Open Access Article
Open Access ArticleSustainable biotransformation of microalgae via probiotic fermentation for enhanced functional, nutritional, and sensory properties
Po-Hsiang
Wang†
 *,
Zann Yi
Qi Tan†
*,
Zann Yi
Qi Tan†
 ,
Choy Eng
Nge
,
Nurhidayah
Basri
,
Lina Xian
Yu Lee
,
Choy Eng
Nge
,
Nurhidayah
Basri
,
Lina Xian
Yu Lee
 ,
Aaron
Thong
,
Aaron
Thong
 ,
Mario
Wibowo
,
Elaine Jinfeng
Chin
,
Mario
Wibowo
,
Elaine Jinfeng
Chin
 ,
Sharon
Crasta
,
Geraldine
Chan
,
Sharon
Crasta
,
Geraldine
Chan
 ,
Yoganathan
Kanagasundaram
and
Siew Bee
Ng
*
,
Yoganathan
Kanagasundaram
and
Siew Bee
Ng
*
Singapore Institute of Food and Biotechnology Innovation (SIFBI), Agency for Science, Technology and Research (A*STAR), 31 Biopolis Way, Nanos, Singapore 138669, Singapore. E-mail: tommy_wang@a-star.edu.sg; ngsb@a-star.edu.sg
First published on 7th November 2025
Abstract
Microalgae represent a sustainable food source with exceptional CO2 fixation efficiency; however, their integration into the food chain is hindered by undesirable organoleptic properties. This study establishes a green biotransformation platform using Generally Recognized As Safe (GRAS) bacterium Lactiplantibacillus plantarum to ferment Chlorella vulgaris biomass. This fermentation process operates without the use of harsh chemicals and organic solvents, enabling the full utilization of the biomass while improving sensory quality. Notably, the L. plantarum fermentation maintained dried biomass weight, in contrast to ∼15–40% loss seen with Bacillus spp., further enhancing the carbon-negative profile of microalgae. Tiered olfactory analysis and gas chromatography-mass spectrometry revealed selective reduction of polyunsaturated fatty acid-derived aldehydes and accumulation of flavor-active volatiles, including pyrazines and phenylethyl derivatives. Electronic tongue and liquid chromatography-mass spectrometry confirmed elevated umami taste, via increased glutamate and nucleotide levels. Additionally, the fermentation of microalgae with L. plantarum converted aromatic amino acids into antioxidant aromatic lactates, exemplifying catalytic, rather than stoichiometric efficiency. Overall, this renewable fermentation strategy converts photoenergy-fuelled, CO2-derived microalgal biomass into direct functional food ingredients under mild, organic solvent-free conditions, while bypassing conventional downstream extraction and purification steps.
Sustainability spotlightThis work establishes a transformative probiotic fermentation platform using GRAS-certified Lactiplantibacillus plantarum to bio-transform CO2-derived Chlorella vulgaris biomass into functional food ingredients without organic solvents, harsh chemicals, or energy-intensive processing. Operating under mild conditions (37 °C, 72 hours), the platform achieves net-zero waste generation while maintaining biomass integrity—contrasting with 15–40% losses from alternative methods. The biotransformation catalytically converts aromatic amino acids into antioxidant lactates while enhancing umami compounds and eliminating off-flavors, producing ready-to-use functional foods without downstream purification. This sustainable approach directly advances UN SDG 2 (zero hunger) by creating scalable alternative protein sources, SDG 12 (responsible consumption) through complete biomass utilization and waste minimization, and SDG 13 (climate action) by amplifying the carbon-negative impact of microalgal CO2 fixation within circular bioeconomy frameworks. |
Introduction
Traditional protein sources and food production methods face significant sustainability challenges, including land usage limitations, excessive water consumption, and substantial carbon footprints.1 As such, edible microalgae have emerged as promising candidates for sustainable food production, owing to their exceptional photo-mixotrophic capabilities, high resource efficiency, minimal land requirements, and remarkable carbon capture potential.2,3 Historically, microalgae have been extensively utilized in industrial and environmental contexts, including bioremediation of wastewater, CO2 capture, biofuel production, and as sources for commodity biochemicals and high-value compounds.4,5 However, their direct application as food ingredients remains comparatively underexplored despite their considerable nutritional value, notably their richness in protein, complex carbohydrates, valuable lipids, and diverse micronutrients. To date, the targeted implementation of microbial fermentation as a method to specifically transform microalgae biomass for direct food production constitutes a significant, yet largely unaddressed, research frontier. The biotransformation of microalgae by carefully selected Generally Recognized As Safe (GRAS) probiotic bacteria can enhance nutritional bioavailability and improve sensory characteristics, unlocking tremendous value in alternative protein systems.Microalgae such as Chlorella can fix CO2 while utilizing solar energy, with photosynthetic efficiency rates substantially higher than terrestrial crops (10–20% versus 1–2%).6 Their rapid life cycles, minimal land footprint, and high fertility, coupled with their rich biochemical composition, render them valuable for producing a wide array of chemicals.7 Additionally, although differences between and within species exist, microalgae generally produce nutrient-dense biomass containing complete protein profiles and valuable bioactive compounds, such as ω-3 polyunsaturated fatty acids (PUFAs), polysaccharides, carotenoids, vitamins, phenolics, and phycobiliproteins.8 Despite their nutritional and environmental merits, challenges related to consumer acceptance hinder the integration of microalgae into mainstream food systems. A key factor among these barriers is the undesirable sensory attributes of microalgae, including intense colour and unpleasant flavours which are commonly described as “fishy,” “grassy,” or “pond-like”, significantly impeding palatability.9 Additionally, the rigid cell walls of many microalgae species limit nutrient bioavailability and digestibility, further reducing their nutritional efficiency.10 These factors have restricted the widespread adoption of microalgae as primary food ingredients despite their exceptional nutritional profiles and environmental credentials.
Biotransformation via microbial fermentation presents an elegant green chemistry solution to these challenges. While microalgae-microbial consortia have been explored for industrial applications, their use for direct food enhancement through probiotic fermentation remains underexplored. By employing carefully selected GRAS edible microorganisms as biocatalysts, the biotransformation of microalgae biomass proceeds without requiring harsh chemicals, high temperatures, or energy-intensive processes. Furthermore, microbial metabolism has the potential to modify the negative organoleptic qualities of microalgae while simultaneously enhancing their nutritional aspects by producing additional bioactive compounds. In this study, we investigate the biotransformation of Chlorella vulgaris microalgal biomass using selected GRAS probiotic bacteria to address the primary limitations hindering the mainstream adoption of microalgal food through a comprehensive platform approach. We employed an enzymatic-based systematic screening pipeline to identify bacterial strains capable of reducing unpleasant sensory attributes while enhancing functional properties, nutrient bioavailability, and bioactive compound profiles. The selected probiotic strain increased the dried biomass weight by 5%, contrasting with the 15–40% weight reduction observed with Bacillus species. Comprehensive analyses using an electronic tongue (E-tongue), olfactory evaluations, liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) confirmed the attenuation of unpleasant flavors and the generation of desirable umami taste and aroma compounds. Collectively, these findings establish a foundation for developing microalgae-based functional foods with improved sensory and nutritional qualities via sustainable biotransformation strategies.
Results and discussion
Establishment of a GRAS microbial fermentation platform for microalgae
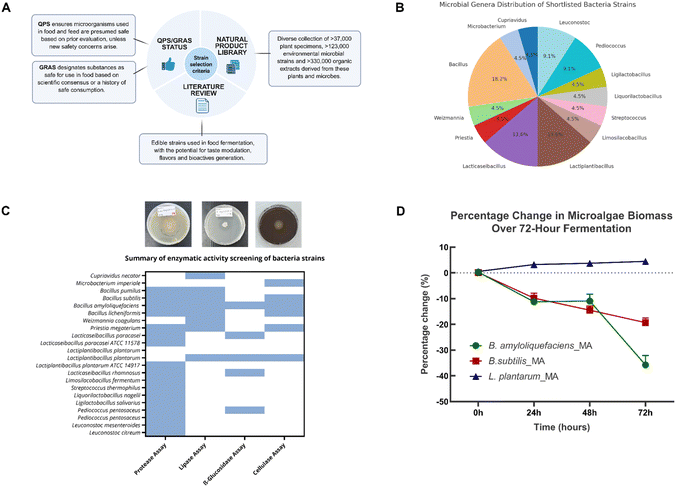
To unlock the nutritional and sensory potential of microalgae through fermentation, we developed a modular bacterial fermentation platform using food-grade bacterial isolates (Fig. 1). The workflow commenced by inoculating microalgae with preselected GRAS strains from the Agency for Science, Technology, and Research's Natural Product Library (A*STAR NPL), a comprehensive resource comprising over 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 plant specimens and 123
000 plant specimens and 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 microbial strains.11 Fermentations were conducted under submerged conditions at 37 °C, followed by freeze-drying and sample extraction for comprehensive chemical and sensory profiling. Analytical outputs were generated using a combination of colorimetric enzyme activity assays, olfactory evaluations, E-tongue measurements, GC-MS and LC-MS. This integrated approach enabled rapid dereplication and high-resolution phenotyping of bacterial fermentates, facilitating the identification of strains and resulting fermentates with desirable nutritional and sensory attributes within the microalgal matrix.
000 microbial strains.11 Fermentations were conducted under submerged conditions at 37 °C, followed by freeze-drying and sample extraction for comprehensive chemical and sensory profiling. Analytical outputs were generated using a combination of colorimetric enzyme activity assays, olfactory evaluations, E-tongue measurements, GC-MS and LC-MS. This integrated approach enabled rapid dereplication and high-resolution phenotyping of bacterial fermentates, facilitating the identification of strains and resulting fermentates with desirable nutritional and sensory attributes within the microalgal matrix.
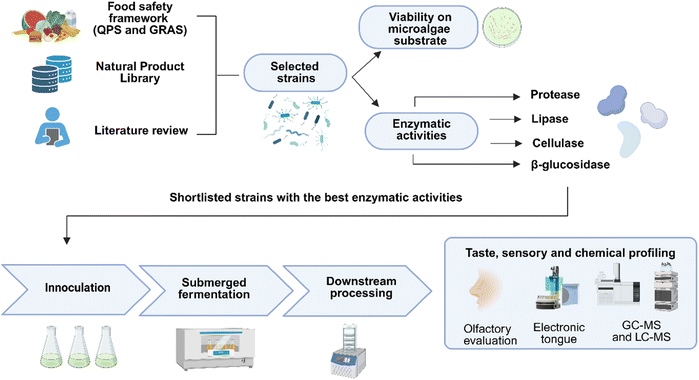 | ||
| Fig. 1 Integrated biotransformation platform showcasing the pipeline for food-grade strain selection, microalgae fermentation, and multi-modal sensory and chemical profiling. | ||
Bacterial strain selection based on bioactivity assays and dried weight yield
Building on the platform's goal to enhance C. vulgaris's suitability for food applications, a systematic screening pipeline was implemented to select bacterial strains capable of improving sensory and nutritional profiles through fermentation. Candidate strains were shortlisted based on food safety status (GRAS), literature evidence supporting their use in food fermentation, enzymatic potential relevant to flavour generation (Fig. 2A), and preliminary sensory profiling. We referred to established safety frameworks to assess food-grade applicability, sourcing shortlisted candidate strains from the A*STAR NPL, a diverse repository for identifying strains with the potential for flavour enhancement and bioactive compound production. Alongside this, literature reviews were conducted to identify strains employed in food fermentations with known abilities to generate desirable flavour profiles and functional metabolites. Based on these criteria, a total of 22 strains spanning 13 microbial genera were selected. The most represented genera include Bacillus (18.2%), Lacticaseibacillus (13.6%), and Lactiplantibacillus (13.6%). The remaining strains belonged to Cupriavidus, Microbacterium, Weizmannia, and Priestia. The shortlisted panel also included other lactic acid bacteria (LAB) genera such as Leuconostoc, Pediococcus, Ligilactobacillus, Liquorilactobacillus, Streptococcus, and Limosilactobacillus (Fig. 2B).The selected strains were subsequently tested in several enzymatic activity assays, including protease, lipase, β-glucosidase, and cellulase assays. Among the tested strains, Bacillus amyloliquefaciens, Bacillus subtilis, and Lactiplantibacillus plantarum demonstrated the highest levels of enzymatic activities (Fig. 2C and Table S1). Even though B. amyloliquefaciens demonstrated superior enzymatic assay results across the board it also produced strong, undesirable odours that would significantly limit its inclusion rate and consumer appeal in a food matrix. Similarly, B. subtilis showed high enzymatic activity but was associated with biomass degradation and potential off flavours. On the other hand, while the selected L. plantarum strain displayed moderate enzymatic activity and did not show strong protease activity, it excelled in sensory attributes and well-documented health benefits crucial for functional food application. Furthermore, the absence of strong proteolytic activity is fairly advantageous, as excessive protein hydrolysis can lead to bitter peptide formation and off-flavors that compromise product acceptability. Moreover, microalgal proteins—such as those from Chlorella and Spirulina—are highly digestible and exhibit favourable amino acid profiles comparable to conventional protein sources.12 Crucially, this allows greater incorporation of the microalgal ingredient into food products, providing nutritional and functional health benefits without compromising sensory quality.
Given that the enzymatic activity profiles and preliminary sensory evaluation (in-house sniff tests) yielded divergent results regarding optimal strain performance, we decided to use dried weight yield analysis to evaluate the scalability and economic viability of the biotransformation platform by the shortlisted strains. We analysed the variations in dried weight during fermentation, a critical parameter often overlooked in microbial fermentation strategies, where substrate loss can undermine sustainability gains. The dry biomass yield of Chlorella vulgaris was monitored over 72 hours of fermentation with the three bacterial strains. Remarkably, L. plantarum fermentation resulted in a progressive biomass increase, gaining ∼5% by 72 hours, while B. subtilis and B. amyloliquefaciens caused substantial biomass losses of ∼15% and ∼40%, respectively (Fig. 2D). This contrasting behaviour likely reflects fundamental differences in metabolic strategies: while Bacillus species catabolize algal biomass as the electron donor for respiration, L. plantarum demonstrates biomass-preserving metabolism for biosynthetic processes during aerobic fermentation, as previously reported for phylogenetically close Lactobacilli.13 The 45% differential in final biomass between L. plantarum (+5%) and B. amyloliquefaciens (−40%) represents a substantial economic advantage for industrial implementation, where feedstock costs typically dominate operational expenses. This biomass enhancement by L. plantarum reflects multiple synergistic mechanisms: (i) accumulation of non-volatile fermentation metabolites including lactic acid, exopolysaccharides,27,28 and aromatic lactates that are retained during lyophilization; (ii) biomass-preserving facultatively heterofermentative metabolism that preferentially utilizes simple nutrients from the medium rather than catabolizing structural microalgal components; and (iii) catalytic biotransformation of aromatic amino acids into bioactive compounds without degrading the bulk biomass as an energy source. Under aerobic fermentation conditions, L. plantarum maintains redox balance through NADH oxidase-mediated NAD+ regeneration, where oxygen serves as an electron acceptor to produce water or hydrogen peroxide rather than supporting complete oxidative catabolism of the biomass. This contrasts fundamentally with Bacillus species, which employ aerobic respiratory metabolism that oxidizes algal biomass to CO2, resulting in substantial weight losses. The dried weight enhancement by L. plantarum thus transforms the fermentation from a degradative process to an accumulative one, where the microalgal biomass serves as a scaffold for functional metabolite production while maintaining its structural integrity. This effectively amplifies the carbon-negative impact of the microalgae feedstock while generating value-added bioactive compounds.
Olfactory profile transformation of microalgae through bacterial fermentation
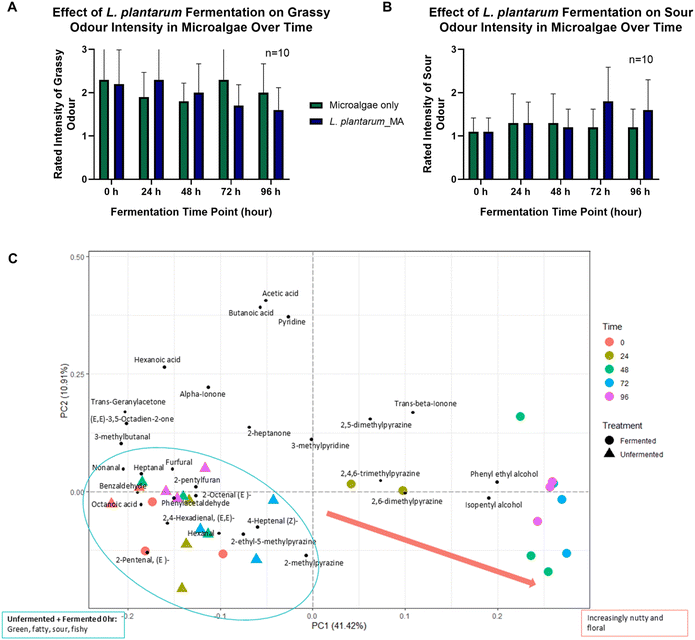
Subsequently, given the critical role of sensory attributes in consumer acceptance of microalgae-based foods, tiered olfactory profile tests and GC-MS analyses were designed to assess L. plantarum's ability to transform C. vulgaris's unpalatable sensory profile, building on its selection for enzymatic and sustainability benefits. We ran a time-course sniff test (n = 10 panellists) on L. plantarum-fermented microalgae samples (Fig. 3A and B). At baseline (0 hours), unfermented microalgae had a strong grassy odour, a known barrier to consumer acceptance. However, fermentation with L. plantarum notably shifted this profile. By 96 hours, the grassy odour had decreased markedly compared to unfermented controls.To confirm this, we used a two-way repeated measures ANOVA—a statistical test that examines how two factors (here, fermentation treatment and time) influence an outcome (odour intensity), while accounting for repeated measurements from the same panellists. There was a significant interaction between treatment and time (P = 0.0188) for grassy odour between the fermented and unfermented samples (declining more in fermented ones). There were no significant main effects for time alone (P = 0.0845) or treatment alone (P = 0.6531), indicating neither factor independently drove the changes without the other. Panellists showed notable differences in their ratings (P < 0.0001), reflecting natural variability in smell perception. Since the data violated the sphericity assumption (a requirement for ANOVA accuracy), we applied the Geisser–Greenhouse correction (ε = 0.8204) to adjust the results. Fermentation also increased sour notes over time. The same ANOVA test also revealed a significant interaction between treatment and time (P = 0.0287) for sour notes, showing that sourness evolved differently based on whether samples were fermented. No significant main effects appeared for time (P = 0.0607) or treatment (P = 0.3431) in isolation (Tables S8–S11). Again, panellist variability was high (P < 0.0001), and we used Geisser–Greenhouse correction for sphericity violation (ε = 0.5751). As negative controls, we assessed fishy and earthy odours, which showed no significant differences (P > 0.05) (Tables S12–S15) between fermented and unfermented samples (Fig. S3), ruling out non-specific effects and strengthening the specificity of grassy and sour changes.
GC-MS analysis corroborated the sensory findings, providing molecular insights into the aroma transformation (Fig. 3C). The volatile profile of microalgae fermented with L. plantarum revealed a significant decrease in polyunsaturated fatty acids (PUFAs)-derived aldehydes such as hexanal, 2-octenal, and 4-heptanol, which are typically associated with lipid oxidation and the undesirable “fishy” and “grassy” odours characteristic of microalgae.14 This reduction in off-flavours was accompanied by an increase in more desirable aroma compounds. Fermentation led to the accumulation of flavour-active compounds, including benzaldehyde, phenylethyl alcohol, pyrazines, and ionone derivatives. The increased production of pyrazines contributed to nutty and roasted notes,15 while phenylethyl compounds introduced floral aromas.16 Notably, pyrazines and ionone derivatives have also been identified as key volatiles in matcha, where they contribute to its roasted and floral-violet-like notes, respectively,17 highlighting their potential role in enhancing the sensory appeal of fermented microalgae. By 96 hours of fermentation, however, the aroma began to develop tangy sour notes of L. plantarum-fermented microalgae. PCA analysis, a technique that visualizes data clustering to show differences, of the volatile compounds showed a clear separation between control and fermented samples over time, indicating a dynamic shift in the volatile metabolome induced by bacterial activity. This separation became more pronounced with extended fermentation, highlighting the progressive nature of the sensory transformation. Together, these beneficial volatile profile modifications substantiate L. plantarum's role in enhancing the sensory qualities of Chlorella, providing the foundation for subsequent taste and metabolite analyses.
Taste modulation and metabolite profile evolution (E-tongue and LC-MS)
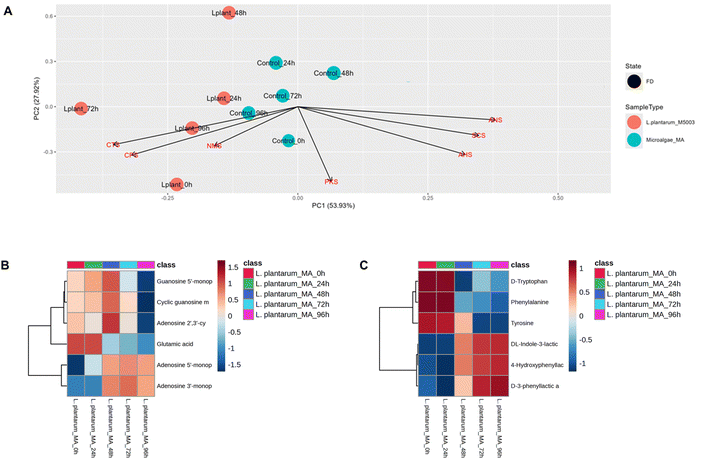
To further enhance C. vulgaris's palatability and characterize its functional benefits, building on the sensory improvements achieved through L. plantarum fermentation, E-tongue and LC-MS analyses were conducted to evaluate taste modulation and metabolite changes. E-tongue analysis revealed a clear divergence between fermented samples and unfermented controls over the 96-hour fermentation period, particularly along principal component axes associated with umami and mild sourness (Fig. 4A). The unfermented controls remained tightly clustered throughout, indicating a relatively stable taste profile and minimal variation in the absence of bacterial activity. In contrast, L. plantarum-fermented samples exhibited progressive shifts in the PCA space, particularly along PC1, reflecting dynamic bacterial-induced changes in taste-active metabolite composition. The distinct clustering of L. plantarum-fermented samples at later time points (48, 72, and 96 hours) suggests an active fermentation-driven metabolic transformation process that continuously alters the taste-active compound profile, while the unfermented controls remain compositionally stable. Overall, the E-tongue findings demonstrated that bacterial fermentation of microalgae leads to substantial shifts in taste properties. LC-MS analysis provided deeper insights into the specific metabolite transformations occurring during bacterial fermentation of microalgae. Fermentation with L. plantarum led to increased levels of umami-enhancing compounds, including glutamic acid and nucleotides such as adenosine monophosphate (AMP) and guanosine monophosphate (GMP) which are well-documented contributors to savoury taste perception18 (Fig. 4B).Additionally, the accumulation of three aromatic acids—indole lactic acid, phenyl lactic acid, and hydroxyphenyl lactic acid—was observed, with concurrent decreases in their amino acid precursors phenylalanine, L-tyrosine, and D-tryptophan (Fig. 4C). These aromatic acids are known for their antioxidant and antimicrobial properties,19 suggesting that L. plantarum fermentation may confer ancillary functional benefits alongside sensory improvements. The metabolite profiles revealed by LC-MS analysis provided mechanistic explanations for the taste changes detected by the E-tongue. The increase in umami compounds in L. plantarum-fermented microalgae likely contributed to the umami-associated PCA shifts, while the production of organic acids possibly influenced the sourness dimension. Altogether, the taste and metabolite enhancements validate the ability of the probiotic-fermented microalgae to produce health-promoting, palatable food ingredients, supporting its sustainability evaluation.
Conclusions
The biotransformation of microalgae via LAB fermentation offers a process-intensified, green alternative to conventional microalgae valorisation strategies. This study demonstrates probiotic fermentation by GRAS-certified Lactiplantibacillus plantarum as a sustainable, scalable biotransformation strategy for edible microalgae. Sensory and metabolomic profiling confirmed significant improvements in aroma, taste, and bioactive compound profiles. Compared to conventional microalgae valorisation methods such as solvent extraction, acid/base hydrolysis, and isoelectric precipitation—each associated with high E-factors (5–30), chemical inputs, and significant energy demands—the probiotic fermentation platform developed in this study offers a substantial green advancement (Table 1). Operating under mild, aerobic conditions without solvents or pH-altering agents, the platform enables biomass preservation (+5%) while generating functional food ingredients directly from CO2-derived biomass. Crucially, the process is catalytic rather than stoichiometric: aromatic amino acids are converted into antioxidant lactates without degrading the bulk biomass. This fermentation avoids post-processing steps, maintains a carbon-negative profile, and valorises all biomass components. These attributes align with Green Chemistry principles 1 (waste prevention), 6 (energy efficiency), 9 (catalysis), and 10 (design for degradation), positioning this strategy as a model for sustainable food biomanufacturing.| Green chemistry principle | Probiotic fermentation | Chemical processing | Enzymatic processing | Green advance & literature support |
|---|---|---|---|---|
| a Data adapted from ref. 20–26. | ||||
| (1) Prevention | Minimal organic waste | High chemical waste (e.g., acidic/alkaline streams, salts) | Moderate enzyme waste | Reduced waste streams |
| (2) Atom economy | >95% theoretical yield | ∼60–70% yields (with losses due to byproducts) | 80–85% yields | Higher atom incorporation |
| (3) Less hazardous synthesis | Water and minerals only | Strong acids/bases (HCl, NaOH, KOH) | Mild enzyme solutions | Eliminates corrosive chemicals |
| (4) Safer solvents | Water-based system | Organic solvents required | Aqueous systems | Zero organic solvents |
| (5) Energy efficiency | 25–45 °C | Often 100–370 °C for thermal–chemical methods | 40–60 °C | Mild temperature operation |
| (6) Catalysis | Whole-cell biocatalyst | Chemical/thermal treatment | Isolated enzymes | Self-contained biocatalysis |
The demonstrated biomass enhancement improves process economics by eliminating typical feedstock losses, while mild fermentation conditions (37 °C, 72 hours) are compatible with existing food fermentation infrastructure. Key scale-up considerations include maintaining consistent inoculum quality and optimizing aeration strategies to preserve the observed biomass-enhancing metabolism across different microalgae batches. In conclusion, through microalgae fermentation by L. plantarum, we achieved a reduction in off-flavour aldehydes, enhancement in umami compounds, and generation of bioactive metabolites—all without chemical additives or complicated downstream processing—thereby advancing microbial fermentation as both a sustainable bioprocessing platform and a practical route to circular food innovation.
Author contributions
Po-Hsiang Wang: conceptualization, writing original draft, manuscript editing and supervision; Zann Yi Qi Tan: lab work, writing original draft and manuscript editing; Choy Eng Nge: lab work, manuscript writing, review & editing; Nurhidayah Basri: lab work, manuscript writing, review & editing; Lina Xian Yu Lee: manuscript writing, review & editing; Aaron Thong: manuscript review and editing; Elaine Chin: manuscript review & editing; Sharon Crasta: manuscript review and editing; Geraldine Chan: manuscript review & editing; Mario Wibowo: manuscript review and editing; Yoganathan Kanagasundaram: manuscript review & editing, supervision; Siew Bee Ng: manuscript review & editing, supervision. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no competing interests.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: supplementary materials and methods, supplementary figures. See DOI: https://doi.org/10.1039/d5fb00492f.Acknowledgements
This research is supported by the National Research Foundation, Prime Minister's Office, Singapore, under its Campus for Research Excellence and Technological Enterprise (CREATE) programme [Proteins4Singapore] and Agency for Science, Technology and Research (A*STAR). We also thank Ryan Soh Han Wei for the use of his custom R script for the E-tongue analysis and Naedia Loh Xuan Yi for assisting in sample extraction and LCMS operations.References
- C. L. Lumsden, J. Jägermeyr, L. Ziska and J. Fanzo, One Earth, 2024, 7, 1187–1201 Search PubMed.
- E. Daneshvar, R. J. Wicker, P.-L. Show and A. Bhatnagar, Chem. Eng. J., 2022, 427, 130884 Search PubMed.
- M. Chen, Y. Chen and Q. Zhang, Sci. Total Environ., 2024, 948, 174462 Search PubMed.
- F. Khavari, M. Saidijam, M. Taheri and F. Nouri, Mol. Biol. Rep., 2021, 48, 4757–4765 CAS.
- Y. Zhao, Q. Wang, D. Gu, F. Huang, J. Liu, L. Yu and X. Yu, Bioresour. Technol., 2024, 393, 130093 Search PubMed.
- C. Chen, T. Tang, Q. Shi, Z. Zhou and J. Fan, Trends Food Sci. Technol., 2022, 126, 99–112 CrossRef CAS.
- C. Xue and I.-S. Ng, Bioresour. Technol., 2022, 343, 126089 CrossRef CAS.
- F. Sandgruber, A. Gielsdorf, A. C. Baur, B. Schenz, S. M. Müller, T. Schwerdtle, G. I. Stangl, C. Griehl, S. Lorkowski and C. Dawczynski, Mar. Drugs, 2021, 19, 310 CrossRef CAS PubMed.
- M. L. Olsen, K. Olsen and P. E. Jensen, Physiol. Plant., 2024, 176, e14337 CrossRef CAS PubMed.
- L. Machado, G. Carvalho and R. N. Pereira, Biomass, 2022, 2, 80–102 CrossRef CAS.
- F. L. Sirota, F. Goh, K.-N. Low, L.-K. Yang, S. C. Crasta, B. Eisenhaber, F. Eisenhaber, Y. Kanagasundaram and S. B. Ng, J. Genomics, 2018, 6, 63–73 CrossRef.
- S. L. Ma, S. Sun, T. Z. Li, Y. J. Yan and Z. K. Wang, Crit. Rev. Food Sci. Nutr., 2024, 1–24 Search PubMed.
- M. J. A. Stevens, A. Wiersma, W. M. de Vos, O. P. Kuipers, E. J. Smid, D. Molenaar and M. Kleerebezem, Appl. Environ. Microbiol., 2008, 74, 4776–4778 CrossRef CAS PubMed.
- B. Coleman, C. Van Poucke, B. Dewitte, A. Ruttens, T. Moerdijk-Poortvliet, C. Latsos, K. De Reu, L. Blommaert, B. Duquenne, K. Timmermans, J. van Houcke, K. Muylaert and J. Robbens, Future Foods, 2022, 5, 100139 CrossRef CAS.
- F. Wang, H. Shen, T. Liu, X. Yang, Y. Yang and Y. Guo, Foods, 2021, 10, 273 CrossRef CAS PubMed.
- H. Issa-Issa, G. Guclu, L. Noguera-Artiaga, D. López-Lluch, R. Poveda, H. Kelebek, S. Selli and Á. A. Carbonell-Barrachina, Food Chem., 2020, 316, 126353 CrossRef CAS.
- Y. Luo, Y. Zhang, F. Qu, P. Wang, J. Gao, X. Zhang and J. Hu, Foods, 2022, 11, 2964 CrossRef CAS PubMed.
- J. Diepeveen, T. C. W. Moerdijk-Poortvliet and F. R. van der Leij, J. Food Sci., 2022, 87, 1449–1465 CrossRef CAS PubMed.
- A. Bensid, N. El Abed, A. Houicher, J. M. Regenstein and F. Özogul, Crit. Rev. Food Sci. Nutr., 2020, 62, 2985–3001 CrossRef PubMed.
- A. Kose and S. S. Oncel, Biotechnol. Rep., 2015, 6, 137–143 CrossRef CAS.
- F. Almomani, H. Hosseinzadeh-Bandbafha, M. Aghbashlo, A. Omar, S.-W. Joo, Y. Vasseghian, H. Karimi-Maleh, S. Shiung Lam, M. Tabatabaei and S. Rezania, Chem. Eng. J., 2023, 455, 140588 CrossRef CAS.
- G. Karabulut, A. Purkiewicz and G. Goksen, Compr. Rev. Food Sci. Food Saf., 2024, 23, e13372 CrossRef CAS.
- L. Soto-Sierra, L. R. Wilken and C. K. Dixon, Bioresour. Bioprocess., 2020, 7, 1–14 CrossRef.
- O. Babich, S. Ivanova, P. Michaud, E. Budenkova, E. Kashirskikh, V. Anokhova and S. Sukhikh, Biotechnol. Rep., 2025, 41, e00827 CrossRef PubMed.
- P. S. Corrêa, W. G. Morais Júnior, A. A. Martins, N. S. Caetano and T. M. Mata, Processes, 2020, 9, 10 CrossRef.
- S. Bleakley and M. Hayes, Foods, 2017, 6, 33 CrossRef PubMed.
- S. V. Dilna, H. Surya, R. G. Aswathy, K. K. Varsha, D. N. Sakthikumar, A. Pandey and K. M. Nampoothiri, LWT--Food Sci. Technol., 2015, 64, 1179–1186 CrossRef CAS.
- Y. M. Abouelela, M. A. Hassan and A. M. Abdelaziz, BMC Microbiol., 2023, 23, 361 CrossRef.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2026 |