In vitro and in vivo anti-inflammatory activity of oenothein B from Eucalyptus leaves and its amelioration mechanism on colitis in mice by regulating fecal microbiota and metabolism
Wei
Li†
c,
Xiaoying
Zhang†
b,
Lu
Xu
b,
Yunjiao
Chen
 b,
Yong
Cao
*b and
Ziyin
Li
b,
Yong
Cao
*b and
Ziyin
Li
 *a
*a
aSchool of Public Health, Guangdong Pharmaceutical University, Guangzhou 510006, China. E-mail: 13751322925@163.com
bCollege of Food Science, South China Agricultural University, Guangdong Provincial Key Laboratory of Nutraceuticals and Functional Foods, Guangdong Research Center for Engineering Technology in Bioactive Natural Products, Guangzhou 510642, China. E-mail: caoyong2181@scau.edu.cn
cInstitute of Biopharmaceutical and Health Engineering, Tsinghua Shenzhen International Graduate School, Shenzhen 518055, China
First published on 26th November 2025
Abstract
Nonvolatile extracts from Eucalyptus leaves possess diverse bioactivities; however, their anti-inflammatory potential and key active components remain insufficiently characterized. In this study, we demonstrated that antioxidant polyphenols extracted from Eucalyptus grandis × E. urophylla (EPEGU) using low-temperature continuous phase transformation extraction (LCPTE) exhibited significant anti-inflammatory effects by suppressing the secretion of nitric oxide (NO), tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6). Furthermore, oenothein B (OEB), isolated from EPEGU, markedly reduced pro-inflammatory cytokine levels and their corresponding mRNA expression in LPS-stimulated RAW264.7 macrophages. In vivo, OEB administration alleviated ulcerative colitis (UC) symptoms in mice, evidenced by attenuation of body weight loss, prevention of colon shortening, reduction of pro-inflammatory mediator secretion, and improvement in spleen weight, disease activity index (DAI), histopathological damage, and oxidative stress markers. Gut microbiota analysis revealed that OEB mitigated dysbiosis by increasing the abundance of beneficial taxa such as Firmicutes, Akkermansia, Lactobacillus, and Ruminococcus, while reducing potentially pathogenic genera including Proteobacteria, Bacteroides, and Escherichia–Shigella. These microbial shifts were associated with alterations in colonic metabolites, primarily involving arachidonic acid and bile acid metabolism. Collectively, these findings indicate that OEB is a promising natural anti-inflammatory agent and potential adjuvant for the prevention and management of inflammatory bowel diseases.
1 Introduction
Ulcerative colitis (UC) is a chronic inflammatory disorder of the colon with a multifactorial etiology involving genetic predisposition, environmental influences, lifestyle factors, and gut microbiota imbalance.1 Clinically, UC is characterized by bloody stools, diarrhea, weight loss, and compromised intestinal health. Alarmingly, both the incidence and prevalence of UC have continued to rise globally.2 Conventional treatments, including immunosuppressants, interferons, and glucocorticoids,3 provide symptomatic relief but fail to achieve a definitive cure. Prolonged use of these agents often results in significant adverse effects and a high risk of disease relapse. Consequently, there is an urgent need for safer and more effective therapeutic strategies for both the prevention and management of UC.Emerging evidence indicates that gut microbiota dysbiosis and associated metabolic disturbances play a pivotal role in UC progression. Dysbiosis is marked by altered microbial composition and abundance,4 including an increased presence of pathogenic bacteria such as Proteobacteria and Bacteroides, alongside a significant reduction in beneficial bacteria such as Lactobacillus and Bifidobacterium.5 This imbalance contributes to metabolic disruptions, notably elevated levels of lipopolysaccharides and reduced short-chain fatty acids, which exacerbate intestinal oxidative stress and inflammation. Maintaining microbial homeostasis is thus considered a promising therapeutic target in UC. Recent studies have shown that natural polyphenols can alleviate UC symptoms by modulating gut microbiota composition.6,7 Identifying polyphenol-rich natural compounds may therefore offer a novel and effective approach for UC prevention and therapy.
The genus Eucalyptus, native to Australia, is widely distributed across tropical and subtropical regions. Various Eucalyptus species have long been used in traditional medicine systems in Australia, China, India, and parts of Europe for treating diverse ailments.8 Notably, several species indigenous to the Dharawal region of Australia have been traditionally employed to manage inflammatory conditions, as documented in the Dharawal Pharmacopeia.9,10 Modern pharmacological studies have extensively demonstrated the anti-inflammatory activity of volatile oils extracted from Eucalyptus leaves.11,12 More recently, research has shifted toward the investigation of nonvolatile extracts, which are rich in bioactive compounds such as polyphenols, flavonoids, terpenoids, and tannins.13,14 These nonvolatile constituents have shown promising antioxidant, antimicrobial, and anti-aging properties. However, the anti-inflammatory potential of these nonvolatile fractions, as well as the identification of specific active components responsible for these effects, remains underexplored. In recent work, our team developed a nonvolatile Eucalyptus leaf extract enriched in phenolic compounds, referred to as Eucalyptus Polyphenol Extract (EPE). EPE has demonstrated strong antioxidant activity, modulation of gut microbiota, and improvements in meat quality.15,16 Despite these promising properties, its anti-inflammatory effects have yet to be evaluated.
To address this gap, the present study aimed to assess the anti-inflammatory potential of EPE and identify its key active components. Initially, the optimal Eucalyptus resource was selected from three economically important species using various extraction methods, with antioxidant capacity as the primary selection criterion. The anti-inflammatory activity of EPE was then evaluated in lipopolysaccharide (LPS)-stimulated RAW264.7 macrophages. Subsequently, bioactivity-guided fractionation was performed to isolate the principal anti-inflammatory constituents, whose efficacy and mechanisms of action were further examined in a dextran sulfate sodium (DSS)-induced colitis mouse model. This study provides new insights into the anti-inflammatory potential of Eucalyptus leaf polyphenols and supports the development of EPE as a promising natural agent for the prevention and management of inflammatory bowel diseases.
2 Materials and methods
2.1 Reagents
Eucalyptus leaves from E. grandis × E. urophylla, E. tereticornis, and E. urophylla, representing economically significant forest species, were collected from the Leizhou Forestry Bureau, Zhanjiang, Guangdong Province, China. RAW264.7 murine macrophages were obtained from the Kunming Cell Bank of the Chinese Academy of Sciences. Dulbecco's modified Eagle's medium (DMEM; cat. no. 11965092), fetal bovine serum (FBS; cat. no. A5670201), dexamethasone (DXM; cat. no. D4902), phosphate-buffered saline (PBS, pH 7.4; cat. no. 10010023), and antibiotics (Streptomycin–Penicillin; cat. no. 15140122) were purchased from Gibco (Waltham, MA, USA). LPS (cat. no. SMB00610), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; cat. no. 475989), and ascorbic acid (cat. no. 1043003) were obtained from Sigma-Aldrich (St Louis, MO, USA). A nitric oxide (NO) assay kit (cat. no. S0021S) was provided by Beyotime Biotechnology (Shanghai, China). Mouse ELISA kits for interleukin-6 (IL-6) (cat. no. EMC004.96), interleukin-1β (IL-1β) (cat. no. EMC001b.96), and tumor necrosis factor-α (TNF-α) (cat. no. EMC102a.96) were purchased from Neobioscience Technology Co., Ltd (Shenzhen, China). Assay kits for superoxide dismutase (SOD; cat. no. A001-3-2), catalase (CAT; cat. no. A007-1-1), glutathione peroxidase (GPx; cat. no. A005-1-2), and malondialdehyde (MDA; cat. no. A003-1-2) were obtained from the Jiancheng Biotechnology Institute (Nanjing, China). DSS (cat. no. MFCD00081551) was sourced from MP Biomedicals (Santa Ana, CA, USA). All other reagents were of analytical grade and used as received.2.2 Sample preparation and isolation of bioactive components
After drying, eucalyptus leaves were pulverized and passed through a 40-mesh sieve. Three extraction methods—solvent extraction (SE), ultrasonic wave-assisted extraction (UWE), and low-temperature continuous phase transformation extraction (LCPTE)—were carried out using 70% (v/v) ethanol to evaluate their effects on extraction efficiency, total polyphenol content, and antioxidant activity.17 For SE, the leaf powder was extracted twice with 70% ethanol at 50 °C for 2 h using magnetic stirring. For UAE, two extraction cycles were performed with 70% ethanol at 50 °C for 2 h using ultrasonic equipment. For LCPTE, the extraction was conducted using 70% ethanol under a pump flow rate of 35 L h−1 at 50 °C for a total of 2 h. All resulting extracts were vacuum-concentrated and spray-dried under the following conditions: inlet temperature 180 °C, outlet temperature 80 °C, and atomizer speed 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm.
000 rpm.
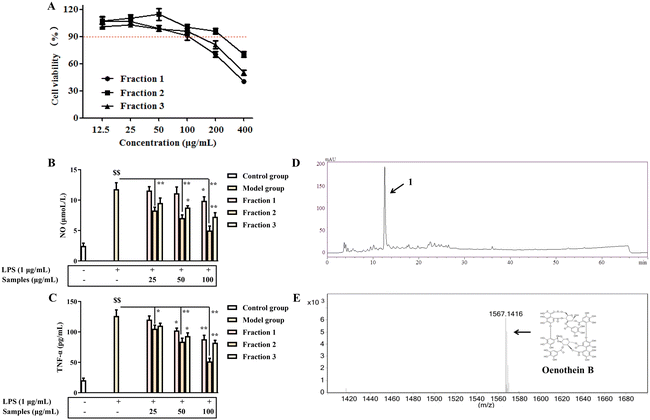
The optimal eucalyptus leaf polyphenol extract was dissolved in 70% ethanol at a concentration of 200 mg mL−1 and stored at 4 °C for 24 h. Following sedimentation, the supernatant was sequentially partitioned with petroleum ether (fraction 1) and ethyl acetate (fraction 2); the remaining aqueous phase was designated as fraction 3. After assessing in vitro anti-inflammatory activity across all fractions, fraction 2 was identified as optimal and subsequently analyzed by high-performance liquid chromatography (HPLC) and further purified using preparative HPLC (LC-8A, Shimadzu). All lyophilized extracts and fractions were stored at −20 °C for subsequent experiments.
2.3 Cell culture
RAW 264.7 macrophages were maintained in DMEM supplemented with 10% fetal bovine serum and 1% antibiotics at 37 °C in a humidified incubator under 5% CO2.2.4 Animal experiment design
A total of 40 male C57BL/6J mice (6–8 weeks old, 18–21 g) were obtained from Zhejiang Weitong Lihua Laboratory Animal Technology Co., Ltd (Zhejiang, China). All animals were housed under standard conditions (25 ± 1 °C, 50 ± 5% humidity) for one week prior to the experiment. The study protocol was approved by the Animal Ethics Committee of South China Agricultural University (SYXK-2019-0136).The mice were randomly assigned to four groups: control, model, low-dose OEB (L-OEB, 25 mg kg−1), and high-dose OEB (H-OEB, 100 mg kg−1). As illustrated in Fig. 7A, OEB was administered via oral gavage once daily for 27 days in the treatment groups, while the control and model groups received sterile water. From day 22 to day 27, 2% DSS was added to the drinking water of all groups except the control to induce ulcerative colitis. Body weight, stool consistency, and the presence of blood in feces were recorded daily during DSS administration. On day 27, the mice were weighed and euthanized. Spleens were harvested and weighed to calculate the spleen index. Colon tissues and fecal samples were collected and stored at −80 °C for further analysis.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C. The resulting supernatants were transferred to new 1.5 mL centrifuge tubes and centrifuged again under the same conditions. The final supernatants were subjected to metabolomic analysis using an LC-MS/MS system (HPLC: Shim-pack UFLC SHIMADZU CBM30A; MS/MS: QTRAP® 6500+ tandem mass spectrometer), following previously described methods.21 Briefly, the ion transport capillary temperature was set to 450 °C, with curtain gas, ion source gas 1, and ion source gas 2 maintained at 25 psi, 55 psi and 60 psi, respectively. The UPLC mobile phase consisted of solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). The optimized gradient elution program was as follows: 0 min, 5% B; 11 min, 90% B; 12 min, 90% B; 12.1 min, 5% B; 14 min, 5% B. The flow rate was set at 0.4 mL min−1, with an injection volume of 2 μL, and a column temperature of 40 °C. All metabolomic analyses were conducted in accordance with standard operating procedures provided by Wuhan MetWare Biotechnology (Wuhan, China). Raw data were processed on the Majorbio Cloud Platform (https://cloud.majorbio.com). Filtered data were used to generate orthogonal partial least squares discriminant analysis (OPLS-DA) score plots and volcano plots to visualize metabolic differences. Venn diagrams and heatmaps were created using OmicStudio tools (https://www.omicstudio.cn/tool). KEGG pathway enrichment analysis was performed using the MetaboAnalyst 6.0 platform (https://www.metaboanalyst.ca/).
000 rpm for 10 min at 4 °C. The resulting supernatants were transferred to new 1.5 mL centrifuge tubes and centrifuged again under the same conditions. The final supernatants were subjected to metabolomic analysis using an LC-MS/MS system (HPLC: Shim-pack UFLC SHIMADZU CBM30A; MS/MS: QTRAP® 6500+ tandem mass spectrometer), following previously described methods.21 Briefly, the ion transport capillary temperature was set to 450 °C, with curtain gas, ion source gas 1, and ion source gas 2 maintained at 25 psi, 55 psi and 60 psi, respectively. The UPLC mobile phase consisted of solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). The optimized gradient elution program was as follows: 0 min, 5% B; 11 min, 90% B; 12 min, 90% B; 12.1 min, 5% B; 14 min, 5% B. The flow rate was set at 0.4 mL min−1, with an injection volume of 2 μL, and a column temperature of 40 °C. All metabolomic analyses were conducted in accordance with standard operating procedures provided by Wuhan MetWare Biotechnology (Wuhan, China). Raw data were processed on the Majorbio Cloud Platform (https://cloud.majorbio.com). Filtered data were used to generate orthogonal partial least squares discriminant analysis (OPLS-DA) score plots and volcano plots to visualize metabolic differences. Venn diagrams and heatmaps were created using OmicStudio tools (https://www.omicstudio.cn/tool). KEGG pathway enrichment analysis was performed using the MetaboAnalyst 6.0 platform (https://www.metaboanalyst.ca/).
2.5 Statistical analysis
All graphs were generated using GraphPad Prism (version 8.0), OmicStudio tools, MetaboAnalyst 6.0, R software (version 2.15.3), and QIIME software (version 1.9.1). Statistical analyses were performed using one-way ANOVA in SPSS (version 18.0), followed by Duncan's or LSD post hoc tests to assess differences among groups. Differences were considered statistically significant at p < 0.05 or p < 0.01. Data are expressed as mean ± standard deviation (SD).3 Results
3.1 Optimization of eucalyptus leaf extracts
Three extraction methods were evaluated to obtain Eucalyptus leaf polyphenol extracts with optimal extraction efficiency and total polyphenol content. As shown in Table 1, the LCPTE method yielded the highest extraction efficiency, followed by UWE and then SE. Among the species tested, Eucalyptus grandis × E. urophylla consistently produced higher extraction yields across all methods compared to the other two species (p < 0.05). Regarding total polyphenol content, both LCPTE and UWE significantly outperformed SE (p < 0.05). Notably, E. grandis × E. urophylla exhibited the highest total polyphenol concentration (300.95 ± 16.37 mg g−1), while E. tereticornis showed the lowest (223.26 ± 14.18 mg g−1) (p < 0.05). These findings indicate that LCPTE is the most effective method in terms of both yield and polyphenol concentration.| Botanical name | Extraction methods | Extraction efficiency (%) | Total polyphenols content (mg g−1) |
|---|---|---|---|
| Different letters among data for extraction efficiency and total content of eucalyptus leaf polyphenols by three extraction methods indicate values that were significantly different (p < 0.05). | |||
| E. grandis × E. urophylla | SE | 25.86 ± 1.33a | 241.80 ± 11.22b |
| UWE | 26.48 ± 0.39a | 272.38 ± 15.58a | |
| LCPTE | 27.01 ± 2.58a | 300.95 ± 16.36a | |
| E. tereticornis | SE | 23.21 ± 2.01b | 223.26 ± 14.18b |
| UWE | 25.35 ± 0.86ab | 251.97 ± 8.24a | |
| LCPTE | 26.46 ± 1.35a | 266.93 ± 8.89a | |
| E. urophylla | SE | 22.61 ± 1.05a | 229.93 ± 23.01b |
| UWE | 24.10 ± 1.83a | 272.38 ± 17.35a | |
| LCPTE | 25.15 ± 1.87a | 290.06 ± 9.20a | |
To further identify the extract with the strongest antioxidant capacity, hydroxyl radical (˙OH) scavenging activity and ABTS+ radical scavenging ability were assessed. As shown in SI Fig. S1A–C, antioxidant activity followed the same trend across all methods: LCPTE > UWE > SE. SI Fig. S1D shows that LCPTE-extracted samples exhibited the lowest IC50 values, indicating stronger antioxidant capacity, while SE extracts had the highest IC50 values. Specifically, EPEGU demonstrated the most potent ˙OH scavenging activity, with the lowest IC50 value of 0.159 ± 0.004 mg mL−1. ABTS+ scavenging assays confirmed these results. As shown in SI Fig. S2A–C, all samples showed concentration-dependent activity, again in the order LCPTE > UWE > SE. EPEGU by LCPTE displayed the lowest ABTS+ IC50 value (0.178 ± 0.01 mg mL−1), further supporting its superior antioxidant potential (SI Fig. S2D). Based on these results, EPEGU extracted via LCPTE was selected for subsequent investigations.
3.2 Anti-inflammatory activity of EPEGU and identification of its anti-inflammatory components
3.3 Anti-inflammatory activity of OEB from EPEGU
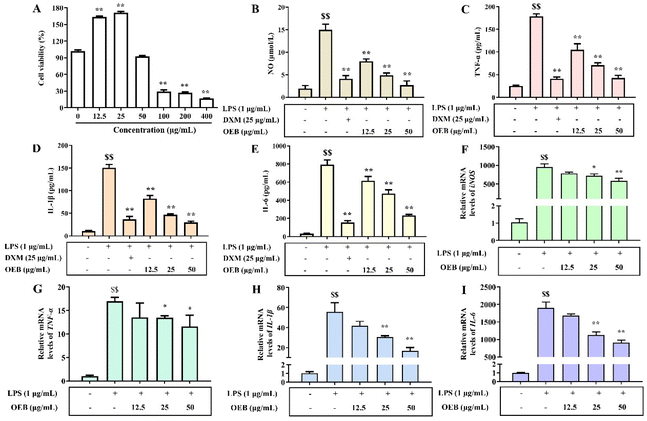
The anti-inflammatory effects of OEB were assessed using an LPS-induced RAW264.7 macrophage model. As shown in Fig. 3A, OEB at concentrations of 12.5–50 μg mL−1 did not exhibit significant cytotoxicity compared to the control group (p > 0.05), and these concentrations were therefore selected for subsequent experiments. As presented in Fig. 3B, OEB significantly and dose-dependently inhibited NO production in LPS-stimulated macrophages. At 50 μg mL−1, OEB reduced NO levels to 2.69 ± 1.62 μmol L−1—an 82.01% decrease compared to the LPS group (14.96 ± 2.20 μmol L−1; p < 0.01)—and showed no significant difference from the control group (1.94 ± 1.12 μmol L−1; p > 0.05). Fig. 3C–E demonstrates that OEB also markedly suppressed the secretion of pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 in a dose-dependent manner. At 50 μg mL−1, the maximum inhibition rates for TNF-α, IL-1β, and IL-6 were 76.15%, 80.16%, and 70.65%, respectively (p < 0.01). To confirm these findings at the transcriptional level, the mRNA expression of iNOS, TNF-α, IL-1β, and IL-6 was measured via RT-qPCR. As shown in Fig. 3F–I, LPS significantly upregulated the expression of all four genes compared to the control (p < 0.05), confirming successful induction of inflammation. OEB treatment led to a significant, dose-dependent reduction in the mRNA levels of iNOS, TNF-α, IL-1β, and IL-6 (p < 0.05). These results collectively demonstrate that OEB exerts potent anti-inflammatory effects in LPS-induced RAW264.7 macrophages by inhibiting both the secretion and gene expression of key inflammatory mediators.3.4 The amelioration effect of OEB on DSS-induced ulcerative colitis
To further characterize the effects of OEB on gut microbiota, taxonomic profiles at the phylum and genus levels were analyzed. At the phylum level, Firmicutes, Verrucomicrobiota, Bacteroidetes, and Proteobacteria were predominant across all groups (Fig. 6E). Compared with the control group, DSS treatment significantly increased the relative abundance of Bacteroidetes, Proteobacteria, Deferribacteres, Desulfobacterota, and Campilobacterota, while reduced Firmicutes and Actinobacteriota levels (p < 0.05). OEB treatment reversed these alterations by elevating Firmicutes abundance, increasing the Firmicutes/Bacteroidetes (F/B) ratio, and suppressing Proteobacteria, Deferribacteres, Desulfobacterota, and Campilobacterota (p < 0.05). At the genus level, DSS exposure led to significant increases in Bacteroides, Escherichia–Shigella, Odoribacter, Vibrio, Prevotella, Agathobacter, Collinsella, Utterella, and Ruegeria (p < 0.05; Fig. 6F). In contrast, OEB supplementation mitigated these changes by decreasing the abundance of Bacteroides, Escherichia–Shigella, and Mucispirillum, while enriching beneficial genera such as Streptococcus and [Eubacterium]_fissicatena_group (p < 0.05). Moreover, the levels of Akkermansia, Lactobacillus, and Ruminococcus, which were reduced in DSS-treated mice, were at least partially restored by OEB treatment (p > 0.05). These results suggest that OEB alleviates dysbiosis by restoring the balance and structural integrity of the gut microbiota.
LEfSe analysis further identified specific microbial biomarkers across groups (Fig. 6G). The control group was enriched in taxa such as p_Firmicutes, o_Lactobacillales, f_Lactobacillaceae, g_Lactobacillus, o_Corynebacteriales, f_Corynebacteriaceae, g_Corynebacterium, and s_Corynebacterium_stationis. In the DSS group, 19 discriminatory taxa were identified, with p_Bacteroidota, p_Proteobacteria, g_Bacteroides, f_Enterobacteriaceae, g_Escherichia–Shigella, s_Escherichia_coli, and g_Schlegelella showing LDA scores above 4.0, indicating strong associations with colitis. In contrast, the OEB-treated group exhibited higher relative abundances of f_Muribaculaceae, g_Dubosiella, and g_Eubacterium_fissicatena_group. Overall, these results demonstrate that DSS-induced colitis significantly disrupts gut microbial composition, while OEB supplementation effectively mitigates dysbiosis by reducing harmful bacteria and promoting beneficial microbial taxa.
4 Discussion
Nonvolatile constituents of Eucalyptus leaves have been reported to exhibit various bioactivities, including antioxidant, antitumor, antibacterial, and antifungal effects.16,26,27 In this study, polyphenols extracted from EPEGU using LCPTE were obtained, yielding a high total polyphenol content with strong antioxidant activity. Notably, antioxidant and anti-inflammatory properties of natural polyphenols are often interrelated.28 Traditionally, Eucalyptus has been used to treat inflammatory conditions, and recent studies suggest that its polyphenols contribute significantly to these effects.29,30 However, the anti-inflammatory potential of Eucalyptus leaf polyphenol extracts remains underexplored. To address this, we preliminarily assessed the anti-inflammatory activity of EPEGU using RAW264.7 macrophages, which are central to initiating and sustaining the inflammatory response in UC. As expected, EPEGU significantly and dose-dependently suppressed NO production and the release of pro-inflammatory cytokines (TNF-α, IL-1β, IL-6) in LPS-stimulated macrophages. These findings suggest that EPEGU holds promise as a natural anti-inflammatory agent. However, EPEGU comprises a complex mixture of phenolics, and the specific compounds responsible for its activity remain unclear. To further clarify the active components, OEB—a unique hydrolyzable tannin—was isolated and identified as a major phenolic compound in E. grandis × E. urophylla leaves. OEB exhibited stronger anti-inflammatory activity than the crude EPEGU extract, indicating that it may be a key contributor to the extract's bioactivity. OEB also significantly downregulated mRNA expression of iNOS, TNF-α, IL-1β, and IL-6 in LPS-stimulated macrophages. Previous studies indicated that OEB from Onagraceae species inhibits TLR/NF-κB-dependent inducible nitric oxide and cytokine synthesis, an anti-inflammatory activity that relied on its intact molecular structure, thereby highlighting its potential as a functional ingredient for dietary use.31 While OEB's in vitro anti-inflammatory effects are well-documented, its in vivo efficacy remains less understood. Therefore, investigating OEB's function in vivo is essential to fully elucidate its therapeutic potential. In summary, OEB derived from Eucalyptus leaves demonstrates potent in vitro anti-inflammatory activity and represents a promising candidate for further development.UC is a chronic inflammatory disease with a multifactorial etiology involving genetic, environmental, and dietary factors.32 Plant-derived phenolic compounds have been reported to alleviate UC symptoms by reducing colonic inflammation and modulating gut microbiota dysbiosis.33,34 Although emerging evidence highlights the therapeutic potential of Eucalyptus-derived polyphenols in inflammatory bowel diseases, including UC,35 their efficacy and underlying mechanisms remain poorly understood due to limited in vivo research. In this study, we identified OEB as a key anti-inflammatory component from Eucalyptus polyphenols. Notably, no prior studies have reported the effects of OEB in a DSS-induced UC mouse model. Our findings demonstrate that OEB significantly alleviated general UC symptoms, including weight loss, splenomegaly, colon shortening, and bloody stools. Histopathological analysis further confirmed that OEB mitigated colonic damage, including mucosal erosion, crypt loss, goblet cell depletion, and inflammatory cell infiltration. These results are consistent with previous reports showing the beneficial effects of polyphenols on colitis symptoms, such as body weight, colon length, and histological integrity.7,36 Given that oxidative stress and inflammation are central to UC pathogenesis,37 the antioxidant and anti-inflammatory properties of OEB are particularly relevant. Excessive production of reactive oxygen species (ROS) impairs the intestinal barrier and promotes inflammation, exacerbating disease progression. Mounting evidence suggests that reducing oxidative stress and inflammation can ameliorate UC symptoms.38 Our data show that OEB enhances antioxidant defenses and suppresses inflammatory responses in the colon. These effects may be attributed to OEB's polyhydroxylated structure, previously reported to contribute to its bioactivity,39 which supported the potential use of OEB as a natural anti-inflammatory agent. In addition, these results hinted that although the leaves of E. grandis × E. urophylla are not conventionally consumed as food, the isolated compound OEB exhibited strong potential as a functional ingredient for nutraceutical or dietary supplement applications in inflammatory bowel diseases. In summary, OEB derived from Eucalyptus leaves effectively alleviates DSS-induced UC in mice, likely by reducing oxidative stress and inflammation. These findings support OEB as a promising natural compound for further investigation in UC therapy.
Accumulating evidence indicates that gut microbiota dysbiosis plays a critical role in the pathogenesis of UC, characterized by a reduction in beneficial microbes and an increase in opportunistic and pathogenic bacteria.40,41 Notably, Firmicutes abundance is negatively correlated with UC incidence. A higher Firmicutes level has been shown to reduce intestinal sensitivity to inflammation, potentially minimizing organ damage caused by endotoxin leakage through a compromised gut barrier.42 In contrast, Bacteroidetes abundance is often elevated in UC, and its enrichment has been associated with pro-inflammatory effects.43 A reduced Firmicutes/Bacteroidetes (F/B) ratio is a common feature in both UC mouse models and human IBD patients, reflecting gut barrier dysfunction and heightened inflammatory response.44,45 In this study, OEB treatment significantly increased Firmicutes abundance and restored the F/B ratio in DSS-induced UC mice, suggesting its potential to protect the intestinal barrier and suppress colonic inflammation by correcting microbial imbalances. Additionally, OEB reduced the relative abundance of several phyla elevated in the DSS group, including Proteobacteria, Deferribacteres, Desulfobacterota, and Campilobacterota, which were previously linked to UC pathology.46,47Proteobacteria comprises numerous pathogenic genera such as Escherichia, Salmonella, and Vibrio, whose expansion is a hallmark of gut dysbiosis and a known driver of IBD.46 Similarly, Deferribacteres, Desulfobacterota, and Campilobacterota are pro-inflammatory taxa associated with increased production of inflammatory mediators and exacerbation of UC symptoms.48 Collectively, these findings suggest that OEB alleviates DSS-induced gut microbiota dysbiosis at the phylum level by promoting the growth of anti-inflammatory taxa like Firmicutes while suppressing inflammation-associated phyla. This microbial modulation may contribute to its protective effects against UC.
Increased abundances of Bacteroides, Escherichia–Shigella, Odoribacter, Vibrio, Prevotella, Agathobacter, Collinsella, and Utterella Ruegeria were observed at the genus level in DSS-induced UC mice, consistent with previous reports.49 Notably, Bacteroides is known to secrete a 20 kDa zinc-dependent metalloprotease toxin that exacerbates colonic inflammation.50 Elevated Bacteroides levels have been detected in both DSS-induced colitis models and UC patients, suggesting its potential as a biomarker for disease onset and progression.51Escherichia–Shigella, Odoribacter, and Vibrio are recognized pathogens that contribute to intestinal inflammation and microbial dysbiosis. Although the precise roles of Prevotella, Agathobacter, Collinsella, and Utterella Ruegeria in UC remain unclear, Prevotella has been associated with pro-inflammatory microbiota and may promote disease severity in certain microbial contexts.52 OEB treatment significantly reduced the abundance of Bacteroides, Escherichia–Shigella, and Mucispirillum, while increased levels of Streptococcus and [Eubacterium]_fissicatena_group, in line with previous findings.53Mucispirillum, a member of the phylum Deferribacteres, has been proposed as a microbial indicator of DSS-induced colitis.54 Importantly, OEB also elevated the relative abundance of Akkermansia, Lactobacillus, and Ruminococcus—genera typically regarded as beneficial due to their roles in modulating inflammation, producing short-chain fatty acids, and maintaining gut barrier integrity.55 Further support came from LEfSe analysis, which revealed that harmful taxa such as p_Bacteroidota, p_Proteobacteria, g_Bacteroides, f_Enterobacteriaceae, g_Escherichia–Shigella, s_Escherichia coli, and g_Schlegelella were enriched in the DSS group. In contrast, beneficial taxa including f_Muribaculaceae, g_Dubosiella, and g_Eubacterium_fissicatena_group were more abundant in the OEB-treated group, reflecting a favorable microbial shift. Collectively, these findings confirm that DSS-induced UC results in pronounced gut microbiota dysbiosis, which can be partially reversed by OEB treatment through the suppression of pathogenic bacteria and enrichment of beneficial taxa.
Colitis-induced dysbiosis of the intestinal microbiota can lead to metabolic disturbances, as evidenced by significant alterations in metabolites observed in both colitis mouse models and patients.56,57 In this study, we found that OEB treatment notably modulated AA metabolism, which plays a key role in inflammatory processes. AA, an ω-6 polyunsaturated fatty acid primarily stored in membrane phospholipids under physiological conditions, is rapidly converted into various bioactive metabolites—such as prostaglandins (PGs), epoxyeicosatrienoic acids (EETs), and thromboxanes (TXs)—in response to external stimuli.58 These metabolites, including PGF2α, TXB2, 5,6-EET, and 8,9-EET, are recognized as proinflammatory mediators and have been positively correlated with the severity of inflammation in UC mouse models.59 Disruption of AA metabolism has been implicated in the exacerbation of UC symptoms.60 Consistent with previous reports, our results showed significant increases in PGF2α, TXB2, 5,6-EET, and 8,9-EET in the DSS-induced colitis group, indicating marked perturbation of AA metabolism. Notably, OEB treatment reversed these alterations. Similar findings have been reported in studies where natural polyphenols ameliorated UC by restoring AA metabolic balance and reducing the levels of these proinflammatory metabolites.61,62 Such interventions are believed to maintain microbial homeostasis, reinforce intestinal barrier integrity, and suppress the release of inflammatory mediators, thereby mitigating DSS-induced colonic inflammation.63 Our data further demonstrated that OEB treatment improved the gut microbiota composition, characterized by an increase in Firmicutes and a reduction in Bacteroidetes and Proteobacteria. These microbial shifts were associated with decreased expression of proinflammatory cytokines (TNF-α, IL-1β, IL-6) and amelioration of colonic inflammation. Additionally, normalization of AA metabolism and a corresponding decrease in proinflammatory metabolites contributed to symptom relief in UC. Collectively, these results suggest that metabolic disturbances in UC are downstream consequences of microbiota dysbiosis. OEB effectively reversed these changes, supporting its therapeutic potential. Moreover, DSS-induced colitis disrupted bile acid metabolism, as evidenced by significantly reduced levels of tauroursodeoxycholic acid, hyocholic acid, and hododeoxycholic acid compared with controls. Previous studies have similarly reported that several bile acids—including taurocholic acid, taurochenodeoxycholic acid, chenodeoxycholic acid, and ursodeoxycholic acid—are significantly reduced in UC mouse models compared to healthy controls.64 In the present study, OEB treatment effectively restored these bile acids in DSS-induced UC mice, suggesting a modulatory effect on bile acid metabolism. This restoration may be partly attributed to the influence of OEB on gut microbiota composition, particularly the increased abundance of Firmicutes and reduced levels of Proteobacteria. Wu et al. demonstrated that suppression of Proteobacteria, especially Escherichia–Shigella, facilitates the recovery of microbial homeostasis and enhances bile acid biosynthesis, notably of taurochenodeoxycholic acid.65 Additionally, members of the Firmicutes phylum—particularly Clostridium species—express key enzymes such as bile salt hydrolase (BSH) and 7α-dehydroxylase, which catalyze the conversion of primary bile acids into secondary bile acids.66 These secondary bile acids function as signaling molecules that help suppress intestinal inflammation and maintain epithelial barrier integrity via activation of the FXR/TGR5 pathways.67 This, in turn, promotes a stable and health-supporting microbial environment. Therefore, the observed improvement in bile acid profiles may result from OEB-mediated microbial modulation. In summary, a balanced gut microbiota supports bile acid metabolism, which in turn contributes to intestinal barrier preservation and inflammation reduction. Our findings suggest that OEB alleviates DSS-induced metabolic disturbances in UC, primarily through the regulation of arachidonic acid and bile acid metabolism.68
5 Conclusions
In conclusion, OEB, derived from Eucalyptus grandis × E. urophylla, demonstrated potent anti-inflammatory effects both in vitro and in vivo. Specifically, OEB treatment significantly alleviated DSS-induced ulcerative colitis in mice by suppressing proinflammatory mediators, enhancing antioxidant defenses, and modulating gut microbiota and related colonic metabolites. Given its origin from natural plant polyphenols, OEB may be a promising dietary functional ingredient used in foods or in dietary supplements for the management of inflammatory diseases.Author contributions
Wei Li: conceptualization, writing – original draft. Xiaoying Zhang: methodology, software. Lu Xu: resources, visualization. Yunjiao Chen: methodology, resources. Yong Cao: project administration, supervision, writing – review & editing. Ziyin Li: supervision, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Data availability
The data supporting this study are available within the article.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5fo03016a.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 82304178), the Shenzhen Science and Technology Program (JCYJ20240813112052067), Dongguan Social Development Science and Technology Project (No. 20231800900292) and the Haday Visionary Science Advancement Fund.References
- H. Lu, M. Shen, Y. Chen, Q. Yu, T. Chen and J. Xie, Alleviative effects of natural plant polysaccharides against DSS-induced ulcerative colitis via inhibiting inflammation and modulating gut microbiota, Food Res. Int., 2023, 167, 112630 CrossRef CAS PubMed.
- D. M. Tshikudi, C. N. Bernstein, S. Mishra, J. Ghia and H. K. Armstrong, Influence of biological sex in inflammatory bowel diseases, Nat. Rev. Gastroenterol. Hepatol., 2025, 22, 415–437 CrossRef CAS PubMed.
- R. Sun, D. Jin, F. Fei, Z. Xu, B. Cao and J. Li, Mushroom polysaccharides from Grifola frondosa (Dicks.) Gray and Inonotus obliquus (Fr.) Pilat ameliorated dextran sulfate sodium-induced colitis in mice by global modulation of systemic metabolism and the gut microbiota, Front. Pharmacol., 2023, 14, 1172963 CrossRef CAS PubMed.
- C. Herrera-deGuise, E. Varela, G. Sarrabayrouse, M. Pozuelo Del Río, V. R. Alonso, N. B. Sainz, F. Casellas, L. F. Mayorga, C. Manichanh and F. A. Vidaur, Gut microbiota composition in long-remission ulcerative colitis is close to a healthy gut microbiota, Inflamm. Bowel Dis., 2023, 29, 1362–1369 CrossRef PubMed.
- Z. Li, T. Chu, X. Sun, S. Zhuang, D. Hou, Z. Zhang, J. Sun, Y. Liu, J. Li and Y. Bian, Polyphenols-rich Portulaca oleracea L.(purslane) alleviates ulcerative colitis through restiring the intestinal barrier, gut microbiota and metabolites, Food Chem., 2025, 468, 142391 CrossRef CAS PubMed.
- C. Yang, W. Yang, Y. Wang, Y. Du, T. Zhao, H. Shao, D. Ren and X. Yang, Nonextractable polyphenols from Fu Brick Tea alleviates ulcerative colitis by controlling colon microbiota-targeted release to inhibit intestinal inflammation in mice, J. Agric. Food Chem., 2024, 72, 7397–7410 CrossRef CAS PubMed.
- Z. Zhou, W. He, H. Tian, P. Zhan and J. Liu, Thyme (Thymus vulgaris L.) polyphenols ameliorate DSS-induced ulcerative colitis of mice by mitigating intestinal barrier damage, regulating gut microbiota, and suppressing TLR4/NF-κB-NLRP3 inflammasome pathways, Food Funct., 2023, 14, 1113–1132 RSC.
- B. Salehi, J. Sharifi-Rad, C. Quispe, H. Llaique, M. Villalobos, A. Smeriglio, D. Trombetta, S. M. Ezzat, M. A. Salem and A. Zayed, Insights into Eucalyptus genus chemical constituents, biological activities and health-promoting effects, Trends Food Sci. Technol., 2019, 91, 609–624 CrossRef CAS.
- P. H. Hart, C. Brand, C. F. Carson, T. V. Riley, R. H. Prager and J. J. Finlay-Jones, Terpinen-4-ol, the main component of the essential oil of Melaleuca alternifolia (tea tree oil), suppresses inflammatory mediator production by activated human monocytes, Inflammation Res., 2000, 49, 619–626 CrossRef CAS PubMed.
- U. R. Juergens, Anti-inflammatory properties of the monoterpene 1.8-cineole: current evidence for co-medication in inflammatory airway diseases, Drug Res., 2014, 64, 638–646 CrossRef CAS PubMed.
- B. Arooj, S. Asghar, M. Saleem, S. H. Khalid, M. Asif, T. Chohan, I. U. Khan, H. M. Zubair and H. S. Yaseen, Anti-inflammatory mechanisms of eucalyptol rich Eucalyptus globulus essential oil alone and in combination with flurbiprofen, Inflammopharmacology, 2023, 31, 1849–1862 CrossRef CAS PubMed.
- H. Belkhodja, B. Meddah, K. Sidelarbi, D. Bouhadi, B. Medjadel and A. Brakna, In vitro and in vivo anti-inflammatory potential of eucalyptus globulus essential oil, J. Appl. Biol. Sci., 2022, 16, 80–88 CrossRef.
- T. Almeida, P. Moreira, F. J. Sousa, C. Pereira, A. J. Silvestre, C. Vilela and C. S. Freire, Bioactive bacterial nanocellulose membranes enriched with Eucalyptus globulus Labill. leaves aqueous extract for anti-aging skin care applications, Materials, 2022, 15, 1982 CrossRef CAS PubMed.
- D. Bencheikh, A. Gueddah, K. Soualat, H. Ben-aissi, A. Benslama, A. Harrar and S. Khennouf, Polyphenolic contents, antioxidant and antibacterial activities of aqueous extracts of Eucalyptus globulus L. and Trigonella foenum-greacum L., J. Appl. Biol. Sci., 2021, 15, 53–63 Search PubMed.
- Y. Chen, H. Chen, W. Li, J. Miao, N. Chen, X. Shao and Y. Cao, Polyphenols in Eucalyptus leaves improved the egg and meat qualities and protected against ethanol-induced oxidative damage in laying hens, J. Anim. Physiol. Anim. Nutr., 2018, 102, 214–223 CrossRef CAS PubMed.
- W. Li, X. Zhang, Z. He, Y. Chen, Z. Li, T. Meng, Y. Li and Y. Cao, In vitro and in vivo antioxidant activity of eucalyptus leaf polyphenols extract and its effect on chicken meat quality and cecum microbiota, Food Res. Int., 2020, 136, 109302 CrossRef CAS PubMed.
- Y. Yuan, J. Li, S. He, Q. Zeng, L. Dong, R. Zhang, D. Su and M. Zhang, Composition of phenolic and antioxidant activity of water chestnut peel during digestion in vitro as affected by blanching time, Int. J. Food Prop., 2019, 22, 71–83 CrossRef CAS.
- E. C. Giese, J. Gascon, G. Anzelmo, A. M. Barbosa, M. A. A. Da Cunha and R. F. H. Dekker, Free-radical scavenging properties and antioxidant activities of botryosphaeran and some other β-D-glucans, Int. J. Biol. Macromol., 2015, 72, 125–130 CrossRef CAS PubMed.
- L. Smolskaitė, P. R. Venskutonis and T. Talou, Comprehensive evaluation of antioxidant and antimicrobial properties of different mushroom species, LWT – Food Sci. Technol., 2015, 60, 462–471 CrossRef.
- W. Yin, M. Liu, Z. Jin, Z. Hao, C. Liu, J. Liu, H. Liu, M. Zheng and D. Cai, Ameliorative effects of insoluble dietary fiber and its bound polyphenols from adzuki bean seed coat on acute murine colitis induced by DSS: The inflammatory response, intestinal barrier and gut microbiota, Int. J. Biol. Macromol., 2025, 286, 138343 CrossRef CAS PubMed.
- W. Li, Z. Li, M. Peng, X. Zhang, Y. Chen, Y. Yang, X. Zhai, G. Liu and Y. Cao, Oenothein B boosts antioxidant capacity and supports metabolic pathways that regulate antioxidant defense in Caenorhabditis elegans, Food Funct., 2020, 11, 9157–9167 RSC.
- Y. Chen, B. Onken, H. Chen, X. Zhang, M. Driscoll, Q. Huang and Y. Cao, Healthy lifespan extension mediated by Oenothein B isolated from Eucalyptus grandis×Eucalyptus urophylla GL9 in Caenorhabditis elegans, Food Funct., 2020, 11, 2439–2450 RSC.
- T. Hatano, T. Yasuhara and M. Matsuda, Oenothein B, a dimeric, hydrolysable tannin with macrocyclic structure, and accompanying tannins from Oenothera erythrosepala, J. Chem. Soc., Perkin Trans., 1990, 37, 2735–2743 RSC.
- I. A. Schepetkin, L. N. Kirpotina, L. Jakiw, A. I. Khlebnikov, C. L. Blaskovich, M. A. Jutila and M. T. Quinn, Immunomodulatory activity of oenothein B isolated from Epilobium angustifolium, J. Immunol., 2009, 183, 6754–6766 CrossRef CAS PubMed.
- Y. Zhao, X. Tian, Y. Yan, S. Tian, D. Liu and J. Xu, Lithospermic acid alleviates oxidative stress and inflammation in DSS-induced colitis through Nrf2, Eur. J. Pharmacol., 2025, 995, 177390 CrossRef CAS PubMed.
- H. Elkolli, M. Elkolli, F. S. Ataya, M. M. Salem-Bekhit, S. A. Zahrani, M. W. Abdelmageed, B. Ernst and Y. Benguerba, In vitro and in Silico activities of E. radiata and E. cinerea as an enhancer of antibacterial, antioxidant, and anti-Inflammatory agents, Molecules, 2023, 28, 7153 CrossRef CAS PubMed.
- A. S. Taha, I. H. Ibrahim, W. A. Abo-Elgat, A. Abdel-Megeed, M. Z. Salem and M. S. A. El-Kareem, GC–MS, quantum mechanics calculation and the antifungal activity of river red gum essential oil when applied to four natural textiles, Sci. Rep., 2023, 13, 18214 CrossRef CAS PubMed.
- T. Hussain, B. Tan, Y. Yin, F. Blachier, M. C. B. Tossou and N. Rahu, Oxidative stress and inflammation: What polyphenols can do for us?, Oxid. Med. Cell. Longevity, 2016, 2016, 1–9 Search PubMed.
- D. E. Ali, R. A. E. Gedaily, S. M. Ezzat, M. A. E. Sawy, M. R. Meselhy and E. Abdel-Sattar, In silico and in vitro anti-inflammatory study of phenolic compounds isolated from Eucalyptus maculata resin, Sci. Rep., 2023, 13, 2093 CrossRef CAS PubMed.
- Y. Qiujian, F. Zongcai, H. Liping, H. Jingwei, Z. Zhongliu and L. Fang, Ellagic acid (EA), a tannin was isolated from Eucalyptus citriodora leaves and its anti-inflammatory activity, Med. Chem. Res., 2021, 30, 2277–2288 CrossRef.
- D. Schmid, M. Gruber, C. Piskaty, F. Woehs, A. Renner, Z. Nagy, A. Kaltenboeck, T. Wasserscheid, A. Bazylko, A. K. Kiss and T. Moeslinger, Inhibition of NF-kappaB-dependent cytokine and inducible nitric oxide synthesis by the macrocyclic ellagitannin oenothein B in TLR-stimulated RAW 264.7 macrophages, J. Nat. Prod., 2012, 75, 870–875 CrossRef CAS PubMed.
- B. Gros and G. G. Kaplan, Ulcerative colitis in adults: a review, J. Am. Med. Assoc., 2023, 330, 951–965 CrossRef CAS PubMed.
- R. Cao, X. Wu, H. Guo, X. Pan, R. Huang, G. Wang and J. Liu, Naringin exhibited therapeutic effects against DSS-induced mice ulcerative colitis in intestinal barrier–dependent manner, Molecules, 2021, 26, 6604 CrossRef CAS PubMed.
- W. Li, L. Zhang, Q. Xu, W. Yang, J. Zhao, Y. Ren, Z. Yu and L. Ma, Taxifolin alleviates DSS-induced ulcerative colitis by acting on gut microbiome to produce butyric acid, Nutrients, 2022, 14, 1069 CrossRef CAS PubMed.
- D. M. Syukri and S. Singh, Medicinal and nutritional importance of Eucalyptus camaldulensis in human health, In Springer, 2024, pp. 185–199 Search PubMed.
- H. Zhang, Z. Hao, R. Zhang, J. Tong, X. Wang, J. Liu, Y. Gao, X. Wang, Q. Su and H. Wen, Artemisia argyi polyphenols attenuates DSS-induced colitis in mice by regulating the structural composition of gut microbiota, Phytomedicine, 2024, 132, 155897 CrossRef CAS PubMed.
- Y. Y. Ren, Y. Geng, Y. Du, W. Li, Z. Lu, H. Xu, G. Xu, J. Shi and Z. Xu, Polysaccharide of Hericium erinaceus attenuates colitis in C57BL/6 mice via regulation of oxidative stress, inflammation-related signaling pathways and modulating the composition of the gut microbiota, J. Nutr. Biochem., 2018, 57, 67–76 CrossRef CAS PubMed.
- Y. Xiong, Z. Cheng, Y. Zhang, T. Liu, Z. Wan, C. Xia, B. Zhou, C. Shan, D. Song and F. Miao, Ellagic acid alleviates DSS–induced ulcerative colitis by inhibiting ROS/NLRP3 pathway activation and modulating gut microbiota in mice, Eur. J. Nutr., 2025, 64, 1–17 CrossRef PubMed.
- T. Yoshida, M. Yoshimura and Y. Amakura, Chemical and biological significance of oenothein B and related ellagitannin oligomers with macrocyclic structure, Molecules, 2018, 23, 552 CrossRef PubMed.
- H. Yang, Y. Zhao, R. Zhang, L. Zhao, H. Yang and X. Liao, CiLi (Rosa roxburghii Tratt.) polyphenols improve colitis via gut microbiota-lipid mediator-immunity axis, Food Res. Int., 2025, 209, 116257 CrossRef CAS PubMed.
- X. Zeng, J. Li, X. Wang, L. Liu, S. Shen, N. Li, Z. Wang, Y. Yuan and T. Yue, Regulation of gut microbiota and microbial metabolome of kefir supernatant against Fusobacterium nucleatum and DSS-coinduced colitis, J. Agric. Food Chem., 2024, 72, 3536–3548 CrossRef CAS PubMed.
- M. Rühlemann, T. Liwinski, F. A. Heinsen, C. Bang, R. Zenouzi, M. Kummen, L. Thingholm, M. Tempel, W. Lieb and T. Karlsen, Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis, Aliment. Pharmacol. Ther., 2019, 50, 580–589 CrossRef PubMed.
- J. Miyoshi and E. B. Chang, The gut microbiota and inflammatory bowel diseases, Transl. Res., 2017, 179, 38–48 CrossRef PubMed.
- X. Dou, N. Gao, D. Yan and A. Shan, Sodium butyrate alleviates mouse colitis by regulating gut microbiota dysbiosis, Animals, 2020, 10, 1154 CrossRef PubMed.
- Y. Yin, T. Yang, Z. Tian, C. Shi, C. Yan, H. Li, Y. Du and G. Li, Progress in the investigation of the Firmicutes/Bacteroidetes ratio as a potential pathogenic factor in ulcerative colitis, J. Med. Microbiol., 2025, 74, 1966 CAS.
- S. Umeda, T. Sujino, K. Miyamoto, Y. Yoshimatsu, Y. Harada, K. Nishiyama, Y. Aoto, K. Adachi, N. Hayashi and K. Amafuji, D-amino acids ameliorate experimental colitis and cholangitis by inhibiting growth of Proteobacteria: potential therapeutic role in inflammatory bowel disease, Cell. Mol. Gastroenterol. Hepatol., 2023, 16, 1011–1031 CrossRef CAS PubMed.
- R. Yu, Q. Zhou, T. Liu, P. Liu, H. Li, Y. Bian and Z. Liu, Kaempferol relieves the DSS-induced chronic colitis in C57BL/6J mice, alleviates intestinal angiogenesis, and regulates colonic microflora structure, J. Funct. Foods, 2023, 107, 105646 CrossRef CAS.
- C. Li, Y. Huang, M. Qin, Q. He, Z. Lin, X. Zhang, H. Ren, D. Xu, X. Liao and Y. Liu, Enocyanin alleviates experimental colitis and restores gut microbiota homeostasis as functional foods, Food Biosci., 2024, 57, 103546 CrossRef CAS.
- X. Feng, Y. Chen, L. Luo, Z. Fang, S. Ma, Z. Li, J. Huang, Y. Pan, H. Lv and S. Gong, Liubao insect tea polyphenols ameliorate DSS-induced experimental colitis by protecting intestinal barrier and regulating intestinal microbiota, Food Chem., 2025, 467, 142156 CrossRef CAS PubMed.
- B. Arif, S. Yasir, M. Saeed and M. Q. Fatmi, Natural products can be potential inhibitors of metalloproteinase II from Bacteroides fragilis to intervene colorectal cancer, Heliyon, 2024, 10, e32838 CrossRef CAS PubMed.
- H. K. M. Leung, E. K. K. Lo and H. El-Nezami, Theabrownin Alleviates colorectal tumorigenesis in murine AOM/DSS model via PI3K/Akt/mTOR pathway suppression and gut microbiota modulation, Antioxidants, 2022, 11, 1716 CrossRef CAS PubMed.
- A. Iljazovic, U. Roy, E. Galvez, T. R. Lesker, B. Zhao, A. Gronow, L. Amend, S. E. Will, J. D. Hofmann, M. C. Pils, K. Schmidt-Hohagen, M. Neumann-Schaal and T. Strowig, Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation, Mucosal Immunol., 2021, 14, 113–124 CrossRef CAS PubMed.
- Y. Liu, H. Zhang, A. Xie, J. Sun, H. Yang, J. Li, Y. Li, F. Chen, Y. Mei and Y. Liang, Lactobacillus rhamnosus L. and plantarum combination treatment ameliorated colitis symptoms in a mouse model by altering intestinal microbial composition and suppressing inflammatory response, Mol. Nutr. Food Res., 2023, 67, 2200340 CrossRef CAS PubMed.
- D. Lin, Y. Jin, D. Hu, S. Ying and Y. Jiang, Influence of TRAIL deficiency on Th17 cells and colonic microbiota in experimental colitis mouse model, Am. J. Med. Sci., 2021, 362, 188–197 CrossRef PubMed.
- Y. Lee, K. Sugihara, M. G. Gillilland, S. Jon, N. Kamada and J. J. Moon, Hyaluronic acid–bilirubin nanomedicine for targeted modulation of dysregulated intestinal barrier, microbiome and immune responses in colitis, Nat. Mater., 2020, 19, 118–126 CrossRef CAS PubMed.
- Y. Chen, K. Li, M. Zhang, J. Liu, M. Wang, Y. Xiu, Q. Zhang and D. Zhao, Amentoflavone ameliorates DSS-induced ulcerative colitis via gut microbiota-bile acid metabolism axis modulation, J. Funct. Foods, 2025, 126, 106706 CrossRef CAS.
- Q. Yang, J. Liu, T. Li, S. Lyu, X. Liu, Z. Du, X. Shang and T. Zhang, Integrated microbiome and metabolomic analysis reveal the repair mechanisms of ovalbumin on the intestine barrier of colitis mice, J. Agric. Food Chem., 2023, 71, 8894–8905 CrossRef CAS PubMed.
- T. Wang, X. Fu, Q. Chen, J. K. Patra, D. Wang, Z. Wang and Z. Gai, Arachidonic acid metabolism and kidney inflammation, Int. J. Mol. Sci., 2019, 20, 3683 CrossRef CAS PubMed.
- T. Murata, K. Aritake, Y. Tsubosaka, T. Maruyama, T. Nakagawa, M. Hori, H. Hirai, M. Nakamura, S. Narumiya and Y. Urade, Anti-inflammatory role of PGD2 in acute lung inflammation and therapeutic application of its signal enhancement, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5205–5210 CrossRef CAS PubMed.
- Z. Yuan, L. Yang, X. Zhang, P. Ji, Y. Hua and Y. Wei, Mechanism of Huang-lian-Jie-du decoction and its effective fraction in alleviating acute ulcerative colitis in mice: Regulating arachidonic acid metabolism and glycerophospholipid metabolism, J. Ethnopharmacol., 2020, 259, 112872 CrossRef CAS PubMed.
- S. Li, A. Zhuge, K. Wang, L. Lv, X. Bian, L. Yang, J. Xia, X. Jiang, W. Wu, S. Wang, Q. Wang and L. Li, Ketogenic diet aggravates colitis, impairs intestinal barrier and alters gut microbiota and metabolism in DSS-induced mice, Food Funct., 2021, 12, 10210–10225 RSC.
- Z. Niu, X. Li, X. Yang and Z. Sun, Protective effects of sinomenine against dextran sulfate sodium-induced ulcerative colitis in rats via alteration of HO-1/Nrf2 and inflammatory pathway, Inflammopharmacology, 2024, 32, 2007–2022 CrossRef CAS PubMed.
- W. Zhong, J. Gong, Q. Su, M. A. Farag, J. Simal-Gandara, H. Wang and H. Cao, Dietary polyphenols ameliorate inflammatory bowel diseases: advances and future perspectives to maximize their nutraceutical applications, Phytochem. Rev., 2025, 24, 1227–1259 CrossRef CAS.
- F. Xiao, F. Dong, X. Li, Y. Li, G. Yu, Z. Liu, Y. Wang and T. Zhang, Bifidobacterium longum CECT 7894 improves the efficacy of infliximab for DSS-Induced colitis via regulating the gut microbiota and bile acid metabolism, Front. Pharmacol., 2022, 13, 902337 CrossRef CAS PubMed.
- Y. Wu, Y. Zheng, X. Wang, P. Tang, W. Guo, H. Ma, A. Zhang, D. Li, Y. Xie and C. Wang, Ginseng-containing sijunzi decoction ameliorates ulcerative colitis by orchestrating gut homeostasis in microbial modulation and intestinal barrier integrity, Am. J. Chin. Med., 2023, 51, 677–699 CrossRef PubMed.
- S. Bamba, O. Inatomi, A. Nishida, M. Ohno, T. Imai, K. Takahashi, Y. Naito, J. Iwamoto, A. Honda and N. Inohara, Relationship between the gut microbiota and bile acid composition in the ileal mucosa of Crohn's disease, Intest. Res., 2022, 20, 370–380 CrossRef PubMed.
- J. Wang, J. Zang, Y. Yu, Y. Liu, H. Cao, R. Guo, L. Zhang, M. Liu, Z. Zhang and X. Li, Lingguizhugan oral solution alleviates MASLD by regulating bile acids metabolism and the gut microbiota through activating FXR/TGR5 signaling pathways, Front. Pharmacol., 2024, 15, 1426049 CrossRef CAS PubMed.
- W. Jia, G. Xie and W. Jia, Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis, Nat. Rev. Gastroenterol. Hepatol., 2018, 15, 111–128 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to the work of this article. |
| This journal is © The Royal Society of Chemistry 2026 |