Open Access Article
Open Access ArticleSustainable enhancement of conjugated linoleic acid (CLA) production in lactic acid bacteria cocultures via ethanol permeabilization
Lan
Cheng
,
Miao
Wang
,
Yingying
Wang
,
Yumeng
Chang
,
Yan
Ding
and
Shuhong
Ye
 *
*
State Key Laboratory of Marine Food Processing & Safety Control, School of Food Science and Technology, Dalian Polytechnic University, Dalian, Liaoning 116034, China. E-mail: shuhongye@dlpu.edu.cn
First published on 9th December 2025
Abstract
Conjugated linoleic acid (CLA) is a highly value added active lipid that could be utilized as a functional food ingredient. This study introduces an innovative, low-energy biocatalytic approach utilizing lactic acid bacteria (LAB) to enhance CLA production in a sustainable manner. A synergistic coculture of Lactobacillus acidophilus (La) and Lactobacillus plantarum (Lp) was developed, wherein La's rapid acidification (pH < 5.5) effectively activated Lp's linoleate isomerase (LAI). Notably, a food-grade ethanol-based permeabilization method—a non-invasive and environmentally friendly processing technique—was applied to modify cell membranes (confirmed via SEM), thereby facilitating efficient uptake and intracellular conversion of linoleic acid (LA) by LAI, resulting in both cis → trans and Δ12 → Δ11 isomerization. The coculture of La![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lp in MRS media containing 500 µg mL−1 LA exhibited a CLA conversion of 41.3%. When permeabilized cells were used in skim milk—an underutilized dairy byproduct—the CLA yield increased to 220.7 µg mL−1 (44.1% conversion), demonstrating the potential for direct integration into dairy fermentation processes. This research establishes ethanol permeabilization as a promising tool in circular bioengineering, enabling energy-efficient CLA production (<50 °C, no toxic reagents) with minimal process waste, offering new opportunities for the production of CLA-enhanced milk products.
Lp in MRS media containing 500 µg mL−1 LA exhibited a CLA conversion of 41.3%. When permeabilized cells were used in skim milk—an underutilized dairy byproduct—the CLA yield increased to 220.7 µg mL−1 (44.1% conversion), demonstrating the potential for direct integration into dairy fermentation processes. This research establishes ethanol permeabilization as a promising tool in circular bioengineering, enabling energy-efficient CLA production (<50 °C, no toxic reagents) with minimal process waste, offering new opportunities for the production of CLA-enhanced milk products.
Sustainability spotlightThis study develops a low-energy biocatalytic platform (operating below 50 °C) through food-grade ethanol permeabilization of lactic acid bacteria cocultures, enabling the conversion of dairy sidestreams—specifically skim milk—into conjugated linoleic acid (CLA)-enriched functional foods with a yield of 44.1%. By eliminating the use of toxic solvents, reducing thermal energy consumption by over 30% compared to conventional methods, and promoting the utilization of underutilized resources, this approach directly supports the objectives of zero-waste biomanufacturing (SDG 12), sustainable nutrition security (SDG 2), and green industrial innovation (SDG 9). |
1. Introduction
Conjugated linoleic acid (CLA) comprises LA isomers differing in double bond location (e.g., 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, 9
9, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11, 10
11, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12, and 11
12, and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13) and geometry (i.e., cis or trans).1 Due to its many health promoting activities such as anticarcinogenic,2 antiatherosclerotic, antidiabetogenic,3 body mass enhancing, antioxidative, immunomodulatory, antimicrobial, hypocholesterolemic, and anabolic attributes,4 there has been increasing scientific interest in this mixture. Amongst them, cis-9, trans-11 (rumenic acid) and trans-10, cis-12 are the more bioactive ones, which exhibit potent anticancer activity and cause reduction in body fat, respectively.5
13) and geometry (i.e., cis or trans).1 Due to its many health promoting activities such as anticarcinogenic,2 antiatherosclerotic, antidiabetogenic,3 body mass enhancing, antioxidative, immunomodulatory, antimicrobial, hypocholesterolemic, and anabolic attributes,4 there has been increasing scientific interest in this mixture. Amongst them, cis-9, trans-11 (rumenic acid) and trans-10, cis-12 are the more bioactive ones, which exhibit potent anticancer activity and cause reduction in body fat, respectively.5
CLA exists naturally in animal foods having ruminants' origin (meat, milk, etc.) as an intermediate product of ruminal biohydrogenation (BH).6–9 This process, mediated by rumen microorganisms, is a primary pathway for CLA formation.10,11 Additionally, mammary Δ9-desaturase-mediated transformation of VA (trans-11 C18:1) elevates milk VA concentration.12,13 Beyond natural sources, microbial synthesis of CLA in vitro has emerged as a promising strategy. Specific bacteria including LAB, bifidobacteria and propionibacteria catalyze the conversion of LA to CLA by linoleate isomerase (LAI).14,15 Notably, LAB strains such as Lactobacillus acidophilus, Lactobacillus plantarum, and Lactobacillus casei have demonstrated CLA-producing capabilities in both culture media and milk matrices.16–18
Despite these promising health benefits and production pathways, a critical challenge in optimizing microbial CLA synthesis lies in overcoming the physical barriers posed by intact cell walls and membranes, which restrict substrate access to intracellular LAI. Coculture systems leverage microbial synergy to enhance metabolic outputs, offering advantages over monocultures in biotechnological applications.19,20 While LAB cocultures are widely employed in dairy fermentation, their potential for CLA production remains underexplored. The enhanced CLA production is attributed to a dual mechanism: (i) synergistic interaction in coculture where La's acidification activates Lp's LAI, and (ii) ethanol permeabilization that increases membrane permeability to facilitate substrate access to intracellular enzymes. To address this, cell permeabilization – a process that increases membrane permeability while preserving enzymatic activity – was implemented as a key innovation in this study. By disrupting cell integrity through ethanol treatment, permeabilized whole cells enable direct interaction between LA and LAI, thereby maximizing catalytic efficiency.21
This work investigates the synergistic effects of LAB cocultures on CLA production, with a focus on the mechanistic role of cell permeabilization. We evaluate how strain ratios, LA concentration, and fermentation duration influence CLA isomer profiles and yields. Furthermore, we demonstrate that permeabilized whole-cell systems significantly enhance LAI activity and CLA conversion rates, particularly in skim milk – a finding with critical implications for developing CLA-enriched functional foods.
2. Materials and methods
2.1. Chemicals
Lipid standards were sourced from Sigma Chemical Co. in St. Louis, MO. For the culture medium, we used MRS medium, which we got from Difco out of Detroit, MI. Skim milk was also used. All other chemicals needed for the fatty acid analysis were of analytical grade and came from Fisher in Springfield, NJ.2.2. Strains and growth conditions
Lactobacillus plantarum, Lactobacillus acidophilus, Streptococcus thermophilus, Lactobacillus bulgaricus, Lactobacillus fermentum, Lactobacillus casei, and Lactobacillus breris (preserved in the laboratory) were used.All seven strains were subcultured twice in a glass tube with 5 mL of MRS medium at 37 °C, 24 h, except for S. thermophilus, which was grown at 42 °C, while other growth conditions were similar to those for other LAB strains. The active culture was split for increased production and testing via fermentation.
2.3. Preparation of LA and LA emulsion
LA and Tween-80 at a ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (m/m) were dissolved in deionized water. To form a stable LA emulsion, the mixtures were treated with an ultrasound cell disruptor (SCIENTZ-IID).
1 (m/m) were dissolved in deionized water. To form a stable LA emulsion, the mixtures were treated with an ultrasound cell disruptor (SCIENTZ-IID).
2.4. Preparation of whole cells
Two percent (v/v) of pre-cultured LAB strains was inoculated in MRS medium containing 0.75 mg mL−1 LA and incubated statically at 37 °C for 24 h. Cells were harvested by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 minutes, it was washed twice with physiological saline and treated with (70% v/v) ethanol at 37 °C for 5 min to permeabilize the cell membrane.21 Ethanol was selected as a food-grade, environmentally benign permeabilizing agent for this study. The permeabilization parameters (70% v/v ethanol, 37 °C, 5 min) were adopted from an established protocol to ensure effective membrane disruption while preserving enzymatic activity.21 The cells were centrifuged again at 10
000 × g for 15 minutes, it was washed twice with physiological saline and treated with (70% v/v) ethanol at 37 °C for 5 min to permeabilize the cell membrane.21 Ethanol was selected as a food-grade, environmentally benign permeabilizing agent for this study. The permeabilization parameters (70% v/v ethanol, 37 °C, 5 min) were adopted from an established protocol to ensure effective membrane disruption while preserving enzymatic activity.21 The cells were centrifuged again at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 minutes and then suspended in phosphoric acid–citric acid buffer (pH 7.0). This permeabilization step disrupts the cell wall and membrane barriers, enabling direct access of LA to intracellular LAI, thereby enhancing enzymatic activity.
000 × g for 10 minutes and then suspended in phosphoric acid–citric acid buffer (pH 7.0). This permeabilization step disrupts the cell wall and membrane barriers, enabling direct access of LA to intracellular LAI, thereby enhancing enzymatic activity.
2.5. Scanning Electron Microscopy (SEM) analysis
In order to observe morphological changes in LAB cells before and after permeabilization, we have used Scanning Electron Microscopy (SEM). Cells from both untreated and permeabilized La–Lp coculture groups were centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 10 min), washed twice with 0.1 M phosphate buffer solution at pH 7.0, fixed with 2.5% glutaraldehyde solution and incubated at 4 °C for 12 hours. They were then dehydrated through different concentrations of ethanol solutions: 30%, 50%, 70%, 90%, and 100%. The dehydration steps mentioned above were repeated until all remaining liquids evaporated on the test slides. The specimens were critical-point dried using carbon dioxide, vacuum transferred to stainless steel rings and sputter coated with gold-palladium (5 nm thick). Finally, imaging was carried out at a 5 kV accelerating voltage with a Hitachi SU8010 Scanning Electronic Microscope (Hitachi High-Tech., Japan) operated at a working distance of 8 mm. Three independent replicates were analyzed for each group.
000 × g, 10 min), washed twice with 0.1 M phosphate buffer solution at pH 7.0, fixed with 2.5% glutaraldehyde solution and incubated at 4 °C for 12 hours. They were then dehydrated through different concentrations of ethanol solutions: 30%, 50%, 70%, 90%, and 100%. The dehydration steps mentioned above were repeated until all remaining liquids evaporated on the test slides. The specimens were critical-point dried using carbon dioxide, vacuum transferred to stainless steel rings and sputter coated with gold-palladium (5 nm thick). Finally, imaging was carried out at a 5 kV accelerating voltage with a Hitachi SU8010 Scanning Electronic Microscope (Hitachi High-Tech., Japan) operated at a working distance of 8 mm. Three independent replicates were analyzed for each group.
2.6. Determination of pH and microbial growth
The pH of the culture media was measured at room temperature (20 ± 2 °C) using a digital pH meter (Hanna, Padova, Italy) with a combined electrode.Bacterial growth was checked during fermentation according to the values obtained by measuring OD600 and cell counts in CFU mL−1.
2.7. Fermentation experiments
All fermentations were conducted as batch processes. Fermentations with seven LAB strains were carried out in MRS medium. Inocula were prepared as follows: all 7 strains were transferred from 4 °C stock or 80 °C stock culture and subcultured twice in MRS medium at 37 °C, except for S. thermophilus, which was cultured at 42 °C. Subcultured cultures were grown again to activate them; each LAB strain sub-cultured separately at OD = 0.1 into 10 mL MRS containing 300 µg mL−1 of LA as the substrate for evaluation of conjugation of unsaturated fatty acids in fermented products such as yogurt. For the coculture study, each of the 21 pairs consisted of one strain sub-cultured individually into 5 mL MRS, and its OD adjusted before inoculating 10 mL of MRS medium supplemented with 300 µg mL−1 of LA, thereby having an activation culture. They were then combined in a sterile vessel, all at the same time and used only once, so that no transfer of cells between each pair of bacteria takes place. For the focused co-culture experiments with La and Lp, fresh pre-cultures of each strain were adjusted to a standard optical density (OD600) and then mixed at defined ratios (e.g., 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) prior to inoculation to ensure a consistent starting population. The stability of the co-culture during fermentation was monitored by tracking the total viable cell counts (CFU mL−1) over time. The cultures formed 21 groups consisting of different strain combinations. Cocultures of strains were activated for 24 h, respectively. Only the mixed strains showing the highest CLA production were subsequently tested in MRS medium.
1) prior to inoculation to ensure a consistent starting population. The stability of the co-culture during fermentation was monitored by tracking the total viable cell counts (CFU mL−1) over time. The cultures formed 21 groups consisting of different strain combinations. Cocultures of strains were activated for 24 h, respectively. Only the mixed strains showing the highest CLA production were subsequently tested in MRS medium.
Fermentation experiments using the above-mentioned selected strains in MRS + 0.3% LA were performed for 24 h to observe the influence caused due to ratio variations on the proportion of CLA isomers. Fermentation experiments were carried out with the selected strains at an appropriate ratio with different LA concentrations and different incubation times to investigate conditions for CLA production between MRS medium and skim milk medium. In the comparative study of single strains and coculture of strains on the CLA-producing activity, sampling was performed at 12, 24, 36, 48, 60 and 72 h of incubation under appropriate conditions.
2.8. Determination of CLA
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) chloroform
1 (v/v) chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol solution. This mixture was then spun down at 3800 × g for 20 minutes at 4 °C. Following centrifugation, the organic layer was carefully separated, dried using anhydrous sodium sulfate to eliminate any water, and concentrated under vacuum at 30 °C. Finally, the resulting sample was brought up to a final volume of 10 mL with hexane in a volumetric flask for subsequent quantification.20
methanol solution. This mixture was then spun down at 3800 × g for 20 minutes at 4 °C. Following centrifugation, the organic layer was carefully separated, dried using anhydrous sodium sulfate to eliminate any water, and concentrated under vacuum at 30 °C. Finally, the resulting sample was brought up to a final volume of 10 mL with hexane in a volumetric flask for subsequent quantification.20
The LA and CLA extract was collected and dried under a stream of nitrogen gas at 70 °C using an evaporator. The dried stuff was then redissolved in 500 µL of hexane and derivatized into methyl esters by reacting it with 1 mL of 5.0% (v/v) HCl in methanol at 100 °C for an hour.23 We analyzed the methyl esters of CLA using a gas chromatography (GC) system. It is important to note that these extraction and derivatization steps were performed for the analytical quantification and identification of CLA within the fermentation matrix. The produced CLA was not isolated as a separate, purified compound, as the study focused on in situ yield and isomer distribution.
2.9. GC-MS analysis
Following the method outlined by Nakao et al.,22 the methyl esters of CLA and LA were analyzed using an Agilent Technologies HP7890 GC system (Wilmington, Delaware, USA). This setup included an automatic injector and a flame ionization detector (FID). We used an HP-50 capillary column (60 m × 0.25 mm inner diameter × 0.20 µm film thickness) to separate the compounds. We injected 1 µL of each sample. The GC conditions were as follows: the injector and detector were both set to 250 °C. The oven temperature was programmed to start at 90 °C, hold for five minutes, and then increase the temperature to 180 °C at a rate of 10 °C per minute. It was held at this temperature for an additional 10 minutes. After that, the temperature was increased to 220 °C at 5 °C per minute and held for 10 minutes before finally reaching 250 °C at 5 °C per minute, with a final hold of 10 minutes. Hydrogen was used as the carrier gas to move everything along.2.10. Statistical analysis
We made sure to run each experiment at least three times. The data are presented as mean values, along with their corresponding standard deviations (SDs). We crunched all the numbers and processed the data using specialized software.3. Results and discussion
3.1. Conversion of LA to CLA by coculturing of LAB
Employing diverse food-safe bacteria for the production of CLA isomers during food processing, notably as starter cultures, offers potential for enhanced nutritional profiles. The synthesis of CLA from free LA by LAB strains has been documented in numerous studies.25 It was found that the addition of probiotic bacteria—specifically Lactobacillus rhamnosus and strains of Propionibacterium freudenreichii (subsp. shermanii 56, shermanii 51, and subsp. freudenreichii 23)—to yogurt starter cultures (Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus salivarius subsp. thermophilus) synergistically increased the CLA-producing ability of fermented milk. What's more, when L. rhamnosus is combined with standard yogurt cultures, the resulting fermented milk boasts the highest levels of CLA.26 This indicated the feasibility of coculturing LAB strains. However, few studies concentrate on production of free LA by coculture of LAB strains. In the present study, the mixed LAB strains were evaluated on unbound LA's transformation into bound CLA. We found that they all have the ability to convert LA to CLA, and CLA production was almost higher than that of single strains (Table 1). CLA production by coculture of different LAB strains was shown for the first time in this study. La cocultured with other LAB strains showed the higher CLA-producing ability than the others, and La and Lp gave the highest CLA production of 107.9 µg mL−1 (LA conversion rate 35.97%) (Table 1). Compared to other LAB strains, such as Lactobacillus acidophilus CRL730 (23.8%), L. acidophilus Q42 (20.0%), Lactobacillus casei CRL87 (17%), and Lactobacillus plantarum NCUL005 (27.6%), the coculture exhibited a relatively high LA conversion rate. Meanwhile, this observation is in agreement with the study that the co-culture of La and Lp (La–Lp) produced more CLA than the co-culture of La and St (La–St).27 It is possible that La has a good compatibility with other LAB strains, which can be used as a starter culture in fermented dairy products. Therefore, further mechanistic studies will focus on La and Lp, which were selected for coculture.Count (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU mL−1) CFU mL−1) |
pH | CLA (µg mL−1) | |
|---|---|---|---|
| LAB strains | |||
| La | 9.11 | 4.53 | 60.3 ± 2.3e |
| Lp | 8.97 | 4.73 | 69.2 ± 1.8e |
| Lf | 10.45 | 4.68 | 80.6 ± 2.2c |
| Lc | 8.86 | 4.77 | 60.7 ± 2.7e |
| Lb | 10.19 | 4.76 | 75.6 ± 2.4d |
| Lbu | 8.73 | 4.69 | 23.1 ± 2.1g |
| St | 9.05 | 4.74 | 51.2 ± 1.5f |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Coculture of LAB strains | |||
| La and Lp | 9.22 | 4.62 | 107.9 ± 2.2a |
| La and Lf | 10.34 | 4.61 | 87.4 ± 2.9c |
| La and Lc | 9.33 | 4.63 | 98.6 ± 2.4b |
| La and Lb | 8.99 | 4.57 | 81.3 ± 2.2c |
| La and Lbu | 8.57 | 4.61 | 73.7 ± 1.9d |
| La and St | 9.47 | 4.59 | 89.6 ± 2.5c |
| Lp and Lf | 10.67 | 4.51 | 67.2 ± 1.7e |
| Lp and Lc | 9.41 | 4.70 | 79.4 ± 3.0d |
| Lp and Lb | 9.89 | 4.72 | 69.9 ± 2.3e |
| Lp and Lbu | 9.14 | 4.77 | 53.2 ± 1.8f |
| Lp and St | 8.97 | 4.81 | 60.5 ± 1.4e |
| Lf and Lc | 10.18 | 4.55 | 86.4 ± 1.6c |
| Lf and Lb | 9.77 | 4.68 | 75.2 ± 2.7d |
| Lf and Lbu | 8.49 | 4.69 | 63.0 ± 1.9e |
| Lf anf St | 9.56 | 4.72 | 60.3 ± 1.6e |
| Lc and Lb | 9.38 | 4.77 | 76.9 ± 2.5d |
| Lc and Lbu | 8.63 | 4.75 | 66.1 ± 2.8e |
| Lc and St | 8.85 | 4.71 | 55.2 ± 2.1f |
| Lb and Lbu | 9.12 | 4.75 | 64.9 ± 1.5e |
| Lb and St | 9.74 | 4.74 | 79.1 ± 2.0d |
| Lbu and St | 9.09 | 4.69 | 59.9 ± 2.4f |
3.2. Permeabilization enhances LAI activity and CLA production
SEM analysis provided direct visual evidence of the impact of ethanol-based permeabilization on LAB cell integrity. The untreated cells had smooth, intact surfaces and typical rod shape (Lactobacillus) morphologies (Fig. 1a). In contrast, permeabilized cells displayed pronounced structural modifications, including surface roughness and partial collapse of the cellular architecture (Fig. 1b). These observations align with previous studies demonstrating that ethanol permeabilization disrupts lipid bilayers and increases membrane fluidity, thereby facilitating substrate access to intracellular enzymes.28 | ||
| Fig. 1 SEM image of LAB cells (La–Lp co-culture). Untreated cells show intact membranes and smooth surfaces (a). Washed cells after permeabilization (b). | ||
Such structural changes likely reduce diffusion barriers for LA, enabling efficient interaction with LAI and subsequent CLA biosynthesis. Following permeabilization treatment, the cellular morphology remained intact, with no visible pores or cytoplasmic leakage observed. Although structural alterations occurred in the membrane of permeabilized cells—manifested as increased permeability—the overall cellular architecture was preserved—a critical feature for maintaining enzymatic activity during prolonged fermentation.29
These findings underscore the mechanistic basis for the 2.3-fold increase in CLA production in permeabilized cocultures (Table 4). By bridging SEM-derived structural insights with biochemical data, this work establishes a direct link between cell permeability and catalytic efficiency in microbial CLA synthesis. The ethanol permeabilization treatment, performed under conditions (70% v/v, 37 °C, 5 min) selected based on prior literature,21 proved to be a critical step for enhancing substrate access to intracellular LAI, thereby significantly contributing to the elevated CLA production observed in this study.
3.3. Combined UV spectrophotometry and GC-MS for quantitative and qualitative analysis of conjugated linoleic acid (CLA)
The gas chromatogram of CLA standards (Fig. 2a) and LA methyl ester (Fig. 2b) revealed the following retention times: 26.044 min for c9,t11-CLA, 26.183 min for t10,c12-CLA, 26.824 min for the t, t mixture (t,t-CLA) and 25.080 min for LA, respectively. The peak of the conjugated double bond's extinction spectrum is 232–234 nm. A linear relationship was observed between the absorption at 233 nm and the standard CLA concentration within the range of 0–12 µg mL−1. The standard curve of CLA UV absorption at 233 nm at room temperature is expressed by the following formula: y = 0.0791x + 0.014 (R2 = 0.999). | ||
| Fig. 2 Gas chromatogram for the conjugated linoleic acid isomer standards (a), LA (b) and the fatty acids of fermentation broth (c). | ||
There are many methods of analysis, including gas–liquid chromatography, silver-ion high-performance liquid chromatography, nuclear magnetic resonance (NMR), and gas chromatography-mass spectrometry (GC-MS). The UV method is used more commonly than other methods; however, it only determines the total CLA production and does not give the distribution of the content of CLA isomers. So we combined GC-MS with UV to conduct the quantitative and qualitative determination of CLA in the study.
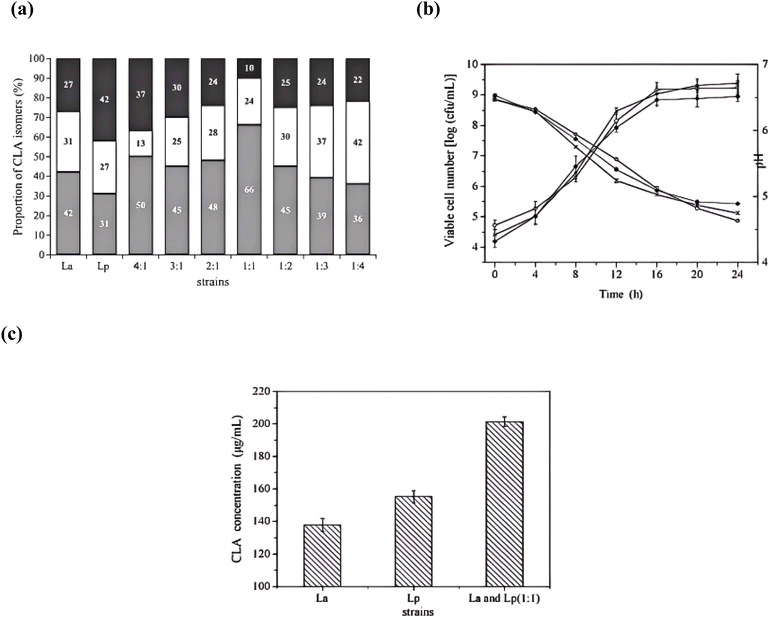
3.4. The effect of different proportions of La and Lp on the proportion of CLA isomers
The effect of different proportions of La and Lp on the total CLA production was investigated, and the CLA production reached the maximum rates when the coculture was inoculated at 37 °C for 48 h in a skim milk medium containing 300 µg mL−1 of LA, with a specific strain ratio of La![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lp = 1
Lp = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 v/v.27 However, the CLA isomer distribution was not studied. In the present study, qualitative analysis revealed that the c9,t11-CLA, t10,c12-CLA, and t,t-CLA isomers were the three main isomers (Fig. 2c) (Table 2). The total CLA production reached a maximum of 116.6 µg mL−1 when La was in coculture with Lp, with specific strain ratios of 1
4 v/v.27 However, the CLA isomer distribution was not studied. In the present study, qualitative analysis revealed that the c9,t11-CLA, t10,c12-CLA, and t,t-CLA isomers were the three main isomers (Fig. 2c) (Table 2). The total CLA production reached a maximum of 116.6 µg mL−1 when La was in coculture with Lp, with specific strain ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (v/v) in the MRS medium containing 300 µg mL−1 of LA (Table 2), and quantitative analysis revealed that the c9,t11-CLA, t10,c12-CLA, and t,t-CLA isomers comprised about 50% (w/w), 13% (w/w) and 37% (w/w) of total CLA produced by the coculture, respectively (Fig. 3a). When La was in coculture with Lp at a ratio of 1
4 (v/v) in the MRS medium containing 300 µg mL−1 of LA (Table 2), and quantitative analysis revealed that the c9,t11-CLA, t10,c12-CLA, and t,t-CLA isomers comprised about 50% (w/w), 13% (w/w) and 37% (w/w) of total CLA produced by the coculture, respectively (Fig. 3a). When La was in coculture with Lp at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, quantitative analysis revealed that the c9,t11-CLA, t10,c12-CLA, and t,t-CLA isomers comprised about 66% (w/w), 24% (w/w) and 10% (w/w) of total CLA produced by the coculture, respectively (Fig. 3a). The different proportions of La and Lp did not affect CLA production. However, they did affect the relative composition of isomers. There were obvious differences in CLA isomer distribution. We discovered that t,t-CLA produced by coculture of La an Lp had a relative reduction in comparison to Lp, and it seemed to favour the formation of t10,c12-CLA when La took up a larger proportion in the coculture. We can conclude that the coculture is able to change different kinds of CLA isomer contents. This result is similar to the research showing that the addition of 0.1% LA to a mixture of yogurt cultures and L. acidophilus significantly increased the content of c9,t11-CLA in nonfat set yogurt.30 The proportion of CLA isomers formed by La or Lp was studied previously, and it showed that c9,t11-CLA, t10,c12-CLA and t,t-CLA isomers were the primary compositions, and t,t-CLA isomers took up the higher proportion.31 It has been reported that c9,t11-CLA shows anticancer activity32 and t10,c12-CLA shows activities that decrease body fat,33 which is beneficial to health, while the bioactivity of t,t-CLA is lower than c9,t11-CLA.33 Thus t,t-CLA isomers were not expected to enrich in the fermented diary food. The coculture of La and Lp reduced the t,t-CLA content obviously, causing a desirable enrichment in c9,t11-CLA with specific strain ratios of 1
1, quantitative analysis revealed that the c9,t11-CLA, t10,c12-CLA, and t,t-CLA isomers comprised about 66% (w/w), 24% (w/w) and 10% (w/w) of total CLA produced by the coculture, respectively (Fig. 3a). The different proportions of La and Lp did not affect CLA production. However, they did affect the relative composition of isomers. There were obvious differences in CLA isomer distribution. We discovered that t,t-CLA produced by coculture of La an Lp had a relative reduction in comparison to Lp, and it seemed to favour the formation of t10,c12-CLA when La took up a larger proportion in the coculture. We can conclude that the coculture is able to change different kinds of CLA isomer contents. This result is similar to the research showing that the addition of 0.1% LA to a mixture of yogurt cultures and L. acidophilus significantly increased the content of c9,t11-CLA in nonfat set yogurt.30 The proportion of CLA isomers formed by La or Lp was studied previously, and it showed that c9,t11-CLA, t10,c12-CLA and t,t-CLA isomers were the primary compositions, and t,t-CLA isomers took up the higher proportion.31 It has been reported that c9,t11-CLA shows anticancer activity32 and t10,c12-CLA shows activities that decrease body fat,33 which is beneficial to health, while the bioactivity of t,t-CLA is lower than c9,t11-CLA.33 Thus t,t-CLA isomers were not expected to enrich in the fermented diary food. The coculture of La and Lp reduced the t,t-CLA content obviously, causing a desirable enrichment in c9,t11-CLA with specific strain ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which has significances to fermented dairy products. So we selected a specific strain ratio of 1
1, which has significances to fermented dairy products. So we selected a specific strain ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to carry out the next experiment.
1 to carry out the next experiment.
| c9,t11-CLA (µg mL−1) | t10,c12-CLA (µg mL−1) | t,t-CLA (µg mL−1) | Total CLA (µg mL−1) | Microbial counts (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU g−1) CFU g−1) |
|
|---|---|---|---|---|---|
| La and Lp | |||||
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
58.3 ± 1.6b | 15.1 ± 1.9d | 43.2 ± 1.4a | 116.6 ± 4.9a | 9.35 ± 0.23 |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
45.2 ± 1.3c | 25.0 ± 1.5c | 30.1 ± 2.4b | 100.3 ± 4.2c | 9.22 ± 0.20 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
46.3 ± 1.6c | 26.7 ± 1.2c | 22.8 ± 1.3c | 95.8 ± 4.1c | 9.11 ± 0.21 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
70.9 ± 1.3a | 25.7 ± 1.0c | 10.9 ± 1.5d | 107.5 ± 3.8b | 9.38 ± 0.22 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
39.5 ± 1.4d | 26.3 ± 1.7c | 21.9 ± 1.2c | 87.7 ± 4.3d | 9.23 ± 0.16 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
37.9 ± 0.8d | 35.9 ± 1.4b | 23.3 ± 1.6c | 97.1 ± 3.8c | 9.06 ± 0.25 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
35.8 ± 1.2e | 41.9 ± 0.7a | 22.1 ± 1.6c | 99.8 ± 3.5c | 9.14 ± 0.27 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Single strains | |||||
| La | 27.3 | 20.2 | 17.4 | 64.9 | 9.19 |
| Lp | 22.4 | 19.5 | 30.5 | 72.4 | 8.95 |
These alterations in the isomer profile suggest that the La–Lp co-culture may influence the isomerization pathway. Specifically, it potentially inhibits the further reduction of the more bioactive c9,t11-CLA and t10,c12-CLA isomers to the less desirable t,t-CLA or other saturated fatty acids, a step in the biohydrogenation pathway. It is plausible that one strain influences the enzymatic activity or metabolic flux of the other, leading to a redirected isomerization pathway that favors the accumulation of specific CLA isomers. The exact mechanism—whether through cross-talk, altered gene expression, or metabolic complementation—warrants further investigation.
When La and Lp were incubated in the MRS media containing 300 µg mL−1 of LA, the total plate count increased dramatically, from 4.73 to 9.11 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CFU mL−1 over the 24 hour incubation period (Fig. 3b). The consistent and robust growth patterns observed for the co-culture (Fig. 3b and 4a) confirm the stable co-existence of both strains throughout the fermentation. The presence of La in the MRS medium, in the presence of other microorganisms, led to slightly higher levels of microbial growth when compared to the MRS medium that contained only Lp and the MRS medium with only La after 24 h of fermentation. The CLA production by the whole cell of the mixed strains is significantly higher than the CLA production by the whole cell of La or Lp (Fig. 3c). Since the coculture of LAB strains shows a higher-producing CLA ability than single LAB, we conducted two speculations: (i) the coculture increases the total microbial growth; this is because microbial counts is important in CLA production.34 (ii) The coculture increases the enzyme activity of strains; this is based on the fact that microorganisms can produce CLA from LA through linoleate isomerase (LAI) activity.35 To verify the above speculations, the growth rate of strains was tested, and we found that there was little difference among the total microbial counts in the MRS medium. Then we carried out thewhole cell experiment. Linoleate isomerase is an intracellular enzyme. Therefore, this study focused exclusively on the activity of the intracellular LAI enzyme. The experimental approach, utilizing permeabilized whole cells, was specifically designed to assess this intracellular activity by overcoming membrane barriers to substrate access. Extracellular enzymatic activity was not investigated. Thus, the cell membrane and cell wall prevent the enzyme from coming into contact with its substrates.21 The drawbacks of using whole cells were overcome by adopting permeabilization technology,36 which can excellently exhibit the LAI activity. Because the whole cell of mixed strains showed a relatively higher conversion activity with LA as compared to the whole cell of La or Lp at the same cell concentration, we draw a conclusion that the higher production by coculture of strains can be due to the improvement of enzyme activity, but it is still unclear why the coculture can improve the LAI activity. We surmised it is possible that some kind of organic chemical composition was produced by coculture, leading to the enhancement of LAI activity.
CFU mL−1 over the 24 hour incubation period (Fig. 3b). The consistent and robust growth patterns observed for the co-culture (Fig. 3b and 4a) confirm the stable co-existence of both strains throughout the fermentation. The presence of La in the MRS medium, in the presence of other microorganisms, led to slightly higher levels of microbial growth when compared to the MRS medium that contained only Lp and the MRS medium with only La after 24 h of fermentation. The CLA production by the whole cell of the mixed strains is significantly higher than the CLA production by the whole cell of La or Lp (Fig. 3c). Since the coculture of LAB strains shows a higher-producing CLA ability than single LAB, we conducted two speculations: (i) the coculture increases the total microbial growth; this is because microbial counts is important in CLA production.34 (ii) The coculture increases the enzyme activity of strains; this is based on the fact that microorganisms can produce CLA from LA through linoleate isomerase (LAI) activity.35 To verify the above speculations, the growth rate of strains was tested, and we found that there was little difference among the total microbial counts in the MRS medium. Then we carried out thewhole cell experiment. Linoleate isomerase is an intracellular enzyme. Therefore, this study focused exclusively on the activity of the intracellular LAI enzyme. The experimental approach, utilizing permeabilized whole cells, was specifically designed to assess this intracellular activity by overcoming membrane barriers to substrate access. Extracellular enzymatic activity was not investigated. Thus, the cell membrane and cell wall prevent the enzyme from coming into contact with its substrates.21 The drawbacks of using whole cells were overcome by adopting permeabilization technology,36 which can excellently exhibit the LAI activity. Because the whole cell of mixed strains showed a relatively higher conversion activity with LA as compared to the whole cell of La or Lp at the same cell concentration, we draw a conclusion that the higher production by coculture of strains can be due to the improvement of enzyme activity, but it is still unclear why the coculture can improve the LAI activity. We surmised it is possible that some kind of organic chemical composition was produced by coculture, leading to the enhancement of LAI activity.
3.5. Conditions for CLA production
The analysis of potential factors leading to CLA production identified two critical variables to test. The concentration of LA was considered an important factor because long-chain FFAs are known to have an inhibitory effect on microbial growth.36 Incubation time was considered another potential factor affecting CLA production. Reports indicating that Lactobacillus acidophilus produces CLA only after reaching the stationary growth phase served as the basis for this hypothesis.37The microbial growth of the coculture of La and Lp proportionally decreased as LA concentration increased to 700 µg mL−1 (Fig. 4a). After a 24 hour period of incubation, the maximal viable cell growth at the free LA level of 500 µg mL−1 was approximately three-quarter that of bacteria grown with 200 µg mL−1. Furthermore, the total CLA production by cocultures of La–Lp with the addition of the LA (substrates) from 200 µg mL−1 to 500 µg mL−1. Then, it decreased due to the addition of 500 µg mL−1. When the LA addition level was 500 µg mL−1, the total CLA production reached its maximum value of 158.6 µg mL−1 (Fig. 4a), and the c9,t11-CLA content was 107.7 µg mL−1 (Table 3). The variation trend in the content of individual isomers was consistent with the total CLA production. As reported, LA has inhibitory effects on bacterial growth38 and the level of tolerance to LA differs between different strains.39 Some have suggested that converting free LA to CLA may serve as a detoxification mechanism in bacteria and that a greater tolerance to LA implies a higher CLA productivity.40 The present study tested the tolerance levels of mixed strains to LA by adding various concentrations of LA to the MRS medium. So this result might be due to a relatively high tolerance to LA for the coculture of La and Lp. But the CLA production was still restrained at over-loading concentrations. This pattern was similar to those observed in previous Lactobacillus strain studies.41 Adding LA increased CLA production during fermentation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in the MRS medium supplemented with 200, 300, 400, 500, 600 and 700 µg mL−1 LA for 24 h of incubation at 37 °C (p < 0.05)
1 ratio in the MRS medium supplemented with 200, 300, 400, 500, 600 and 700 µg mL−1 LA for 24 h of incubation at 37 °C (p < 0.05)
| LA concentration (µg mL−1) | c9,t11-CLA (µg mL−1) | t10,c12-CLA (µg mL−1) | t,t-CLA (µg mL−1) | Total CLA (µg mL−1) |
|---|---|---|---|---|
| 200 | 47.2 ± 0.8e | 16.5 ± 1.5e | 11.1 ± 1.0d | 74.8 ± 3.3f |
| 300 | 68.2 ± 1.6d | 23.1 ± 1.4d | 13.6 ± 0.8c | 104.9 ± 3.8e |
| 400 | 81.1 ± 1.7c | 31.4 ± 1.6bc | 18.3 ± 1.3b | 130.8 ± 4.6c |
| 500 | 107.7 ± 1.6a | 34.4 ± 1.7a | 16.5 ± 0.8b | 158.6 ± 4.1a |
| 600 | 93.4 ± 1.4b | 32.7 ± 1.6ab | 23.0 ± 1.3a | 149.1 ± 4.3b |
| 700 | 72.3 ± 1.7d | 29.6 ± 2.3c | 16.7 ± 1.5b | 118.6 ± 5.5d |
First, we analyzed samples of MRS broth supplemented with 500 µg mL−1 of LA and inoculated with a culture of LA and Lp. The samples were then incubated at 37 °C for different periods of time (6, 12, 24, 36, 48, 60, and 72 hours) to determine the growth stage at which CLA was produced. As fermentation progresses, the total content of CLA increases, reaching a maximum of 206.5 µg mL−1 after 48 hours (Fig. 4b), and the corresponding conversion rate from LA to CLA was 41.3%. The similar tendency was observed in a previous study.27 We also discovered that most CLA was produced at the stationary stage. This is similar to the findings of P. Liu et al.42
Moreover, we discovered that CLA production by coculture of La and Lp did not decline significantly after 72 h of incubation (Fig. 5a), so we investigated the CLA-producing activity of La and Lp under same conditions for 72 h at 37 °C. When La and Lp were each incubated in MRS, both of them reached the highest production levels after 48 h, but then the CLA production levels declined sharply (Fig. 5b). Previously, the production of CLA by Lactobacillus plantarum IP15 increased with incubation time, ranging from 0 to 48 hours. A rapid decrease in LA conversion was observed at incubation times above 48 hours.21 The reduction to the total CLA production by La was mainly attributed to the reduction to the proportion of the c9,t11-CLA isomer and the t10,c12-CLA isomer, and the reduction to the total CLA production by Lp was mainly ascribed to reduction to the proportion of the c9,t11-CLA isomer and t,t-CLA (Table 4). Compared with La or Lp, the c9,t11-CLA produced by the coculture decreased slightly, and t10,c12-CLA content and t,t-CLA content were basically unchanged (Table 4). Studies have shown that CLA is an intermediate product in the BH process and that rumen BH involves sequential yet distinct enzymatic reactions that produce saturated fatty acids. These processes include the spontaneous isomerization of LA to CLA, as well as the reduction of CLA to VA and stearic acid. Studies have shown that CLA is an intermediate product in the BH process. Ruminal BH involves sequential, yet distinct, enzymatic processes that yield saturated fatty acids. First, LA spontaneously isomerizes into CLA. Then, CLA is reduced into VA and stearic acid,12 so we deduced that La in coculture with Lp may decrease the BH activity compared to the single LAB strain. And it has been found that this characteristic of coculture of La and Lp made it possible to maintain the CLA production during fermentation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), La and Lp in the MRS medium supplemented with 500 µg mL−1 LA after 48, 60, and 72 h of incubation at 37 °C (p < 0.05)
1), La and Lp in the MRS medium supplemented with 500 µg mL−1 LA after 48, 60, and 72 h of incubation at 37 °C (p < 0.05)
| Time (h) | Lp | La | La and Lp | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 48 | 60 | 72 | 48 | 60 | 72 | 48 | 60 | 72 | |
| c9,t11-CLA (µg mL−1) | 35.6 ± 0.7a | 26.6 ± 1.3b | 19.4 ± 1.1c | 38.9 ± 2.2a | 26.3 ± 1.2b | 20.7 ± 0.8c | 128.0 ± 1.6a | 124.3 ± 1.9b | 121.4 ± 2.0c |
| t10,c12-CLA (µg mL−1) | 30.9 ± 1.2a | 26.3 ± 1.4b | 22.7 ± 1.7c | 29.7 ± 1.4a | 21.6 ± 0.8b | 15.6 ± 1.4c | 51.6 ± 1.7a | 49.4 ± 1.6ab | 48.3 ± 1.7b |
| t,t-CLA (µg mL−1) | 44.2 ± 1.8a | 24.6 ± 1.9b | 17.6 ± 2.0c | 24.3 ± 1.5a | 21.4 ± 0.9b | 19.1 ± 1.3c | 26.9 ± 1.3a | 25.0 ± 1.8a | 24.8 ± 1.9a |
| Total CLA (µg mL−1) | 110.7 ± 3.7a | 77.5 ± 4.6b | 59.7 ± 4.8c | 92.7 ± 5.1a | 69.3 ± 2.9b | 55.4 ± 3.5c | 206.5 ± 4.6a | 198.7 ± 5.3ab | 194.2 ± 5.6b |
We found that the CLA isomer ratio was not impacted by LA concentration and incubation time, which indicated that the distribution of CLA isomers is mainly influenced by the different proportions of coculture of La and Lp (Fig. 5a and c). And this needs to be further validated in future trials.
3.6. Application of the coculture of La and Lp in the skim milk medium
Further investigation is needed to determine the availability of La and Lp cultures, so we conducted the same experiments in the skim milk including varying LA concentrations and incubation time. The results are similar to the fermentation in the MRS medium. CLA production increased proportionally before the LA concentration reached 500 µg mL−1 (Fig. 4a), and the optimal incubation time for producing CLA was 48 hours (Fig. 4b). When La was in coculture with Lp in the skim milk medium containing 500 µg mL−1 of LA for 48 h, and other culture conditions were the same as those in MRS medium, the total CLA production reached 220.7 µg mL−1 (Fig. 4b) and the corresponding conversion rate from LA to CLA was 44.1%. We found that the CLA production in skim milk is higher than it in MRS medium, but the CLA isomer ratio was nearly unchanged compared with the CLA isomer ratio in the MRS medium. An increase in cell numbers during the late log phase may have been caused by the nutrients in skim milk. Actually, the environmental factors that lead to an increase in cell numbers are related to the production of CLA,34 and this should be kept in mind when making yogurt with CLA.4. Conclusions
The enhanced CLA production is primarily driven by a dual mechanism: the synergistic interaction in the La–Lp coculture, where La's acidification activates Lp's LAI, combined with the significantly improved substrate access afforded by ethanol permeabilization of cell membranes. This strategy proved robust and effective, not only yielding higher total CLA compared to monocultures but also favorably altering the isomer profile by enriching the more bioactive c9,t11-CLA isomer. Optimal production was achieved with a La![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lp ratio of 1
Lp ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, a LA concentration of 500 µg mL−1, and an incubation time of 48 hours. The co-culture exhibited high LA tolerance and, notably, maintained stable CLA production without significant decline during extended fermentation, unlike the monocultures. Validation in skim milk fermentation confirmed the practical applicability of this approach, resulting in even higher CLA yields. This work establishes the La–Lp co-culture combined with ethanol permeabilization as a promising, sustainable method for producing CLA-enriched fermented foods.
1, a LA concentration of 500 µg mL−1, and an incubation time of 48 hours. The co-culture exhibited high LA tolerance and, notably, maintained stable CLA production without significant decline during extended fermentation, unlike the monocultures. Validation in skim milk fermentation confirmed the practical applicability of this approach, resulting in even higher CLA yields. This work establishes the La–Lp co-culture combined with ethanol permeabilization as a promising, sustainable method for producing CLA-enriched fermented foods.
Future research will focus on elucidating the mechanisms underpinning: (i) the enhanced LAI activity in co-culture and the role of increased membrane permeability, (ii) the sustained CLA stability during prolonged in-process fermentation in the La–Lp consortium, and (iii) the stability of CLA isomers in the final product under various storage and food processing conditions. Concurrently, we will explore strategies to further augment CLA yield, including refined cell preparation techniques (such as optimized washing protocols), culture condition optimization, mutagenesis, and genetic engineering approaches. Expanding the application of this co-culture system incorporating the permeability-enhancing step to diverse fermented food matrices, including various milk types and plant-based (vegetable) fermentations, will be prioritized to maximize its impact on sustainable and nutritious food innovation. Furthermore, investigating the potential for extracellular LAI activity or enzyme leakage, particularly under permeabilization conditions, will be crucial to fully delineate the biocatalytic mechanism and optimize the process. Additionally, a comprehensive analysis of the nutritional composition and sensory properties of the CLA-enriched fermented products is essential for their development as viable functional foods.
Author contributions
Lan Cheng and Shuhong Ye conceived and designed the experiments. Lan Cheng, Miao Wang, and Yingying Wang performed the experiments; Lan Cheng and Shuhong Ye wrote the original manuscript; Yumeng Chang and Yan Ding validated and reviewed the final manuscript.Conflicts of interest
The authors of this article declare that they have no conflicts of interest.Data availability
All data supporting the findings of this article are also presented in the manuscript's tables and figures. There are no restrictions on the availability of these data.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5fb00425j.
Acknowledgements
This study was supported by the Basic Research Funds for Undergraduate Universities in Liaoning Province in 2024 (LJ212410152067) and National Key R & D Program of China (2021YFD2100100).References
- M. Iorizzo, G. Paventi and C. Di Martino, Curr. Issues Mol. Biol., 2024, 46, 200–220 CrossRef CAS.
- M. J. Du, M. Y. Gong, G. C. Wu, J. Jin, X. G. Wang and Q. Z. Jin, J. Agric. Food Chem., 2024, 72, 5503–5525 CrossRef CAS.
- Y. Park, K. J. Albright, J. M. Storkson, W. Liu and M. W. Pariza, J. Funct. Foods, 2010, 2, 54–59 CrossRef CAS.
- L. J. den Hartigh, Nutrients, 2019, 11, 29 CrossRef PubMed.
- P. G. Toral, G. Hervás and P. Frutos, J. Dairy Sci., 2019, 102, 1213–1223 CrossRef CAS PubMed.
- K. Wang, Z. M. Xin, Z. Chen, H. A. Li, D. M. Wang and Y. Yuan, Animals, 2023, 13, 18 Search PubMed.
- I. C. A. Balcazar, L. D. G. Rivera, J. S. Chavira, B. E. Drouaillet, M. R. Albarran and Y. B. Martínez, Animals, 2022, 12, 15 Search PubMed.
- Y. R. Yanza, M. Szumacher-Strabel, D. Lechniak, S. Slusarczyk, P. Kolodziejski, A. K. Patra, Z. Váradyová, D. Lisiak, M. Vazirigohar and A. Cieslak, J. Anim. Sci. Biotechnol., 2022, 13, 19 Search PubMed.
- L. P. Thanh, P. T. T. Kha, J. J. Loor and T. T. T. Hang, J. Anim. Sci., 2022, 100, 9 Search PubMed.
- T. Aziz, A. Sarwar, M. Naveed, M. Shahzad, M. A. Shabbir, A. S. Dablool, J. U. Din, A. A. Khan, S. Naz, H. Y. Cui and L. Lin, Food Res. Int., 2022, 154, 9 Search PubMed.
- F. L. Zhang, Y. M. Fu, J. F. Wang, L. M. Lang, S. Y. Liang, S. L. Zhang, L. N. Wang, P. Gao, G. Shu, C. J. Zhu, R. F. Wu, Q. Y. Jiang and S. B. Wang, Food Funct., 2024, 15, 5000–5011 Search PubMed.
- L. Chamekh, M. Caivo, T. Khorchani, P. Castro-Gómez, M. Hammadi, J. Fontecha and M. H. Yahyaoui, J. Food Compos. Anal., 2020, 87, 9 Search PubMed.
- P. G. Toral, G. Hervás and P. Frutos, J. Dairy Sci., 2022, 105, 255–268 Search PubMed.
- A. S. Salsinha, L. L. Pimentel, A. L. Fontes, A. M. Gomes and L. M. Rodríguez-Alcalá, Microbiol. Mol. Biol. Rev., 2018, 82, 21 Search PubMed.
- I. Carafa, D. Masuero, U. Vrhovsek, G. Bittante, E. Franciosi and K. M. Tuohy, Proc. Nutr. Soc., 2020, 79, E628 Search PubMed.
- P. Simatende, M. Siwela and T. H. Gadaga, S. Afr. J. Sci., 2019, 115, 7 Search PubMed.
- L. Marras, M. Caputo, S. Bisicchia, M. Soato, G. Bertolino, S. Vaccaro and R. Inturri, Microorganisms, 2021, 9, 21 Search PubMed.
- S. Martini, L. Sola, A. Cattivelli, M. Cristofolini, V. Pizzamiglio, D. Tagliazucchi and L. Solieri, Front. Microbiol., 2024, 15, 21 Search PubMed.
- A. Burmeister, Q. Akhtar, L. Hollmann, N. Tenhaef, F. Hilgers, F. Hogenkamp, S. Sokolowsky, J. Marienhagen, S. Noack, D. Kohlheyer and A. Grünberger, ACS Synth. Biol., 2021, 10, 1308–1319 CAS.
- A. Alanzi, E. A. Elhawary, M. L. Ashour and A. Y. Moussa, Arch. Pharmacal Res., 2023, 46, 273–298 Search PubMed.
- T. Li, Z. L. Gu and G. Q. Zhou, Anal. Chem., 2024, 96, 19155–19159 CAS.
- T. Nakao, T. Yuzuriha and H. Kakutani, Microchem. J., 2025, 208, 10 Search PubMed.
- M. Fiastru-Irimescu and D. Margina, Stud. Univ. Babes-Bolyai, Chem., 2024, 69, 175–186 Search PubMed.
- L. M. Rodríguez-Alcalá, T. Braga, F. X. Malcata, A. Gomes and J. Fontecha, Food Chem., 2011, 125, 1373–1378 Search PubMed.
- K. Zongo, F. Yazici and A. H. Con, Indian J. Biochem. Biophys., 2022, 59, 741–750 Search PubMed.
- Y. Jang, A. G. Elnar, S. J. Hur and G. B. Kim, Crit. Rev. Food Sci. Nutr., 2025, 65, 3911–3927 CrossRef CAS.
- S. H. Ye, T. Yu, H. Yang, L. Li, H. Wang, S. Xiao and J. H. Wang, Ann. Microbiol., 2013, 63, 707–717 CrossRef CAS.
- L. Y. Li, R. Zhang, L. Chen, X. T. Tian, T. Li, B. C. Pu, C. H. Ma, X. Y. Ji, F. Ba, C. W. Xiong, Y. F. Shi, X. Q. Mi, J. Li, J. D. Keasling, J. W. Zhang and Y. F. Liu, Adv. Sci., 2022, 9, 11 Search PubMed.
- Z. H. Gao, E. B. M. Daliri, J. Wang, D. H. Liu, S. G. Chen, X. Q. Ye and T. Ding, J. Food Prot., 2019, 82, 441–453 CrossRef CAS PubMed.
- N. Baliyan, A. K. Maurya, A. Kumar, V. K. Agnihotri and R. Kumar, LWT-Food Sci. Technol., 2023, 176, 9 CrossRef.
- T. Malovrh, E. Melkic, D. Kompan, A. Levart and L. Kompan, Eur. J. Nutr., 2014, 53, 989–993 CrossRef CAS.
- E. K. Paulin, E. Leung, L. I. Pilkington and D. Barker, Int. J. Mol. Sci., 2023, 24, 12 Search PubMed.
- C. G. Padilha, C. Ribeiro and D. E. Oliveira, J. Anim. Physiol. Anim. Nutr., 2023, 107, 1028–1034 CrossRef CAS.
- M. F. Peng, Z. Tabashsum, P. Patel, C. Bernhardt and D. Biswas, Front. Microbiol., 2018, 9, 14 CrossRef PubMed.
- M. Iorizzo, C. Di Martino, F. Letizia, T. W. Crawford and G. Paventi, Foods, 2024, 13, 23 CrossRef.
- C. Borreby, E. M. S. Lillebaek and B. H. Kallipolitis, FEMS Microbiol. Rev., 2023, 47, 14 CrossRef PubMed.
- Z. Y. Wang, C. L. Su, Y. S. Zhang, S. F. Shangguan, R. M. Wang and J. Su, Front. Microbiol., 2024, 14, 14 Search PubMed.
- P. Poudel, Y. Tashiro and K. Sakai, Biosci., Biotechnol., Biochem., 2016, 80, 642–654 CrossRef CAS.
- C. Wu, H. Q. Chen, Y. C. Mei, B. Yang, J. X. Zhao, C. Stanton and W. Chen, Prog. Lipid Res., 2024, 93, 15 CrossRef PubMed.
- M. H. A. El-Salam, K. El-Shafei, O. M. Sharaf, B. A. Effat, F. M. Asem and M. El-Aasar, Int. J. Dairy Technol., 2010, 63, 62–69 CrossRef.
- C. S. Chen, F. Tong, R. H. Sun, Y. Zhang, Z. H. Pang and X. Q. Liu, Foods, 2024, 13, 20 Search PubMed.
- P. Liu, S. R. Shen, H. Ruan, Q. Zhou, L. L. Ma and G. Q. He, J. Zhejiang Univ., Sci., B, 2011, 12, 923–930 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |