Open Access Article
Open Access ArticleResonant acoustic mixing enables solvent-less amide coupling in solid-phase peptide synthesis
Alice
Nanni
 ,
Panayiotis
Bilalis
,
Panayiotis
Bilalis
 and
Magnus
Rueping
and
Magnus
Rueping
 *
*
KAUST Catalysis Center (KCC), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia. E-mail: magnus.rueping@kaust.edu.sa
First published on 30th October 2025
Abstract
Solid-phase peptide synthesis (SPPS) is the backbone of modern peptide production. However, it relies heavily on relatively toxic solvents and generates significant waste, limiting its sustainability and scalability. To address these limitations, we report the first fully solvent-less peptide coupling protocol for SPPS enabled by Resonant Acoustic Mixing (RAM), representing a step toward greener peptide manufacturing. This method eliminates bulk solvent use, reagent pre-dissolution, and pre-activation during coupling by using mechanical agitation to drive efficient amide bond formation. Optimized conditions (95g acceleration, 5 min, 1.5 equiv. Fmoc-amino acids) afford rapid and clear reactions with high conversion and purity. Notably, no external solvent is added during coupling; instead, residual solvent retained from resin pre-swelling creates a localized microenvironment sufficient for in situ activation. Compared to conventional SPPS, this protocol significantly reduces solvent and reagent use, reaction time, and waste. Process Mass Intensity (PMI) calculations show clear improvements, highlighting the method's environmental and economic benefits. This approach was validated by synthesizing two bioactive peptides (IKVAV and Angiotensin 1–7) in high yield and purity, and further demonstrated excellent scalability in a tenfold scale-up.
Green foundation1. Our work introduces a solvent-less amide coupling for solid-phase peptide synthesis (SPPS), addressing one of the most environmentally burdensome aspects of peptide production. By utilizing Resonant Acoustic Mixing (RAM), a non-contact, high-efficiency mechanical mixing technique we eliminate the need for: (a) bulk solvent use, (b) reagent pre-dissolution and preactivation and (c) excess amino acid and reagent consumption typically required in conventional SPPS. This reduces the environmental footprint, chemical waste, and process mass intensity (PMI). In doing so, we establish a scalable platform that maintains the efficiency and versatility of SPPS while significantly improving its sustainability.2. Our most significant green chemistry achievement lies in the development of a solvent-less amide coupling protocol for solid-phase peptide synthesis (SPPS) in RAM. Quantitatively, this method demonstrated a 5-fold reduction in Process Mass Intensity (PMI). This dramatic improvement reflects substantial savings in solvents and reagents. Notably, the coupling steps require no added solvent whatsoever; only the small amount of DMF retained from resin swelling is sufficient to facilitate in situ activation. Furthermore, reagent use was minimized by optimizing the protocol to require 1.5 equivalents of amino acids, down from the 3 to 5 equivalents commonly used, without sacrificing reaction efficiency. Each coupling cycle was reduced to only five minutes, delivering products in excellent crude purity (up to 99%) and high yields. These results were consistently reproduced across short (KAV), medium (IKVAV), and longer peptide sequences (Angiotensin 1–7), as well as successfully scaled up to 10 mmol, underscoring both the robustness and scalability of the approach. 3. The RAM-enabled protocol marks a major step toward greener peptide synthesis. However, several avenues remain to further enhance its sustainability. Currently, solvent-less conditions are limited to the coupling steps, whereas deprotection and washing still require some solvent. Developing alternative, greener reagents or solvent-free protocols for these steps would greatly reduce overall solvent dependency. Overall, continued innovation in these areas holds the potential to establish a more comprehensive green platform for peptide manufacturing. |
Introduction
Peptides have emerged as an essential class of bioactive molecules, playing a key role in therapeutics, diagnostics, and materials science.1–3 Their unique ability to target biological systems with high affinity and selectivity has driven the rapid growth of peptide-based medicines, with more than one hundred approved drugs and many in development.4 By bridging the gap between small molecules and biologics, peptides offer tunable structures with strong biological activity, making them attractive candidates for modern healthcare solutions.5–7 However, the production of peptides often carries a significant environmental burden, largely due to solvent-intensive synthetic methods and high reagent consumption.8Since its introduction by Merrifield in 1963,9 solid-phase peptide synthesis (SPPS) has remained the gold standard for peptide production for both academic research and industrial manufacturing. This technique allows efficient stepwise chain elongation on an insoluble resin, enabling rapid synthesis, straightforward purification, and broad compatibility with complex sequences, and has therefore become the backbone of modern peptide synthesis.10–12 Despite its advantages, however, SPPS presents serious sustainability challenges. The method relies heavily on more hazardous polar aprotic solvents, which increasingly face regulatory pressure due to their higher toxicity and difficulty of disposal safety.13 In addition, SPPS typically requires large excesses of amino acids and coupling reagents, resulting in high waste generation, elevated process mass intensities (PMIs), and increased production costs.14,15
In recent years, various strategies have been developed to make SPPS more sustainable. Microwave-assisted SPPS synthesis has reduced reaction times, while alternative solvent systems have aimed to lower toxicity without compromising solubility or resin swelling.16–23 Technologies such as ultrasound-assisted synthesis, continuous-flow processes,24 and mechanochemical methods such as ball milling have introduced new ways to reduce solvent use and improve reaction efficiency.25–32 Progress has also been made in reducing solvent consumption during washing and deprotection, the most solvent-intensive stages of SPPS.33,34 However, many of these approaches still rely on solvent-intensive coupling steps, require reagent pre-dissolution and activation, and face scalability limitations or resin compatibility issues.35–39
To address these limitations, we introduce Resonant Acoustic Mixing (RAM) as a new platform for solvent-less peptide coupling under mild and scalable conditions. RAM is a non-contact mixing technology that uses low-frequencies acoustic energy (∼60 Hz) to generate efficient bulk motion and localized micro-mixing zones throughout the reaction vessel.40 Unlike ball milling, RAM operates without grinding media, offering precise control over mixing intensity through adjustable acceleration (up to 100g).41–46 Although RAM has shown great potential in other areas of mechanochemistry, including cocrystal formation, metal–organic framework synthesis, cross-coupling and depolymerisation reactions,47–53 its application in SPPS remains unexplored.
Herein, we report the first practical and scalable RAM-based approach for solvent-less peptide coupling in SPPS. The method eliminates the need for bulk solvent use, reagent pre-dissolution and pre-activation, relying solely on the residual swelling medium retained in the resin to support in situ activation and diffusion. We developed a custom RAM-compatible reaction setup, validating resin integrity, and optimized reaction parameters to achieve rapid and efficient coupling, even for longer sequences up to seven amino acids. We further demonstrate scalability up to 10 mmol without compromising performance. This work establishes RAM as a rubust platform for greener solid-phase peptide synthesis, offering substantial reductions in solvent use, chemical waste, production costs, and processing time, while maintaining the reliability and efficiency of SPPS. Moreover, the future integration of RAM with an automated platform holds promise for future automation of SPPS processes.54,55
Results and discussion
Experimental setup design and evaluation of resin bead integrity under RAM conditions
A critical requirement for any SPPS-compatible mixing technique is preserving the integrity of the resin beads during agitation. Excessive mechanical stress can damage the beads, compromising synthesis efficiency and reproducibility.52,56To assess whether RAM preserves resin morphology, commercially available Fmoc-Rink Amide MBHA resin (loading: 0.664 mmol g−1, scale: 0.3 mmol) was swollen in 1.5 mL of N,N-dimethylformamide (DMF) and subjected to RAM at 100g for 30 minutes. Polypropylene fritted syringes used in these reactions are incompatible with the standard RAM vial holder. Therefore, a custom-made sample holder was designed in CAD and 3D printed to securely and reproducibly mount the 10 mL disposable polypropylene fritted syringe into the LabRAM II instrument. The syringe was sealed with a rubber stopper at the top and a cap at the bottom, to create a closed, RAM-compatible system (Fig. 1a–c). This holder can also be adapted and used of non-standard reaction vessels (e.g., syringes) and allows multiple samples to be processed simultaneously, increasing the method's throughput and flexibility (Fig. 1d). The resin morphology was examined using high-speed digital microscopy and compared to untreated control samples (Fig. 1e). RAM-treated beads retained their spherical shape with no observable surface damage (Fig. 1f), indicating that RAM preserves resin integrity under these conditions. In contrast, when the same resin was treated under ball milling conditions (30 Hz, 30 min) the obtained resin revealed notable micro-pitting and surface degradation, attributed to the compressive forces of the grinding media (Figure S2). These results confirm that RAM provides efficient, non-destructive mixing without abrasive or compressive stress, extending resin bead lifetimes and offering a robust, scalable alternative fully compatible with SPPS.
Process optimization using a benchmark tripeptide
Efficient coupling in solid-phase peptide synthesis depends on the diffusion of reagents into the swollen resin matrix and their penetration through its porous network.56 While reagent design and protocol improvements have enhanced SPPS over time, poor mixing remains a key limitation. To overcome this, conventional methods often rely on large reagent excesses and pre-activation or pre-dissolution of coupling agents.57–59 These practices significantly increase chemical waste, especially when using high-value amino acids and polar aprotic solvents.35,60,61Microwave-assisted SPPS protocols frequently employ up to five equivalents of amino acids and coupling reagents in 0.5 M solutions to achieve good yield and purity.62,63 Even on small scales (e.g., 0.3 mmol), this requires substantial solvent volumes and generates a high process mass intensity (PMI) per coupling cycle.15
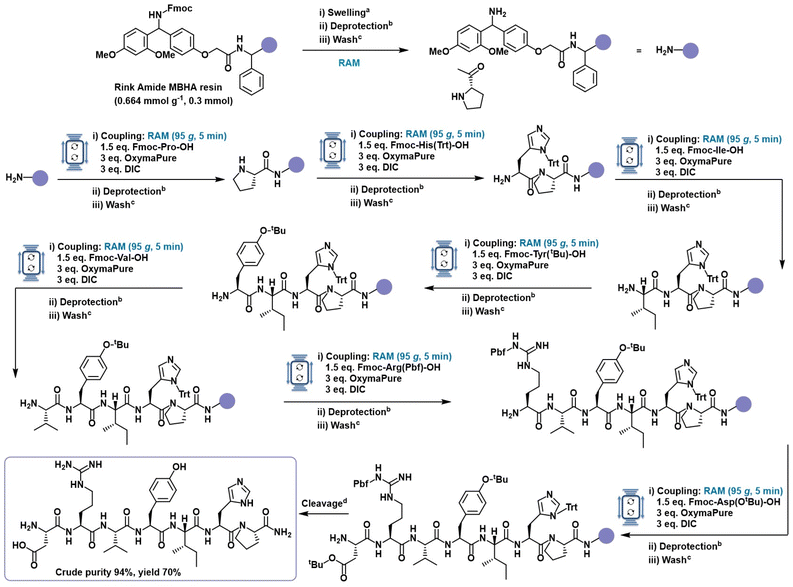
To evaluate and optimize the performance of Resonant Acoustic Mixing for solid-phase peptide synthesis, the tripeptide KAV (Lys–Ala–Val) was selected as a model system. Its combination of hydrophilic, positively charged (Lys), neutral (Ala), and hydrophobic, branched (Val) amino acids, provided a suitable challenge for benchmarking. All couplings were performed using the widely adopted Fmoc (9-fluorenylmethyloxycarbonyl) strategy for temporary N-terminal protection, combined with tert-butyl-based protecting groups for the side-chains. This orthogonal protection scheme allows selective deprotection under basic conditions for the Fmoc removal and acidic conditions for the side chain deprotection and final cleavage.64 The coupling system of N,N′-diisopropylcarbodiimide (DIC) and ethylcyanohydroxyiminoacetate (OxymaPure) was selected for its high coupling efficiency and minimal racemization risk,65–69 ensuring reliable peptide bond formation under all tested conditions (Fig. 2). After each coupling and deprotection step, the resin was washed with DMF to remove the excess reagents and by-products. Detailed synthetic procedure provided in the SI.
Initial coupling experiments were performed on a 0.3 mmol scale using Fmoc-Rink Amide MBHA resin (loading: 0.664 mmol g−1) with four equivalents of Fmoc-amino acids, and coupling reagents in 500 μL of DMF per coupling cycle, in RAM at 80g per 30 minutes. Notably, all reagents were added directly to the pre-swollen resin in solid or neat form without any pre-activation or pre-dissolution, allowing for in situ activation during RAM. This afforded the target tripeptide in 85% yield and 99% crude purity (Table 1, entry 1). When the solvent volume was reduced to 250 μL, the reactions still proceeded efficiently, maintaining the same crude purity (99%) and yielding 84% (Table 1, entry 2). The couplings were next performed under fully solvent-less conditions. Remarkably, despite the absence of externally added solvent, the reaction proceeded efficiently, delivering the target tripeptide in 80% yield and 98% crude purity (Table 1, entry 3). This outcome was enabled by the small amount of DMF retained in the pre-swollen resin, experimentally quantified as ≈500 μL per 0.1 mmol of resin (see the SI). This residual solvent provided a localized, sufficiently solvated microenvironment that, under RAM, facilitated efficient reagent diffusion and in situ activation through high-intensity mixing and enhanced mass transfer. While traditional SPPS protocols rely on solvent not only for resin swelling but also to dissolve reagents and promote diffusion,70–73 these results demonstrate that RAM enables efficient peptide coupling even in the absence of added solvent.
| Entry | DMF [μL] | Time [min] | Acceleration [g] | X equivalent | Y equivalent | Isolated yield (%) | Purity (%) |
|---|---|---|---|---|---|---|---|
| 1 | 500 | 30 | 80 | 4 | 4 | 85 | 99 |
| 2 | 200 | 30 | 80 | 4 | 4 | 84 | 99 |
| 3 | — | 30 | 80 | 4 | 4 | 80 | 98 |
| 4 | — | 10 | 80 | 4 | 4 | 80 | 97 |
| 5 | — | 5 | 80 | 4 | 4 | 79 | 97 |
| 6 | — | 5 | 60 | 4 | 4 | 64 | 84 |
| 7 | — | 5 | 95 | 4 | 4 | 84 | 99 |
| 8 | — | 5 | 95 | 3 | 3 | 81 | 99 |
| 9 | — | 5 | 95 | 1.5 | 1.5 | 61 | 81 |
| 10 | — | 5 | 95 | 1.5 | 3 | 84 | 99 |
Having demonstrated that coupling can proceed under solvent-less conditions, we next explored whether RAM could also accelerate the process without compromising efficiency. When the coupling time was reduced from 30 to 10 minutes, and further to just 5 minutes, using 4 equivalents of each reagent under solvent-less conditions, crude purity remained high (97–97%) and yields were comparable (80–79%, entries 4 and 5). The influence of mixing intensity was next examined by performing the coupling at 60g and 95g under the same solvent-less, 5-minute conditions. At 60g, the reaction gave incomplete conversion (64%, entry 6) and formation of a Lys–Ala deletion product (see the SI), suggesting inefficient mixing and poor reagent diffusion. Increasing the acceleration to 95g restored complete conversion and yielded 84% of the desired tripeptide with 99% crude purity (entry 7). These results highlight the importance of tuning the mechanical energy input to enhance reagent–resin interactions by rapid mixing and in situ activation. Lastly, the reagent stoichiometry was also optimized. Reducing all components to 3 equivalents still resulted in high conversion (81%) and 99% crude purity (entry 8). However, lowering the equivalents of both amino acids and coupling reagents to 1.5 led to incomplete coupling and the formation of deletion sequence (Lys–Ala) as a side product (entry 9). Interestingly, when only Fmoc-amino acids were reduced to 1.5 equivalents while maintaining 3 equivalents of DIC and OxymaPure, coupling proceeded efficiently, affording the tripeptide in 84% yield and 99% crude purity (entry 10).
Based on this evaluation, the optimized conditions for RAM-assisted SPPS were defined as 1.5 equivalent of Fmoc-amino acid, 3 equivalents of DIC and OxymaPure, 5 minutes of mixing at 95g, and no added solvent or pre-activation. This optimized protocol maintains high yield and crude purity while reducing solvent use, reaction time, and reagent input up to 60% less of amino acid consumption compared to the conventional SPPS protocol,74 underscoring the capability of RAM to achieve efficient coupling using only the residual swelling medium. These findings demonstrate that even the traditionally resource-intensive coupling step in SPPS can be effectively streamlined without compromising performance, marking a significant advance toward greener and more sustainable peptide manufacturing.
Protocol extension to bioactive peptides: synthesis of IKVAV and Angiotensin (1–7)
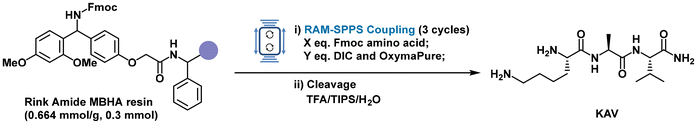
To further demonstrate the validity of the optimized coupling protocol under RAM conditions, the methodology was extended to the synthesis of more complex bioactive peptides. The first target was the pentapeptide IKVAV (Ile–Lys–Val–Ala–Val), a laminin-derived sequence known for its role in cell adhesion, neurite outgrowth, and tissue engineering applications (Fig. 3).75–79 Synthesis was performed on a 0.3 mmol scale using Fmoc-Rink Amide MBHA resin (loading: 0.664 mmol g−1). Each coupling step used 1.5 equivalents of Fmoc-amino acids and 3 equivalents of DIC and OxymaPure, added directly to the resin in solid or neat form without pre-activation or dissolution. In situ activation was enabled by the residual solvent retained in the pre-swollen resin. Each coupling was carried out for 5 minutes at 95g using the LabRAM II instrument. Fmoc deprotection was performed with 20% piperidine in DMF under RAM at 80g for 5 minutes, followed by standard steps. After cleavage and precipitation, the crude peptide was analysed by LC-MS and NMR, confirming excellent crude purity (95%) and isolated yield (81%). Importantly, no deletion sequences were observed, highlighting the method's ability to deliver full-length sequences with high fidelity under solvent-less conditions (see the SI).To further test the robustness of the method with a longer and more complex sequence, the protocol was applied to the heptapeptide Angiotensin 1–7 (Asp–Arg–Val–Tyr–Ile–His–Pro), a bioactive hormone of the renin-angiotensin system with therapeutic potential in cardiovascular and pulmonary diseases, including complications related to COVID-19.80,81 This sequence presents synthetic challenges, including the risk of aspartimide formation from Asp, the potential for His epimerization, and the steric inderance of Pro at the C-terminus.82,83 It also includes side-chain protected residues such as Arg(Pbf), His(Trt), and Tyr(tBu), which further test the efficiency of coupling and the method's compatibility with diverse protecting groups. Using the same RAM protocol employed for IKVAV, identical conditions for coupling, deprotection, and cleavage, Angiotensin (1–7) was synthesized in high crude purity (94%) and good isolated yield (70%) (Fig. 4). LC-MS and NMR analyses confirmed the absence of deletion sequences or side products, demonstrating the method's ability to deliver full-length peptides with high sequence integrity even for longer and more sensitive sequences (see the SI).
Scalable coupling of model tripeptide via RAM
To demonstrate the scalability of the optimized RAM-assisted SPPS protocol, the synthesis of the model tripeptide Lys–Ala–Val (KAV) was scaled up from 0.3 mmol to 10 mmol, representing a 30-fold increase. Notably, this scale exceeds the typical capacity of most automated microwave-assisted synthesizers, which are usually limited to a batches of up to 2 mmol, and only up to 5 mmol on specialized systems.29 The synthesis was carried out using the same optimized conditions developed at small scale: 1.5 equivalents of each Fmoc-amino acid, 3 equivalents of DIC and OxymaPure, 5 minutes of coupling time, and 95g acoustic acceleration using the LabRAM II instrument. For this larger batch, a 250 mL Pyrex jar was used as the reaction vessel to provide adequate space and ensure efficient mixing across the full resin bed. Unlike the 10 mL syringe setup used in small-scale experiments, our current set-up involves a Pyrex jar lacking an integrated filtration system (see the SI). Importantly, the coupling steps remained fully solvent-less throughout the process. All reagents were added directly in solid or neat form, with deprotection and washing steps carried out manually between cycles. Despite the increased volume, the synthesis proceeded smoothly with no signs of compromised mixing or reaction performance. After cleavage and precipitation, the crude peptide was analysed by LC-MS and NMR, confirming a high isolated yield of 71% and crude purity of 99%, consistent with the results obtained at the 0.3 mmol scale. This successful scale-up demonstrate the ability of RAM to translate efficient solvent-less coupling to gram-scale production. In contrast to the microwave-assisted system, which often face challenges in scaling due to limited heating uniformity and irradiation penetration in handling larger volumes,37 the RAM provides uniform energy distribution and effective mass transfer regardless the scale. Together, these results support the potential of RAM as a scalable, sustainable, and industrially relevant platform for solid-phase peptide synthesis.Environmental impact assessment: process mass intensity
To evaluate the environmental performance of the optimized RAM-assisted SPPS protocol, the Process Mass Intensity (PMI) was calculated. PMI, a widely used green chemistry metric, representing the total mass of all input materials used in the process relative to the mass of the final product, serving as a direct indicator of material efficiency.84,85 For the synthesis of the model tripeptide Lys–Ala–Val (KAV) on a 0.3 mmol scale, the RAM-based SPPS process was calculated to be 953. This value includes all the reagents, solvent used for resin swelling, washing, and deprotection, but excludes materials used during cleavage and final isolation (see the SI). For comparison, a conventional microwave-assisted SPPS process using the same amino acids and coupling reagents produced a considerably higher PMI of 4695. This five-fold improvement clearly demonstrates the reduced material consumption achieved with RAM-SPPS. These results highlight the enhanced efficiency and lower environmental footprint of the RAM approach, particularly in terms of solvent and reagent use. Even at the early stages of process development, RAM offers a more sustainable alternative to traditional SPPS methods and supports its potential greener, scalable peptide manufacturing.Conclusions
This study presents the first solvent-less peptide coupling protocol for solid-phase peptide synthesis (SPPS) using Resonant Acoustic Mixing (RAM). By eliminating the need for added solvent in the coupling, reagent pre-dissolution, and pre-activation, the method achieved efficient peptide bond formation through mechanical mixing alone. Under optimized peptide coupling conditions, RAM delivers high conversion and purity using reduced reagent amounts and no external solvent, relying on the residual solvent in the swollen resin to support in situ activation. The protocol was successfully applied to the synthesis of bioactive peptides IKVAV and Angiotensin (1–7), and scaled up to 10 mmol for the model tripeptide Lys–Ala–Val, maintaining high product quality throughout the full process. The environmental benefits of the RAM-SPPS technique was quantified by using the PMI green chemistry metric, showing a five-fold reduction compared to traditional methods. This work establishes a strong foundation for solvent-less SPPS and supports further development toward more sustainable peptide synthesis.Author contributions
A. N. performed all experiments, analyzed the data, and prepared the initial draft of the manuscript. P. B. and M. R. provided guidance during manuscript preparation and contributed to reviewing and editing the final version. All authors approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data that support the findings of this study are available in the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5gc04067a.Acknowledgements
This work was financially supported by the King Abdullah University of Science and Technology (KAUST), Saudi Arabia, Office of Sponsored Research (URF/1/4405 and URF/1/4726). We acknowledge Dr Srinivas Banala for helpful discussions.References
- L. Wang, N. Wang, W. Zhang, X. Cheng, Z. Yan, G. Shao, X. Wang, R. Wang and C. Fu, Signal Transduction Targeted Ther., 2022, 7, 1–27 CrossRef PubMed.
- W. Xiao, W. Jiang, Z. Chen, Y. Huang, J. Mao, W. Zheng, Y. Hu and J. Shi, Signal Transduction Targeted Ther., 2025, 10, 74 CrossRef CAS PubMed.
- X. Ji, A. L. Nielsen and C. Heinis, Angew. Chem., Int. Ed., 2024, 63, e202308251 CrossRef CAS PubMed.
- V. D'Aloisio, P. Dognini, G. A. Hutcheon and C. R. Coxon, Drug Discovery Today, 2021, 26, 1409–1419 CrossRef PubMed.
- B. G. De La Torre and F. Albericio, Molecules, 2020, 25, 2293 CrossRef CAS PubMed.
- D. J. Drucker, Nat. Rev. Drug Discovery, 2020, 19, 277–289 CrossRef CAS.
- K. Fosgerau and T. Hoffmann, Drug Discovery Today, 2015, 20, 122–128 CrossRef CAS PubMed.
- Y. E. Jad, A. Kumar, A. El-Faham, B. G. de la Torre and F. Albericio, ACS Sustainable Chem. Eng., 2019, 7, 3671–3683 CrossRef CAS.
- R. B. Merrifield, J. Am. Chem. Soc., 1963, 85, 2149–2154 CrossRef CAS.
- A. J. Mijalis, D. A. Thomas III, M. D. Simon, A. Adamo, R. Beaumont, K. F. Jensen and B. L. Pentelute, Nat. Chem. Biol., 2017, 13, 464–468 CrossRef CAS PubMed.
- A. Isidro-Llobet, M. N. Kenworthy, S. Mukherjee, M. E. Kopach, K. Wegner, F. Gallou, A. G. Smith and F. Roschangar, J. Org. Chem., 2019, 84, 4615–4628 CrossRef CAS PubMed.
- B. L. Bray, Nat. Rev. Drug Discovery, 2003, 2, 587–593 CrossRef CAS.
- V. Martin, P. H. G. Egelund, H. Johansson, S. T. Le Quement, F. Wojcik and D. S. Pedersen, RSC Adv., 2020, 10, 42457–42492 RSC.
- M. H. A. Somehsaraie, V. F. Vavsari, M. Kamangar and S. Balalaie, Iran. J. Pharm. Res., 2022, 21, e123879 CAS.
- I. Kekessie, K. Wegner, I. Martinez, M. E. Kopach, T. D. White, J. K. Tom, M. N. Kenworthy, F. Gallou, J. Lopez, S. G. Koenig, P. R. Payne, S. Eissler, B. Arumugam, C. Li, S. Mukherjee, A. Isidro-Llobet, O. Ludemann-Hombourger, P. Richardson, J. Kittelmann, D. S. Pedersen and L. J. van den Bos, J. Org. Chem., 2024, 89, 4261–4282 CrossRef CAS PubMed.
- S. B. Lawrenson, R. Arav and M. North, Green Chem., 2017, 19, 1685–1691 RSC.
- D. S. MacMillan, J. Murray, H. F. Sneddon, C. Jamieson and A. J. B. Watson, Green Chem., 2013, 15, 596–600 RSC.
- K. Wegner, D. Barnes, K. Manzor, A. Jardine and D. Moran, Green Chem. Lett. Rev., 2021, 14, 153–164 CrossRef CAS.
- G. Vivenzio, M. C. Scala, G. Auriemma, C. Sardo, P. Campiglia and M. Sala, Green Chem. Lett. Rev., 2024, 17, 2404234 CrossRef.
- Y. E. Jad, G. A. Acosta, T. Govender, H. G. Kruger, A. El-Faham, B. G. de la Torre and F. Albericio, ACS Sustainable Chem. Eng., 2016, 4, 6809–6814 CrossRef CAS.
- L. Ferrazzano, D. Corbisiero, G. Martelli, A. Tolomelli, A. Viola, A. Ricci and W. Cabri, ACS Sustainable Chem. Eng., 2019, 7, 12867–12877 CrossRef CAS.
- A. Kumar, Y. E. Jad, J. M. Collins, F. Albericio and B. G. de la Torre, ACS Sustainable Chem. Eng., 2018, 6, 8034–8039 CrossRef CAS.
- C. Loffredo, N. A. Assunção, J. Gerhardt and M. T. M. Miranda, J. Pept. Sci., 2009, 15, 808–817 CrossRef CAS PubMed.
- D. Daurio, C. S. Jacobsen, K. Nagapudi, R. Saw, M. V. S. Elipe, O. Thiel, R. Balgley, S. P. Chamarthy and F. Alvarez-Nunez, J. Pharm. Sci., 2025, 103941 CrossRef PubMed.
- A. Wróblewska, I. I. Bak-Sypien, P. Paluch, E. Wielgus, J. Zając, A. Jeziorna, S. Kaźmierski and M. J. Potrzebowski, Chem. – Eur. J., 2024, 30, e202400177 CrossRef PubMed.
- A. Rios-Echeverri, C. E. Puerto-Galvis, K. J. Ardila-Fierro and J. G. Hernández, ChemSusChem, 2025, 18, e202402446 CrossRef CAS PubMed.
- V. Declerck, P. Nun, J. Martinez and F. Lamaty, Angew. Chem., Int. Ed., 2009, 48, 9318–9321 CrossRef CAS PubMed.
- Y. Yeboue, M. Jean, G. Subra, J. Martinez, F. Lamaty and T.-X. Métro, Org. Lett., 2021, 23, 631–635 CrossRef CAS.
- G. Sabatino, A. D'Ercole, L. Pacini, M. Zini, A. Ribecai, A. Paio, P. Rovero and A. M. Papini, Org. Process Res. Dev., 2021, 25, 552–563 CrossRef CAS.
- S. Mottola, A. D. Bene, V. Mazzarella, R. Cutolo, I. Boccino, F. Merlino, S. Cosconati, S. D. Maro and A. Messere, Ultrason. Sonochem., 2025, 114, 107257 CrossRef CAS PubMed.
- C. Bolm and J. G. Hernández, ChemSusChem, 2018, 11, 1410–1420 CrossRef CAS PubMed.
- E. Juaristi and C. G. Avila-Ortiz, Synthesis, 2023, 55, 2439–2459 CrossRef CAS.
- J. M. Collins, S. K. Singh, T. A. White, D. J. Cesta, C. L. Simpson, L. J. Tubb and C. L. Houser, Nat. Commun., 2023, 14, 8168 CrossRef CAS PubMed.
- A. Kumar, A. Sharma, B. G. de la Torre and F. Albericio, Green Chem., 2022, 24, 4887–4896 RSC.
- L. Ferrazzano, M. Catani, A. Cavazzini, G. Martelli, D. Corbisiero, P. Cantelmi, T. Fantoni, A. Mattellone, C. D. Luca, S. Felletti, W. Cabri and A. Tolomelli, Green Chem., 2022, 24, 975–1020 RSC.
- O. Maurin, P. Verdié, G. Subra, F. Lamaty, J. Martinez and T.-X. Métro, Beilstein J. Org. Chem., 2017, 13, 2087–2093 CrossRef CAS.
- A. D. L. Hoz, J. Alcázar, J. Carrillo, M. A. Herrero, J. D. M. Muñoz, P. Prieto, A. D. Cózar, A. Diaz-Ortiz, A. D. L. Hoz, J. Alcázar, J. Carrillo, M. A. Herrero, J. D. M. Muñoz, P. Prieto, A. D. Cózar and A. Diaz-Ortiz, in Microwave Heating, IntechOpen, 2011 Search PubMed.
- Y. F. Varela, M. Vanegas-Murcia and M. E. Patarroyo, Molecules, 2018, 23, 2877 CrossRef.
- S. L. Pedersen, A. P. Tofteng, L. Malik and K. J. Jensen, Chem. Soc. Rev., 2012, 41, 1826–1844 RSC.
- I. U. Khan, R. Guo, U. Farooq, S. Adhikari and H. Zhou, Processes, 2023, 11, 266 CrossRef.
- K. Nagapudi, E. Y. Umanzor and C. Masui, Int. J. Pharm., 2017, 521, 337–345 CrossRef CAS PubMed.
- A. A. L. Michalchuk, K. S. Hope, S. R. Kennedy, M. V. Blanco, E. V. Boldyreva and C. R. Pulham, Chem. Commun., 2018, 54, 4033–4036 RSC.
- H. M. Titi, J.-L. Do, A. J. Howarth, K. Nagapudi and T. Friščić, Chem. Sci., 2020, 11, 7578–7584 RSC.
- F. Effaty, L. Gonnet, S. G. Koenig, K. Nagapudi, X. Ottenwaelder and T. Friščić, Chem. Commun., 2023, 59, 1010–1013 RSC.
- L. Gonnet, T. H. Borchers, C. B. Lennox, J. Vainauskas, Y. Teoh, H. M. Titi, C. J. Barrett, S. G. Koenig, K. Nagapudi and T. Friščić, Faraday Discuss., 2023, 241, 128–149 RSC.
- M. Wohlgemuth, S. Schmidt, M. Mayer, W. Pickhardt, S. Grätz and L. Borchardt, Chem. – Eur. J., 2023, 29, e202301714 CrossRef CAS PubMed.
- A. Nanni, D. Kong, C. Zhu and M. Rueping, Green Chem., 2024, 26, 8341–8347 RSC.
- A. Dey, R. Kancherla, K. Pal, N. Kloszewski and M. Rueping, Commun. Chem., 2024, 7, 295 CrossRef CAS PubMed.
- J.-L. Do and T. Friščić, ACS Cent. Sci., 2017, 3, 13–19 CrossRef CAS PubMed.
- A. S. Makarov and M. Rueping, Green Chem., 2025, 27, 716–721 RSC.
- D. Kong, L. Yi, A. Nanni and M. Rueping, Nat. Commun., 2025, 16, 3983 CrossRef CAS PubMed.
- I. Alshanski, M. Bentolila, A. Gitlin-Domagalska, D. Zamir, S. Zorsky, S. Joubran, M. Hurevich and C. Gilon, Org. Process Res. Dev., 2018, 22, 1318–1322 CrossRef CAS.
- E. Hamzehpoor, F. Effaty, T. H. Borchers, R. S. Stein, A. Wahrhaftig-Lewis, X. Ottenwaelder, T. Friščić and D. F. Perepichka, Angew. Chem., Int. Ed., 2024, 63, e202404539 CrossRef CAS PubMed.
- M. Seifrid, F. Strieth-Kalthoff, M. Haddadnia, T. C. Wu, E. Alca, L. Bodo, S. Arellano-Rubach, N. Yoshikawa, M. Skreta, R. Keunen and A. Aspuru-Guzik, Digital Discovery, 2024, 3, 1319–1326 RSC.
- M. C. Martin, G. M. Goshu, J. R. Hartnell, C. D. Morris, Y. Wang and N. P. Tu, Org. Process Res. Dev., 2019, 23, 1900–1907 CrossRef CAS.
- M. Amblard, J.-A. Fehrentz, J. Martinez and G. Subra, Mol. Biotechnol., 2006, 33, 239–254 CrossRef CAS PubMed.
- D. Grunhaus, E. R. Molina, R. Cohen, T. Stein, A. Friedler and M. Hurevich, Org. Process Res. Dev., 2022, 26, 2492–2497 CrossRef CAS PubMed.
- J. Dasan, Amino Acids, 2018, 50, 39–68 CrossRef.
- J. M. Palomo, RSC Adv., 2014, 4, 32658–32672 RSC.
- J. N. Tawara, A. Blokhin, T. A. Foderaro, F. R. Stermitz and H. Hope, J. Org. Chem., 1993, 58, 4813–4818 CrossRef CAS.
- A. Banerjee and H. Onyuksel, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2012, 4, 562–574 CAS.
- A. Öhlander, C. Lüdtke, A. Sahakjan and R. E. Johnsson, J. Pept. Sci., 2024, 30, e3612 CrossRef PubMed.
- P. Hivare, A. Gangrade, G. Swarup, K. Bhavsar, A. Singh, R. Gupta, P. Thareja, S. Gupta and D. Bhatia, Nanoscale, 2022, 14, 8611–8620 RSC.
- O. A. Musaimi, B. G. de la Torre and F. Albericio, Green Chem., 2020, 22, 996–1018 RSC.
- K. E. Ruhl, M. J. Di Maso, H. B. Rose, D. M. Schultz, F. Lévesque, S. T. Grosser, S. M. Silverman, S. Li, N. Sciammetta and U. F. Mansoor, Org. Process Res. Dev., 2024, 28, 2896–2905 CrossRef CAS.
- D. Anguraj, S. Chaudhary, M. Pasupuleti, L. R. Singh and A. Sharma, Int. J. Pept. Res. Ther., 2025, 31, 63 CrossRef CAS.
- R. Subirós-Funosas, R. Prohens, R. Barbas, A. El-Faham and F. Albericio, Chem. – Eur. J., 2009, 15, 9394–9403 CrossRef PubMed.
- S. R. Manne, A. Sharma, A. Sazonovas, A. El-Faham, B. G. de la Torre and F. Albericio, ACS Omega, 2022, 7, 6007–6023 CrossRef CAS PubMed.
- A. Orlandin, I. Guryanov, L. Ferrazzano, B. Biondi, F. Biscaglia, C. Storti, M. Rancan, F. Formaggio, A. Ricci and W. Cabri, Molecules, 2022, 27, 4235 CrossRef CAS PubMed.
- S. B. H. Kent, J. Pept. Sci., 2025, 31, e70013 CrossRef CAS PubMed.
- S. Lawrenson, M. North, F. Peigneguy and A. Routledge, Green Chem., 2017, 19, 952–962 RSC.
- V. Martin, S. Jadhav, P. H. G. Egelund, R. Liffert, H. J. Castro, T. Krüger, K. F. Haselmann, S. T. L. Quement, F. Albericio, F. Dettner, C. Lechner, R. Schönleber and D. S. Pedersen, Green Chem., 2021, 23, 3295–3311 RSC.
- C. Amadi-Kamalu, H. Clarke, M. McRobie, J. Mortimer, M. North, Y. Ran, A. Routledge, D. Sibbald, M. Tickias, K. Tse and H. Willway, ChemistryOpen, 2020, 9, 431–441 CrossRef CAS PubMed.
- I. M. Mándity, B. Olasz, S. B. Ötvös and F. Fülöp, ChemSusChem, 2014, 7, 3172–3176 CrossRef PubMed.
- M. Nomizu, B. S. Weeks, C. A. Weston, W. H. Kim, H. K. Kleinman and Y. Yamada, FEBS Lett., 1995, 365, 227–231 CrossRef CAS PubMed.
- R. Patel, M. Santhosh, J. K. Dash, R. Karpoormath, A. Jha, J. Kwak, M. Patel and J. H. Kim, Polym. Adv. Technol., 2019, 30, 4–12 CrossRef CAS.
- Y. Yin, W. Wang, Q. Shao, B. Li, D. Yu, X. Zhou, J. Parajuli, H. Xu, T. Qiu, A. K. Yetisen and N. Jiang, Biomater. Sci., 2021, 9, 2887–2892 RSC.
- F.-Y. Wu and H.-C. Lin, Molecules, 2022, 27, 4115 CrossRef CAS PubMed.
- Z. B. Y. ÇeviK, B. Köken and O. Karaman, Int. Adv. Res. Eng. J., 2023, 7, 97–102 CrossRef.
- M. Gheblawi, K. Wang, A. Viveiros, Q. Nguyen, J.-C. Zhong, A. J. Turner, M. K. Raizada, M. B. Grant and G. Y. Oudit, Circ. Res., 2020, 126, 1456–1474 CrossRef CAS PubMed.
- R. A. S. Santos, A. C. Simões-e-Silva, C. Maric, D. M. R. Silva, R. P. Machado, I. de Buhr, S. Heringer-Walther, S. V. B. Pinheiro, M. T. Lopes, M. Bader, E. P. Mendes, V. S. Lemos, M. J. Campagnole-Santos, H.-P. Schultheiss, R. Speth and T. Walther, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 8258–8263 CrossRef CAS.
- Y. Zhou, H. Li, Y. Huang, J. Li, G. Deng, G. Chen, Z. Xi and C. Zhou, Nat. Commun., 2023, 14, 5324 CrossRef CAS PubMed.
- T. Fantoni, A. Orlandin, I. D. Stefano, M. Macis, A. Tolomelli, A. Ricci, W. Cabri and L. Ferrazzano, Green Chem., 2024, 26, 10929–10939 RSC.
- R. A. Sheldon, ACS Sustainable Chem. Eng., 2018, 6, 32–48 CrossRef CAS.
- E. R. Monteith, P. Mampuys, L. Summerton, J. H. Clark, B. U. W. Maes and C. R. McElroy, Green Chem., 2020, 22, 123–135 RSC.
| This journal is © The Royal Society of Chemistry 2026 |


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) swelling: 4 mL DMF, RAM (80
swelling: 4 mL DMF, RAM (80