Open Access Article
Open Access ArticleComputer vision for high-throughput materials synthesis: a tutorial for experimentalists
Madeleine A.
Gaidimas†
 a,
Abhijoy
Mandal†
b,
Pan
Chen
b,
Shi Xuan
Leong
cd,
Gyu-Hee
Kim
a,
Akshay
Talekar
e,
Kent O.
Kirlikovali
a,
Abhijoy
Mandal†
b,
Pan
Chen
b,
Shi Xuan
Leong
cd,
Gyu-Hee
Kim
a,
Akshay
Talekar
e,
Kent O.
Kirlikovali
 a,
Kourosh
Darvish
bf,
Omar K.
Farha
a,
Kourosh
Darvish
bf,
Omar K.
Farha
 *ag,
Varinia
Bernales
*cef and
Alán
Aspuru-Guzik
*ag,
Varinia
Bernales
*cef and
Alán
Aspuru-Guzik
 *bcfhijkl
*bcfhijkl
aDepartment of Chemistry and International Institute for Nanotechnology, Northwestern University, Evanston, IL 60208, USA. E-mail: o-farha@northwestern.edu
bDepartment of Computer Science, University of Toronto, Toronto, ON M5S 2E4, Canada. E-mail: alan@aspuru.com
cDepartment of Chemistry, University of Toronto, Toronto, ON M5S 2E4, Canada. E-mail: varinia@bernales.org
dSchool of Chemistry, Chemical Engineering and Biotechnology, Nanyang Technological University, Singapore 637371, Singapore
eMaterials Discovery Research Institute, UL Research Institutes, Skokie, IL 60077, USA
fAcceleration Consortium, Toronto, ON M5S 3H6, Canada
gDepartment of Chemical and Biological Engineering, Northwestern University, Evanston, IL 60208, USA
hVector Institute for Artificial Intelligence, Toronto, ON M5G 1M1, Canada
iDepartment of Chemical Engineering and Applied Chemistry, University of Toronto, Toronto, ON M5S 3E5, Canada
jDepartment of Materials Science and Engineering, University of Toronto, Toronto, ON M5S 3E4, Canada
kSenior Fellow, Canadian Institute for Advanced Research (CIFAR), Toronto, ON M5G 1M1, Canada
lNVIDIA, Toronto, ON M5V 1K4, Canada
First published on 23rd December 2025
Abstract
Advances in high-throughput instrumentation and laboratory automation are revolutionizing materials synthesis by enabling the rapid generation of large libraries of novel materials. However, efficient characterization of these synthetic libraries remains a significant bottleneck in the discovery of new materials. Traditional characterization methods are often limited to sequential analysis, making them time-intensive and cost-prohibitive when applied to large sample sets. In the same way that chemists interpret visual indicators to identify promising samples, computer vision (CV) is an efficient approach to accelerate materials characterization across varying scales when visual cues are present. CV is particularly useful in high-throughput synthesis and characterization workflows, as these techniques can be rapid, scalable, and cost-effective. Although there is a set of growing examples in the literature, we have found a lack of resources where newcomers interested in the field could get a hold of a practical way to get started. Here, we aim to fill that identified gap and present a structured tutorial for experimentalists to integrate computer vision into high-throughput materials research, providing a detailed roadmap from data collection to model validation. Specifically, we describe the hardware and software stack required for deploying CV in materials characterization, including image acquisition, annotation strategies, model training, and performance evaluation. As a case study, we demonstrate the implementation of a CV workflow within a high-throughput materials synthesis and characterization platform to investigate the crystallization of metal–organic frameworks (MOFs). By outlining key challenges and best practices, this tutorial aims to equip chemists and materials scientists with the necessary tools to harness CV for accelerating materials discovery.
1 Introduction
Automation and high-throughput (HT) experimentation tools are transforming the discovery of novel materials.1–5 These tools have enabled the rapid exploration of vast chemical and synthetic parameter spaces, allowing researchers to generate more candidate materials than previously possible in an efficient manner. Integrating these automated workflows with artificial intelligence (AI) to create self-driving labs (SDLs) presents further opportunities for autonomous materials discovery by incorporating automated analysis to make decisions based on existing data and proposing subsequent experiments to achieve a certain target material or property.6,7 Enabling real-time reaction monitoring, analysis, and decision-making within SDLs is crucial for accelerating new materials discovery, as well as enhancing the efficiency, precision, and reproducibility of materials synthesis. While HT experimentation has already demonstrated reasonable success in synthesizing molecules and materials, the full potential of autonomous materials discovery is still limited by the specific challenges of HT materials characterization.8 While essential for understanding structure–property relationships and driving discovery, integrating materials characterization steps into automated workflows often requires labor-intensive, time-consuming, and resource-heavy analytical techniques.9 Assessing the progress of the experiment itself is also necessary to ensure synthetic reproducibility and reliability of the results. Therefore, streamlining characterization tasks within materials SDLs is crucial to strike a balance between efficiency, data quality, and resource allocation.AI tools such as computer vision (CV), used to rapidly analyze digital images and videos, have significant potential to enhance automated materials discovery workflows.10 CV analysis can enable automated image classification,11–13 segmentation,14,15 and object detection.16,17 These capabilities have been implemented in a wide range of disciplines, including medical imaging,18,19 self-driving technologies,20 industrial automation,21 and agriculture.22,23 Within research laboratories, CV can be leveraged to monitor visual cues such as color changes,24 morphological changes,25 phase transitions,26 and crystal formation.27 Researchers often rely on such visual cues to make decisions about their synthetic protocol: for instance, determining if a compound is fully dissolved or waiting for a color change before proceeding to the next step. Within materials chemistry SDLs, where human researchers are not present to monitor reactions visually, CV techniques are particularly useful for assessing reaction progress and making decisions on next steps. Utilizing CV analysis and classification reduces researcher time spent on tedious, repetitive work and standardizes outputs to minimize the subjectivity associated with analyses performed by different human researchers. The improved consistency, speed, and scalability of CV analysis methods make them valuable tools within materials synthesis workflows.28–30
Despite these advantages, widespread implementation of CV analysis in the domain of materials discovery is hindered by a lack of publicly available, high-quality data and information on the requirements of integrating CV into specific experimental setups. Materials chemistry encompasses a broad range of subfields, with vastly different synthetic conditions, sample vessel requirements, and characterization procedures.8 Experimental researchers seeking to incorporate CV analysis into their workflows may lack expertise in machine learning and be unfamiliar with constructing a CV pipeline tailored to their unique experimental needs. To enable easier CV tool development and facilitate the broader use of these techniques within the materials discovery domain, standardized practices and instructions for non-experts on how to set up their own CV analysis pipelines are necessary.
As a case study, intended to help the community learn the “ins and outs” of the field, we apply CV techniques to analyze the high-throughput crystallization experiment of nanomaterials, specifically metal–organic frameworks (MOFs). MOFs are self-assembled materials comprised of metal ions or cluster “nodes” and organic “linker” molecules.31–33 The crystallinity, porosity, and tunability of MOFs have enabled their use in applications ranging from carbon dioxide capture34 and hydrogen storage35 to catalysis36 and drug delivery.37 Due to the vast range of nodes and linkers, as well as the diverse arrangements of these components, the synthetic parameter space of MOFs is extremely large. While HT methods have the potential to expedite the synthesis of novel MOFs, HT crystalline materials characterization remains a significant bottleneck and typically requires expensive, specialized instrumental setups. Efficient allocation of such characterization resources requires identifying promising candidate MOFs following a HT synthesis protocol. For instance, an unsuccessful reaction that does not produce solid MOF material should be excluded from further characterization. The development of rapid, cost-effective methods to screen promising candidate MOFs is essential for advancing novel MOF discovery in automated workflows.
A collaboration amongst our research groups has recently integrated CV into an automated, HT synthesis platform for MOF crystallization. From images of sample vials containing MOF precursors, such as metal salts and linkers in solution, we utilize CV analysis to rapidly classify material phases, including solids, liquids, and residues. The implementation of CV as a screening tool facilitates the identification of promising MOF candidates during exploratory synthesis programs that can involve screening hundreds or thousands of reactions per campaign. Considering their scalability, speed, and low implementation cost, CV techniques represent a valuable complement to more advanced characterization methods. While our case study focuses on MOF crystallization, we emphasize that the same procedure for constructing a CV pipeline can be applied to other material synthesis domains that rely on visual cues, such as color or phase changes. In this tutorial, we provide a comprehensive guide for experimental chemists and materials scientists to incorporate CV into their own synthesis workflows. We describe the design and optimization of a CV pipeline for detecting sample vials and classifying their contents based on images acquired during synthesis. In addition, we detail the challenges associated with defining phase labels in our MOF platform and provide recommendations for other researchers to adapt such decisions to their own chemistry tasks. Finally, we evaluate the performance of our classification model in terms of accuracy and speed, benchmarking it against human performance by surveying a cohort of researchers with varying technical familiarity with experimental chemistry and artificial intelligence.
2 High-throughput MOF synthesis and characterization
While the transition from manual techniques to automated MOF synthesis creates unique opportunities to explore larger parameter spaces, adapting traditional synthesis protocols to automated platforms presents significant challenges. A typical MOF synthesis involves the reaction of a metal salt with an organic linker, which is dissolved in an organic solvent, sealed in a glass vial, and transferred to an oven or heating block.38–40 These sample vials often appear as clear or slightly cloudy liquids upon their initial preparation. As the reaction proceeds, MOFs gradually precipitate from the solution as powders or crystals, forming a layer of solid material that commonly sinks to the bottom of the vial (Fig. 1). The synthetic conditions for MOFs vary considerably, depending on the starting materials, desired product, and optimal crystal size. Some MOFs precipitate rapidly in mild conditions, while others require heating at elevated temperatures for multiple days. Following synthesis, solid products are isolated for characterization using powder X-ray diffraction (PXRD), the primary technique for determining MOF phase identity,41 as well as other methods, including gas sorption isotherms and thermogravimetric analysis (TGA).42 | ||
| Fig. 1 Schematic depicting MOF self-assembly. Initially, sample vials contain organic linker molecules and metal ions dissolved in solution. Following synthesis, solid product MOFs typically appear as a layer of powder or crystals within the vial. See Fig. 2 for computer vision (CV) images corresponding to this schematic. | ||
Some aspects of the MOF synthesis process are more easily translated into automated HT setups. For instance, liquid handlers can transfer stock solutions of reagents, automated capping tools can seal vials, and integrated shaker plates and heating blocks can agitate samples and control reaction temperature.43–45 MOF researchers have employed these techniques in large screening protocols to determine optimal synthetic parameters for specific target MOFs,46–48 as well as the discovery of new framework materials.49 Despite advancements in HT MOF synthesis, automated characterization of the solid products remains challenging. Efforts to increase the throughput of PXRD characterization have included the use of motorized multi-sample holders,50 robotic sample changers,51 and articulated robotic arms to interact with existing diffraction instrumentation.52 However, these methods remain expensive and time-consuming, further complicated by the challenge of transferring MOF powders from their synthesis vessels into suitable sample holders for characterization.9
Besides HT characterization, another difficult aspect of automating MOF synthesis is acquiring feedback on the progress and results of the crystallization reaction. In a common manual MOF synthesis, human researchers rely on visual feedback to make decisions about the synthetic procedure and necessary characterization tasks. For instance, a researcher may check samples to confirm the starting materials are fully dissolved before heating, and alternatively, continue agitating or sonicating the sample if reagent dissolution is incomplete. Following a synthesis procedure, a researcher will inspect samples to determine if MOF powder or crystals have been formed. Adapting MOF synthesis to automated high-throughput platforms makes these visual feedback steps more challenging to implement, as human researchers are not present to monitor the status of each reaction. While a well-established synthetic protocol may not require visual assessment of reaction progress, synthesis campaigns aimed at discovering novel materials will likely involve screening a range of synthetic conditions with unknown outcomes. Many trial conditions may be unsuccessful: for instance, forming no solid material at all instead of MOF powder or crystals. Visually assessing the results of a synthesis campaign provides the researcher with valuable information about potential products and aids in selecting promising samples for further characterization.
To restore real-time visual feedback—unavailable in the absence of human researchers during automated syntheses—we incorporated computer vision (CV) analysis into our automated synthesis platform to monitor reaction progress and complement traditional materials characterization techniques. We sought to use CV as a filter to identify which sample vials contained solid MOF products and required further characterization by PXRD. For MOF synthesis, CV offers a scalable tool to streamline the discovery process, particularly when synthesis campaigns involve screening hundreds or thousands of reactions with unknown outcomes. Although PXRD analysis is ultimately needed to confirm crystallinity, an initial pass with CV rapidly identifies which conditions yield solid precipitate and merit further analysis, enabling more efficient allocation of time-intensive characterization resources. While CV is not a replacement for advanced characterization techniques, it is a cost-effective and scalable tool that can rapidly expedite materials characterization in a HT setting.
3 Computer vision techniques for materials synthesis image analysis
To plan and implement a CV workflow effectively, it is essential to first understand the key components that influence the model's performance, from defining the right analytical task to selecting suitable training data and model architecture. In this section, we provide a high-level overview of the main considerations that guide the development of CV systems for automated materials characterization, laying the conceptual groundwork for the case study presented in Section 4.Common CV tasks that are relevant to automated materials synthesis and characterization include image classification, object detection, semantic segmentation, and instance segmentation. For example, in MOF image analysis, image classification determines whether specific material phases (e.g., solids, liquid, residue) are present in an image of a sample vial. Object detection goes further to identify the spatial positions of these phases and enclose them within bounding boxes. Semantic segmentation provides a more granular differentiation by assigning each pixel to a specific phase, while instance segmentation distinguishes individual instances within the same phase, such as separate crystals within a vial. The choice of the CV task typically depends on the specific objectives of the HT image analysis.
A critical component in the CV pipeline is the training dataset, which comprises images labeled with objects of interest. This training dataset must be of sufficient quality and quantity to ensure adequate performance of the CV model. While some published datasets of chemistry-related images are available,15,53,54 many experimental use cases require custom, task-specific datasets. Collecting and annotating in-house data can be a resource-intensive process. Data augmentation techniques can be used to expand limited datasets by generating modified versions of existing images through methods such as color modulation, image rotation or scaling, mosaicking multiple images together, and adding pixel noise.55 Increasing data variability through these modifications also reduces the model's sensitivity to minor input variations, such as noise. Beyond dataset size, image quality is critical to ensure the CV model does not learn from unwanted noise, such as glare. Examples of noise reduction techniques include preprocessing approaches such as histogram equalization for glare reduction, and environmental adjustments such as controlling lighting conditions.56–59
Another key component is the model architecture. For many CV models, convolutional neural networks (CNNs) are the benchmark method,60 with seminal CNN architectures such as LeNet61 and AlexNet.62 Deeper, more complex networks, including ResNet,63 You Only Look Once (YOLO),64 and Swin Transformer,65 among others, have achieved exemplary performance in CV tasks such as image classification and object detection. Importantly, CNN-based CV models have shown promise in materials chemistry applications, such as the classification, segmentation, and subsequent quantity estimation of chemically relevant artifacts, including liquids, solids, and residues on vessel walls.66,67 However, these initial models adopted an end-to-end approach that is effective only in highly controlled environments, where external factors such as lighting and environmental noise are minimized.64 When applied to larger or more complex systems where environmental factors are harder to control, model performance can degrade.64 Expanding these models to real-world settings thus requires significantly larger and more diverse labeled training datasets, making scalability technically demanding.
On the other hand, hierarchical models address the data scarcity challenge by breaking down the CV task into multiple stages, allowing different models and processing techniques to handle specific aspects of the task. In the context of MOF image analysis, a hierarchical model would first detect the region of interest (i.e., glass sample vials) in an image before classifying material phases (i.e., solid, liquid) within the detected region. Rather than attempting classification directly from raw images, this approach decouples environmental variability from the task of identifying chemically relevant artifacts. Given the high cost and time-consuming nature of image labelling,60 this decoupling provides a practical way to leverage large external datasets for the first model, which needs only general object detection, while allowing a more task-specific downstream model to be fine-tuned on a smaller, in-house dataset. Hierarchical models have previously been deployed for HT screening,67–69 demonstrating their potential to enhance scalability and adaptability for diverse materials chemistry tasks. In addition to model architecture selection, additional performance gains can be achieved through hyperparameter tuning (e.g. learning rate, batch size etc.), which we did not emphasize in our implementation but may be explored for enhanced model performance.
4 Building a CV pipeline for a HT MOF synthesis workflow
In this section, we provide a technical walkthrough on building an automated screening system using computer vision, with an example of how we implemented a CV workflow within our HT platform to investigate MOF crystallization. We will cover four major steps to build an effective CV system: (1) problem and task definition, (2) hardware setup, (3) data collection and annotation, and (4) building CV models. While each step is relatively straightforward, building a robust and reliable system is an iterative process that often requires refinement at each step to address problems or unwanted behaviors, which are likely specific to the use case. Throughout this walkthrough, we highlight common pitfalls and precautions to speed up the development of such systems.4.1 Problem and task definition
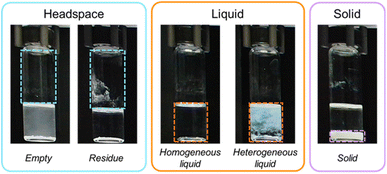
To determine whether a CV system is the appropriate solution, it is essential to clearly define the problem it is intended to solve. This begins with identifying the overarching chemistry task, including the experimental scale and the specific bottlenecks that image analysis could help overcome. Key guiding questions might include: “Do I rely on visual cues to assess outcomes at any stage of this experiment?” and “If I were to run this experiment at scale (e.g., perform hundreds of these experiments in parallel), would I need to visually inspect all samples at regular intervals?”. If the answer to either question is yes, a CV system could provide significant improvements by automating visual inspections, enabling parallel screening, and reducing human bias or inconsistency. Once the need for a CV system is established, the next step is to define the specific visual cues that the system should detect. These cues and the level of granularity are connected to the chemistry task we are trying to solve and how the chemistry task's automated decisions will be made, i.e., how the model output is used in downstream tasks. These cues will guide the data annotation and evaluation metrics throughout the system development.In our case, the defined CV task involves detecting five material phases within MOF sample vials. These phases, commonly observed in MOF synthesis, are distributed across three layers from top to bottom: headspace, liquid, and solid (from left to right in Fig. 2). The headspace layer, located at the top of the vial, is classified as either empty or residue, where the latter indicates visible material deposits on the vial walls, as shown on the left side box Fig. 2 (additional examples are provided in Fig. S1 in the SI). The liquid phase, situated below the headspace (central box in Fig. 2), is classified as either homogeneous, meaning it consists of clear and uniform liquids, or heterogeneous, which appears cloudy or contains suspended particles (additional examples are shown in Fig. S2 and S3, respectively). Finally, the solid phase corresponds to powder or crystallites settled at the bottom of the vial (Fig. S4). To successfully implement a CV pipeline, it is essential to clearly define each class, particularly when handling edge cases that may be subjective. More detailed descriptions of each class can be found in Section S2.1 of the SI.
4.2 Hardware setup
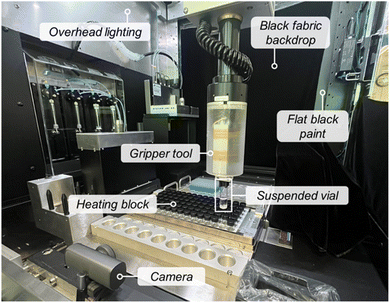
As discussed in Section 3, an effective CV pipeline relies heavily on high-quality image acquisition for training. Good hardware design and setup can significantly reduce the need for image post-processing and lower the amount of training data required by minimizing variability at the source. Some best practices that we identified include (1) ensuring lighting uniformity to minimize shadows and reflections that can interfere with image analysis, (2) maintaining a consistent background to simplify object detection, and (3) capturing all images from a consistent angle and distance to reduce variability. These design considerations directly inform the placement of the camera and light sources. For instance, glare can often be mitigated by positioning lights above or to the side of the vial, using diffused illumination rather than direct beams, and by lining the imaging enclosure with non-reflective materials. During hardware setup and pilot testing, it is helpful to capture diagnostic images under different conditions (e.g., time of day, varying reaction contents, empty vials) to identify unresolved setup-related image quality issues. These suggestions are particularly valuable when developing task-specific models intended for controlled environments and well-defined tasks, rather than for general-purpose foundation models designed for broad applicability.For our MOF crystallization example workflow, we utilized a HT synthesis platform equipped with automated liquid and solid dispensing, screw capping, and a robotic gripper tool to manipulate vials. Our MOF syntheses were performed in glass sample vials housed in a heating block. We installed a USB webcam inside the enclosure, positioning it to capture images of vials suspended above the heating block (Fig. 3). Images were captured when the gripper picked up each vial from the block and briefly held it in place. This process was automated for all vials, enabling the capture of images at specified points throughout the synthesis. To optimize image quality for CV analysis, we made minor modifications to our automated synthesis platform, including creating a consistent background for the vials by adding a black fabric backdrop opposite the camera. To minimize unwanted reflections, flat black paint was used to conceal metallic surfaces in the camera path. We also repositioned the enclosure's LED lights to illuminate the vials from above, reducing glare on the vial surfaces and ensuring uniform lighting regardless of the time of day. This approach was more effective than software-based glare reduction techniques,70 which we found removed critical artifacts of interest, such as falsely detected residue on the vial walls.
4.3 Data collection and annotation
Once a satisfactory hardware setup is implemented, the next step is to collect images that capture a diverse set of scenarios containing multiple examples of each class or visual cue of interest. The images should be as clear and sharp as possible, especially in moving setups where the edges of the frames might be out of focus. Samples should be zoomed in as much as possible to preserve detail and improve the model's accuracy in detecting subtle artifacts. However, this creates a tradeoff between image quality and hardware setup complexity that should be evaluated iteratively (see Section 4.4 for further discussion). Once a suitable dataset is collected, it must be annotated according to the visual cue definitions in Section 4.1. To minimize labeling bias, it is recommended that at least two people annotate the images, with a third independent subject matter expert resolving any discrepancies that may arise. Annotations can be performed using tools such as RoboFlow71 and SuperAnnotate,72 which support direct export in CV model-specific formats for common CV models.In our case, we collected images with varying amounts of liquids and solids at different time points to capture different levels of turbidity and solid levels. Further, we captured images of vials lifted from various positions in the well plate. We created a dataset of 168 images captured from 56 unique sample vials (representing 3 replicate images of each vial taken at different timepoints throughout the synthesis reaction). The images were annotated with 5 classes: empty, residue, homogeneous liquid, heterogeneous liquid, and solid, as defined in Section 4.1. We used RoboFlow to annotate our datasets and export them in YOLO format. The solid materials in our images are exclusively white in color, though similar datasets can be constructed with a wider chromatic range.
4.4 Building CV models
After data collection and annotation, we can start training the CV model. For experimental setups involving individual reaction vessels, such as vials, flasks, and containers, a hierarchical CV approach is often helpful. This strategy involves first training a model to detect and isolate the container itself, followed by a second model that analyzes the contents of the container. Existing datasets can often be leveraged for the initial stage and should be augmented with images collected from the current hardware setup to improve robustness. Hierarchical CV systems are more robust to changes in hardware setup and provide more stable detection of chemistry-related visual cues, as environmental distractors are cropped out. However, if the hardware configuration already captures close-up images with minimal background noise, such as when a camera follows and zooms in on individual samples, a single-stage CV model may suffice. Thus, selecting between a hierarchical and single-stage architecture involves balancing hardware complexity against CV architecture complexity, and should be carefully evaluated before opting for either approach. The specific constraints and goals of the experimental workflow should guide this decision.Regardless of the model design, data augmentation can improve model performance, especially in low-data regimes. Most existing CV libraries offer on-demand augmentation, generating augmented images and annotations during training without increasing data storage burden. When the model is trained, it should also be qualitatively evaluated for any undesirable or wrong behavior by visually inspecting outputs. While quantitative metrics, such as mean Average Precision (mAP), F1 score, and precision, are good measures for monitoring training progress, these can be difficult to interpret accurately; hence, qualitative analysis becomes important. Finally, the visual cue detection model must be trained using inputs that match its deployment conditions. In a hierarchical pipeline, this means training on cropped container images produced by the first-stage detector. In contrast, a single-stage system can be trained directly on raw images from the camera with some optional pre-processing.
In our example workflow, we employ a hierarchical model architecture, consisting of two sequential YOLO models. We chose YOLO models for their balance of accuracy, speed, and ease of training. These models are particularly well-suited for non-specialists, thanks to the availability of pre-trained weights, built-in support for data augmentation, evaluation metrics, and hyperparameter tuning. Additionally, YOLO models are compatible with both high-performance GPUs and less powerful machines, making them broadly accessible. The first model in our hierarchy is designed to detect vessels of interest, specifically glass sample vials. Since the vial position and distance from the camera may vary, the model needs to be robust to change in viewpoint and placement. To train the vessel detection model, we used the Vector-LabPics dataset, which includes a collection of 7900 images of laboratory equipment, such as beakers and flasks.15 We further augmented this dataset with 168 images of vials collected from our automated platform. The model was trained using default hyperparameters defined in YOLOv5 for 60 epochs, with an 80/20 train/validation split.
We assessed model performance using mAP, a standard metric in object detection that measures the overlap between predicted bounding boxes and ground-truth objects. This overlap is quantified using the Intersection over Union (IoU) metric, which ranges from 0 to 1. We assess both mAP50, which considers the detection correct if the IoU is at least 0.5 (50%), and mAP50-95, which provides a stricter measure by averaging across multiple IoU thresholds from 0.50 to 0.95. Our vial detection model achieved a mAP50-95 of 0.826 (Table S1), demonstrating reliable detection performance within our automated synthesis platform. Additionally, we measured the model's precision and recall, which represent the proportion of predicted detections that are correct and the proportion of actual objects that are successfully detected, respectively (see Section S3.1 in the SI for the equations). The vial detection model achieved perfect scores of 1.00 for both metrics (Table S1). The vial detection model outputs bounding box coordinates for each detected vial in an image, which are then used to crop the image and isolate the vial from the background. These cropped vial regions serve as inputs for the second phase detection model (Fig. 4).
The downstream phase detection model is a YOLOv8 model trained to detect five material classes (empty, residue, homogeneous liquid, heterogeneous liquid, and solid) described above. The training dataset consisted of 168 images captured within our HT synthesis platform from 56 unique MOF sample vials. The vials encompassed a representative range of chemical artifacts and instances of each material phase (Table S2). Each phase was manually annotated using RoboFlow71 to create the labelled dataset. The phase detection model was similarly trained using an 80/20 train/validation split, with the default YOLOv8 hyperparameters for 25 epochs. In this case, fewer epochs were required compared to the training of the vial detection model, as the LabPics dataset contained greater image diversity. To enhance dataset diversity, we utilized YOLOv8's built-in data augmentation techniques in the model training function.73 The phase detection model achieved an overall mAP50-95 of 0.851 across all five phases (Table 1). Table 1 also reports precision and recall, calculated according to eqn (S1) and (S2), respectively. To further assess model performance, we analyzed the normalized confusion matrix shown in Fig. 5. Among all classes, ‘empty’ was the most challenging to predict, with the lowest proportion of correct predictions (0.73). For all classes, most errors stemmed from missed detections, where the model failed to identify any object. Other notable misclassifications include ‘residue’ frequently predicted as ‘homogeneous liquid’, and ‘empty’ often mistaken as ‘heterogeneous liquid’ (with values of 0.12 and 0.27 in the confusion matrix, respectively, as shown in Fig. 5). The phase detection model outputs bounding box coordinates for each detected region in the image and classifies it as one of the defined phases. These predictions and coordinates are then utilized in downstream post-processing methods, including visualization and annotation, as well as decision-making in subsequent characterization tasks.
| Phase (N) | mAP50 | mAP50-95 | Precision | Recall |
|---|---|---|---|---|
| Empty (11) | 0.964 | 0.851 | 0.877 | 0.948 |
| Residue (11) | 0.951 | 0.927 | 0.818 | 0.909 |
| Homogeneous liquid (16) | 0.995 | 0.940 | 0.881 | 1.000 |
| Heterogeneous liquid (8) | 0.995 | 0.822 | 0.898 | 1.000 |
| Solid (12) | 0.934 | 0.684 | 0.907 | 0.833 |
| Overall average performance | 0.964 | 0.851 | 0.877 | 0.948 |
If the trained model does not meet the desired performance levels, this may indicate a need for further refinement. We recommend interpreting the evaluation metrics in the context of your specific objectives to identify appropriate next steps. For example, a low recall score suggests the model is missing true objects, and collecting more representative labelled data may help. Readers are encouraged to consult additional resources for more detailed guidance on interpreting evaluation metrics.74 However, we would like to emphasize that what is considered ‘satisfactory’ model performance highly depends on the specific use case. For example, a less precise model may be sufficient for prioritizing promising samples in early-stage exploratory screening, while higher-stakes scenarios (such as discarding expensive materials) may require stricter thresholds and more iterative model refinements. In some cases, it may also be appropriate to prioritize performance on specific classes of interest.
5 Software tutorial
To enable experimentalists to build their own YOLO-based computer vision models, we provide a GitHub repository as part of this tutorial (https://github.com/AccelerationConsortium/CV-HTE-Tutorial). The tutorial outlines the training and application of our phase detection model as a working example. In brief, we provide four core Python scripts, designed to be executed in the following order:1. Dataset.py – This script prepares the annotated dataset that, in our case, is exported from Roboflow (Section S2.4) into YOLO format for subsequent model training. While there are no strict rules, we recommend a minimum of 100 instances per class. For more robust models, particularly when working with images featuring diverse backgrounds or subtle differences between classes, we recommend increasing the dataset size to over 500 instances per class. As described in Section 3, data augmentation techniques can be strategically employed to enhance model performance.
2. ProcessLabPics.py – This script creates a vessel detection dataset from the Vector-LabPics dataset by grouping labels corresponding to vessels such as flasks, beakers, vials etc. into one class called “vessel”.15 The resulting dataset is then randomly split into 80% training and 20% testing sets. Models trained on just this dataset can detect a variety of vessels in general lab settings. However, in more specialized setups or setups not seen in the LabPics dataset, such models can be less successful. To address this, the dataset is enhanced by adding images of the unseen/new setup, with vessels of interest labelled. These images can then be added into the dataset obtained from the LabPics dataset to improve detection accuracy.
3. Train.py – This script splits the dataset into training and validation sets, then initiates training using the YOLOv8 framework. Key training parameters include: (i) batch size, which determines the number of images processed in a single iteration; (ii) image size, which defines the input image resolution; and (iii) number of epochs, which refers to the number of complete training cycles performed over the entire dataset. In most cases, the default YOLO hyperparameters perform well; however, advanced users may choose to explore hyperparameter tuning (e.g., adjusting the learning rate) to further optimize model performance. From these training parameters, the number of epochs is particularly important: too few epochs may lead to model underperformance, while too many can cause the model to “memorize” the training data, resulting in poorer performance on unseen images. Other hyperparameters, such as batch size, can be adjusted to optimize memory usage during training. Once training is complete, the script returns the model weights, saved in a format compatible with YOLO inference (best.pt for YOLOv8), along with evaluation metrics to assess model training performance.
4. Test.py – This script performs inference on new images using the trained YOLO model. For each detected object, the model outputs bounding box coordinates, class predictions, and confidence scores. By default, YOLO returns normalized bounding box coordinates in the format: [x_center, y_center, width, height], where each value is scaled between 0 and 1 relative to the image dimensions. In our case, we convert these normalized coordinates back to absolute pixel values using the original image's width and height. These coordinates are then used to overlay bounding boxes on the vial images for visualization. The model outputs can also be utilized in downstream tasks such as region-based quantification or cropping.
6 User study
Anticipating the widespread adoption of CV-based automation tools in the chemistry and materials science communities, we conducted a user study to evaluate users' perceptions of the model's effectiveness in improving the accuracy and speed of phase classification. This study aims to clarify how and where such AI models can best support HT experimentation by scientists in the laboratory. A total of 143 researchers from 16 institutions participated via a web-based Qualtrics survey (Section S4.8). Participation was voluntary, and data were collected only after receiving informed consent. The survey consisted of four sections: (1) scientific training background, followed by an accuracy-focused labelling task (2) and a speed labelling task (3) in which response time was measured, and finally, (4) a post-task survey. We only analyzed the responses from participants who attempted at least one of the two labelling tasks, resulting in a total of 111 participants (see Section S4.1 for more details). As participants were not required to complete all sections or questions and could skip or exit the survey at any time, the number of responses received varied across sections (Table S3). We observed a well-distributed scientific training background among the participants in terms of their technical familiarity with experimental chemistry and artificial intelligence. Of the 90 participants who responded to this section, the largest group (47.8%) identified as chemists without experience in AI; 22.2% identified as chemists with an AI background; 15.6% had an AI background but no formal training in chemistry; and 14.4% reported no background in either field (Table S3 and Fig. S5). This broad distribution enabled us to capture diverse perspectives on the model's utility and perceived performance.Both the accuracy and speed tasks utilized a dataset comprising 378 images of 42 unique MOF sample vials, captured using our automated setup, as described in the previous sections (a detailed phase distribution among these images is presented in Table S4). This dataset is unique to the user study; none of the images were used in model training or validation. Ground truths were established through independent annotations by two domain experts, with any disagreements resolved by a third expert (for more details, see Section S4.2, Table S5 and Fig. S6).
For the accuracy task, participants were shown definitions and examples of five phase categories: empty, residue, homogeneous liquid, heterogeneous liquid, and solid (Fig. S7). They were then asked to label five randomly selected images by drawing bounding boxes around each vial and entering the corresponding phase name (Fig. S8). A Flask-based server was used to randomly select images. Accuracy was measured by comparing participant labels to ground truth labels, and F1 scores were computed for each phase to assess per-class performance (eqn (S3)). We observe that the model outperformed human participants in overall F1 scores across all phases, with notable improvements in detecting empty, residue, and solid phases (Fig. 6). In addition, the model achieved higher accuracy than human participants, regardless of the number of phases present in the image (Fig. S12). For additional discussion, see Section S4.3 in the SI.
For the speed task, participants were shown five randomly selected vial images and asked a yes-or-no question about the presence of a specific phase (e.g., “Is the solid phase present?”), as shown in Section S4.4 of the SI. The time each participant spent observing the image and answering the question was recorded. On average, the model was over 50 times faster at identifying phases than the human participants, processing each image in 0.062 seconds when run on two NVLINK Nvidia RTX A6000 GPUs (48 GB VRAM). In comparison, participants took an average of about 3.65 seconds per image (Fig. S14). Importantly, this speed advantage did not compromise phase identification performance. During the speed task, the model outperformed human participants in terms of accuracy across all phases, by 31%, 11%, 17%, and 10% for residue, homogeneous liquid, heterogeneous liquid and solid phases, respectively (Fig. S15).
To evaluate user perspectives, participants were asked to respond to a series of general statements about conducting parallel experiments (Fig. S16) and using CV analysis for phase labeling (Fig. S17). Responses were recorded on a three- and five-point Likert scale to gauge perceptions of the model's utility, considering responses ranging from “strongly disagree” to “strongly agree” for the latter. Overall, responses reflected a strong recognition of the model's practical benefits: 84% agreed that manual labeling is tedious (calculated by combining 31% “agree” and 53% “strongly agree”), and 82% agreed that the model could accelerate the identification of samples requiring further characterization (questions a and b, Fig. S17). Additionally, 80% agreed that the model would facilitate conducting parallel experiments (question g, in Fig. S17).
However, perceptions of trust and accuracy were more divided. Only 41% indicated they would trust the model for their experiments (question f, Fig. S17), and just 17% preferred it over human annotators (question e, Fig. S17). Moreover, only 34% of participants believed the model labelled phases more accurately than humans, while 28% disagreed (question j, Fig. S17). These responses suggest that while users appreciate the efficiency gains, many remain cautious about entirely deferring to the model's judgment. While 69% of participants reported understanding the model's logic, the same proportion indicated they had never known or used similar AI tools before (questions d and h, Fig. S17). This lack of previous interaction and user experience with such AI models may hinder trust and broader adoption. These results highlight a valuable opportunity: although the model is seen as helpful in reducing workload and enabling scalability, its broader acceptance will depend on increasing transparency, interpretability, and user training. Notably, more than 61% of the 33 participants who regularly conduct parallel experiments reported that doing so compromises the time they can devote to each experiment (Fig. S16), further underscoring the potential value of automated tools like the one introduced here.
Finally, we included two open-ended questions to survey participants on how this AI model could benefit their own experimental workflows, as well as to identify any potential concerns (Table S12). A word cloud generated from responses to the first question highlighted ‘time’ and ‘help’ as prominent keywords (Fig. S18). Qualitative analyses of the 49 responses to the first question revealed that 22 participants explicitly mentioned timesaving as a key advantage, along with related terms such as “efficiency” and “faster” (see Table S12 for all responses). These responses underscore that participants recognize the model's potential and value in streamlining experimental processes and improving overall efficiency.
In addition, we analyzed the results from the experimental chemists separately (Fig. S19–S21), and we observed that phase identification is a difficult task generally, rather than a task for which a specific experimental background significantly boosts performance.
Overall, the user study suggests that employing CV models for phase detection and progress tracking in chemistry tasks can make these processes more quantifiable and objective. The higher error rates observed in human subject annotations indicate that CV models could contribute to establishing more standardized interpretations of visual cues relevant to chemical processes. Moreover, while such models have the potential to reduce cognitive load on scientists and enhance their performance, responses to the user trust questionnaire highlight the importance of maintaining human oversight in automated systems. Human oversight is not only relevant for the accuracy of the computer vision system but also for related safety considerations.75,76
7 Discussion and outlook
A collaborative, multidisciplinary approach was crucial to developing our CV workflow. Our team was an international collaboration spanning three institutions, drawing on expertise from researchers with diverse backgrounds in experimental and computational chemistry, as well as data and computer science. Throughout our collaboration, we developed a deeper understanding of the challenges facing this multidisciplinary problem, and the unique perspective of each researcher allowed us to improve our final workflow based on contributions from both the experimental and computational sides. A benefit of this approach was the ability to leverage the specific expertise of experimentalists during the process of developing our model, resulting in a highly customized tool that non-CV experts could confidently apply to their chemistry tasks. While the involvement of multiple subject matter experts in the generation and annotation of training datasets may seem trivial, one challenge we encountered was subjective bias in phase labeling. For this reason, we emphasize the importance of clearly defining each artifact to be identified, with particular attention to edge cases. For instance, in our study we classify liquid phases as either “homogeneous” or “heterogeneous,” though the cloudiness of a real sample is not necessarily binary. Clearly defining the CV task is therefore crucial to ensure that samples are labeled consistently. This subjectivity of different human labelers also underscores the utility of an adequately trained CV model, which can minimize bias compared to multiple human experimenters who may vary in their assessment of the results.An observation shared by multiple user study participants related to the challenges of identifying phases from a single-viewpoint image compared to handling the actual sample vial. One respondent noted, “In a real lab setting, a human would jiggle the flask and change their viewing angle to ensure they identify the vial contents correctly; this dramatically increases the accuracy of a human chemist's identifications.” Accurate classification from a single image is more challenging, particularly in cases involving more ambiguous phase labeling. For instance, a sample containing colorless crystals in solution is difficult to distinguish from a sample containing a similar clear liquid without close examination of the real vial. Capturing multiple images from different angles could potentially increase the robustness of a CV model in such cases. For our MOF crystallization workflow, we aimed to use CV to complement additional characterization techniques rather than a standalone analysis method. HT materials characterization necessarily involves a tradeoff between throughput and data quality and accuracy. We believe that the speed and scalability of CV techniques make them a valuable counterpart to traditional characterization methods. CV analysis is particularly useful when manual inspection of every sample of a high-throughput campaign is not feasible. Unlike human researchers, CV models can consistently handle large image datasets without fatigue.
CV analysis is clearly not applicable to situations where successful and failed syntheses cannot be visually distinguished, such as homogeneous chemical reactions that require characterization with spectroscopic methods. An exception perhaps would be if imaging beyond the visible ranges (infrared, ultraviolet, etc.) is available via hyperspectral cameras. However, many tasks relevant to materials synthesis can benefit from incorporating CV tools. Beyond MOF crystallization, user survey participants offered a range of chemical tasks where CV may be useful, including solubility testing, extractions, liquid level detection, and precipitation reactions. The hierarchical model architecture employed in this tutorial enables generalization of our approach to additional chemistry tasks through the fine-tuning of each individual model. For instance, the vessel detection model can be retrained to include different sample vessel sizes, positions, or lighting conditions. Similarly, the phase detection model can be fine-tuned to distinguish other relevant artifacts, such as solution color. By creating custom, fine-tuned models, chemists and materials scientists can integrate their experimental expertise directly into the model itself, without relying on previous datasets that may not apply to their specific use case.
In our example MOF crystallization workflow, we utilized CV to distinguish promising samples following synthesis, based on the appearance of solid material from initially clear solutions. Beyond this simple case of product identification, we anticipate that this CV workflow will provide additional insights into MOF crystallization kinetics by comparing phases identified from photos captured at multiple timepoints. A CV approach enables rapid and inexpensive kinetic information, in contrast to traditional time-resolved materials characterization techniques such as in situ PXRD, which is not feasible in a HT setting. Many further opportunities exist to apply CV techniques within materials synthesis workflows, including real-time sample analysis, integration into automated platforms to guide decision-making, and handling potential errors from automated protocols without human intervention. We emphasize that the tunability of a hierarchical model approach enables the implementation of customized CV models tailored to specific chemistry goals while minimizing the amount of training data required. Considering the range of chemistry and materials science tasks where visual analysis is relevant, we hope this approach empowers experimentalists to incorporate CV tools within their own synthesis workflows. A revolution is happening in science by the advent of agentic systems.77–80 We believe that the integration of these computer vision workflows in agentic self-driving lab experiments81–83 will help in further expanding the toolset of agentic science by providing further “eyes” to automated AI science agents.
8 Recommended computer vision learning resources
• You only look once: unified, real-time object detection.64• ImageNet classification with deep convolutional neural networks.62
• No-code computer vision with RoboFlow.84
• OpenCV Bootcamp85
• IBM: introduction to computer vision and image processing.86
• The State University of New York: computer vision basics.87
Author contributions
Conceptualization: M. A. G., A. M., K. D., O. K. F., V. B., A. A.-G.; methodology: M. A. G., A. M., K. D., O. K. F., V. B., A. A.-G.; investigation: M. A. G., A. M., P. C., G.-H. K., K. D., V. B.; data curation: M. A. G., A. M., P. C., G.-H. K, K. D., V. B.; software: A. M., P. C., S.-X. L.; formal analysis: A. M., P. C., S.-X. L., K. D., V. B.; validation: M. A. G., A. M., S.-X. L., G.-H. K.; visualization: M. A. G., A. M., P. C., S.-X. L., G.-H. K., V. B.; writing (original draft): M. A. G., A. M., P. C., S.-X. L., G.-H. K., K. D., V. B.; writing (review and editing): all authors; supervision and project administration: K. O. K., K. D., O. K. F., V. B., A. A.-G.; resources and funding acquisition: O. K. F., V. B., A.A.-G.Conflicts of interest
O. K. F. has a financial interest in Numat Technologies, a startup company seeking to commercialize metal–organic frameworks.Data availability
The image datasets are publicly available in the following repository: https://zenodo.org/records/4736111,88 and https://zenodo.org/records/16209653.89 The machine learning methods used in this study are open-source. The object detection method YOLOv8 is accessible at https://github.com/ultralytics/ultralytics. The utilization in this paper and the command example are shared at https://github.com/AccelerationConsortium/CV-HTE-Tutorial/.Supplementary information (SI): hardware information, model training and performance details, and user study methodology and results. See DOI: https://doi.org/10.1039/d5dd00384a.
Acknowledgements
A. A.-G. thanks Anders G. Frøseth for his generous support. A. A.-G. also acknowledges the generous support of Natural Resources Canada and the Canada 150 Research Chairs program. This research is part of the University of Toronto's Acceleration Consortium, which receives funding from the Canada First Research Excellence Fund (CFREF) via CFREF-2022-00042. O. K. F. gratefully acknowledges support from the Underwriters Laboratories' Materials Discovery Research Institute and the Trienens Institute for Sustainability and Energy at Northwestern University. M. A. G. gratefully acknowledges support from the Ryan Fellowship and the International Institute for Nanotechnology (IIN) at Northwestern University. S. X. L. acknowledges support from Nanyang Technological University, Singapore, and the Ministry of Education, Singapore, for the International Postdoctoral Fellowship. P. C. acknowledges support from Google NSERC Industrial Research Chair Grants.References
- P. Nikolaev, D. Hooper, N. Perea-López, M. Terrones and B. Maruyama, ACS Nano, 2014, 8, 10214–10222 CrossRef CAS PubMed.
- C. W. Coley, D. A. Thomas, J. A. M. Lummiss, J. N. Jaworski, C. P. Breen, V. Schultz, T. Hart, J. S. Fishman, L. Rogers, H. Gao, R. W. Hicklin, P. P. Plehiers, J. Byington, J. S. Piotti, W. H. Green, A. J. Hart, T. F. Jamison and K. F. Jensen, Science, 2019, 365, eaax1566 CrossRef CAS PubMed.
- R. W. Epps, M. S. Bowen, A. A. Volk, K. Abdel-Latif, S. Han, K. G. Reyes, A. Amassian and M. Abolhasani, Adv. Mater., 2020, 32, 2001626 CrossRef CAS PubMed.
- J. Li, J. Li, R. Liu, Y. Tu, Y. Li, J. Cheng, T. He and X. Zhu, Nat. Commun., 2020, 11, 2046 CrossRef CAS PubMed.
- B. P. MacLeod, F. G. L. Parlane, T. D. Morrissey, F. Häse, L. M. Roch, K. E. Dettelbach, R. Moreira, L. P. E. Yunker, M. B. Rooney, J. R. Deeth, V. Lai, G. J. Ng, H. Situ, R. H. Zhang, M. S. Elliott, T. H. Haley, D. J. Dvorak, A. Aspuru-Guzik, J. E. Hein and C. P. Berlinguette, Sci. Adv., 2020, 6, eaaz8867 CrossRef CAS PubMed.
- R. W. Epps, A. A. Volk, M. Y. S. Ibrahim and M. Abolhasani, Chem, 2021, 7 Search PubMed.
- M. Abolhasani and E. Kumacheva, Nat. Synth., 2023, 2, 483–492 CrossRef CAS.
- R. Potyrailo, K. Rajan, K. Stoewe, I. Takeuchi, B. Chisholm and H. Lam, ACS Comb. Sci., 2011, 13, 579–633 CrossRef CAS PubMed.
- I. G. Clayson, D. Hewitt, M. Hutereau, T. Pope and B. Slater, Adv. Mater., 2020, 32, 2002780 CrossRef CAS PubMed.
- R. Sasaki, M. Fujinami and H. Nakai, Digital Discovery, 2024, 3, 2458–2464 RSC.
- I. Singh, S. K. Singh, S. Kumar and K. Aggarwal, in Congress on Intelligent Systems: Proceedings of CIS 2021, Springer Nature, Singapore, 2021, vol. 1, pp. 247–261 Search PubMed.
- M. Dyrmann, H. Karstoft and H. S. Midtiby, Biosyst. Eng., 2016, 151, 72–80 CrossRef.
- F. L. C. D. Santos, M. Paci, L. Nanni, S. Brahnam and J. Hyttinen, Biosyst. Eng., 2015, 138, 11–22 CrossRef.
- M. Vardhana, N. Arunkumar, S. Lasrado, E. Abdulhay and G. Ramirez-Gonzalez, Cogn. Syst. Res., 2018, 50, 10–14 CrossRef.
- S. Eppel, H. Xu, M. Bismuth and A. Aspuru-Guzik, ACS Cent. Sci., 2020, 6, 1743–1752 CrossRef CAS PubMed.
- A. Mhalla, T. Chateau, S. Gazzah and N. E. B. Amara, IEEE Trans. Intell. Transport. Syst., 2019, 20, 4006–4018 Search PubMed.
- X. Cheng, S. Zhu, Z. Wang, C. Wang, X. Chen, Q. Zhu and L. Xie, Artif. Intell. Chem., 2023, 1, 100016 CrossRef.
- A. Schmidt, O. Mohareri, S. DiMaio, M. C. Yip and S. E. Salcudean, Med. Image Anal., 2024, 94, 103131 CrossRef PubMed.
- E. Elyan, P. Vuttipittayamongkol, P. Johnston, K. Martin, K. McPherson, C. F. Moreno-García, C. Jayne and M. M. K. Sarker, Artif. Intell. Surg., 2022, 2, 24–45 Search PubMed.
- É. Zablocki, H. Ben-Younes, P. Pérez and M. Cord, Int. J. Comput. Vis., 2022, 130, 2425–2452 CrossRef.
- L. Zhou, L. Zhang and N. Konz, IEEE Trans. Syst. Man Cybern. Syst., 2023, 53, 105–117 Search PubMed.
- H. Tian, T. Wang, Y. Liu, X. Qiao and Y. Li, Inf. Process. Agric., 2020, 7, 1–19 Search PubMed.
- D. I. Patrício and R. Rieder, Comput. Electron. Agric., 2018, 153, 69–81 CrossRef.
- Y. Kosenkov and D. Kosenkov, J. Chem. Educ., 2021, 98, 4067–4073 CrossRef CAS.
- N. Taherimakhsousi, M. Fievez, B. P. MacLeod, E. P. Booker, E. Fayard, M. Matheron, M. Manceau, S. Cros, S. Berson and C. P. Berlinguette, npj Comput. Mater., 2021, 7, 190 CrossRef CAS.
- Y. Suh, A. Chandramowlishwaran and Y. Won, npj Comput. Mater., 2024, 10, 65 CrossRef.
- S.-J. Burgdorf, T. Roddelkopf and K. Thurow, Chem. Ing. Tech., 2024, 96, 1107–1115 CrossRef CAS.
- R. El-khawaldeh, M. Guy, F. Bork, N. Taherimakhsousi, K. N. Jones, J. M. Hawkins, Lu Han, R. P. Pritchard, B. A. Cole, S. Monfette and J. E. Hein, Chem. Sci., 2024, 15, 1271–1282 RSC.
- C. Yan, C. Fyfe, L. Minty, B. Henry, J. Craig and M. Reid, Chem. Sci., 2023, 14, 11872–11880 RSC.
- K. X. Chong, Q. Alsabia, Z. Ye, A. McDaniel, D. Baumgardner, D. Xiao and S. Sun, ChemRxiv, 2025, preprint, chemrxiv-2025-nq31g, DOI:10.26434/chemrxiv-2025-nq31g.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- H.-C. J. Zhou and S. Kitagawa, Chem. Soc. Rev., 2014, 43, 5415–5418 RSC.
- O. M. Yaghi, M. O'Keeffe, N. W. Ockwig, H. K. Chae, M. Eddaoudi and J. Kim, Nature, 2003, 423, 705–714 CrossRef CAS PubMed.
- Z.-M. Ye, Y. Xie, K. O. Kirlikovali, S. Xiang, O. K. Farha and B. Chen, J. Am. Chem. Soc., 2025, 147, 5495–5514 CrossRef CAS PubMed.
- L. Jimenez-Lopez, R. M. Ospino, L. G. d. Araujo, A. Celzard and V. Fierro, Nanoscale, 2025, 17, 6390–6413 RSC.
- K. E. McCullough, D. S. King, S. P. Chheda, M. S. Ferrandon, T. A. Goetjen, Z. H. Syed, T. R. Graham, N. M. Washton, O. K. Farha, L. Gagliardi and M. Delferro, ACS Cent. Sci., 2023, 9, 266–276 CrossRef CAS PubMed.
- H. D. Lawson, S. P. Walton and C. Chan, ACS Appl. Mater. Interfaces, 2021, 13, 7004–7020 CrossRef CAS PubMed.
- A. J. Howarth, A. W. Peters, N. A. Vermeulen, T. C. Wang, J. T. Hupp and O. K. Farha, Chem. Mater., 2017, 29, 26–39 CrossRef CAS.
- N. Stock and S. Biswas, Chem. Rev., 2012, 112, 933–969 CrossRef CAS PubMed.
- R. S. Forgan, Chem. Sci., 2020, 11, 4546–4562 RSC.
- C. F. Holder and R. E. Schaak, ACS Nano, 2019, 13, 7359–7365 CrossRef CAS PubMed.
- N. Saadatkhah, A. C. Garcia, S. Ackermann, P. Leclerc, M. Latifi, S. Samih, G. S. Patience and J. Chaouki, Can. J. Chem. Eng., 2020, 98, 34–43 CrossRef CAS.
- S. Lo, S. G. Baird, J. Schrier, B. Blaiszik, N. Carson, I. Foster, A. Aguilar-Granda, S. V. Kalinin, B. Maruyama, M. Politi, H. Tran, T. D. Sparks and A. Aspuru-Guzik, Digital Discovery, 2024, 3, 842–868 RSC.
- M. Politi, F. Baum, K. Vaddi, E. Antonio, J. Vasquez, B. P. Bishop, N. Peek, V. C. Holmberg and L. D. Pozzo, Digital Discovery, 2023, 2, 1042–1057 RSC.
- B. G. Pelkie and L. D. Pozzo, Digital Discovery, 2023, 2, 544–556 RSC.
- S. M. Moosavi, A. Chidambaram, L. Talirz, M. Haranczyk, K. C. Stylianou and B. Smit, Nat. Commun., 2019, 10, 539 CrossRef CAS PubMed.
- N. P. Domingues, S. M. Moosavi, L. Talirz, K. M. Jablonka, C. P. Ireland, F. M. Ebrahim and B. Smit, Commun. Chem., 2022, 5, 170 CrossRef CAS PubMed.
- K. Sumida, S. Horike, S. S. Kaye, Z. R. Herm, W. L. Queen, C. M. Brown, F. Grandjean, G. J. Long, A. Dailly and J. R. Long, Chem. Sci., 2010, 1, 184–191 RSC.
- R. Banerjee, A. Phan, B. Wang, C. Knobler, H. Furukawa, M. O'Keeffe and O. M. Yaghi, Science, 2008, 319, 939–943 CrossRef CAS PubMed.
- E. Biemmi, S. Christian, N. Stock and T. Bein, Microporous Mesoporous Mater., 2009, 117, 111–117 CrossRef CAS.
- Y. Yotsumoto, Y. Nakajima, R. Takamoto, Y. Takeichi and K. Ono, Digital Discovery, 2024, 2523–2532, 10.1039/D4DD00190G.
- A. M. Lunt, H. Fakhruldeen, G. Pizzuto, L. Longley, A. White, N. Rankin, R. Clowes, B. Alston, L. Gigli, G. M. Day, A. I. Cooper and S. Y. Chong, Chem. Sci., 2024, 15, 2456–2463 RSC.
- L. C. O. Tiong, H. J. Yoo, N. Kim, C. Kim, K.-Y. Lee, S. S. Han and D. Kim, npj Comput. Mater., 2024, 10, 42 CrossRef.
- R. Sasaki, M. Fujinami and H. Nakai, Data Brief, 2024, 52, 110054 CrossRef CAS PubMed.
- C. Shorten and T. M. Khoshgoftaar, J. Big Data, 2019, 6, 60 CrossRef.
- S. F. Tan and N. A. M. Isa, IEEE Access, 2019, 7, 70842–70861 Search PubMed.
- W. A. Mustafa and M. M. M. Abdul Kader, J. Phys.: Conf. Ser., 2018, 1019, 012026 CrossRef.
- M. Wu and Q. Zhong, Syst. Soft. Comput., 2024, 6, 200169 CrossRef.
- Y. Xie, L. Ning, M. Wang and C. Li, J. Phys.: Conf. Ser., 2019, 1314, 012161 CrossRef.
- X. Zhao, L. Wang, Y. Zhang, X. Han, M. Deveci and M. Parmar, Artif. Intell. Rev., 2024, 57, 99 CrossRef.
- Y. Lecun, L. Bottou, Y. Bengio and P. Haffner, Proc. IEEE, 1998, 86, 2278–2324 CrossRef.
- A. Krizhevsky, I. Sutskever and G. Hinton, Adv. Neural Inf. Process. Syst., 2012, 25, 1 Search PubMed.
- R. Wightman, H. Touvron and H. Jégou, NeurIPS 2021 Workshop on ImageNet: Past, Present, and Future, 2021, https://openreview.net/forum?id=NG6MJnVl6M5 Search PubMed.
- J. Redmon, S. Divvala, R. Girshick and A. Farhadi, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 779–788 Search PubMed.
- Z. Liu, Y. Lin, Y. Cao, H. Hu, Y. Wei, Z. Zhang, S. Lin and B. Guo, in Proceedings of the IEEE/CVF international conference on computer vision, 2021, pp. 10012–10022 Search PubMed.
- R. El-khawaldeh, M. Guy, F. Bork, N. Taherimakhsousi, K. N. Jones, J. M. Hawkins, L. Han, R. P. Pritchard, B. A. Cole, S. Monfette and J. E. Hein, Chem. Sci., 2024, 15, 1271–1282 RSC.
- R. El-khawaldeh, A. Mandal, N. Yoshikawa, W. Zhang, R. Corkery, P. Prieto, A. Aspuru-Guzik, K. Darvish and J. E. Hein, Device, 2024, 2, 100404 CrossRef.
- G. Pizzuto, J. De Berardinis, L. Longley, H. Fakhruldeen and A. I. Cooper, 2022 International Joint Conference on Neural Networks (IJCNN), 2022, pp. 1–7, DOI:10.1109/IJCNN55064.2022.9892533.
- IEEE/SICE International Symposium on System Integration (SII), 2025, DOI:10.1109/SII59315.2025.10870899.
- M. Z. Alam, Z. Kaleem and S. Kelouwani, IEEE Trans. Intell. Veh., 2024, 9, 7030–7044 Search PubMed.
- B. Dwyer, J. Nelson and T. Hansen, Roboflow (Version 1.0), 2024, https://roboflow.com/ Search PubMed.
- I. SuperAnnotate AI, SuperAnnotate Streamline AI Data Workflows, https://www.superannotate.com/, accessed June 25, 2025.
- G. Jocher, J. Qiu and A. Chaurasia, YOLO by Ultralytics, 2023 Search PubMed.
- YOLO Performance Metrics Guide, https://docs.ultralytics.com/guides/yolo-performance-metrics/.
- S. X. Leong, C. E. Griesbach, R. Zhang, K. Darvish, Y. Zhao, A. Mandal, Y. Zou, H. Hao, V. Bernales and A. Aspuru-Guzik, Nat. Rev. Chem., 2025, 9, 707–722 CrossRef PubMed.
- F. Munguia-Galeano, Z. Zhou, S. Veeramani, H. Fakhruldeen, L. Longley, R. Clowes and A. I. Cooper, arXiv, 2025, preprint arXiv:2508.05148, DOI:10.48550/arXiv.2508.05148.
- Y. Zou, A. H. Cheng, A. Aldossary, J. Bai, S. X. Leong, J. A. Campos-Gonzalez-Angulo, C. Choi, C. T. Ser, G. Tom, A. Wang, Z. Zhang, I. Yakavets, H. Hao, C. Crebolder, V. Bernales and A. Aspuru-Guzik, Matter, 2025, 8, 102263 CrossRef CAS.
- A. Aspuru-Guzik and V. Bernales, Polyhedron, 2025, 281, 117707 CrossRef CAS.
- A. Ghafarollahi and M. J. Buehler, 2025, arXiv, 2025, preprint arXiv:2504.19017, DOI:10.48550/arXiv.2504.19017.
- K. Kawaharazuka, J. Oh, J. Yamada, I. Posner and Y. Zhu, IEEE Access, 2025, 13, 162467–162504 Search PubMed.
- S. Cao, Z. Zhang, M. Alghadeer, S. D. Fasciati, M. Piscitelli, M. Bakr, P. Leek and A. Aspuru-Guzik, arXiv, 2025, preprint arXiv:2412.07978, 2024, DOI:10.48550/arXiv.2412.07978.
- F. Strieth-Kalthoff, H. Hao, V. Rathore, J. Derasp, T. Gaudin, N. H. Angello, M. Seifrid, E. Trushina, M. Guy, J. Liu, X. Tang, M. Mamada, W. Wang, T. Tsagaantsooj, C. Lavigne, R. Pollice, T. C. Wu, K. Hotta, L. Bodo, S. Li, M. Haddadnia, A. Wołos, R. Roszak, C. T. Ser, C. Bozal-Ginesta, R. J. Hickman, J. Vestfrid, A. Aguilar-Granda, E. L. Klimareva, R. C. Sigerson, W. Hou, D. Gahler, S. Lach, A. Warzybok, O. Borodin, S. Rohrbach, B. Sanchez-Lengeling, C. Adachi, B. A. Grzybowski, L. Cronin, J. E. Hein, M. D. Burke and A. Aspuru-Guzik, Science, 2024, 384, eadk9227 CrossRef CAS PubMed.
- T. Song, M. Luo, X. Zhang, L. Chen, Y. Huang, J. Cao, Q. Zhu, D. Liu, B. Zhang, G. Zou, G. Zhang, F. Zhang, W. Shang, Y. Fu, J. Jiang and Y. Luo, J. Am. Chem. Soc., 2025, 147, 12534–12545 CrossRef CAS PubMed.
- Roboflow 101: Learn Computer Vision, https://roboflow.com/learn, accessed March 2025.
- OpenCV Bootcamp, https://opencv.org/university/free-opencv-course/?utm_source=opcvu%26utm_medium=menu%26utm_campaign=obc, accessed March 2025.
- J. Santarcangelo and A. Egwaikhide, IBM: Introduction to Computer Vision and Image Processing, https://www.coursera.org/learn/introduction-computer-vision-watson-opencv, accessed March 2025.
- R. Dasari and J. Yuan, https://www.coursera.org/learn/computer-vision-basics, accessed March 2025.
- S. Eppel, H. Xu, A. Aspuru-Guzik and M. Bismuth, LabPics dataset for visual understanding of Medical and Chemistry Labs (2.0) [Data set], Zenodo, 2021, DOI:10.5281/zenodo.4736110.
- M. A. Gaidimas, A. Mandal, P. Chen, S. Xuan, G.-H. Kim, A. Talekar, K. Kirlikovali, K. Darvish, O. Farha, V. Bernales and A. Aspuru-Guzik, Tutorial for computer vision for high throughput experimentation (1.0) [Data set], Zenodo, 2025, DOI:10.5281/zenodo.16209653.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2026 |