Open Access Article
Open Access ArticlePersistent effects of early-life arsenic exposure in Caenorhabditis elegans
Kathleen A.
Hershberger
 *abc,
Shaza
Gaballah
a,
Britney Jiayu
He
*abc,
Shaza
Gaballah
a,
Britney Jiayu
He
 c,
Luoyu
Zhang
c,
Emily
Barefoot-Gautier
a,
Caroline B.
Reed
a,
Nelson A.
Rivera
Jr
d,
Heileen
Hsu-Kim
c,
Luoyu
Zhang
c,
Emily
Barefoot-Gautier
a,
Caroline B.
Reed
a,
Nelson A.
Rivera
Jr
d,
Heileen
Hsu-Kim
 d and
Joel N.
Meyer
d and
Joel N.
Meyer
 *a
*a
aNicholas School of the Environment, Duke University, Durham, NC, USA. E-mail: joel.meyer@duke.edu
bDepartment of Biology, Case Western Reserve University, Cleveland, OH, USA. E-mail: kathleen.hershberger@case.edu
cDepartment of Biology, Grinnell College, Grinnell, IA, USA
dDepartment of Civil & Environmental Engineering, Duke University, Durham, NC, USA
First published on 16th January 2026
Abstract
Arsenic exposure is a major global health challenge. In addition to well-documented toxic effects in exposed people and animals, there is evidence that exposure to arsenic may lead to transgenerational effects. Transgenerational effects of low levels of exposure are challenging to study in species with long generation times. The model organism Caenorhabditis elegans offers the ability to quickly carry out transgenerational experiments with very large sample sizes of isogenic animals, reducing variation, and numerous biological replicates, to increase statistical rigor. An important challenge historically associated with this species for such work is uncertainty about internal dosimetry and toxicokinetics. Here, we report a 4-generation experiment in which C. elegans were exposed during larval development to sodium arsenite concentrations in the parental generation at concentrations resulting in no or mild growth inhibition up to significant growth inhibition. These exposures resulted in internal concentrations between 0.4 and 6.7 nM and rapid excretion (t1/2 = 3 hours), despite the lack of arsenic methylation in this species. These exposures had strong and significant effects on the exposed generation later in life, but no transgenerational effects were detected. We discuss possible reasons for this “negative” result. We also report strong similarity of the nematode transcriptomic, metabolomic, and fat accumulation responses in the exposed generation to responses reported in other organisms, including persistent alterations in cysteine and fatty acid metabolism, phase II and III metabolic processes, and increased adiposity. Finally, we discuss ways to take advantage of this species difference in arsenic metabolism for the use of C. elegans in toxicology testing.
Environmental significanceChronic arsenic exposure through consumption of contaminated food and drinking water affects millions of people worldwide and is associated with severe outcomes, including various cancers, diabetes, and cardiovascular disease. However, the long-term consequences of in utero arsenic exposure remain poorly understood, largely due to challenges in conducting such studies. We developed and validated C. elegans as a simple and efficient model to provide physiological and molecular insight as to the effects of prenatal arsenic exposure. Establishing such models is essential for understanding how environmental toxicants affect human health and for developing strategies to mitigate their effects. |
Introduction
Arsenic exposure is a global problem, with an estimated 90 million to 220 million people exposed to groundwater containing arsenic at levels above the WHO guidelines of 10 µg L−1(1). Over 94% of people exposed to high levels of arsenic in groundwater reside in Asia, though significant numbers of people in South America, North America, and Africa are also at risk of persistent exposure to arsenic.1 While the primary route of exposure to arsenic globally is via drinking water or using arsenic contaminated water in cooking, exposure due to industrial processes also occurs. The toxicokinetic processes of arsenic exposure in people are well documented. Absorption of inorganic arsenic species (arsenate or arsenite) occurs primarily in the gastrointestinal tract and arsenic is rapidly distributed to other organs. The half-life of inorganic arsenic in the blood is relatively short (4–6 hours), with rapid urinary excretion in humans facilitated by methylation mediated by the arsenic methyltransferase enzyme.2,3Chronic arsenic exposure can result in severe diseases, ranging from skin lesions and changes in pigmentation to cancers affecting various organ systems, diabetes, and heart disease.4,5 Importantly, there is often a long latency period between the exposure to arsenic and the development of the associated diseases.6 In the past few decades, much research has focused on the effects of arsenic exposure in utero. Recent reviews7,8 highlight key findings of the impacts of in utero exposures to arsenic, so it is not discussed in depth here. Human and animal studies suggest that in utero exposure to arsenic increases immediate risks to the infant such as increased adverse birth outcomes and increased risk of infection and cardiovascular mortality, among other adverse health effects in childhood.8 Additionally, in utero exposure to arsenic is associated with an increased risk of cancer mortality, renal disease mortality, and cardiovascular disease mortality in adulthood.8 The mechanism of the long latency periods of diseases caused by exposure to inorganic arsenic (iAs) and the effects that extend into later life after in utero exposure are not well characterized but may be epigenetic. One early study demonstrated that prenatal iAs exposure in mice resulted in a 90% decrease in methylation of the ER-α promoter and a 3-fold increase in ER-α mRNA expression in hepatocellular tumors.9 In another study examining epigenetic mechanisms of disease, 40 newborn blood samples were characterized to identify 12 miRNAs that increased with maternal arsenic exposure and were involved in signaling pathways related to known outcomes of iAs exposure.10 In addition to exposure to toxic compounds in utero, there remains the further question of whether effects of toxic exposures can be passed through multiple generations, or even transgenerationally via epigenetic mechanisms.11
Epidemiological studies in humans have characterized the adverse effects of early-life arsenic exposure and advanced our current understanding of potential mechanisms of life-long and inherited effects.5,7,12–14 However, the challenges and timelines of undertaking these studies to provide a clear mechanistic picture motivated our interest in developing a complementary model to ask how early life exposure to arsenic leads to persistent effects, and if these effects are transgenerational. Caenorhabditis elegans is a well-characterized genetic and developmental biology model as well as an established model to study environmental toxicology.15 Previous work in C. elegans has elucidated mechanisms of arsenic toxicity including shifts in metabolism to a reliance on glycolysis16 and effects on mitochondrial dynamics.17 Research in cell culture suggests that mitochondrial impacts of arsenic18 can drive epigenetic memory formation, but possible transgenerational impacts of arsenic on mitochondrial function in vivo have not been tested. Given the short life cycle and ease of culturing large populations of C. elegans, this model organism is ideal for investigating transgenerational effects of exposures. Indeed, research has suggested that arsenic exposure has transgenerational effects on brood side19 and behavior.20 Despite identifying transgenerational phenotypes of arsenic exposure in C. elegans, researchers have not systematically characterized arsenic toxicokinetics, although it has been determined that C. elegans lack the AS3MT gene which encodes an enzyme that methylates As, promoting excretion.21 The goal of this study is to provide characterization of early-life arsenic exposure in C. elegans and identify phenotypes of arsenic exposure that persist into adulthood and transgenerationally. Metabolomics and transcriptomics were used to understand mechanisms of persistent effects of early-life exposure to arsenic.
Methods
C. elegans strains and culture conditions and transgenerational methods
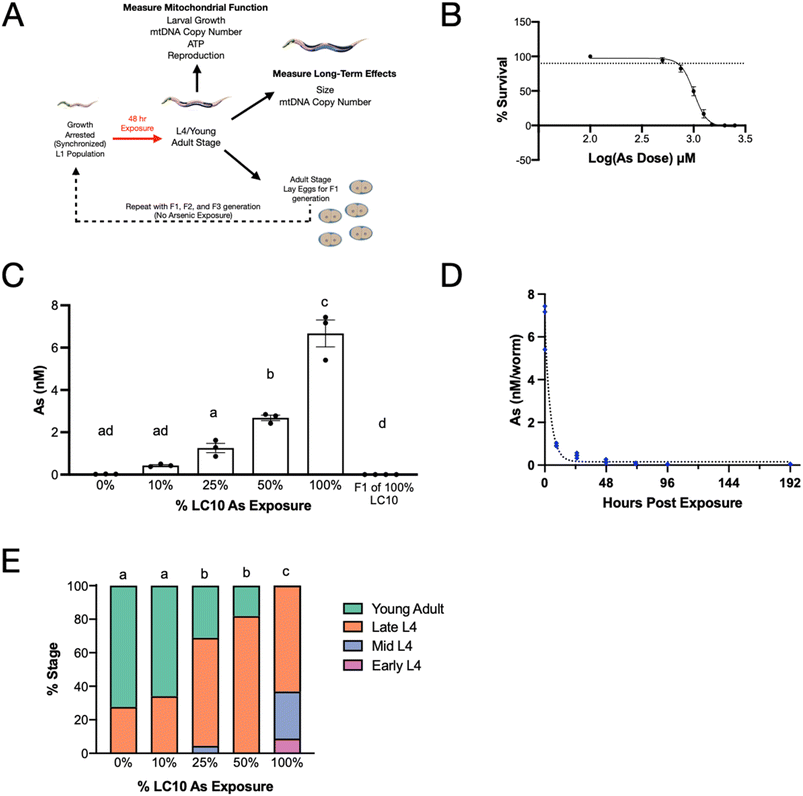
Wild-type (N2, Bristol) and LIU2 (ldrls2) nematodes were purchased from the Caenorhabditis Genetics Center (CGC, University of Minnesota). Synchronized populations of larval stage (L1) nematodes were generated via sodium hydroxide bleach treatment as previously described,22 followed by overnight incubation in a 25 cm2 tissue culture flask on an orbital shaker in complete K-medium (150 µl 1 M CaCl2, 150 µl 1 M MgSO4, 25 µl 10 mg mL−1 cholesterol, 50 mL sterile K medium (2.36 g KCl, 3 g NaCl, 1 L ddH2O)) at 20 °C. Synchronous populations of nematodes were exposed in liquid for 48 hours then maintained on K-agar plates seeded with Escherichia coli OP50 at 20 °C as previously described.23 Nematodes in the exposed parental generation are referred to as the F0 generation. For transgenerational assays, nematodes were maintained on the K-agar plates for approximately 30 hours post-exposure. The gravid adults were bleached and F1 eggs were incubated overnight and arrested in the L1 stage as for the F0 generation. L1 larvae were plated on K-agar plates seeded with OP50 and incubated at 20 °C for 48 hours and prepared for assays as described below. The same protocol was followed to generate the F2 and F3 populations.Exposure to sodium arsenite
C. elegans were exposed to sodium arsenite in 96-well flat bottom cell culture plates. Wells on the outside of the plate were not used for exposures. 50 L1 nematodes were exposed in each well. The total well volume was 100 µl and contained 25 µl of 4× UVC-irradiated UvrA (UVC sensitive E. coli strain due to a lack of nucleotide excision repair24), sodium arsenite, and synthetic moderately hard freshwater “EPA water”25 (96 mg NaHCO3, 60 mg MgSO4·7H2O, 60 mg CaSO4·2H2O, 4 mg KCl, 500 µl 10 mg mL−1 cholesterol, 1 L ddH2O). Plates were incubated at 20 °C for 48 hours on an orbital shaker. Sodium arsenite exposure concentrations were based on the LC10 as calculated by the ECanything (4 parameter logistic model with a variable slope) model in Prism (0%, 10%, 25%, 50%, and 100%).Lethality
C. elegans were exposed to various concentrations (100 µM, 500 µM, 750 µM, 1 mM, 1.5 mM, 2 mM, and 2.5 mM) of sodium arsenite as described above, with only 8–15 nematodes per well. Nematodes were added to wells in 10 µl of complete EPA water and counted. 10 µl of 4× UvrA E. coli and an appropriate amount of sodium arsenite were added, and well volumes were brought to 100 µl. Number of live nematodes were counted after 48 hours of exposure.Measurement of in vivo arsenic concentrations
C. elegans were exposed to various concentrations of sodium arsenite for 48 hours as described above. Immediately after exposures, C elegans of the same exposure concentration were combined in a 15 mL conical, spun down, and rinsed with K-medium. Nematodes were incubated on an orbital shaker at 20 °C for twenty minutes to allow contents of their guts to clear. Nematodes were spun down again, counted, and brought to a concentration of 2 nematodes per µl. 700 µl of nematodes were flash frozen in liquid nitrogen. For later time points, nematodes were plated on K-agar plates seeded with OP50 and maintained at 20 °C. Nematodes were transferred daily by washing nematodes off plates with K-medium, allowing to gravity settle, and removing progeny. Rinsing was performed 3 times, and the adults were plated onto new K-agar OP50 plates. For later time points collected, nematodes were brought to a concentration of 2 nematodes per µl. 700 µl of nematodes were flash frozen in liquid nitrogen. Nematodes were stored at −80 °C for further downstream analysis.C. elegans samples were analyzed for As content by ICP-MS (Agilent 7900). For analysis, samples were thawed on ice and 500 µL of each sample was transferred to a clean vial. Samples were digested with 1.0 mL of nitric acid (trace metal grade, Fisher Scientific) per 0.5 mL of sample. The mixture was heated on a hot block (Environmental Express) at 95 °C for two hours. Aliquots of the digestates were diluted with 2% nitric acid/0.5% hydrochloric acid (v/v) diluent spiked with a 89Y internal standard prior to instrumental analysis. Arsenic quantification was performed in a He collision gas mode to reduce polyatomic interferences. The instrument calibration was performed with a multi-element standard (Spex Certiprep) and verified by a second source reference (CRM-TMDW-A, High Purity Standards). Trace metal grade acids and 18.2 MΩ water (Millipore MilliQ) were used to prepare reagents and dilutions.
Developmental staging
C. elegans were exposed to various concentrations of sodium arsenite for 48 hours as described above. Immediately after exposures, C elegans of the same exposure concentration were combined in a 15 mL conical, spun down, and rinsed with K-medium. Worms were plated onto OP50 and groups were blinded. Approximately 30 worms per group were picked into a 96 well imaging plate. Each well contained 100 µL K-medium and 6 µL of 1 M sodium azide to paralyze the worms. Worms were viewed with a Keyence BZ-X700 using a 40× objective and developmental stage was assessed by viewing vulval morphology.26 Stages were binned as follow: young adult, late L4 (L4.7–L4.9), mid L4 (L4.3–L4.6), and early L4 (L4.0–L4.2).COPAS biosort
After exposure to sodium arsenite from the L1 to L4 stage, nematode length was measured using the COPAS Biosort (Union Biometrica Inc., Somerville, MA) as previously described.17 Time of Flight (TOF), a surrogate for worm length, was normalized to the non-exposed control (control-ratio-based approach) of the F0 generation prior to statistical analysis.Reproduction
C. elegans were exposed to various concentrations of sodium arsenite for 48 hours as described above. Immediately after exposures, C elegans of the same exposure concentration were combined in a 15 mL conical, spun down, and rinsed with K-medium. Single worms were picked onto K-agar plates seeded with OP50 and incubated at 20 °C for the duration of the experiment. Single worms were moved to new plates daily for their reproductive lifespan (4 days) and progeny on the plates were counted and combined to determine the brood size of a single nematode. Brood size was normalized to the non-exposed control of the F0 generation prior to statistical analysis.mtDNA copy number
Post sodium arsenite exposure, mitochondrial DNA (mtDNA) copy number was determined according to methods previously published.27 Briefly, DNA was extracted from 6 L4 nematodes per condition.28 mtDNA copy number was measured using a plasmid-based standard curve and real-time PCR as described.27 mtDNA copy number was normalized to number of worms in the sample and to average size of the worm (extinctoin coefficient or “EXT” from COPAS Biosort data) because arsenic exposure affected worm size. Finally, this value was normalized to the non-exposed control (control-ratio-based approach) of the F0 generation prior to statistical analysis.ATP quantification
ATP was quantified using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) and adapted from previously published methods.29 Nematodes were rinsed in K-medium and brought to a concentration of 2 worms per µL. 100 µL of the worm solution was flash frozen in liquid nitrogen. Samples were heated at 95 °C for 15 minutes, placed on ice for 5 minutes, and spun down at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 minutes at 4 °C. Supernatant was moved to a clean 1.7 mL tube and diluted 1
000 × g for 10 minutes at 4 °C. Supernatant was moved to a clean 1.7 mL tube and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6. Standards ranging from 10 to 1000 nM ATP were created. 50 µl of standards or sample were added to a 96-well white plate and 50 µl of the prepared reagent was added to the wells. After 10 minutes of incubation, the luminescence was recorded every minute for 3 minutes. The average of the three reads and technical duplicates was used as the sample value. Amount of ATP was calculated based on the standard curve. ATP was averaged to the protein content of each sample (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific). This value was normalized to the non-exposed control (control-ratio-based approach) of the F0 generation prior to statistical analysis.
6. Standards ranging from 10 to 1000 nM ATP were created. 50 µl of standards or sample were added to a 96-well white plate and 50 µl of the prepared reagent was added to the wells. After 10 minutes of incubation, the luminescence was recorded every minute for 3 minutes. The average of the three reads and technical duplicates was used as the sample value. Amount of ATP was calculated based on the standard curve. ATP was averaged to the protein content of each sample (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific). This value was normalized to the non-exposed control (control-ratio-based approach) of the F0 generation prior to statistical analysis.
Sodium arsenite Re-challenge experiment
Worms were exposed to no sodium arsenite or the LC10 of sodium arsenite for 48 hours beginning from a synchronized L1 population as described. F1 progeny were collected as described for the transgenerational experiments. Synchronized L1s were exposed to a range of sodium arsenite concentrations (0–15 mM) in a 96-well plate for 24 hours and counted according to the lethality methods. Survival curves were generated and the LC50 was determined using GraphPad Prism ECAnything equation.Worm sizer
Worms were exposed to sodium arsenite for 48 hours as described above. After rinsing, the worms (up to 500) were plated on K-agar plates without OP50 and allowed to dry. After the plates were dry, a Keyence BZ-X700 was used to take images across the entire plate using brightfield. Images were uploaded to FIJI and analyzed using the WormSizer macro.30 Data was exported from WormSizer into Excel and Prism for statistical analysis.Extraction of metabolites from C. elegans
Worms were exposed to various concentrations of sodium arsenite for 48 hours and rinsed immediately after exposure as described above. For the day zero timepoint, worms were counted and approximately 1000 worms per condition were added to pre-weighed 1.5 mL Eppendorf tube. Worms were gently spun down, and liquid was removed. Tubes were weighed and immediately flash frozen in liquid nitrogen. The rest of the worm population was plated on K-agar plates with OP50 food. The worms were transferred daily for 8 days to avoid starvation and contamination of the population from progeny. Worms were rinsed 3× with K-medium, counted, and approximately 1000 worms were added to pre-weighed 1.5 mL Eppendorf tubes. Worms were gently spun down, liquid was removed, and tubes were weighed and flash frozen.Worm samples were shipped to the NYU Metabolomics Core Resource Laboratory on dry ice for metabolomic analysis. Prior to extraction, samples were moved from −80 °C storage to wet ice and thawed. Extraction buffer, consisting of 80% methanol (Fisher Scientific) and 500 nM metabolomics amino acid mix standard (Cambridge Isotope Laboratories, Inc.), was prepared and placed on dry ice. Samples were extracted by mixing pre weighed C. elegans with extraction buffer (10 mg mL−1) in 2.0 mL screw cap vials containing ∼100 µL of disruption beads (Research Products International, Mount Prospect, IL). Each sample was homogenized for 10 cycles on a bead blaster homogenizer (Benchmark Scientific, Edison, NJ). Cycling consisted of a 30 s homogenization time at 6 m s−1 followed by a 30 s pause. Samples were subsequently spun at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 3 min at 4 °C. A set volume of each (450 µL) was transferred to a 1.5 mL tube and dried down by SpeedVac (Thermo Fisher, Waltham, MA). Samples were reconstituted in 50 µL of Optima LC/MS grade water (Fisher Scientific, Waltham, MA). Samples were sonicated for 2 min, then spun at 21
000 g for 3 min at 4 °C. A set volume of each (450 µL) was transferred to a 1.5 mL tube and dried down by SpeedVac (Thermo Fisher, Waltham, MA). Samples were reconstituted in 50 µL of Optima LC/MS grade water (Fisher Scientific, Waltham, MA). Samples were sonicated for 2 min, then spun at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 3 min at 4 °C. 20 µL were transferred to LC vials containing glass inserts for analysis. The remaining sample was placed in −80 °C for long term storage.
000 g for 3 min at 4 °C. 20 µL were transferred to LC vials containing glass inserts for analysis. The remaining sample was placed in −80 °C for long term storage.
LC-MS/MS with the hybrid metabolomics method
Samples were subjected to an LCMS analysis to detect and quantify known peaks. A metabolite extraction was carried out on each sample based on a previously described method.31 The LC column was a MilliporeTM ZIC-pHILIC (2.1 ×150 mm, 5 µm) coupled to a Dionex Ultimate 3000 TM system and the column oven temperature was set to 25 °C for the gradient elution. A flow rate of 100 µL min−1 was used with the following buffers; A) 10 mM ammonium carbonate in water, pH 9.0, and B) neat acetonitrile. The gradient profile was as follows; 80–20%B (0–30 min), 20–80%B (30–31 min), 80–80%B (31–42 min). Injection volume was set to 2 µL for all analyses (42 min total run time per injection).MS analyses were carried out by coupling the LC system to a Thermo Q Exactive HFTM mass spectrometer operating in heated electrospray ionization mode (HESI). Method duration was 30 min with a polarity switching data-dependent Top 5 method for both positive and negative modes. Spray voltage for both positive and negative modes was 3.5 kV and capillary temperature was set to 320 °C with a sheath gas rate of 35, aux gas of 10, and max spray current of 100 µA. The full MS scan for both polarities utilized 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolution with an AGC target of 3 × 106 and a maximum IT of 100 ms, and the scan range was from 67–1000 m/z. Tandem MS spectra for both positive and negative mode used a resolution of 15
000 resolution with an AGC target of 3 × 106 and a maximum IT of 100 ms, and the scan range was from 67–1000 m/z. Tandem MS spectra for both positive and negative mode used a resolution of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, AGC target of 1 × 105, maximum IT of 50 ms, isolation window of 0.4 m/z, isolation offset of 0.1 m/z, fixed first mass of 50 m/z, and 3-way multiplexed normalized collision energies (nCE) of 10, 35, 80. The minimum AGC target was 1 × 104 with an intensity threshold of 2 × 105. All data were acquired in profile mode.
000, AGC target of 1 × 105, maximum IT of 50 ms, isolation window of 0.4 m/z, isolation offset of 0.1 m/z, fixed first mass of 50 m/z, and 3-way multiplexed normalized collision energies (nCE) of 10, 35, 80. The minimum AGC target was 1 × 104 with an intensity threshold of 2 × 105. All data were acquired in profile mode.
Relative quantification of metabolites
The resulting ThermoTM RAW files were converted to mzXML format using ReAdW.exe version 4.3.1 to enable peak detection and quantification. The centroided data were searched using an in-house python script Mighty_skeleton version 0.0.2 and peak heights were extracted from the mzXML files based on a previously established library of metabolite retention times and accurate masses adapted from the Whitehead Institute,32 and verified with high-resolution MS/MS spectral curated against the NIST14MS/MS33 and METLIN (2017)34 tandem mass spectral libraries. Metabolite peaks were extracted based on the theoretical m/z of the expected ion type e.g., [M + H]+, with a ±5 part-per-million (ppm) tolerance, and a ±7.5 second peak apex retention time tolerance within an initial retention time search window of ±0.5 min across the study samples. The resulting data matrix of metabolite intensities for all samples and blank controls was processed with an in-house statistical pipeline Metabolyze version 1.0 and final peak detection was calculated based on a signal to noise ratio (S/N) of 3× compared to blank controls, with a floor of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (arbitrary units). For samples where the peak intensity was lower than the blank threshold, metabolites were annotated as not detected, and the threshold value was imputed for any statistical comparisons to enable an estimate of the fold change as applicable. The resulting blank corrected data matrix was then used for all group-wise comparisons, and t-tests were performed with the Python SciPy (1.1.0)35 library to test for differences and generate statistics for downstream analyses. Any metabolite with p-value <0.05 was considered significantly regulated (up or down).
000 (arbitrary units). For samples where the peak intensity was lower than the blank threshold, metabolites were annotated as not detected, and the threshold value was imputed for any statistical comparisons to enable an estimate of the fold change as applicable. The resulting blank corrected data matrix was then used for all group-wise comparisons, and t-tests were performed with the Python SciPy (1.1.0)35 library to test for differences and generate statistics for downstream analyses. Any metabolite with p-value <0.05 was considered significantly regulated (up or down).
Transcriptomics
Worms for transcriptomics were taken from the same population as the worms for metabolomic analysis. Immediately after exposure, worms were counted and approximately 500 worms were added to a 1.5 mL Eppendorf tube. Worms were spun down, liquid was aspirated, 1 mL of RLT buffer (from Qiagen RNeasy Kit) with β-mercaptoethanol (BME) was added to the samples, and the samples were flash frozen and stored at −80 °C. The rest of the worm population was plated on K-agar plates with OP50 food. The worms were transferred daily for 8 days to avoid starvation and contamination of the population from progeny. Worms were rinsed 3× with K-medium, counted, and approximately 500 worms were added to 1.5 mL Eppendorf tubes. Worms were spun down, liquid was aspirated, 1 mL of RLT with BME was added to the samples, and the samples were flash frozen and stored at −80 °C.For RNA extraction, samples were thawed on ice and zirconia/silica beads were added to the thawed samples. Samples were homogenized using a beadbeater (spun for 1 minute intervals at maximum speed for 4 minutes total in the cold room). RNA extraction was performed using an RNeasy Kit (Qiagen, Germany) and according to manufacturer's instructions. RNA was eluted in 50 µL of molecular grade water. RNA concentration and purity was measured using a Thermo Scientific NanoDrop One Microvolume UV-Vis Spectrophotomer (Thermo Fisher Scientific) and samples were brought to a concentration of 30 ng µl−1 and were stored at −20 °C until sequencing.
The Sequencing and Genomics Technologies Core Facility at Duke University performed mRNA-sequencing. mRNA was enriched from total RNA and reversed transcribed into cDNA to build sequencing libraries using the Kapa stranded mRNA Kit from Roche (Code: KR0960). Libraries were pooled to equimolar concentration and sequenced on the NovaSeq 6000 S-Prime flow cell to produce 50 bp paired-end reads.
Bioinformatics processing of mRNA sequencing data
Quality of FASTQ files was evaluated using FastQC and ends were trimmed to remove adapters using Trimmomatic (V0.32) default parameters. Trimmed FASTQ files were uploaded to Galaxy (version 21.01) for further processing. Reads were aligned to the C. elegans reference genome (WBcel235) using HISAT2 (default parameters) and quantification was performed using featureCounts (default parameters) with the WS282 C. elegans genome annotation. Read count files were downloaded and analyzed in R (version 4.1.0) using DESeq2.Fat staining
Synchronized LIU2 L1 nematodes were exposed to various concentrations of sodium arsenite for 48 hours and rinsed immediately after exposure as described above. Nematodes were plated on K-agar plates seeded with OP50 and maintained at 20 °C. Nematodes were transferred daily by washing nematodes off plates with K-medium, allowing to gravity settle, and removing progeny. Rinsing was performed 3 times, and the adults were plated onto new K-agar OP50 plates. After 8 days post exposure, approximately 30 adult nematodes per condition were moved to agar pads on glass slides for imaging. 5 µl of 0.5 µM sodium azide was added to anesthetize the nematodes and a coverslip was added. A Keyence BZ-X700 was used to take fluorescent images of the nematodes and quantification of fluorescence was performed in FIJI. Mean fluorescence was normalized to worm area.Statistical analysis
Statistical analysis was performed in GraphPad Prism10 or in R as stated in figure legends. Outliers were removed in experiments with n > 100 using the two-sided Grubbs' test. Graphs were generated in GraphPad Prism10.Results
Development and characterization of experimental system to study long term effects of developmental exposure
First, to identify exposure scenarios that would permit us to interrogate the long-term (later-life and transgenerational) effects of developmental exposure to arsenite, we characterized the uptake of arsenic and its effects on nematode development and viability. Populations of L1 nematodes that were age-synchronized by growth arrest were exposed to various concentrations of arsenic for 48 hours in liquid cultures. After 48 hours of exposure, worms (that had reached a mix of L4 and adult stages that varied by treatment: Fig. 1E) were washed and plated on agar plates for downstream assays (Fig. 1A).Sodium arsenite exposure concentrations for subsequent experiments were determined by generating a lethality dose response curve (Fig. 1B). Percent survival was calculated immediately after the 48-exposure. Although worm size and developmental stage were not characterized at the highest arsenic doses used to generate the dose–response curve, worms were clearly smaller and developmentally delayed at these doses. The LC10 was determined to be approximately 735 µM. Exposures for subsequent experiments were based on percentages of this value. Due to the cuticle of C. elegans, the actual dose of sodium arsenic was likely much less than the exposure concentration.36 To determine the concentration of sodium arsenite inside the nematode after exposure, worms were exposed to arsenic and the internal concentration of arsenic was measured by ICP-MS (Fig. 1C). With increasing exposure concentrations of arsenic, the amount of arsenic measured in the nematodes also increased. Nematodes exposed to 73.5 µM sodium arsenite (10% LC10) had an average arsenic concentration (internal dose) of 0.43 ± 0.04 nM per worm, while nematodes exposed to 735 µM (100% LC10) sodium arsenite had an average concentration of 6.67 ± 0.64 nM per worm arsenic, an approximate 15-fold increase between the lowest and highest sodium arsenite exposures. The progeny of nematodes exposed to the highest concentration of sodium arsenite were also collected to measure free arsenic. No detectable arsenic was measured in the progeny of worms exposed to the highest concentration of arsenic (limit of detection: 9.4 nM; 300–1500 worms per sample). In addition to measuring the amount of sodium arsenite present immediately after the 48 hours exposure, we also measured the elimination of arsenic from the nematodes exposed to 735 µM sodium arsenite (Fig. 1D). Arsenic was eliminated rapidly, with a half-life of 3 hours (95% CI [2, 3.9]). After 24 hours, arsenic levels were 0.45 ± 0.05 nM, which is comparable to the amount of arsenic measured in the worms exposed to 73.5 µM sodium arsenite immediately post exposure (Fig. 1C). Free arsenic measured was not detectably different than the non-exposed control by 96 hours post exposure.
To begin to characterize the sub-lethal effects of these exposures, we next determined the effect of the sodium arsenite exposure on worm development. 48 hours of growth from the L1 stage should result in a population of worms in the late L4 to young adult stage under these culture conditions. Our results demonstrate that worms exposed to the lowest concentration of sodium arsenite are in the same developmental stage as the non-exposed controls following the 48 hours liquid exposure (Fig. 1E). However, exposure to sodium arsenite above 73.5 µM (10% of the LC10) resulted in a statistically significant delay in development. About 70% of the populations of worms exposed to 10% of the LC10 and the non-exposed controls were young adults, with the remainder of the worms in late L4 stages. However, the 25% and 50% of the LC10 exposures had only approximately 30% and 20%, respectively, of the populations reach the young adult stage. Finally, the group exposed to 735 µM sodium arsenite had no young adults present in the population and instead had 63% in the late L4 stage, 28% in the mid L4 stage, and 9% in the early L4 stage.
Transgenerational phenotypes
Previous studies have reported that sodium arsenite exposure in C. elegans had transgenerational effects including decreased reproduction19 and development of behavioral alterations.20 We examined five potential transgenerational phenotypes that represented both physiological and molecular phenotypes: worm length, reproduction, mitochondrial DNA copy number, ATP levels, and resilience to subsequent arsenite challenge. Based on the results shown in Fig. 1, we chose exposure levels that varied from no or mild to significant effects on nematode development. The highest concentration was the LC10 for 48 h viability (735 µM).Worm length was measured at the young adult stage using the COPAS Biosort time of flight (TOF) value as a surrogate. In the F0 generation, worms exposed to all concentrations of sodium arsenite were shorter than the non-exposed controls (Fig. 2A). Length decreased in a concentration-dependent manner, which is consistent with previously reported studies.17,19 Worm length was assessed in the same manner for three generations post arsenic exposure. In all the subsequent generations, there was no difference in worm length between the progeny of non-exposed and exposed parental worms within a generation. However, we did observe a decrease in length of the offspring of the control and low-exposure groups in the F1 compared to the F0 generation. Specifically, compared to the F0 group of the same exposure, the F1 populations of worms exposed to 0%, 10%, and 25% of the LC10 were statistically significantly shorter. This may be attributed to worms in the F1 generation growing only on solid K-agar plates while the F0 generation was grown for 48 hours in a liquid culture, which has been shown to increase worm length.37 The F2 and F3 generations showed no difference in worm length within generations, with some statistically significant differences compared to the F0 population of the same exposure level. Again, this was likely due to differences in liquid versus solid culture. Together, these data confirm that exposure to sodium arsenite in early life stunted growth as measured by length and suggests that molecular mechanisms causing a decrease in length were not transferred or inherited across generations.
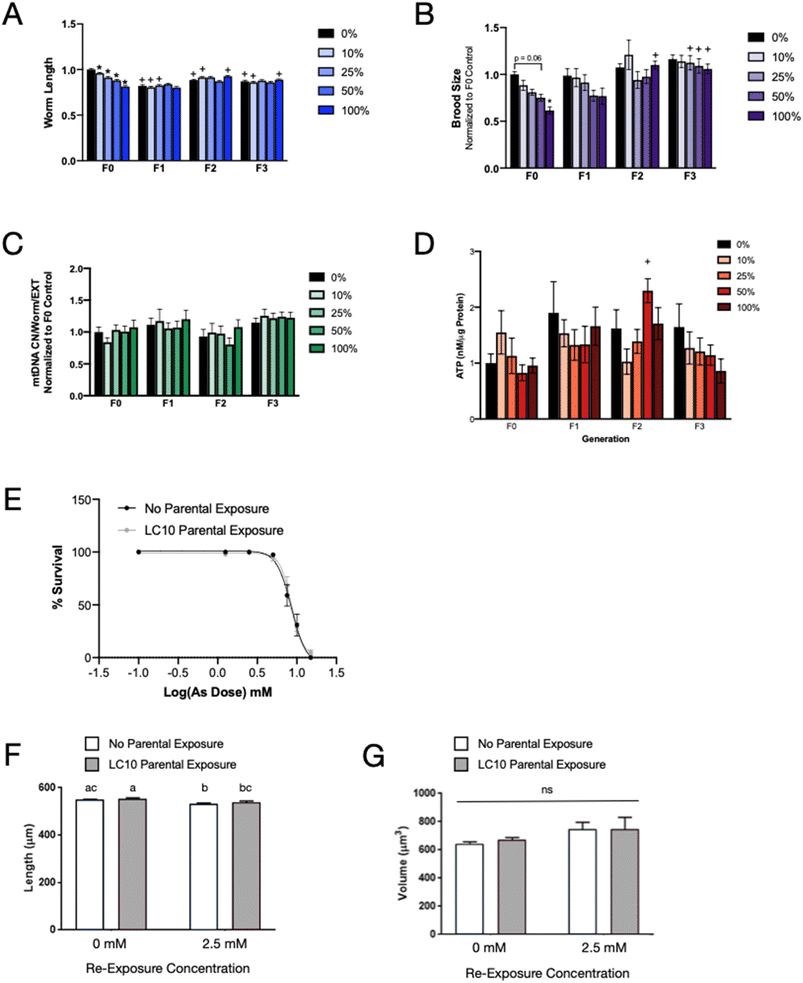 | ||
| Fig. 2 Transgenerational effects of exposure to sodium arsenite are minimal. (A) Worm length (TOF) normalized to F0 control. n > 100 per group per experiment. 3 independent experiments. Two-way ANOVA with Bonferroni's correction for multiple comparisons.31 * p ≤ 0.05 compared to control within generation. + p ≤ 0.05 compared to F0 of same exposure concentration. (B) Brood size normalized to F0 control. n = 7–22 per group. 6 independent experiments. Two-way ANOVA with Bonferroni's correction for multiple comparisons.31 * p ≤ 0.05 compared to control within generation. + p ≤ 0.05 compared to F0 of same exposure concentration. (C) mtDNA copy number normalized to worm number, EXT and F0 control. n = 3 per group per experiment. Two-way ANOVA with Bonferroni's correction for multiple comparisons.31 (D) ATP (nM µg−1 protein) normalized to F0 control. n = 3 per group per experiment. 2 independent experiments (50% F1 group has only one independent experiment). Two-way ANOVA with Bonferroni's correction for multiple comparisons.31 + p ≤ 0.05 compared to F0 of same exposure concentration. (E) Sodium arsenite re-challenge. Worms were exposed to the LC10 of sodium arsenite or control for 24 hours. F1 progeny were exposed to a range of concentrations (0–15 mM) to determine the LC50 for each group. There was no significant difference between the groups. (F) Worm length and (G) volume was measured using WormSizer ref. 25. Worms were exposed to the LC10 of sodium arsenite or control for 24 hours. F1 progeny were exposed to control or 2.5 mM sodium arsenite for 48 hours. n = 140–190. 2 independent experiments. Two-way ANOVA with Tukey's post hoc test for multiple comparisons. Different letters represent statistically significant differences (p < 0.05). | ||
We next measured individual nematode brood. In the exposed parental generation, we saw a dose-dependent decline in brood size with increased exposure to sodium arsenite (Fig. 2B). The highest sodium arsenite exposure resulted in a significantly decreased brood size compared to the non-exposed control. This was the only same-generation effect that was seen across the four generations assayed. Although F1 progeny of the highest doses of sodium arsenite exposure in the parental generation (50% and 100% of LC10) appear to have a smaller brood size (approximately 20% fewer progeny per worm) compared to F1 progeny of non-exposed nematodes, this difference was not statistically significant (p = 0.35 and 0.26, respectively). This observation is not present in the F2 and F3 generations, suggesting that the trend of decreased brood size with high sodium arsenite exposure in the parental generation, even if biologically real despite not reaching statistical significance, was not persistent across generations. Indeed, in the F3 generation, the brood size of all exposure groups was about the same as the brood size of the non-exposed controls in the F1 generation, providing further evidence that this is not an inherited effect of sodium arsenite exposure.
Because we did not see any evidence of transgenerational physiological phenotypes, we looked at molecular phenotypes that could be altered by sodium arsenite exposure and might be more sensitive to perturbation. We have previously reported that worms with mitochondrial deficiencies are more susceptible to the toxic effects of sodium arsenite exposure.17 Therefore, we quantified two readouts of mitochondrial health—mitochondrial DNA (mtDNA) content and ATP levels—post arsenic exposure and for three subsequent non-exposed generations. At these exposure concentrations, neither mtDNA (Fig. 2C) nor ATP levels (Fig. 2D) were different in the parental generation across any level of sodium arsenite exposure. Additionally, no transgenerational phenotypes were observed.
Lastly, we decided to conduct a sodium arsenite re-challenge experiment to determine if progeny of exposed nematodes were more resistant or more susceptible to sodium arsenite toxicity. A shift in the LC50 of the progeny of exposed nematodes could indicate the presence of a molecular phenotype that altered the response to sodium arsenite in subsequent generations. However, comparing the LC50 of F1 progeny from exposed (8.22 ± 1.43 mM) versus control non-exposed nematodes (8.50 ± 0.18 mM) indicated that there were no differences in sensitivity to arsenic lethality based on parental exposure (Fig. 2E). Additionally, there was no difference in worm length (Fig. 2F) or volume (Fig. 2G) after exposure to arsenic in progeny of non-exposed or LC10-exposed parental generation. There was a small but significant effect of the 2.5 mM re-exposure on worm length, but this was independent of treatment group. These data further support our observations that under these experimental conditions, exposure to sodium arsenic does not have obvious or deleterious multigenerational or transgenerational effects.
Later life effects of early life arsenic exposure
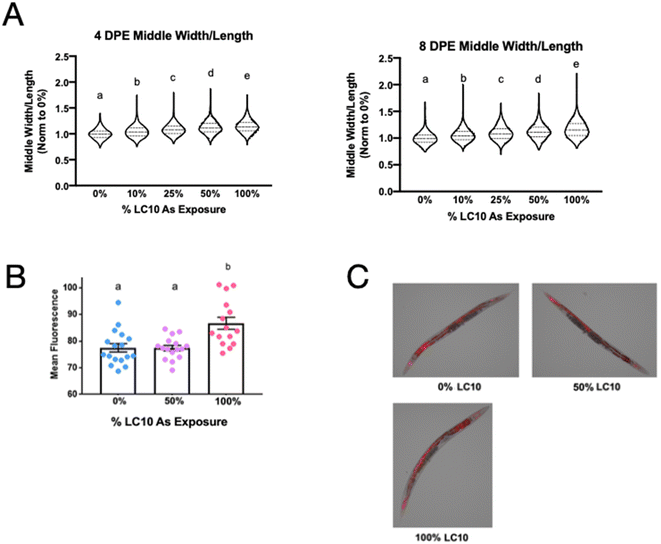
To contextualize the lack of multigenerational and transgenerational effects of sodium arsenite exposure and to elucidate molecular mechanisms of arsenic action in C. elegans compared to other species, given the extensive literature on later-life effects of early-life arsenic exposure, we decided to look at effects that might persist throughout the life of the worm. Worm length (Fig. 3A) and volume (Fig. 3B) showed statistically significant dose-dependent decreases with higher levels of arsenic exposure at both 4- and 8 days post exposure. Immediately after exposure, worms exposed to the highest exposure level (LC10) were approximately 19% shorter compared to control worms (Fig. 2A). Four days post-exposure, worms exposed to the LC10 were about 16% shorter, and were about 22% shorter than controls 8 days post-exposure (Fig. 3A). Similar trends were observed with volume measurements, with worms exposed to the LC10 having a 25% decrease in volume 4 days post-exposure, and a 36.5% decrease in volume 8 days post-exposure compared to non-exposed control worms (Fig. 3B).mtDNA copy number was also assessed at 8 days post-exposure (due to worms actively reproducing at 4 days post-exposure, which involves very extensive mtDNA replication, and the fact that we determined that these exposures affect reproduction, mtDNA copy number was not able to be accurately measured at this time point). mtDNA copy number was normalized to the non-exposed control and calculated per worm. To prevent the mtDNA copy number readout from merely reflecting the size of the worms, the mtDNA copy number was normalized to the average volume of the worms for each dose and independent experiment. The calculated average mtDNA copy number appeared to be approximately 10% lower for both the 50% LC10 and 100% LC10 exposed worms compared to the non-exposed controls (Fig. 3C). However, this decrease was not statistically significant (p = 0.50 and p = 0.57, respectively).
Metabolomics and transcriptomics
To characterize molecular changes that persisted through reproduction and into adulthood, we measured metabolomics and transcriptomics. Worms were exposed to control, low (10% of the LC10), or high (100% of the LC10) concentrations of arsenic for 48 hours beginning at the L1 stage. Samples of worms were flash frozen immediately following exposure, and 8 days after exposure. Samples were processed and prepared for metabolomic or transcriptomic analysis.Overall, 117 unique metabolites were identified across the 24 samples submitted for analysis. Of these, 56 metabolites were identified in all 24 samples, and 23 metabolites were identified in 12 or fewer samples. Metabolites were normalized to weight per worm for each sample. For each comparison (10% LC10 Day 0 vs. Control Day 0, 100% LC10 Day 0 vs. Control Day 0, 10% LC10 Day 8 vs. Control Day 8, and 100% LC10 Day 8 vs. Control Day 8), the average log2FC (if there were less than 2 reads of a metabolite in a sample group, this metabolite was omitted from the analysis) and −log10(p-value) were calculated to generate volcano plots (Fig. 4A–D). Enrichment and pathway analyses were performed in Metaboanalyst38 using lists of upregulated and downregulated metabolites for each of the 4 comparisons. Metabolite lists consisted of metabolites that had a log2FC greater than 1 and an adjusted p-value ≤0.05 for the upregulated metabolites and a log2FC less than −1 and an adjusted p-value ≤0.05 for the downregulated metabolites. The background list was the list of all metabolites identified in this study.
Overall, there were few changes that were individually statistically significant in metabolic profile in the treatment groups. Early life exposure to the LC10 of sodium arsenite resulted in larger changes in metabolite concentrations compared to early life exposure with 10% of the LC10. Additionally, more metabolites were changed immediately after the 48 hours exposure to sodium arsenite compared to 8 days post-exposure. Some trends existed across treatments. Dihydroxyacetone phosphate was significantly decreased at both treatment levels immediately after exposure (Fig. 4A and B), but not at the 8 days post-exposure timepoint. Citrulline appears to be increased in all conditions tested but is not statistically significant. Valerylcarnitine was decreased in worms exposed to the LC10 of sodium arsenite immediately after exposure, and butyrylcarnitine was decreased at both exposure levels 8 days post-exposure. Because not many metabolites were identified and few changed, enrichment analyses results do not have statistical power and are therefore not reported.
Transcriptomic analysis revealed a greater number of changes in mRNA expression across treatment conditions (Fig. 5) compared to metabolomic profiling. Many transcripts were upregulated (log2FC ≥ 1, adjusted p-value ≤ 0.05) immediately after exposure in both the low (175 transcripts) (Fig. 5A) and high (651 transcripts) (Fig. 5B) exposure conditions. 233 transcripts were also downregulated (log2FC ≤ 1, adjusted p-value ≤ 0.05) immediately after the high exposure condition. In the low exposure group, only three transcripts were significantly upregulated 8 days after the exposure ended (Fig. 5C), and none were downregulated. In contrast, 236 transcripts were upregulated 8 days post exposure in the high arsenic exposure groups (Fig. 5D), while only 9 transcripts were downregulated. Venn diagrams were created39 to determine the overlap of transcripts upregulated (Fig. 5E) or downregulated (Fig. 5F) at both concentrations of arsenic exposure and at the two timepoints.39 Interestingly, the transcripts that were upregulated in the high arsenic treatment group immediately after exposure were nearly entirely different than the transcripts upregulated in the high arsenic treatment group 8 days post (Fig. 5E). Only 11 of the transcripts were upregulated both immediately after and 8 days post exposure. This suggests that the response to arsenic exposure is much different than the transcriptional profile of worms in later life that had been exposed to arsenic in early life. However, the set of transcripts upregulated in the low and high arsenic exposures immediately after exposure were more similar. Of the 175 transcripts that were significantly upregulated in the low arsenic treatment immediately after exposure, 127 of these transcripts were also significantly upregulated in the high arsenic treatment immediately after exposure (Fig. 5E). Downregulated transcripts in each treatment group showed similar trends, though only the high exposure condition immediately after exposure had more than ten significantly downregulated transcripts (Fig. 5F).
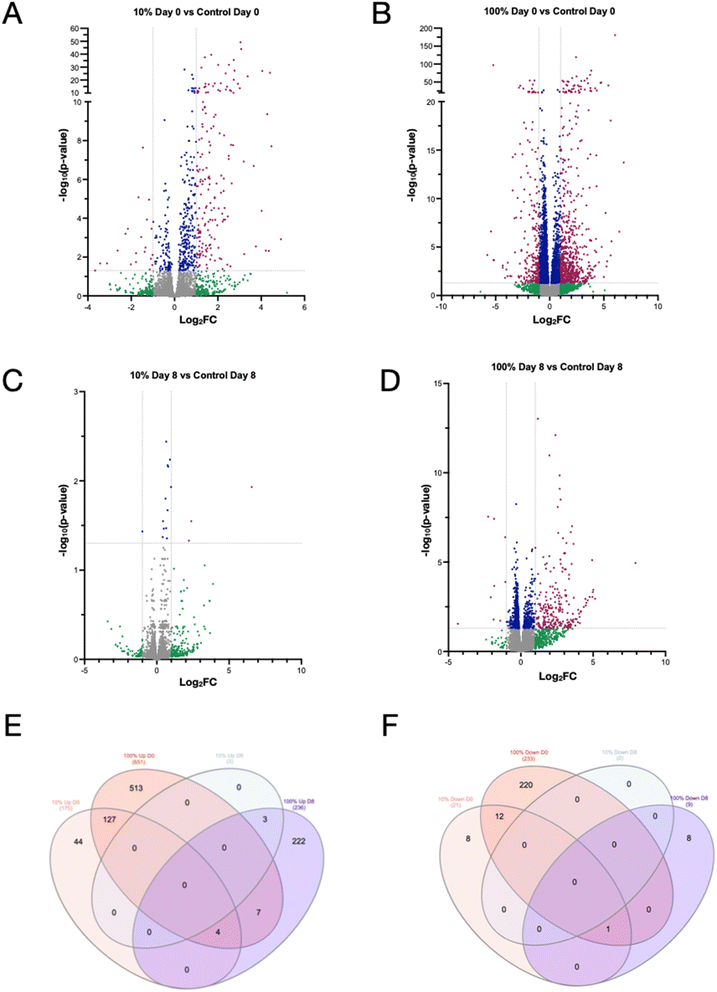 | ||
| Fig. 5 Transcriptomic profiles of worms exposed to sodium arsenite are different immediately after exposure and 8 days post exposure. Volcano plots (−log10(p-value) vs. log2FC) of transcripts identified in (A) 10% LC10 day 0 vs. control day 0, (B) 100% LC10 day 0 vs. control day 0, (C) 10% LC10 day 8 vs. control day 8, and (D) 100% LC10 day 8 vs. control day 8. Red data points represent transcripts with a log2FC ≥ 1 and a p-value ≤ 0.05 compared to control on the same day. Venn diagram comparing overlap of (E) upregulated and (F) downregulated metabolites normalized to control of the same day across all conditions. Venn diagrams were created using InteractiVenn ref. 39. | ||
To gain more insight into transcriptomic changes, lists of significantly upregulated and downregulated transcripts for each of the four treatment groups were uploaded and analyzed in DAVID.40,41 The background list was set to all C. elegans genes. Lists with less than 10 transcripts that met the cutoff criteria (10% Day 8 vs. Control Day 0 upregulated and downregulated, and 100% Day 8 vs. Control Day 0 downregulated) were not included in the analysis. As expected, based on the high degree of overlap between transcripts upregulated immediately after exposure in the 10% LC10 and LC10 groups, there was a high degree of similarity in KEGG pathways and Biological Process (BP) and Molecular Function (MF) Gene Ontology Terms (Table 1). General themes of stress response, metabolism, and signaling/developmental pathways were represented among the upregulated transcripts. To further understand these themes, WormMine42 was used identify the specific genes of upregulated transcripts represented in the KEGG pathways (Table 2). Two major families of genes involved in stress response were evident in the KEGG pathways overrepresented by upregulated transcripts—glutathione S-transferase (GST) genes, and skp-1 related (SKR) genes. Transcripts belonging to the GST family of genes made up at least half of the genes identified in all the significantly overrepresented 10% LC10 KEGG pathways. GST genes also significantly contribute to the upregulated transcripts in the top 6 significantly overrepresented KEGG pathways in the LC10 exposure group.
| Term | 10% LC10 | 100% LC10 | |||
|---|---|---|---|---|---|
| Rank | Adj P-value | Rank | Adj P-value | ||
| KEGG pathways | Metabolism of xenobiotics by cytochrome P450 | 1 | 4.80 × 10−6 | 1 | 1.20 × 10−14 |
| Drug metabolism – cytochrome P450 | 2 | 4.80 × 10−6 | 2 | 3.30 × 10−14 | |
| Glutathione metabolism | 3 | 5.60 × 10−6 | 3 | 6.70 × 10−11 | |
| Drug metabolism – other enzymes | 4 | 2.10 × 10−4 | 4 | 4.50 × 10−9 | |
| Longevity regulating pathway – worm | 5 | 8.60 × 10−4 | 5 | 2.30 × 10−6 | |
| Metabolic pathways | 6 | 4.40 × 10−2 | 6 | 6.16 × 10−6 | |
| Wnt signaling pathway | NA | NA | 7 | 4.10 × 10−4 | |
| Polycomb repressive complex | NA | NA | 8 | 5.80 × 10−3 | |
| TGF-beta signaling pathway | NA | NA | 9 | 7.00 × 10−3 | |
| Lysosome | NA | NA | 10 | 5.30 × 10−2 | |
| BP direct | Innate immune response | 1 | 4.20 × 10−26 | 2 | 3.50 × 10−14 |
| Glutathione metabolic process | 2 | 6.70 × 10−8 | 1 | 5.80 × 10−17 | |
| Defense response to Gram-negative bacteria | 3 | 1.80 × 10−2 | 3 | 9.30 × 10−4 | |
| PERK-mediated unfolded protein response | 4 | 2.10 × 10−2 | NA | NA | |
| Response to gamma radiation | 5 | 5.70 × 10−2 | NA | NA | |
| Defense response to Gram-positive bacteria | NA | NA | 4 | 2.10 × 10−2 | |
| Anatomical structure development | NA | NA | 5 | 2.60 × 10−2 | |
| MF direct | Glutathione transferase activity | 1 | 2.40 × 10−8 | 1 | 2.50 × 10−18 |
| FMN binding | 2 | 3.50 × 10−1 | 7 | 3.20 × 10−2 | |
| UDP-glycosyltransferase activity | 3 | 3.60 × 10−1 | NA | NA | |
| Oxidoreductase activity | 4 | 3.60 × 10−1 | 2 | 5.00 × 10−4 | |
| Structural constituent of cuticle | NA | NA | 3 | 9.20 × 10−4 | |
| Structural constituent of chromatin | NA | NA | 4 | 2.00 × 10−2 | |
| Protein heterodimerization activity | NA | NA | 5 | 2.60 × 10−2 | |
| Cullin family protein binding | NA | NA | 6 | 2.60 × 10−2 | |
| Pathway | 10% LC10 | 100% LC10 | ||||||
|---|---|---|---|---|---|---|---|---|
| Count | Fold enrichment | Genes | Benjamini | Count | Fold enrichment | Genes | Benjamini | |
| Metabolism of xenobiotics by cytochrome P450 | 7 | 24.60 | gst-25, gst-4, gst-7, gst-30, gst-5, gst-38, ugt-18 | 4.80 × 10−6 | 16 | 19.42 | gst-1, gst-10, gst-25, adh-1, gst-4, gst-2, gst-24, gst-7, gst-30, gst-5, gst-38, gst-6, sodh-2, gsto-2, C02D5.4, ugt-18 | 1.19 × 10−14 |
| Drug metabolism – cytochrome P450 | 7 | 22.23 | gst-25, gst-4, gst-7, gst-30, gst-5, gst-38, ugt-18 | 4.80 × 10−6 | 16 | 17.56 | gst-1, gst-10, gst-25, adh-1, gst-4, gst-2, gst-24, gst-7, gst-30, gst-5, gst-38, gst-6, sodh-2, gsto-2, C02D5.4, ugt-18 | 3.32 × 10−14 |
| Glutathione metabolism | 7 | 20.28 | gcs-1, gst-25, gst-4, gst-7, gst-30, gst-5, gst-38 | 5.61 × 10−6 | 14 | 14.01 | gst-1, gcs-1, gst-10, gst-25, gst-4, gst-2, gst-24, gst-7, gst-30, gst-5, gst-38, gst-6, gsto-2, C02D5.4 | 6.69 × 10−11 |
| Drug metabolism – other enzymes | 6 | 15.48 | gst-25, gst-4, gst-7, gst-5, gst-38, ugt-18 | 2.05 × 10−4 | 13 | 11.59 | gst-1, gst-10, gst-25, gst-4, gst-2, gst-24, gst-7, gst-5, gst-38, gst-6, gsto-2, C02D5.4, ugt-18 | 4.49 × 10−9 |
| Longevity regulating pathway – worm | 6 | 11.01 | gcs-1, kgb-2, gst-4, gst-7, gst-5, gst-38 | 8.64 × 10−4 | 12 | 7.61 | gcs-1, kgb-2, mtl-1, gst-4, gsto-2, gst-2, gst-24, gst-7, C02D5.4, gst-5, gst-38, gst-6 | 2.31 × 10−6 |
| Metabolic pathways | 12 | 2.08 | gcs-1, gst-25, ech-9, gst-4, cysl-2, gst-7, nit-1, asm-3, gst-30, gst-5, gst-38, ugt-18 | 0.0435794 | 35 | 2.09 | gst-10, rml-3, gst-25, adh-1, ech-9, gst-4, gst-2, gst-24, gst-7, haly-1, gst-30, gst-5, gst-6, dot-1.4, gsto-2, asah-1, nit-1, pycr-4, hacd-1, gst-1, gcy-23, gcs-1, gana-1, rml-4, cat-4, asm-3, gst-38, sodh-2, cysl-2, pdxk-1, asah-2, R04B5.6, C02D5.4, ugt-18, cbs-1 | 6.13 × 10−6 |
| Wnt signaling pathway | 9 | 6.42 | skr-10, kgb-2, skr-13, cyd-1, skr-15, skr-14, skr-7, vang-1, skr-5 | 4.11 × 10−4 | ||||
| Polycomb repressive complex | 6 | 7.78 | skr-10, skr-13, skr-15, skr-14, skr-7, skr-5 | 0.006 | ||||
| TGF-beta signaling pathway | 6 | 7.28 | skr-10, skr-13, skr-15, skr-14, skr-7, skr-5 | 0.007 | ||||
| Lysosome | 7 | 3.73 | gana-1, lipl-1, cpr-3, asah-2, asah-1, asm-3, crn-6 | 0.053 | ||||
| Ubiquitin mediated proteolysis | 6 | 3.76 | skr-10, skr-13, skr-15, skr-14, skr-7, skr-5 | 0.098 | ||||
| Protein processing in endoplasmic reticulum | 7 | 2.92 | skr-10, kgb-2, skr-13, skr-15, skr-14, skr-7, skr-5 | 0.136 | ||||
Additionally, several KEGG pathways, Biological Process, and Molecular Function terms were related to oxidoreductive metabolism and reactive oxygen species (ROS) including glutathione metabolism and flavin mononucleotide binding. Of interest, within the transcripts upregulated that are categorized in the “metabolic pathways” KEGG pathway, four of the transcripts are related to fatty acid degradation. In the high exposure group, several molecular function terms were related to worm anatomy, specifically components of the cuticle. In conjunction with the observed increase in cysteine and glutathione metabolism genes, this suggests a cuticular stress response. Because the number of downregulated transcripts immediately after arsenic exposure was much lower, fewer KEGG pathways and gene ontology terms were significantly overrepresented in these lists. Terms that did appear also suggested a general stress and metabolic response. Interestingly, a highly significant (log2FC −1.56, p-value 4 × 10−44) transcript downregulated in the high exposure group identified in the metabolic pathways KEGG pathway is pck-1 (phosphoenolpyruvate carboxykinase), potentially suggesting a shift away from oxidative metabolism43 and supporting previous findings of a Warburg-like effect in worms exposed to arsenic.16
Due to a low number of upregulated transcripts in the 10% LC10 group 8 days after exposure (only three), only the upregulated LC10 8 days post-exposure list of transcripts was analyzed in DAVID. Although 236 transcripts were uploaded, few KEGG pathways and gene ontology terms were significantly overrepresented in the list (Table 3). Nearly all significant terms were related to critical components of C. elegans anatomy, including cuticle development and myosin phosphatase activity. The overrepresentation of transcripts in these pathways could suggest a compensatory response to cuticular damage from the arsenic exposure, or a long-term effect of the arsenic exposure. To investigate the signature of transcripts that were upregulated immediately after arsenic exposure and 8 days following the exposure, the list of 11 transcripts that were in both groups was uploaded to DAVID. Even though the list size was small, two biological processes terms were significantly overrepresented in this list of genes: “innate immune response” (genes: clec-67, gst-38, and lys-3) and “PERK-mediated unfolded protein response” (genes: clec-67 and gst-38). This suggests that the initial stress response might be persistent across the time course as these biological process terms were also significantly overrepresented immediately after arsenic exposure.
| Term | 100% LC10 | ||
|---|---|---|---|
| Rank | Adj P-value | ||
| KEGG pathways | mRNA surveillance pathway | 1 | 1.10 × 10−1 |
| BP direct | Anatomical structure development | 1 | 1.00 × 10−59 |
| Cuticle development involved in collage and cuticulin-based cuticle molting cycle | 2 | 7.10 × 10−3 | |
| PERK-mediated unfolded protein response | 3 | 4.30 × 10−2 | |
| Collagen and cuticulin-based cuticle development | 4 | 7.70 × 10−2 | |
| MF direct | Structural constituent of cuticle | 1 | 3.80 × 10−70 |
| Myosin phosphatase activity | 2 | 7.90 × 10−2 | |
To integrate the metabolomics and transcriptomics data, both normalized data sets were uploaded to the Comparative Genome Dashboard of the BioCyc web application.44 All identified metabolites and transcripts were included in the analysis (Fig. 6). Each panel shows the combined data for all gene or metabolite changes related to a specific metabolite or group of metabolites within a pathway or process. The heights of the bars provide the sum of all the fold change values of genes or metabolites related to a metabolite or pathways, while the large dot represents the average fold change. This visualization of the combined omics data sets shows that, in general, comparing metabolomic and transcriptomic data in each group revealed similar trends, providing validation of the pathway signatures. The higher dose of arsenic tends to have a greater effect on pathways compared to the lower dose at the same timepoint, which is consistent with previous results presented here. Interestingly, effects at 8 days post arsenic exposure (yellow and green bars) appear to have a larger magnitude of effect compared to the effects immediately after arsenic exposure despite fewer number of overall transcripts changed.
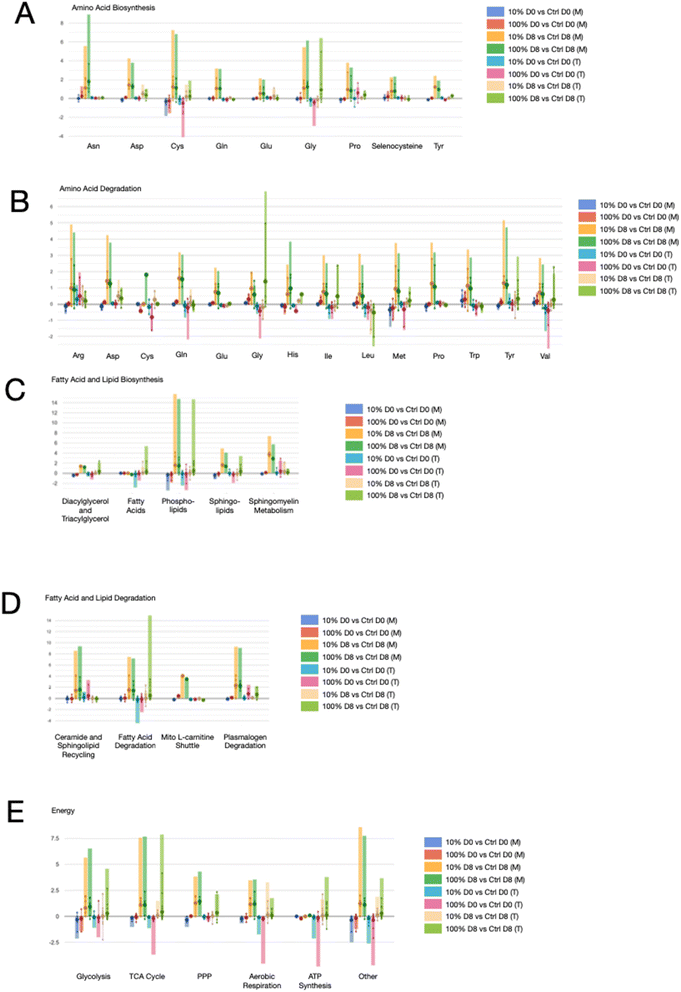 | ||
| Fig. 6 Integration of metabolomic and transcriptomic data validates pathway signatures. Metabolomic and transcriptomic data were uploaded to the Comparative Genome Dashboard of the BioCyc web application ref. 39 for multi-omics analysis. Each panel shows the combined data for all gene or metabolite changes related to a specific metabolite or group of metabolites within a pathway or process. The height of each bar represents the sum of all normalized data for the relative change graphed for a specific metabolite or pathway. The large dot represents the average fold change, while the small dots represent the range of the data. The first four bars for each metabolite or pathway represent the metabolic data, while the second set of four bars represent the transcriptomics data. Each comparison is color matched between metabolomic transcriptomic data sets. (A) Amino acid biosynthesis; (B) amino acid degradation; (C) fatty acid and lipid biosynthesis; (D) fatty acid and lipid degradation; and (E) major energy pathways. M indicates metabolomics data and T indicates transcriptomics data. | ||
Given the transcriptomic analysis that indicated protein degradation and cuticle structure, pathways of amino acid biosynthesis and degradation were further explored (Fig. 6A and B). Glycine synthesis and degradation is high at the 8 days post-exposure timepoint, which could indicate high turnover of the amino acid due to cuticle breakdown as glycine is the most abundant amino acid in the cuticle. The branched chain amino acids (isoleucine, leucine, and valine) are only observed in the amino acid degradation panel. Immediately post arsenic exposure, degradation of the BCAAs is decreased according to the more robust transcriptomics data (Fig. 6B). Recently it was reported that arsenic inhibits BCKDH (branched-chain α-ketoacid dehydrogenase complex) which catalyzes an irreversible step in BCAA catabolism.45 The observation of decreased BCAA degradation supports a decrease in BCAA catabolism. At the 8 days post exposure timepoint, there is an increase in degradation of isoleucine and valine, suggesting that inhibition of BCAA catabolism does not persist. Cysteine biosynthesis is decreased in both the metabolomic and transcriptomic datasets immediately after arsenic exposure, which is consistent with arsenic's affinity for sulfhydryl groups. This effect is not persistent as the 8 days post-exposure group shows an increase in cysteine biosynthesis, suggesting a compensatory effect.
Because we observed an increase in expression of transcripts involved in fatty acid degradation immediately after exposure we looked at the fatty acid and lipid biosynthesis and degradation profiles from the multi-omics analysis. We observed an increase in metabolites and transcripts involved in triacylglycerol synthesis at the 8 days timepoint (Fig. 6C). Fatty acid synthesis and degradation pathways appear to be in flux; however, metabolites associated with the mitochondrial L-carnitine shuttle are increased at the 8 days post-exposure timepoint (Fig. 6D), which could suggest an upregulation of the shuttle to compensate for a decrease in fatty acid oxidation.46 Consistent with previous studies,16 the multi-omics data supports a slight decrease in aerobic respiration and ATP synthesis immediately after arsenic exposure (Fig. 6E). Transcriptomic data suggests a return to baseline, or slightly elevated, aerobic respiration and ATP synthesis 8 days following the arsenic exposure.
Morphology and lipid content
The multi-omics analysis provided evidence of a possible shift in fatty acid degradation and a decrease in oxidative phosphorylation. Therefore, we calculated the middle-width to length ratio of C. elegans 4- and 8 days post arsenic exposure to quantify worm morphology, predicting that a shift in metabolism could lead to differences in worm adiposity. The middle-width to length value increased statistically significantly in a concentration-dependent manner throughout adulthood (Fig. 7A). Worms exposed to the LC10 had an approximate 15% and 17% increase in middle width to length ratio compared to non-exposed controls at 4- and 8 days post-exposure, respectively. To determine if fat content contributed to the difference in worm morphology in later life, fat content was analyzed using LIU2 nematode. This strain expresses mCherry ubiquitously in the intestine, muscle, hypodermis, and embryos, and is localized primarily in lipid droplets. 8 days post-exposure, there was a significant increase in mCherry fluorescence in worms exposed to the high arsenic exposure compared to control (Fig. 7B and C). There was no difference in lipid content between the control and worms exposed to 50% of the LC10. In conjunction with the multi-omics analysis, this phenotype confirms an alteration of fat content and metabolism that persists into adulthood after an early life exposure to arsenic.Discussion
The goals of this study were to characterize the persistent and transgenerational effects of early-life arsenic exposure in C. elegans. Additionally, we sought to define toxicokinetic parameters of arsenic metabolism in worms and explore the utility of C. elegans as a model to study persistent and transgenerational effects of arsenic exposure. We successfully defined the uptake and half-life of sodium arsenite after a 48 hours larval exposure and rigorously examined several transgenerational phenotypes that were relevant to defined effects of arsenic exposure. While transgenerational phenotypes were not robustly observed, we discovered several phenotypes related to worm size and fat content that persisted through mid-adulthood (we did not examine older worms). A multi-omics approach to investigating the molecular signatures of these phenotypes revealed shifts in metabolism immediately after arsenic exposure, and a potential signature of increased fat storage and fatty acid metabolism 8 days post arsenic exposure.Characterization of the elimination of sodium arsenite in C. elegans showed the half-life of sodium arsenite to be 3 hours (Fig. 1D). In humans, the half-life of arsenic species in urine is about 4 days, and on the order of hours in blood.13 The half-life of arsenic varies widely depending on the species, with rat models of arsenic toxicokinetics yielding a half-life on the order of 70 days.47 The rapid elimination of arsenic from the worm appears to be more in line with human elimination of arsenic in the blood. Although C. elegans lack the main enzyme that methylates inorganic arsenic (AS3MT) and promotes excretion of arsenic species in humans, the excretion of arsenic from the worm remains very fast. Our transcriptomic data suggest that the cuticle may be a major target of arsenic toxicity, which suggests an alternate mechanism for the rapid excretion of arsenic. After each larval stage, the worm cuticle is shed and replaced by a new cuticle, including upon entering the young adult stage.48 Most worms (60%) were in the late L4 stage after 48 hours of exposure to the highest concentration of arsenic (Fig. 1E). The amount of time between the late L4 stage and adult stage is approximately 2 hours,26 which is consistent with the half-life observed. Other mechanisms for reducing the total body burden of arsenic may be present, but the shedding of the cuticle could be a major component of arsenic elimination. Future studies examining elimination after arsenic exposure in the adult stage could provide further insight as to additional mechanisms of arsenic elimination; these should include post-reproductive life stages or studies in germline-deficient strains to isolate possible transfer into gametes and eggshells. The absence of arsenic methylation in C. elegans creates the opportunity to separate the toxic effects of inorganic arsenic from its various methylated forms. For example, future studies could dose separately with different arsenic species to study the toxicokinetics and toxicodynamics of the different species without the confounding effect of in vivo methylation. In addition, it may be useful to create a “humanized” C. elegans that expresses human arsenic methyltransferase.
Interestingly, the immediate effects of arsenic exposure on the transcriptome were quite different from the transcriptomic signature in later life (Fig. 5). While immediate effects were centered on stress response, proteasomal degradation, and detoxification, later-life effects of early-life arsenic exposure were consistent with physiological and biochemical effects observed in humans. The cuticle damage described above appears analogous to the effects of chronic arsenic exposure resulting in increased incidence of skin lesion and skin cancer in humans.12 Additionally, we observed a physiological increase in fat deposition (Fig. 7B and C) as well as a molecular signature of increased triacylglycerol synthesis 8 days post-exposure (Fig. 6). This data mirrors epidemiological studies that show an association of increased adiposity (as measured by leptin levels and waist circumference) in people exposed to high levels of arsenic.49 Additionally, our multi-omics results supported a decrease in cellular respiration, consistent with previous findings.16 However, given the differences in experimental design, further studies are needed to definitively prove altered OXPHOS in later life. Together, our data support C. elegans as a model to study the effects of sodium arsenic due to similarities in metabolism and physiological response in humans.
The most striking later-life phenotype observed was the decreased body size (length and volume) (Fig. 3) and increased middle width to length ratio that persisted 8 days after the exposure to arsenic (Fig. 7). Doses as low at 10% of the LC10 resulted in significantly different worm body morphologies. Importantly, although an initial developmental delay (Fig. 1E) was observed in worms exposed to 25% of the LC10 and higher doses, there was no developmental delay observed in the group exposed to 10% of the LC10. The emergence of a persistent phenotype of smaller worm size in this group suggests that there were persistent effects of the arsenic exposure that did not immediately affect worm development. The dramatic change in the transcriptomic profile between immediately after exposure and 8 days post exposure (Fig. 5) suggests that the phenotype is a result of effects of arsenic unrelated to the initial stress response. How this persistent, later-life effect is mediated despite clearance of detectable arsenic is unclear. Mechanistically, most studies in organisms related to the persistent, intergenerational and transgenerational effects of arsenic exposure have been related to DNA methylation.11 Given the atypical or absent DNA methylation patterns in C. elegans,50 other modes of persistently altered gene regulation such as small RNAs may be at play. However, defining precise mechanisms of regulation has proved challenging and remains an open question in the field.51
Multiple transgenerational phenotypes were tested based on reported transgenerational effects in C. elegans19,20 and mice.52 However, in our study, we found no transgenerational phenotypes. Some possible reasons for this lack of congruency include differences in experimental design such as concentrations of arsenic tested, length of exposure, and/or stage of worm during the exposure. In particular, the stress of synchronizing worms with the standard sodium hypochlorite procedure53 as well as exposing worms post embryonically may have reduced our ability to observe transgenerational phenotypes. Despite these potential shortcomings, a large sample size and repeating experiments in triplicate provided a robust data set that clearly showed effects in the exposed generation (Fig. 2) that were absent in three subsequent generations, highlighting the effectiveness of C. elegans as a model organism to study transgenerational phenotypes. While some independent experiments appeared to support a transgenerational phenotype, combining three replicates of an experiment eliminated these observations, suggesting that the observations were noise that washed out with repeated biological replicates.
Conclusions
In conclusion, we have presented a rigorous data set that highlights the utility of C. elegans in studying persistent and transgenerational effects of sodium arsenic exposure. The biochemical and physiological effects observed were in line with effects observed in human populations, including large alterations in cysteine metabolism, phase II and phase III xenobiotic metabolism processes, protein quality control, bioenergetic shifts, innate immune responses, fat deposition, and cuticle damage that appears analogous to effects on skin in people. The fast excretion of sodium arsenic results in a short half-life comparable to the half-life of sodium arsenic in blood in people, albeit likely because of different excretion mechanisms. Finally, although no transgenerational effects were observed, we report a dramatic effect of decreased body size and increased fat accumulation in worms exposed to a low level of sodium arsenite that persists into adulthood.Author contributions
Kathleen A Hershberger: conceptualization, methodology, software, formal analysis, investigation, writing – original draft, visualization. Shaza Gaballah: methodology, formal analysis, investigation, visualization. Britney He: software, formal analysis. Luoyu Zhang: software, formal analysis. Emily S. Barefoot: investigation. Caroline B. Reed: investigation. Nelson A Rivera: methodology, investigation, resources, formal analysis. Heileen Hsu-Kim: supervision. Joel N. Meyer: conceptualization, writing – review and editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: differential metabolite expression (normalized to worm number) and mRNA read counts mapped to WormBase gene IDs. mRNA read count IDs for individual samples are represented as concentration_replicate-day. See DOI: https://doi.org/10.1039/d5em00655d.Acknowledgements
We thank Tori Rodrick and the NYU Metabolomics Core Resource Laboratory for performing the metabolite extraction, LC-MS/MS and raw metabolite quantification for this study. We thank the Duke University School of Medicine for the use of the Sequencing and Genomic Technologies Shared Resource, which provided the mRNA-sequencing service. This research was funded by the National Institute of Environmental Health Sciences, grant number P42ES010356 and T32ES021432.References
- J. Podgorski and M. Berg, Global threat of arsenic in groundwater, Science, 2020, 368(6493), 845–850 CrossRef CAS PubMed.
- C. D. Klaassen, Casarett & Doull's Toxicology: the Basic Science of Poisons, 9th edn, McGraw Hill, 2018, pp. 1115–1119 Search PubMed.
- N. K. Roy, A. Murphy and M. Costa, Arsenic Methyltransferase and Methylation of Inorganic Arsenic, Biomolecules, 2020, 10(9), 1351 CrossRef CAS PubMed.
- A. P. Sage, B. C. Minatel, K. W. Ng, G. L. Stewart, T. J. B. Dummer and W. L. Lam, et al., Oncogenomic disruptions in arsenic-induced carcinogenesis, Oncotarget, 2017, 8(15), 25736–25755 CrossRef PubMed.
- B. C. Minatel, A. P. Sage, C. Anderson, R. Hubaux, E. A. Marshall and W. L. Lam, et al., Environmental arsenic exposure: from genetic susceptibility to pathogenesis, Environ. Int., 2018, 112, 183–197 CrossRef CAS PubMed.
- G. Marshall, C. Ferreccio, Y. Yuan, M. N. Bates, C. Steinmaus and S. Selvin, et al., Fifty-Year Study of Lung and Bladder Cancer Mortality in Chile Related to Arsenic in Drinking Water, J. Natl. Cancer Inst., 2007, 99(12), 920–928 CrossRef CAS PubMed.
- J. L. Young, L. Cai and J. C. States, Impact of prenatal arsenic exposure on chronic adult diseases, Syst. Biol. Reprod. Med., 2018, 64(6), 469–483 CrossRef CAS PubMed.
- S. F. Farzan, M. R. Karagas and Y. Chen, In utero and early life arsenic exposure in relation to long-term health and disease, Toxicol. Appl. Pharmacol., 2013, 272(2), 384–390 CrossRef CAS PubMed.
- M. P. Waalkes, J. Liu, H. Chen, Y. Xie, W. E. Achanzar and Y. S. Zhou, et al., Estrogen Signaling in Livers of Male Mice With Hepatocellular Carcinoma Induced by Exposure to Arsenic In Utero, J. Natl. Cancer Inst., 2004, 96(6), 466–474 CrossRef CAS PubMed.
- J. E. Rager, K. A. Bailey, L. Smeester, S. K. Miller, J. S. Parker and J. E. Laine, et al., Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood, Environ. Mol. Mutagen., 2014, 55(3), 196–208 CrossRef CAS PubMed.
- R. Verdikt, A. A. Armstrong and P. Allard, Transgenerational inheritance and its modulation by environmental cues, Curr. Top. Dev. Biol., 2022, 152, 31–76 Search PubMed.
- M. R. Karagas, A. Gossai, B. Pierce and H. Ahsan, Drinking Water Arsenic Contamination, Skin Lesions, and Malignancies: A Systematic Review of the Global Evidence, Curr. Environ. Health Rep., 2015, 2(1), 52–68 CrossRef CAS PubMed.
- J. Ashley-Martin, M. Fisher, P. Belanger, C. M. Cirtiu and T. E. Arbuckle, Biomonitoring of inorganic arsenic species in pregnancy, J. Exposure Sci. Environ. Epidemiol., 2023, 33(6), 921–932 CrossRef CAS PubMed.
- L. A. Eaves, G. Choi, E. Hall, F. C. M. Sillé, R. C. Fry and J. P. Buckley, et al., Prenatal Exposure to Toxic Metals and Neural Tube Defects: A Systematic Review of the Epidemiologic Evidence, Environ. Health Perspect., 2023, 131(8), 086002 CrossRef CAS PubMed.
- M. C. K. Leung, P. L. Williams, A. Benedetto, C. Au, K. J. Helmcke and M. Aschner, et al., Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology, Toxicol. Sci., 2008, 106(1), 5–28 CrossRef CAS PubMed.
- A. L. Luz, T. R. Godebo, D. P. Bhatt, O. R. Ilkayeva, L. L. Maurer and M. D. Hirschey, et al., From the Cover: Arsenite Uncouples Mitochondrial Respiration and Induces a Warburg-like Effect in Caenorhabditis elegans, Toxicol. Sci., 2016, 152(2), 349–362 CrossRef CAS PubMed.
- A. L. Luz, T. R. Godebo, L. L. Smith, T. C. Leuthner, L. L. Maurer and J. N. Meyer, Deficiencies in mitochondrial dynamics sensitize Caenorhabditis elegans to arsenite and other mitochondrial toxicants by reducing mitochondrial adaptability, Toxicology, 2017, 387, 81–94 CrossRef CAS PubMed.
- A. Cheikhi, C. Wallace, C. S. Croix, C. Cohen, W. Y. Tang and P. Wipf, et al., Mitochondria are a substrate of cellular memory, Free Radical Biol. Med., 2019, 130, 528–541 CrossRef CAS PubMed.
- C. W. Yu and V. H. C. Liao, Transgenerational Reproductive Effects of Arsenite Are Associated with H3K4 Dimethylation and SPR-5 Downregulation in Caenorhabditis elegans, Environ. Sci. Technol., 2016, 50(19), 10673–10681 CrossRef CAS PubMed.
- X. Zhang, H. Q. Zhong, Z. W. Chu, X. Zuo, L. Wang and X. L. Ren, et al., Arsenic induces transgenerational behavior disorders in Caenorhabditis elegans and its underlying mechanisms, Chemosphere, 2020, 252, 126510 CrossRef CAS PubMed.
- I. L. C. Arsenic, Exposure Sources, Health Risks, and Mechanisms of Toxicity, ed. J. W. Sons, 2015, pp. 469–493 Search PubMed.
- J. A. Lewis and J. T. Fleming, Basic Culture Methods, Elsevier, 1995 Search PubMed.
- T. Stiernagle, Maintenance of C. elegans, WormBook: the online review of C. elegans biology, 2006, pp. 1–11 Search PubMed.
- D. L. Croteau, M. J. DellaVecchia, L. Perera and B. V. Houten, Cooperative damage recognition by UvrA and UvrB: identification of UvrA residues that mediate DNA binding, DNA repair, 2008, 7(3), 392–404 CrossRef CAS PubMed.
- Methods for Measuring the AcuteToxicity of Effluents and Receiving Waters to Freshwater and Marine Organisms, US Environmental Protection Agency, 2002 Search PubMed.
- D. Z. L. Mok, P. W. Sternberg and T. Inoue, Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage, BMC Dev. Biol., 2015, 15(1), 26 CrossRef PubMed.
- T. C. Leuthner, J. H. Hartman, I. T. Ryde and J. N. Meyer, Mitochondrial Regulation, Methods and Protocols, Methods Mol. Biol., 2021, 2310, 91–111 CrossRef CAS PubMed.
- C. P. Gonzalez-Hunt, J. P. Rooney, I. T. Ryde, C. Anbalagan, R. Joglekar and J. N. Meyer, PCR-Based Analysis of Mitochondrial DNA Copy Number, Mitochondrial DNA Damage, and Nuclear DNA Damage, Curr. Protoc. Toxicol., 2016, 67, 20 11 1-20 11 25, available from: http://www.ncbi.nlm.nih.gov/pubmed/26828332 Search PubMed.
- K. Palikaras and N. Tavernarakis, Intracellular Assessment of ATP Levels in Caenorhabditis elegans, Bio-Protoc., 2016, 6(23), e22048 Search PubMed.
- B. T. Moore, J. M. Jordan and L. R. Baugh, WormSizer: high-throughput analysis of nematode size and shape, PLoS One, 2013, 8(2), e57142 CrossRef CAS PubMed.
- D. R. Jones, Z. Wu, D. Chauhan, K. C. Anderson and J. Peng, A Nano Ultra-Performance Liquid Chromatography-High Resolution Mass Spectrometry Approach for Global Metabolomic Profiling and Case Study on Drug-Resistant Multiple Myeloma, Anal. Chem., 2014, 86(7), 3667–3675 CrossRef CAS PubMed.
- W. W. Chen, E. Freinkman, T. Wang, K. Birsoy and D. M. Sabatini, Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism, Cell, 2016, 166(5), 1324–1337 CrossRef CAS PubMed.
- Y. Simón-Manso, M. S. Lowenthal, L. E. Kilpatrick, M. L. Sampson, K. H. Telu and P. A. Rudnick, et al., Metabolite Profiling of a NIST Standard Reference Material for Human Plasma (SRM 1950): GC-MS, LC-MS, NMR, and Clinical Laboratory Analyses, Libraries, and Web-Based Resources, Anal. Chem., 2013, 85(24), 11725–11731 CrossRef PubMed.
- C. A. Smith, G. O. Maille, E. J. Want, C. Qin, S. A. Trauger and T. R. Brandon, et al., METLIN, Ther. Drug Monit., 2005, 27(6), 747–751 CrossRef CAS PubMed.
- E. Jones, E. Oliphant and P. Peterson, SciPy: Open Source Scientific Tools for Python, 2001, [Internet], [cited 2019 Aug 19], available from: http://www.scipy.org/ Search PubMed.
- J. H. Hartman, S. J. Widmayer, C. M. Bergemann, D. E. King, K. S. Morton and R. F. Romersi, et al., Xenobiotic metabolism and transport in Caenorhabditis elegans, J. Toxicol. Environ. Health B Crit. Rev., 2021, 24(2), 51–94 CrossRef CAS PubMed.
- I. Lev, R. Bril, Y. Liu, L. I. Ceré and O. Rechavi, Inter-generational consequences for growing Caenorhabditis elegans in liquid, Philos. Trans. R. Soc. London, Ser. B, 2019, 374(1770), 20180125 CrossRef CAS PubMed.
- Y. Lu, Z. Pang and J. Xia, Comprehensive investigation of pathway enrichment methods for functional interpretation of LC–MS global metabolomics data, Briefings Bioinf., 2022, 24(1), bbac553 CrossRef PubMed.
- H. Heberle, G. V. Meirelles, F. R. da Silva, G. P. Telles and R. Minghim, InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams, BMC Bioinf., 2015, 16(1), 169 CrossRef PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc., 2008, 4(1), 44–57 CrossRef PubMed.
- B. T. Sherman, M. Hao, J. Qiu, X. Jiao, M. W. Baseler and H. C. Lane, et al., DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update), Nucleic Acids Res., 2022, 50(W1), W216–W221 CrossRef CAS PubMed.
- T. W. Harris, V. Arnaboldi, S. Cain, J. Chan, W. J. Chen and J. Cho, et al., WormBase: a modern Model Organism Information Resource, Nucleic Acids Res., 2019, 48(D1), D762–D767 Search PubMed.
- Y. Yuan, P. Hakimi, C. Kao, A. Kao, R. Liu and A. Janocha, et al., Reciprocal Changes in Phosphoenolpyruvate Carboxykinase and Pyruvate Kinase with Age Are a Determinant of Aging in Caenorhabditis elegans, J. Biol. Chem., 2016, 291(3), 1307–1319 CrossRef CAS PubMed.
- P. D. Karp, P. E. Midford, R. Billington, A. Kothari, M. Krummenacker and M. Latendresse, et al., Pathway Tools version 23.0 update: software for pathway/genome informatics and systems biology, Briefings Bioinf., 2019, 22(1), 109–126 CrossRef PubMed.
- S. Zdraljevic, B. W. Fox, C. Strand, O. Panda, F. J. Tenjo and S. C. Brady, et al., Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism, eLife, 2019, 8, e40260 CrossRef PubMed.
- M. A. Virmani and M. Cirulli, The Role of l-Carnitine in Mitochondria, Prevention of Metabolic Inflexibility and Disease Initiation, Int. J. Mol. Sci., 2022, 23(5), 2717 CrossRef CAS PubMed.
- V. Diacomanolis, B. N. Noller and J. C. Ng, Bioavailability and pharmacokinetics of arsenic are influenced by the presence of cadmium, Chemosphere, 2014, 112, 203–209 CrossRef CAS PubMed.
- M. Kamal, L. Tokmakjian, J. Knox, P. Mastrangelo, J. Ji and H. Cai, et al., A spatiotemporal reconstruction of the C. elegans pharyngeal cuticle reveals a structure rich in phase-separating proteins, eLife, 2022, 11, e79396 CrossRef CAS PubMed.
- A. M. Jubayar, S. Khan, J. Sadi, N. Uddin, O. Goni and M. Khatun, et al., Novel evidence of arsenic-related excess adiposity and its implication in the risk of cardiometabolic diseases, Environ. Res., 2025, 271, 121059 CrossRef CAS PubMed.
- E. L. Greer, M. A. Blanco, L. Gu, E. Sendinc, J. Liu and D. Aristizábal-Corrales, et al., DNA Methylation on N6-Adenine in C. elegans, Cell., 2015, 161(4), 868–878 CrossRef CAS PubMed.
- L. R. Baugh and T. Day, Nongenetic inheritance and multigenerational plasticity in the nematode C. elegans, eLife, 2020, 9, e58498 CrossRef CAS PubMed.
- L. E. Nava-Rivera, N. D. Betancourt-Martínez, R. Lozoya-Martínez, P. Carranza-Rosales, N. E. Guzmán-Delgado and I. E. Carranza-Torres, et al., Transgenerational effects in DNA methylation, genotoxicity and reproductive phenotype by chronic arsenic exposure, Sci. Rep., 2021, 11(1), 8276 CrossRef CAS PubMed.
- N. O. Burton, A. Willis, K. Fisher, F. Braukmann, J. Price and L. Stevens, et al., Intergenerational adaptations to stress are evolutionarily conserved, stress-specific, and have deleterious trade-offs, eLife, 2021, 10, e73425 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |