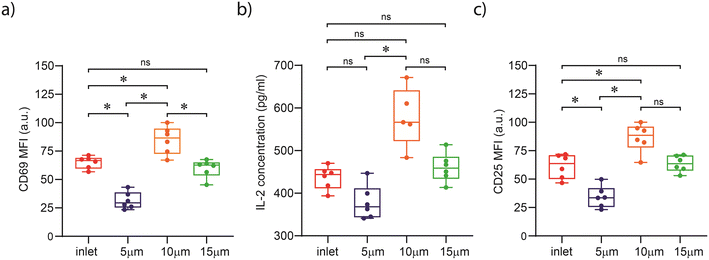
It is known that mechanical stimulation of cells can induce in situ changes in cytoskeleton reorganization, with the cytoskeleton playing a pivotal role in the mechanotransduction of signals into cells.30,38,39 Actin and tubulin are the primary sites of force transmission into the cell from the extracellular environment. Importantly, actin and tubulin play an essential role in T cell activation, since they are used by T cells to exert forces on opposing surfaces, such as antigen-presenting cells.32 Accordingly, we examined the structural organization of the T cell cytoskeleton during activation (after exposure to spatial constraints) by visualizing actin and tubulin using confocal fluorescence microscopy (Fig. 4). Cells collected from the inlet and outlets of the 15 μm and 10 μm channels exhibited the expected cytoskeletal organization, where an actin ring is formed around the cell periphery, and the microtubule-organizing center (MTOC) is directed towards the center of the cell. However, cells that passed through the 5 μm-high channel exhibited a less organized cortical actin structure, characterized by a diminished peripheral ring, while their tubulin networks showed signs of disruption, with fewer extended filaments compared to controls. Such an observation suggests that cytoskeletal organization is disturbed after exposure to constraints that are comparable to the cell size under study (5–8 μm for human primary T cells). It should be noted that Fig. 4 provides representative images highlighting possible cytoskeletal alterations under confinement; quantitative conclusions are drawn from flow cytometry data, with microscopy providing qualitative supporting evidence. Indeed, previous studies have shown that a loss of structural integrity results in impaired T cell activation,33 in line with our observation of compromised T cell activation in the 5 μm high channels (Fig. 3).Confinement within 5 μm channels induces nuclear envelope rupture
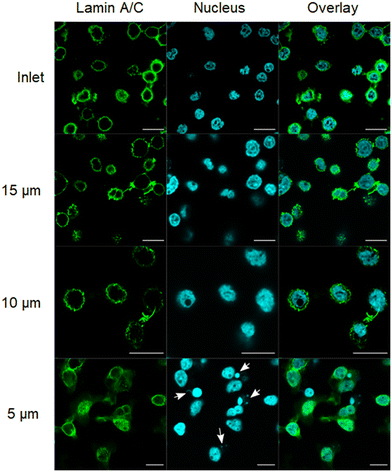
Recent studies have shown that cells experience transient or permanent nuclear envelope rupture during the course of compression from cytoskeletal stress or external forces.40,41 This rupture can lead to DNA damage and an uncontrolled exchange of products between the cytosol and the nucleus.14,15 Nuclear envelope integrity can be visualized by immunostaining of lamin, a major structural component of the nucleus. During nuclear envelope breakdown, lamin disassembles, creating locally depleted areas where chromatin is able to protrude through the lamin structure. Confocal microscopy images of lamin A/C revealed that the lamin structure remains intact in cells that had passed through 10 and 15 μm channels (Fig. 5). However, T cells confined in 5 μm channels exhibited morphological signs of nuclear envelope stress. In many cells, we observed small DAPI bright foci distinct from the main nucleus (arrows in Fig. 5, 5 μm-confinement condition). These structures lacked lamin staining and resembled micronuclei, which form when chromatin becomes separated from the primary nucleus following nuclear envelope rupture.42 Control cells or those confined in 10 and 15 μm channels rarely exhibited such features. The observation of nuclear rupture for cells that had passed through the 5 μm constriction reveals that mechanical confinement can engender enormous stresses on the nucleus. Further analysis is required to determine whether the induced damage is transient or permanent. However, it is evident that the stress induced by 10 and 15 μm channels does not result in significant breakdown of the lamin structure. That said, 5 μm-confined cells showed signs of cytoskeletal disruption (Fig. 4) and nuclear damage (Fig. 5); such stress may impair signal transduction and cytokine secretion, leading to reduced IL-2 despite normal viability. Fig. 3–5 should be interpreted together: flow cytometry provides quantitative evidence of altered activation, while representative imaging reveals cytoskeletal and nuclear perturbations that help explain the reduced responsiveness under extreme confinement.Mechanical stress triggers cell signaling
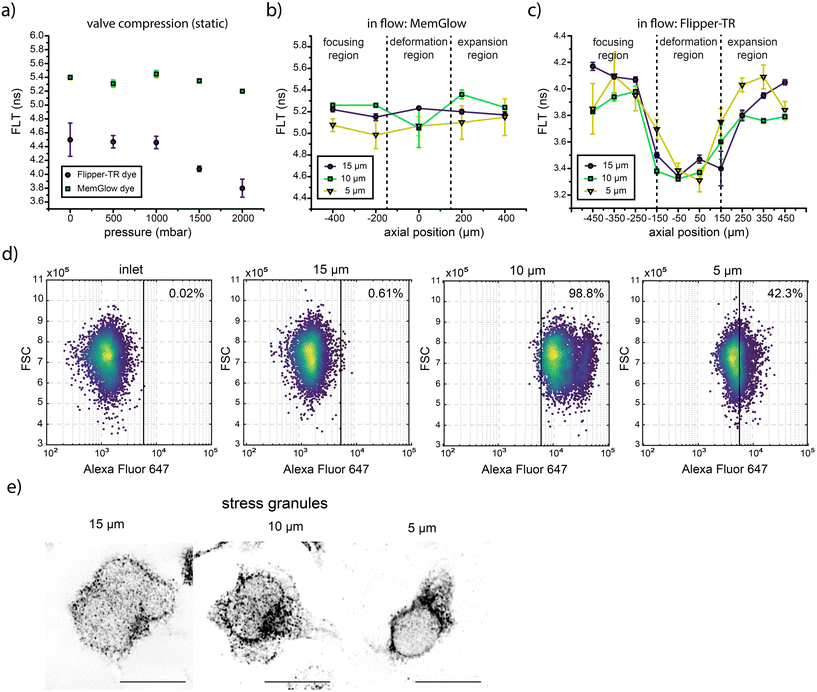
The data reported up until this point confirm that mechanical stress is a cause of alterations in cellular organization and function in primary human T cells translocating through narrow microfluidic channels. More specifically, the compromised activity of T cells after their migration through 5 μm high channels is induced by extensive damage to both the cytoskeleton and the nucleoskeleton. That said, the mechanisms associated with the enhancement of T cell activation after passage through 10 or 15 μm high channels (where cells are not extensively deformed) are yet to be elucidated.Cells can sense membrane tension, which can be used to trigger biological processes such as the phosphorylation of mechanosensitive proteins.43,44 Recent studies have shown that high membrane stresses can activate mechanobiological pathways.45–47 Accordingly, compression of T cells through a microchannel could potentially be used to trigger cell signaling by inducing tension in the cell membrane. To investigate constriction effects on cell membrane tension,48 fluorescence lifetime measurements were performed. First, fluorescence decay times of single cells trapped in a microfluidic chamber were measured before and after pneumatic compression (Fig. 6a and S5), to determine if changes in the fluorescence decay time of Flipper-TR® can be associated to variations in membrane tension. Compression was achieved through the use of a polydimethylsiloxane (PDMS)-based pneumatic valve, with Flipper-TR® being used to assess membrane tension and MemGlow™ being used to visualize the plasma membrane (and act as a control). Pressures of 0, 500, 1000, 1500, and 2000 mbar were used to compress cells. As shown in Fig. 6a, minimal variation in the average fluorescence lifetime (between 5.4 and 5.2 ns) was observed for MemGlow™ between 0 and 2000 mbar. In contrast, the average lifetime of Flipper-TR® decreased from 4.5 ns to 3.8 ns over the same pressure range, demonstrating the effects of dynamic compression on cell membrane tension. Next, variations in membrane tension of cells moving through the deformability platform were investigated (Fig. 6b and c). To do this, the average fluorescence lifetime (for each condition) was measured by sampling at least 100 cells. Specifically, three different geometries with constriction channels of 15 μm × 15 μm, 10 μm × 10 μm and 5 μm × 5 μm in cross section each with a 300 μm length were used. As can be seen in Fig. 6b, the average lifetime of MemGlow™ (in the cell membrane) shows minimal variation (5.0 to 5.4 ns) between the focusing, deformability and expansion regions for all channel heights. This highlights its suitability as a robust internal reference, unaffected by pressure-induced effects. However, the average fluorescence lifetime of Flipper-TR® decreases from 4.1 ns to 3.3 ns as cells enter the constriction region (Fig. 6c), with similar variations being observed for the 10 and 5 μm high channels. These findings directly confirm that the cell membrane is under significant tensile stress as a cell transits the microchannel.
One of the most important mechanobiological pathways linked with T cell activation is the extracellular signal-regulated kinase (ERK) pathway. This is part of the mitogen-activated protein kinase (MAP kinase) pathway, a major signaling cascade in regulating T cell activation and proliferation.49 With this in mind, and to showcase that cell deformation can affect important mechanobiological pathways, we examined the phosphorylation of ERK as a marker for pathway activation by measuring fluorescence immediately after the translocation of primary human T cells through the microchannels. T cells were stained with Alexa Fluor 647 anti-ERK1/2 Phospho Thr202/Tyr204 to monitor ERK phosphorylation via flow cytometry (Fig. 6d). Interestingly, cells that migrated through 10 and 5 μm high microchannels displayed significantly higher ERK phosphorylation, indicating that mechanical compression can induce ERK signaling (Fig. 6d). However, as can be observed, the 5 μm constriction leads to downregulation of the ERK pathway. This result aligns with a recent study demonstrating that ERK can be downregulated by cell compaction, with (in vivo) downregulation of ERK activity triggering cell elimination.50
Another mechanism potentially involved in the mechanotransduction process is the phase transition of stress granules caused by mechanical stresses within the cytoskeletal fibers. Stress granules contain mRNA- and RNA-binding proteins and contribute to cell signaling through their formation and deformation.51 Importantly, the size and distribution of stress granules are controlled by the mechanical stress in their local environment.51 To this end, after confinement, cells were stained with G3BP antibody to allow examination of stress granules. As can be seen in Fig. 6e, the concentration of stress granules increases as microchannel height decreases. After confinement within the 10 and 5 μm high channels, stress granules were primarily distributed close to the nucleus (cortical actin), possibly due to the higher stress at the cortex where actin filaments are tightly cross-linked. These results indicate that cytoskeletal stress induced by mechanical confinement can indeed contribute to cell signaling.
We also performed COMSOL simulations to predict shear stress and deformation behavior of T cells across different confinement sizes (see SI text). Specifically, we estimate that T cells in our device experience peak shear stresses on the order of 0.1–1 kPa for <1 ms (velocity of ∼0.4 m s−1, for additional details see Fig. S6) during passage through 10 μm constrictions. In comparison, during physiological extravasation through blood vessel walls, T cells migrate much more slowly (between 10 and 15 μm min−1)52 and experience wall shear stresses (on the order of 0.1–1 Pa).53 Despite the ultrashort duration of our mechanical stimulus, the fact that significant cellular responses are observed suggests that even brief deformation can trigger molecular signaling cascades. This observation aligns with aspects of natural transmigration, where squeezing through narrow gaps (5–10 μm) provokes cytoskeletal and nuclear adaptations, albeit over longer timescales.
Conclusions
We have introduced a high-throughput viscoelastic fluid-based microfluidic device for transient cell confinement. While related investigations have been conducted on stromal cells,54 our findings underscore for the first time the pivotal role of transient mechanical confinement in regulating T cell behavior. By varying microchannel dimensions, we have demonstrated that T cells respond to transient (millisecond-scale) compression in distinct ways: narrower channels (5 μm) cause severe nuclear deformation and disintegration of the lamin network and cytoskeleton, whereas larger channels (10 and 15 μm) preserve lamin and cytoskeletal integrity while enhancing ERK signaling and T cell activation. These results demonstrate the significance of transient confinement on both immediate and long-term phenotypic changes (≥4 h after confinement) in T cell function, suggesting new strategies for controlling immune responses in vitro. Importantly, the ability to leverage specific channel dimensions for robust T cell expansion has potential implications for adoptive immunotherapy, including CAR T cell manufacturing, where efficient, scalable production of highly functional T cells is essential. Significant ERK phosphorylation is observed in T cells passing through 10 μm channels, highlighting the potency of transient mechanical confinement in activating downstream pathways involved in T cell function. Notably, the mechanical transient confinement used herein (sub-millisecond, high-strain deformation) is very different from physiological T cell transmigration, which unfolds over minutes under far lower shear stresses. Yet, intriguingly, even a millisecond-scale pulse was sufficient to induce signaling changes, underscoring the exquisite mechanosensitivity of T cells. Likewise, the formation and redistribution of stress granules suggest that transient mechanical compression modulates cell signaling by sequestering or releasing key regulatory molecules, impacting protein and gene regulation. We show that immediate responses to transient mechanical stress can produce long-lasting changes in T cell function, either hampering activation in 5 μm channels or enhancing activation in 10 μm channels. Mechanical deformation in 10 μm channels may augment immune synapse formation or mechanosensitive signalling pathways, thereby enhancing T cell activation, whereas 5 μm confinement causes structural damage that interferes with productive activation.Enhanced T cell activation has an obvious impact in immunotherapy. Accordingly, transient confinement of cells within microfluidic constrictions could be used as a method to enhance T cell activation. Our results raise the possibility of mechanically ‘pre-conditioning’ T cells as a novel step in CAR T cell manufacturing. In current protocols, T cells are typically activated using antibody-coated beads or antigen-presenting cell (APC) mimics for days. Introducing a short mechanical stimulation phase (for example, passing cells through a 10 μm constriction) before or during standard activation could potentially enhance signaling pathways (such as ERK phosphorylation) and induce early gene expression. This process could lower the threshold or decrease the time necessary for robust activation once stimulatory antibodies or ligands are provided. This mechanical activation can also complement rather than replace chemical stimulation; for instance, synergizing with suboptimal TCR/CD28 stimulation to boost T cell responsiveness. Such strategies may ultimately reduce culture times or boost cell yields, although further research is needed to optimize these mechanical parameters for clinical-scale manufacturing.
The degree of mechanical confinement and its implications in protein and gene expression may need to be taken into account when designing a therapeutic approach. The potential of adoptive cancer immunotherapy is currently being explored in numerous clinical trials, many of which involve CAR T cells.55,56 To fully realize the potential of such therapies, new manufacturing technologies are required. Our results suggest that mechanobiological regulation of cells through transient viscoelastic microfluidic confinement has the potential to improve current methods for in vitro activation and expansion of CAR T cells. This mechanical pre-conditioning could be integrated as an additional unit operation in CAR T cell production. Rather than replacing classical TCR-mediated activation, a transient mechanical stimulus might enhance it, for example, by increasing ERK phosphorylation and early activation in gene expression, thereby lowering the threshold or shortening the time required for full activation once stimulatory antibodies or ligands are provided. Such a strategy could reduce culture times or improve yields, although future studies are needed to optimize parameters for clinical manufacturing.
Materials and methods
Microfluidic device fabrication
A negative master mold was created using standard photolithography and replicated in polydimethylsiloxane (PDMS).57 The microfluidic device comprises two sections: the focusing and deformability regions, with each having different channel heights. The master mold was fabricated in two steps by depositing and patterning two layers to account for the different heights of the two sections. The focusing region comprises 10 parallel channels, all 3 cm long, 50 μm wide and 50 μm high. The deformability region comprises 10 parallel channels that are 300 μm long, 15 μm wide and either 15 μm, 10 μm or 5 μm high. To fabricate the master mold, two lithographic masks were designed using AutoCAD 2018 (Autodesk, San Francisco, CA, USA) and laser-printed on a 5 inch Cr/fused silica transparency mask (Micro Lithography Services, Chelmsford, UK). The first mask was used to pattern the deformability region, and the second mask to pattern the focusing region. SU8-2010 photoresist (Micro Resist Technology, Berlin, Germany) was spin-coated onto a silicon wafer (Siegert Wafer, Aachen, Germany). The thickness of the coating was controlled by varying the rotation speed (30 seconds at 500 rpm s−1 and then 1500, 2500 and 3000 rpm for the 15, 10 and 5 μm thick layers, respectively). All wafers were prebaked at 65 °C for 3 minutes and then at 95 °C for 9 minutes. Subsequently, the photoresist was exposed to UV light (140 mJ cm−2) and post-baked at 65 °C for 2 minutes and 95 °C for 4 minutes. The entire photoresist was developed in an AZ300 MIF developer (MicroChem, Ulm, Germany) for 3 minutes and hard-baked at 150 °C for 10 minutes. Next, the second mask was aligned with the first layer using a mask aligner (KLOE, St Mathieu de Tréviers, France). The second layer was fabricated as follows: SU-8 2050 photoresist (Micro Resist Technology, Berlin, Germany) was spin-coated (acceleration: 500 rpm s−1, speed: 3000 rpm, time: 30 seconds) on the silicon wafer and pre-baked at 65 °C for 3 minutes and 95 °C for 9 minutes. Subsequently, the photoresist was exposed to UV light (150 mJ cm−2) using the second mask, and post-baked at 65 °C for 2 minutes and 95 °C for 4 minutes. The photoresist was developed for 5 minutes and hard-baked at 150 °C. In this manner, the second layer was created side by side with the first layer. To replicate the microfluidic device from the master mold, a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of PDMS monomer and curing agent (Sylgard 184, Dow Corning Midland, MI, USA) was poured over the master mold and polymerized at 70 °C for 4 hours. Inlet and outlet ports were created using a hole-puncher (SYNEO, West Palm Beach, FL, USA). Subsequently, the structured PDMS substrate was bonded to a 1 mm thick glass substrate after exposing both surfaces to an oxygen plasma (EMITECH K1000X, Quorum Technologies, Lewes, UK) for 60 seconds. A Flow EZ pressure pump (Fluigent, OKABE, France) was used as a flow-generating device. Pressures required to cause appreciable deformability varied between 200 mbar and 2 bar depending on the stiffness of the cells being studied.
1 mixture of PDMS monomer and curing agent (Sylgard 184, Dow Corning Midland, MI, USA) was poured over the master mold and polymerized at 70 °C for 4 hours. Inlet and outlet ports were created using a hole-puncher (SYNEO, West Palm Beach, FL, USA). Subsequently, the structured PDMS substrate was bonded to a 1 mm thick glass substrate after exposing both surfaces to an oxygen plasma (EMITECH K1000X, Quorum Technologies, Lewes, UK) for 60 seconds. A Flow EZ pressure pump (Fluigent, OKABE, France) was used as a flow-generating device. Pressures required to cause appreciable deformability varied between 200 mbar and 2 bar depending on the stiffness of the cells being studied.
Since the microfluidic device used for cell compression studies integrates features of varying height, master molds were fabricated using multi-step resist coating and development techniques.58 In brief, microfluidic channels having semi-circular cross sections were generated by the melting and reflowing of a positive AZ 4620 photoresist (MicroChem, Ulm, Germany), whilst microfluidic channels having rectangular cross sections were fabricated using SU-8 (MicroChem, Ulm, Germany). The fluidic layer comprises 20 μm high microchannels with a rectangular cross section. The fluidic layer mold was created by spin-coating AZ 4620 onto a silicon wafer at a speed of 3750 rpm for 40 seconds. The coated wafer was soft-baked at 126 °C for 8 minutes and then cooled to ambient temperature over a period of 10 minutes. Photolithographic patterning of the photoresist was performed using a UV-KUB 3 mask aligner (KLOE, St Mathieu de Tréviers, France), with the appropriate exposure time being calculated for an incident intensity of 400 mJ cm−2. The exposed wafer was then baked again at 105 °C. Next, the wafer was developed in an AZ300 MIF developer (MicroChem, Ulm, Germany), until the desired microstructures were visible to the naked eye. The wafer was then washed in deionized water and dried under a stream of nitrogen. Semi-circular cross section channel features were generated by melting and reflowing the AZ 4620 photoresist. Specifically, the temperature was increased from ambient to 65 °C, heated up to 150 °C at a rate of 10 °C per hour, and then cooled to ambient temperature. The master mold for the control channel layer was fabricated in SU-8 on a standard silicon wafer following the lithographic procedures described above.
Primary human T cell isolation
Mononuclear cells were isolated from human whole blood (Zurich Blood Donation Center Blutspende SRK, Zurich, Switzerland) by density centrifugation using SepMate-50 tubes (STEMCELL Technologies, Vancouver, Canada). Briefly, density gradient medium (1.077 g mL−1) was added to the tubes through the insert, and whole blood samples, previously diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 with phosphate-buffered saline (PBS) (Sigma-Aldrich, Buchs, Switzerland) supplemented with 2% fetal bovine serum (FBS) (Sigma-Aldrich, Buchs, Switzerland) were added. Density centrifugation was performed at 1200g for 10 minutes at room temperature. Next, the top fraction (containing enriched mononuclear cells) was removed, washed twice with PBS at 300g for 8 minutes, and finally resuspended in PBS. T cells were subsequently selected from this mixture using the EasySep Human Naive Pan T Cell Isolation Kit (STEMCELL Technologies, Vancouver, Canada). Briefly, the sample was mixed with the Isolation cocktail and TCR Gamma/Delta Depletion Cocktail containing combinations of monoclonal antibodies (50 μL mL−1 of sample volume each) and incubated for 5 minutes at room temperature. For the negative depletion of other cell types, magnetic Dextran RapidSpheres (STEMCELL Technologies, Vancouver, Canada) were added to the sample to a concentration of 60 μL mL−1, mixed and incubated for 3 minutes at room temperature prior to being topped up to 2.5 mL with PBS and incubated for another 3 minutes within the EasySep Magnet (STEMCELL Technologies, Vancouver, Canada). Subsequently, the solution containing enriched T cells was decanted into a new tube for a second incubation in the EasySep Magnet, decanted again, and diluted to a final concentration of 5 × 105 cells per mL with RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS and 1% penicillin–streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). Freshly isolated, non-activated T cells were used on the day of isolation without subjecting them to prolonged culture. During the study, cell cultures were maintained for up to 2 days with no observed abnormal cell loss.
2 with phosphate-buffered saline (PBS) (Sigma-Aldrich, Buchs, Switzerland) supplemented with 2% fetal bovine serum (FBS) (Sigma-Aldrich, Buchs, Switzerland) were added. Density centrifugation was performed at 1200g for 10 minutes at room temperature. Next, the top fraction (containing enriched mononuclear cells) was removed, washed twice with PBS at 300g for 8 minutes, and finally resuspended in PBS. T cells were subsequently selected from this mixture using the EasySep Human Naive Pan T Cell Isolation Kit (STEMCELL Technologies, Vancouver, Canada). Briefly, the sample was mixed with the Isolation cocktail and TCR Gamma/Delta Depletion Cocktail containing combinations of monoclonal antibodies (50 μL mL−1 of sample volume each) and incubated for 5 minutes at room temperature. For the negative depletion of other cell types, magnetic Dextran RapidSpheres (STEMCELL Technologies, Vancouver, Canada) were added to the sample to a concentration of 60 μL mL−1, mixed and incubated for 3 minutes at room temperature prior to being topped up to 2.5 mL with PBS and incubated for another 3 minutes within the EasySep Magnet (STEMCELL Technologies, Vancouver, Canada). Subsequently, the solution containing enriched T cells was decanted into a new tube for a second incubation in the EasySep Magnet, decanted again, and diluted to a final concentration of 5 × 105 cells per mL with RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS and 1% penicillin–streptomycin (Thermo Fisher Scientific, Waltham, MA, USA). Freshly isolated, non-activated T cells were used on the day of isolation without subjecting them to prolonged culture. During the study, cell cultures were maintained for up to 2 days with no observed abnormal cell loss.
Sample and reagent preparation for microfluidic confinement
Viscoelastic fluid was prepared by completely dissolving 1 MDa polyethylene oxide (PEO, Sigma-Aldrich, Buchs, Switzerland) in PBS to a concentration of 1% (w/v). Solutions produced in this manner were then aged at 4 °C for 1 week to generate a steady-state viscosity. PEO was diluted in the cell solution to a concentration of 0.1% prior to running experiments. Flasks were stored inside a Galaxy 170S incubator (Eppendorf, Basel, Switzerland) at 37 °C, 5% CO2, and 95% humidity, with media being refreshed every two days. The cell concentration was maintained at 2 million cells per ml for all experiments. In each experiment, cells were centrifuged at 5000 rpm for 5 minutes at 24 °C, filtered through a strainer with a pore size of 40 μm (Corning, New York, NY, USA), centrifuged, and re-suspended in a fresh cell medium.Deformability analysis
Bright-field imaging of cells was performed using a Y5.1 high-speed camera (Motion Pro, Niederoenz, Switzerland) mounted on an inverted Eclipse Ti-E microscope (Nikon, Zurich, Switzerland) using a 30×, 0.45 NA S Plan Fluor objective (Nikon, Zurich, Switzerland), and ultra-high power UHP-T-560-DI-DF LED (Prizmatix, Holon, Israel). Image processing algorithms were implemented in MATLAB. For the selected region of interest, the video is cropped, and the background image calculated by averaging the intensities of the captured images. First, a median filter is used to reduce noise. A binary filter with a specified threshold is used to detect cell contours. The cell area (A) and perimeter length (l) were then calculated for each cell. Deformability, D, is calculated using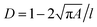 . To capture blur-free images of cells, exposure times were set to 2 μs with a recording rate of 10
. To capture blur-free images of cells, exposure times were set to 2 μs with a recording rate of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames per second. Finally, deformability values and cell sizes from detected cell contours were calculated and plotted as a scatter plot.
000 frames per second. Finally, deformability values and cell sizes from detected cell contours were calculated and plotted as a scatter plot.
T cell activation
The activation of the primary T cells was performed in 24-well plates coated with 400 μL of anti-CD3 and anti-CD28 antibody solution (Thermo Fisher Scientific, Waltham, MA, USA) (both 5 μg mL−1 in PBS). 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded in each well and cultured at 37 °C for 24 hours. The culture medium was supplemented with human IL-2 (Thermo Fisher Scientific, Waltham, MA, USA) at a concentration of 60 U mL−1. The culture medium was composed of RPMI 1640 supplemented with 10% fetal bovine serum (FBS, from Sigma-Aldrich F7524), 1× penicillin/streptomycin, 2-ME (50 mM), nonessential amino acids (Thermo Fisher Scientific, Waltham, MA, USA), sodium pyruvate (Thermo Fisher Scientific, Waltham, MA, USA), HEPES (Thermo Fisher Scientific, Waltham, MA, USA), and glutamate (Thermo Fisher Scientific, Waltham, MA, USA). The collection of peripheral blood mononuclear cells (PBMCs) was approved by the Kantonale Ethikkommission Zürich (KEK-ZH-Nr. 2012-0111), and written consent was obtained from all subjects. A total of 12 separate healthy donors participated in this study.
000 cells were seeded in each well and cultured at 37 °C for 24 hours. The culture medium was supplemented with human IL-2 (Thermo Fisher Scientific, Waltham, MA, USA) at a concentration of 60 U mL−1. The culture medium was composed of RPMI 1640 supplemented with 10% fetal bovine serum (FBS, from Sigma-Aldrich F7524), 1× penicillin/streptomycin, 2-ME (50 mM), nonessential amino acids (Thermo Fisher Scientific, Waltham, MA, USA), sodium pyruvate (Thermo Fisher Scientific, Waltham, MA, USA), HEPES (Thermo Fisher Scientific, Waltham, MA, USA), and glutamate (Thermo Fisher Scientific, Waltham, MA, USA). The collection of peripheral blood mononuclear cells (PBMCs) was approved by the Kantonale Ethikkommission Zürich (KEK-ZH-Nr. 2012-0111), and written consent was obtained from all subjects. A total of 12 separate healthy donors participated in this study.
Flow cytometry
For surface staining, cells were incubated with the following antibodies for 30 minutes at 4 °C: PerCP/Cy5.5 anti-human CD69 (clone FN50), APC anti-human CD25 (clone BC96), Alexa Fluor 647 anti-human TCR α/β (306![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 714), Alexa Fluor 488 anti-human CD45 antibody (368
714), Alexa Fluor 488 anti-human CD45 antibody (368![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 535), CD11a (350
535), CD11a (350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 606), Zombie NIR fixable viability kit for live/dead test (423
606), Zombie NIR fixable viability kit for live/dead test (423![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106), FITC-annexin V apoptosis detection kit with PI (640
106), FITC-annexin V apoptosis detection kit with PI (640![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 914), all from BioLegend, San Diego, CA, USA. Intracellular staining of phosphorylated ERK was performed by fixing cells using 2% formaldehyde (16%, methanol free, Ultra-Pure, Polysciences, Warrington, PA, USA, 18814-10) for 10 minutes at room temperature, and permeabilized with 100% ice-cold methanol on ice dropwise whilst vortexing. pERK was stained for 30 minutes using Alexa Fluor 647 anti-ERK1/2 Phospho Thr202/Tyr204 in PBS 1% bovine serum albumin (BSA) (clone 4B11B69, BioLegend, San Diego, CA, USA). Samples were acquired using a CytoFLEX Flow Cytometer (Beckman Coulter, Brea, CA, USA). All assays were performed using 5000 viable cells per condition. Data analysis was carried out using Python 3 and the Cytoflow package (https://github.com/cytoflow/cytoflow).
914), all from BioLegend, San Diego, CA, USA. Intracellular staining of phosphorylated ERK was performed by fixing cells using 2% formaldehyde (16%, methanol free, Ultra-Pure, Polysciences, Warrington, PA, USA, 18814-10) for 10 minutes at room temperature, and permeabilized with 100% ice-cold methanol on ice dropwise whilst vortexing. pERK was stained for 30 minutes using Alexa Fluor 647 anti-ERK1/2 Phospho Thr202/Tyr204 in PBS 1% bovine serum albumin (BSA) (clone 4B11B69, BioLegend, San Diego, CA, USA). Samples were acquired using a CytoFLEX Flow Cytometer (Beckman Coulter, Brea, CA, USA). All assays were performed using 5000 viable cells per condition. Data analysis was carried out using Python 3 and the Cytoflow package (https://github.com/cytoflow/cytoflow).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated in triplicate for each experimental condition across three independent experiments. Samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 in cell culture medium and then split into 3 aliquots containing approximately 300
5 in cell culture medium and then split into 3 aliquots containing approximately 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells each. The RNeasy Mini Kit (QIAGEN, Hilden, Germany) was used according to the manufacturer's instructions. Briefly, cells were pelleted at 300g for 5 minutes and the supernatant was removed before the cells were lysed in 350 μL l buffer (RNeasy Mini Kit, QIAGEN, Hilden, Germany) supplemented with 1% mercaptoethanol (Sigma-Aldrich, Buchs, Switzerland). The lysate was homogenized by running through a QIAshredder spin column (QIAGEN, Hilden, Germany) in the centrifuge at full speed for 2 minutes, mixed with 350 μL 70% ethanol and transferred into RNeasy spin columns. RNA molecules (>200 nucleotides long) are selectively bound to the membrane during centrifugation at maximum speed and are subsequently washed with the washing buffers provided in the kit. The RNA was eluted into a new collection tube by first adding 50 μL and then 37.5 μL RNase-free water and centrifuged at full speed for 1 minute. Finally, eluate RNA concentrations were measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and frozen at −20 °C.
000 cells each. The RNeasy Mini Kit (QIAGEN, Hilden, Germany) was used according to the manufacturer's instructions. Briefly, cells were pelleted at 300g for 5 minutes and the supernatant was removed before the cells were lysed in 350 μL l buffer (RNeasy Mini Kit, QIAGEN, Hilden, Germany) supplemented with 1% mercaptoethanol (Sigma-Aldrich, Buchs, Switzerland). The lysate was homogenized by running through a QIAshredder spin column (QIAGEN, Hilden, Germany) in the centrifuge at full speed for 2 minutes, mixed with 350 μL 70% ethanol and transferred into RNeasy spin columns. RNA molecules (>200 nucleotides long) are selectively bound to the membrane during centrifugation at maximum speed and are subsequently washed with the washing buffers provided in the kit. The RNA was eluted into a new collection tube by first adding 50 μL and then 37.5 μL RNase-free water and centrifuged at full speed for 1 minute. Finally, eluate RNA concentrations were measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and frozen at −20 °C.
Isolated RNA samples were thawed to synthesize cDNA by reverse transcription using the iScript Advanced cDNA Synthesis Kit for RT-qPCR (Bio-Rad, Hercules, CA, USA). Reactions were assembled on ice and on a nuclease-free bench. A master mix was prepared with 6 μL 5× iScript Advanced Reaction Mix and 1.5 μL iScript Advanced Reverse Transcriptase per 32.5 μL RNA. If RNA contents were higher than 7.5 μg, samples were diluted accordingly with nuclease-free water. For 40 μL reactions, 7.5 μL master mix and 32.5 μL RNA were combined in a PCR tube, loaded onto a Mastercycler (Eppendorf, Basel, Switzerland), and incubated with the custom CDNAADV program, i.e. (1) reverse transcription at 42 °C for 30 minutes, (2) inactivation at 85 °C for 5 minutes, (3) hold at 4 °C. The final cDNA samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 with nuclease-free water and stored at 4 °C. The primer pair candidates for RT-qPCR were based on the complete coding sequence of target genes retrieved from the NCBI Nucleotide Database (https://www.ncbi.nlm.nih.gov/nuccore).
4 with nuclease-free water and stored at 4 °C. The primer pair candidates for RT-qPCR were based on the complete coding sequence of target genes retrieved from the NCBI Nucleotide Database (https://www.ncbi.nlm.nih.gov/nuccore).
RT-qPCR was performed using the SsoAdvanced Universal SYBR Green Supermix (an antibody-mediated hot-start Sso7d fusion polymerase, and the dsDNA-binding dye SYBR Green; Bio-Rad, Hercules, CA, USA) in a CFX Connect Real-Time PCR Machine (Bio-Rad, Hercules, CA, USA). 96-well-plate setups were designed using a sample maximization strategy, such that all samples of a target gene replicate are evaluated in the same run. This facilitates comparison and reduces the effects of inter-run variations. All primers were purchased from Microsynth, Balgach, Switzerland. The reference gene 18S rRNA was chosen for normalization of RT-qPCR data and included on every plate. Each reaction was run in triplicate. Working on a nuclease-free bench, supermixes were created for each target gene containing 1 μL forward primer (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with nuclease-free water), 1 μL reverse primer (diluted 1
10 with nuclease-free water), 1 μL reverse primer (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with nuclease-free water), 3 μL nuclease-free water and 10 μL SsoAdvanced Universal SYBR Green Supermix per reaction well. Subsequently, 15 μL target gene supermix and 5 μL sample cDNA were mixed in Hard-Shell 96-well PCR plate wells (Bio-Rad, Hercules, CA, USA) and sealed with Microseal ‘B’ sealing film (Bio-Rad, Hercules, CA, USA). RT-qPCR was then performed on a CFX Connect (Bio-Rad, Hercules, CA, USA) instrument according to the following protocol. First, polymerase activation was performed at 95 °C for 3 minutes. Denaturation (95 °C for 10 seconds), annealing and extension (62.0 °C for 30 seconds) and optical readout were performed 39 times. Finally, a melt curve was obtained between 65 °C and 95 °C in increments of 0.5 °C. Subsequently, amplification and melt peak curves were visually inspected to evaluate amplification success and detect errors. The statistical significance of differences in gene expression was evaluated using the Wilcoxon rank sum test in Prism 9.4 (GraphPad, La Jolla, CA, USA).
10 with nuclease-free water), 3 μL nuclease-free water and 10 μL SsoAdvanced Universal SYBR Green Supermix per reaction well. Subsequently, 15 μL target gene supermix and 5 μL sample cDNA were mixed in Hard-Shell 96-well PCR plate wells (Bio-Rad, Hercules, CA, USA) and sealed with Microseal ‘B’ sealing film (Bio-Rad, Hercules, CA, USA). RT-qPCR was then performed on a CFX Connect (Bio-Rad, Hercules, CA, USA) instrument according to the following protocol. First, polymerase activation was performed at 95 °C for 3 minutes. Denaturation (95 °C for 10 seconds), annealing and extension (62.0 °C for 30 seconds) and optical readout were performed 39 times. Finally, a melt curve was obtained between 65 °C and 95 °C in increments of 0.5 °C. Subsequently, amplification and melt peak curves were visually inspected to evaluate amplification success and detect errors. The statistical significance of differences in gene expression was evaluated using the Wilcoxon rank sum test in Prism 9.4 (GraphPad, La Jolla, CA, USA).
ELISA
Cultured cell supernatant was obtained 24 hours after activation to measure interleukin-2 (IL-2) content. Human IL-2 ELISA Set (ab48471, Abcam, Cambridge, UK) was used according to the manufacturer's instructions. Supernatants were assessed in triplicate and at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and 1
4 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 dilutions. A Tecan M200 Plate Reader spectrometer (Tecan, Zurich, Switzerland) was used to measure the absorbance of each well at 450 nm. The absolute concentration of IL-2 in each well was calculated by comparison with standards (0–1000 pg mL−1) using Origin 2019 (OriginLab, Northampton, MA, USA).
5 dilutions. A Tecan M200 Plate Reader spectrometer (Tecan, Zurich, Switzerland) was used to measure the absorbance of each well at 450 nm. The absolute concentration of IL-2 in each well was calculated by comparison with standards (0–1000 pg mL−1) using Origin 2019 (OriginLab, Northampton, MA, USA).
Immunostaining
Cells were seeded on microscope coverslips (Fisher Scientific, Reinach, Switzerland) and incubated at 37 °C and 5% CO2 for the duration. All buffers were pre-warmed at 37 °C. For cytoskeletal staining, cells were pre-fixed for 1 minute in 0.5% PFA (formaldehyde, Polysciences GmbH, Warrington, PA, USA) in cytoskeletal buffer (10 mM MES pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM MgCl2). Cells were subsequently fixed and permeabilized with 4% PFA and 0.1% Triton-X (Sigma-Aldrich, Buchs, Switzerland) in cytoskeletal buffer for 7 minutes. Samples were washed with PBS three times and incubated with 0.01% NaCl4 (Sigma-Aldrich, Buchs, Switzerland) in PBS for 10 minutes to reduce autofluorescence. Samples were then washed with PBS three times and blocked with 2% BSA (Sigma-Aldrich, Buchs, Switzerland) in PBS for 1 hour at room temperature prior to staining. No pre-fixation step was used when staining lamin. The following reagents were used according to the manufacturer's instructions over a period of 1 hour at room temperature in the dark: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution of phalloidin (Alexa Fluor 488, 546, 647, Thermo Fisher Scientific, Waltham, MA, USA), 5 μg mL−1 DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride, Thermo Fisher Scientific, Waltham, MA, USA), anti-lamin A + lamin C antibody [131C3] (Abcam, Cambridge, UK), anti-alpha tubulin antibody, Microtubule Marker (ab15246, Abcam, Cambridge, UK) and recombinant anti-G3BP antibody (ab181150, Abcam, Cambridge, UK). 1% BSA in PBS was used as a buffer to dilute the antibodies. The secondary antibodies were added and incubated for 2 hours at room temperature in the dark. The samples were then washed with PBS three times before mounting with Prolong gold anti-fade reagent (Molecular Probes, Eugene, OR, USA).
200 dilution of phalloidin (Alexa Fluor 488, 546, 647, Thermo Fisher Scientific, Waltham, MA, USA), 5 μg mL−1 DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride, Thermo Fisher Scientific, Waltham, MA, USA), anti-lamin A + lamin C antibody [131C3] (Abcam, Cambridge, UK), anti-alpha tubulin antibody, Microtubule Marker (ab15246, Abcam, Cambridge, UK) and recombinant anti-G3BP antibody (ab181150, Abcam, Cambridge, UK). 1% BSA in PBS was used as a buffer to dilute the antibodies. The secondary antibodies were added and incubated for 2 hours at room temperature in the dark. The samples were then washed with PBS three times before mounting with Prolong gold anti-fade reagent (Molecular Probes, Eugene, OR, USA).
Fluorescence lifetime imaging
Fluorescence lifetime measurements were performed using a custom-built setup. A picosecond diode laser with an excitation wavelength of 488 ± 5 nm (PicoQuant, Berlin, Germany) was operated at a repetition rate of 20 MHz. Emission was coupled to a Nikon C2si confocal laser scanner mounted on a Nikon Eclipse Ti2 inverted microscope (Nikon, Egg, Switzerland), collected in an optical fiber (OZ Optics, Ottawa, Canada) and passed through a collimator (F950FC-A, Thorlabs, Bergkirchen, Germany), focused with a doublet lens (AC254-050-A, Thorlabs, Bergkirchen, Germany) and passed through either a 525/39 nm (525/39 BrightLine HC, AHF, Tübingen, Germany) or 590/50 nm (590/50 BrightLine HC, AHF, Tübingen, Germany) wavelength filter. Fluorescence photons were detected by an avalanche photodiode-based single-photon counting module (Excelitas Technologies, Waltham, MA, USA). Both the microscope and the photon detection optics were controlled by two computers communicating over a local network. NIS-Elements C (Nikon, Egg, Switzerland) was used to control the scanner, and SymPhoTime 64 data acquisition software (PicoQuant, Berlin, Germany) was used to collect and analyze the fluorescence lifetime data. The emission filters 525/39 and 590/50 were used for the detection of MemGlow 488 (Cytoskeleton, Denver, CO, USA) and Flipper-TR (LubioScience, Zürich, Switzerland) dyes respectively.Statistical analysis
Data are presented as the mean of replicates, with error bars representing mean ± standard deviation. All statistical analyses were performed with GraphPad Prism 9.4 (GraphPad Software, La Jolla, CA, USA). In all graphs the significance level was defined as p < 0.05 and p values are represented by *p < 0.033, **p < 0.0021, ***p < 0.0002 and ****p < 0.0001. Statistical significance of overall differences of cell size and deformability data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test. Statistical significance of gene and protein expression data was evaluated using a two-sided Wilcoxon signed-rank test. The p values reported from each experiment come from comparison of the given sample to the control condition obtained.Confocal microscopy
Imaging of immunostained samples was conducted using a Zeiss LSM 880 laser scanning confocal microscope with an Airyscan detector (Carl Zeiss AG, Jena, Germany) equipped with a 60× oil objective. A minimum of 10 cells were imaged per condition.Conflicts of interest
There are no conflicts of interest to declare.Data availability
The data that support the findings of this study are available within the article and the supplementary information (SI).Supplementary information: the SI includes simulation data of velocity, shear stress, and deformation for various confinement sizes, detailed fabrication procedures, and additional experimental data that further substantiate the conclusions of this work. See DOI: https://doi.org/10.1039/d5lc00833f.
Acknowledgements
The authors acknowledge partial financial support from ETH Zurich and the Swiss National Science Foundation (Grant number: 205321/176011/1).References
- P. Schineis, P. Runge and C. Halin, Vasc. Pharmacol., 2019, 112, 31–41 CrossRef CAS.
- D. Masopust and J. M. Schenkel, Nat. Rev. Immunol., 2013, 13, 309–320 CrossRef CAS PubMed.
- L. Lei, D. Huang, H. Gao, B. He, J. Cao and N. A. Peppas, Sci. Adv., 2022, 8, eadc8738 CrossRef CAS PubMed.
- A. Chauveau, G. Pirgova, H.-W. Cheng, A. De Martin, F. Y. Zhou, S. Wideman, J. Rittscher, B. Ludewig and T. I. Arnon, Immunity, 2020, 52, 794–807.e7 CrossRef CAS PubMed.
- M. J. Ludford-Menting, J. Oliaro, F. Sacirbegovic, E. T.-Y. Cheah, N. Pedersen, S. J. Thomas, A. Pasam, R. Iazzolino, L. E. Dow, N. J. Waterhouse, A. Murphy, S. Ellis, M. J. Smyth, M. H. Kershaw, P. K. Darcy, P. O. Humbert and S. M. Russell, Immunity, 2005, 22, 737–748 CrossRef CAS.
- J. L. Galeano Niño, S. V. Pageon, S. S. Tay, F. Colakoglu, D. Kempe, J. Hywood, J. K. Mazalo, J. Cremasco, M. A. Govendir, L. F. Dagley, K. Hsu, S. Rizzetto, J. Zieba, G. Rice, V. Prior, G. M. O'Neill, R. J. Williams, D. R. Nisbet, B. Kramer, A. I. Webb, F. Luciani, M. N. Read and M. Biro, eLife, 2020, 9, e56554 CrossRef.
- H. T. Nia, L. L. Munn and R. K. Jain, Science, 2020, 370, eaaz0868 CrossRef CAS.
- J. Renkawitz, A. Kopf, J. Stopp, I. de Vries, M. K. Driscoll, J. Merrin, R. Hauschild, E. S. Welf, G. Danuser, R. Fiolka and M. Sixt, Nature, 2019, 568, 546–550 CrossRef CAS.
- S. Hervé, A. Scelfo, G. Bersano Marchisio, M. Grison, K. Vaidžiulytė, M. Dumont, A. Angrisani, A. Keikhosravi, G. Pegoraro, M. Deygas, G. P. F. Nader, A.-S. Macé, M. Gentili, A. Williart, N. Manel, M. Piel, Y. A. Miroshnikova and D. Fachinetti, Nat. Cell Biol., 2025, 27, 73–86 CrossRef.
- Y. Kalukula, A. D. Stephens, J. Lammerding and S. Gabriele, Nat. Rev. Mol. Cell Biol., 2022, 23, 583–602 CrossRef CAS PubMed.
- M. Le Berre, J. Aubertin and M. Piel, Integr. Biol., 2012, 4, 1406–1414 CrossRef CAS PubMed.
- A. J. Lomakin, C. J. Cattin, D. Cuvelier, Z. Alraies, M. Molina, G. P. F. Nader, N. Srivastava, P. J. Sáez, J. M. Garcia-Arcos, I. Y. Zhitnyak, A. Bhargava, M. K. Driscoll, E. S. Welf, R. Fiolka, R. J. Petrie, N. S. De Silva, J. M. González-Granado, N. Manel, A. M. Lennon-Duménil, D. J. Müller and M. Piel, Science, 2020, 370, eaba2894 CrossRef CAS PubMed.
- M. Raab, M. Gentili, H. de Belly, H. R. Thiam, P. Vargas, A. J. Jimenez, F. Lautenschlaeger, R. Voituriez, A. M. Lennon-Duménil, N. Manel and M. Piel, Science, 2016, 352, 359–362 CrossRef CAS.
- C. M. Denais, R. M. Gilbert, P. Isermann, A. L. McGregor, M. te Lindert, B. Weigelin, P. M. Davidson, P. Friedl, K. Wolf and J. Lammerding, Science, 2016, 352, 353–358 CrossRef CAS.
- J. T. Long and J. Lammerding, Dev. Cell, 2021, 56, 156–158 CrossRef CAS.
- P. Shah, C. M. Hobson, S. Cheng, M. J. Colville, M. J. Paszek, R. Superfine and J. Lammerding, Curr. Biol., 2021, 31, 753–765.e6 CrossRef CAS PubMed.
- M. R. Zanotelli, A. Rahman-Zaman, J. A. VanderBurgh, P. V. Taufalele, A. Jain, D. Erickson, F. Bordeleau and C. A. Reinhart-King, Nat. Commun., 2019, 10, 4185 CrossRef PubMed.
- N. Jain and V. Vogel, Nat. Mater., 2018, 17, 1134–1144 CrossRef CAS.
- I. Bozdag-Turan, R. G. Turan, L. Paranskaya, N. S. Arsoy, C. H. Turan, I. Akin, S. Kische, J. Ortak, H. Schneider, S. Ludovicy, T. Hermann, G. D'Ancona, S. Durdu, A. R. Akar, H. Ince and C. A. Nienaber, J. Transl. Med., 2012, 10, 143 CrossRef PubMed.
- M. P. Nikolova and M. S. Chavali, Bioact. Mater., 2019, 4, 271–292 Search PubMed.
- Y. Qiu, B. Ahn, Y. Sakurai, C. E. Hansen, R. Tran, P. N. Mimche, R. G. Mannino, J. C. Ciciliano, T. J. Lamb, C. H. Joiner, S. F. Ofori-Acquah and W. A. Lam, Nat. Biomed. Eng., 2018, 2, 453–463 CrossRef CAS PubMed.
- C. L. Thompson, S. Fu, H. K. Heywood, M. M. Knight and S. D. Thorpe, Front. Bioeng. Biotechnol., 2020, 8, 602646 CrossRef.
- E. Gasser, E. Su, K. Vaidžiulytė, N. Abbade, H. Cognart, J.-B. Manneville, J.-L. Viovy, M. Piel, J.-Y. Pierga, K. Terao and C. Villard, Lab Chip, 2024, 24, 3930–3944 RSC.
- M. Asghari, S. D. Ivetich, M. K. Aslan, M. Aramesh, O. Melkonyan, Y. Meng, R. Xu, M. Colombo, T. Weiss, S. Balabanov, S. Stavrakis and A. J. deMello, Sci. Adv., 2024, 10, eabj1133 CrossRef CAS PubMed.
- G. Holzner, B. Mateescu, D. van Leeuwen, G. Cereghetti, R. Dechant, S. Stavrakis and A. deMello, Cell Rep., 2021, 34, 108824 CrossRef CAS.
- G. Holzner, S. Stavrakis and A. deMello, Anal. Chem., 2017, 89, 11653–11663 CrossRef CAS.
- O. Otto, P. Rosendahl, A. Mietke, S. Golfier, C. Herold, D. Klaue, S. Girardo, S. Pagliara, A. Ekpenyong, A. Jacobi, M. Wobus, N. Töpfner, U. F. Keyser, J. Mansfeld, E. Fischer-Friedrich and J. Guck, Nat. Methods, 2015, 12, 199–202 CrossRef CAS.
- M. Piergiovanni, V. Galli, G. Holzner, S. Stavrakis, A. DeMello and G. Dubini, Lab Chip, 2020, 20, 2539–2548 RSC.
- M. Urbanska, H. E. Muñoz, J. Shaw Bagnall, O. Otto, S. R. Manalis, D. Di Carlo and J. Guck, Nat. Methods, 2020, 17, 587–593 CrossRef CAS PubMed.
- L. Kaitao, R. William, Y. Zhou and Z. Cheng, Curr. Opin. Biomed. Eng., 2019, 12, 102–110 CrossRef.
- D. L. Harrison, Y. Fang and J. Huang, Front. Phys., 2019, 7, 45 CrossRef PubMed.
- W. Jin, F. Tamzalit, P. K. Chaudhuri, C. T. Black, M. Huse and L. C. Kam, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 19835–19840 CrossRef CAS.
- S. Kumari, S. Curado, V. Mayya and M. L. Dustin, Biochim. Biophys. Acta, 2014, 1838, 546–556 CrossRef CAS PubMed.
- M. Ecker, R. Schregle, N. Kapoor-Kaushik, P. Rossatti, V. M. Betzler, D. Kempe, M. Biro, N. Ariotti, G. M. Redpath and J. Rossy, eLife, 2022, 11, e67550 CrossRef CAS.
- M. Aramesh, D. Stoycheva, I. Sandu, S. J. Ihle, T. Zünd, J.-Y. Shiu, C. Forró, M. Asghari, M. Bernero, S. Lickert, M. Kotowski, S. J. Davis, A. Oxenius, V. Vogel and E. Klotzsch, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2107535118 CrossRef CAS PubMed.
- J. M. Hope, J. A. Dombroski, R. S. Pereles, M. Lopez-Cavestany, J. D. Greenlee, S. C. Schwager, C. A. Reinhart-King and M. R. King, BMC Biol., 2022, 20, 61 CrossRef CAS PubMed.
- M. Adamczyk, J. Bartosińska, D. Raczkiewicz, M. Kowal, A. Surdacka, D. Krasowska, A. Michalak-Stoma and D. Krasowska, Clin. Med., 2023, 12, 6573 CAS.
- M. Huse, Nat. Rev. Immunol., 2017, 17, 679–690 CrossRef CAS.
- J. Hong, C. Ge, P. Jothikumar, Z. Yuan, B. Liu, K. Bai, K. Li, W. Rittase, M. Shinzawa, Y. Zhang, A. Palin, P. Love, X. Yu, K. Salaita, B. D. Evavold, A. Singer and C. Zhu, Nat. Immunol., 2018, 19, 1379–1390 CrossRef CAS.
- E. M. Hatch and M. W. Hetzer, J. Cell Biol., 2016, 215, 27–36 CrossRef CAS PubMed.
- S. Cho, J. Irianto and D. E. Discher, J. Cell Biol., 2017, 216, 305–315 CrossRef CAS.
- C. M. Denais, R. M. Gilbert, P. Isermann, A. L. McGregor, M. te Lindert, B. Weigelin, P. M. Davidson, P. Friedl, K. Wolf and J. Lammerding, Science, 2016, 352, 353–358 CrossRef CAS.
- V. Vogel and M. Sheetz, Nat. Rev. Mol. Cell Biol., 2006, 7, 265–275 CrossRef CAS.
- F. Martino, A. R. Perestrelo, V. Vinarský, S. Pagliari and G. Forte, Front. Physiol., 2018, 9, 824 CrossRef PubMed.
- H. T. McMahon and E. Boucrot, J. Cell Sci., 2015, 128, 1065–1070 CrossRef CAS PubMed.
- H. De Belly, A. Stubb, A. Yanagida, C. Labouesse, P. H. Jones, E. K. Paluch and K. J. Chalut, Cell Stem Cell, 2021, 28, 273–284.e6 CrossRef CAS.
- S. Huang and D. E. Ingber, Cancer Cell, 2005, 8, 175–176 CrossRef CAS.
- A. Colom, E. Derivery, S. Soleimanpour, C. Tomba, M. D. Molin, N. Sakai, M. González-Gaitán, S. Matile and A. Roux, Nat. Chem., 2018, 10, 1118–1125 CrossRef CAS PubMed.
- J. E. Smith-Garvin, G. A. Koretzky and M. S. Jordan, Annu. Rev. Immunol., 2009, 27, 591–619 CrossRef CAS PubMed.
- E. Moreno, L. Valon, F. Levillayer and R. Levayer, Curr. Biol., 2019, 29, 23–34.e8 CrossRef CAS PubMed.
- D. S. W. Protter and R. Parker, Trends Cell Biol., 2016, 26, 668–679 CrossRef CAS PubMed.
- D. J. Fowell and M. Kim, Nat. Rev. Immunol., 2021, 21, 582–596 CrossRef CAS PubMed.
- I. Linares, K. Chen, A. Saffren, M. Mansouri, V. V. Abhyankar, B. L. Miller, S. Begolo, H. A. Awad and J. L. McGrath, Sci. Rep., 2025, 15, 3227 CrossRef CAS PubMed.
- M. T. Doolin, R. A. Moriarty and K. M. Stroka, Front. Physiol., 2020, 11 Search PubMed.
- S. Upadhaya, J. X. Yu, M. Shah, D. Correa, T. Partridge and J. Campbell, Nat. Rev. Drug Discovery, 2021, 20, 503–504 CrossRef CAS PubMed.
- M. Aramesh, D. Yu, M. Essand and C. Persson, Adv. Funct. Mater., 2024, 34, 2400487 CrossRef CAS.
- K. Vijayakumar, S. Gulati, A. J. deMello and J. B. Edel, Chem. Sci., 2010, 1, 447–452 RSC.
- X. Cao, T. Buryska, T. Yang, J. Wang, P. Fischer, A. Streets, S. Stavrakis and A. deMello, Lab Chip, 2023, 23, 2029–2038 RSC.
| This journal is © The Royal Society of Chemistry 2026 |