A computational protocol for 15N NMR parameter prediction in aqueous peptide ensembles using optimized DFT methods
Minji
Kim
 a,
Jung Ho
Lee
a,
Jung Ho
Lee
 *a and
Keunhong
Jeong
*a and
Keunhong
Jeong
 *b
*b
aDepartment of Chemistry, Seoul National University, Seoul 08826, Republic of Korea. E-mail: jungho.lee@snu.ac.kr
bDepartment of Physics and Chemistry, Korea Military Academy, Seoul 01805, Republic of Korea. E-mail: doas1mind@kma.ac.kr
First published on 17th November 2025
Abstract
Accurate prediction of 15N NMR chemical shifts in flexible peptide systems remains challenging. We present an ensemble-based computation protocol combining density functional theory with replica-exchange molecular dynamics. This approach outperformed single-structure predictions with deviations of 2.5–5.6 ppm for most residues and 1.1 ppm for leucine in a model peptide.
Nuclear magnetic resonance (NMR) spectroscopy stands as one of the most powerful analytical techniques for elucidating molecular structure and dynamics at the atomic level. Among the various NMR-active nuclei, nitrogen-15 (15N) provides particularly valuable structural information for biomolecular systems, especially proteins, through its exceptional sensitivity to local electronic environments and hydrogen bonding patterns.1,2
However, experimental acquisition of 15N NMR data faces significant challenges due to low natural abundance (0.365%) and relatively small gyromagnetic ratio, resulting in poor sensitivity compared to proton detection.3 Recent advances in density functional theory (DFT) have enabled increasingly accurate predictions of NMR chemical shifts across diverse molecular systems.4–6 AI-based tools have also demonstrated high accuracy in chemical shift prediction in previous studies.7–10 However, these AI-based approaches face inherent limitations in accurately incorporating specific solvent environments into their predictive models, whereas DFT-based methodologies enable explicit consideration of solvation effects through quantum mechanical calculations.
Computational frameworks specifically designed for nitrogen nuclei in aqueous environments remain largely underexplored. While reliable scaling factors for 15N chemical shift predictions have been established, these calibrations primarily relied on gas-phase calculations or non-aqueous solvation models.11–15 Therefore, the development of a comprehensive methodology specifically validated for aqueous systems is crucial for practical applications in water-based biological systems. The challenge becomes even more complex when dealing with intrinsically disordered proteins (IDPs), which lack rigid three-dimensional structures and exhibit dynamic conformational variability. These proteins exist as ensembles of multiple conformers, with observed NMR signals representing ensemble-averaged value over all accessible conformations.16 Our investigation focuses on the short model peptide, Asp–Gln–Leu–Gly–Lys (DQLGK), corresponding to residues 98–102 of α-synuclein IDP (Fig. 1).
This pentapeptide is expected to form an ensemble of multiple conformers. Boltzmann-weighted averaging is therefore anticipated to improve the accuracy of chemical shift predictions for such flexible systems.
In this study, we first establish an optimal computational protocol by systematically evaluating17 882 functional/basis set combinations across 19 small organic molecules (43 nitrogen datapoints) in water3 to determine accurate scaling factors for aqueous environments. Using this optimized protocol, we predict Boltzmann-averaged 15N chemical shifts for a pentapeptide and validate their accuracy against experimental data. Representative conformers composing the ensemble are derived from replica-exchange molecular dynamics (REMD)18 simulations, and the chemical shifts are calculated with density functional theory (DFT). This approach ensures reliable predictions for flexible peptide systems in biologically relevant conditions.
To obtain experimental 15N chemical shift for the pentapeptide, we performed 1H–15N HSQC, 1H–1H TOCSY, and 1H–1H NOESY experiments. All NMR experiments in this work were performed on a 600 MHz NMR spectrometer equipped with a z-gradient cryoprobe. 15N chemical shifts of backbone amides were measured from the HSQC spectrum (Fig. S1, SI), and the five peaks were assigned to each residue by TOCSY (Fig. S2, SI) and NOESY (Fig. S3, SI) spectra. All samples were 6 mM peptides (without 15N enrichment) in 20 mM sodium phosphate (pH 6.0), 20 mM sodium chloride, 5% D2O, and measured at 20 °C.
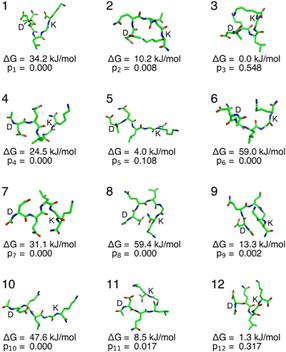
The conformational ensemble of the pentapeptide was derived via REMD simulation in GROMACS 24.02.19–26 The simulation employed the AMBER99SB force field27 and the SPC/E water model.28 Simulations were run for 40 ns with a 2 fs time step. Trajectories were clustered with the GROMOS method,29 yielding 12 clusters. The lowest-energy frame from each cluster was selected as its representative conformer.
Each conformer was first geometry optimized and then subjected to 15N chemical shift calculations, using the same DFT functional/basis set identified as optimal during the scaling factor optimization. Then, Gibbs free energy was calculated using DFT B3LYP30–32/6-31G(d,p).33–35 All DFT calculations were performed using Gaussian16 Rev. A.0336 on the NURION supercomputer (KISTI) using Knights Landing nodes with Intel Xeon Phi 7250 processors (68 cores, 96 GB RAM).
Geometry optimization was the most time-consuming step, with CPU times ranging from 40 to 100 days, and up to 200 days for the representative conformer of the second cluster. In contrast, NMR calculations typically required CPU time about 7 hours, and frequency calculations took approximately 8 days on average.
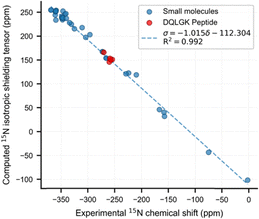
Scaling factor optimization used datasets of small molecules (Table S1, SI). The isotropic shielding tensors were calculated with a broad range of DFT methods. For geometry optimization, 3 functional/basis set were employed, whereas the NMR calculations explored 294 combinations (49 functionals × 6 basis sets) (Tables S2–S4, SI). Subsequently, we carried out linear regressions between the experimental chemical shifts and the computed shielding tensors for each case (Tables S5–S7, SI). The optimal scaling factor were adopted for the peptide study.
Through systematic evaluation, we establish an optimal computational protocol specifically calibrated for aqueous environments (Fig. 2); B3LYP[GD3BJ]30–32,37/6-311++G(d,p)38–46 for geometry optimization and LC-ωHPBE47–50/6-31+G(d,p)33–35,38,39,51–55 for NMR calculations. The linear regression yielded σ = −1.015δ − 112.304 with R2 = 0.992, demonstrating excellent agreement between the fitted slope −1.015 and the theoretical slope −1 in δsample = σref − σsample.56
Thermodynamic analysis using DFT-calculated Gibbs free energies revealed substantial differences in conformer stability. The three most populated conformers represent 54.75%, 31.73%, and 10.76% of the ensemble, respectively, amounting to 97.24% of the total population (Fig. 3).
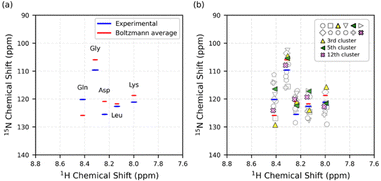
The calculation of ensemble-averaged 15N chemical shifts involved Boltzmann-weighted averaging of individual conformer contributions according to their thermodynamic populations. Comparison between calculated ensemble-averaged chemical shifts and experimental values obtained from 1H–15N HSQC spectroscopy showed good agreement (Fig. 4).
Ensemble averaging improved prediction accuracy compared to using only the dominant 3rd cluster's chemical shifts. (Fig. S6, SI) While the 3rd cluster exhibited an MAE (Mean Absolute Error) of 5.1 ppm, the ensemble-averaged chemical shifts yielded a reduced MAE of 3.6 ppm. This finding supports the view that the peptide exists as a dynamic mixture of interconverting conformers and that the experimentally observed chemical shifts represent an ensemble-averaged property.
The computational predictions showed deviations ranging from 2.5 to 5.6 ppm for most backbone nitrogen atoms (Asp, Gln, Gly, Lys), with all calculated values appearing up-field relative to experimental measurements, except for glutamine. Notably, the leucine residue demonstrated exceptional agreement with only 1.1 ppm deviation from the experimental value. Averaged 15N shift deviations of 3.6 ppm is slightly smaller than those reported in prior 15N DFT study – averaged deviation of 4.0 in ubiquitin and 4.6 ppm in GB3.57
In contrast to the four residues, the central residue exhibits accurate prediction results. The accuracy of quantum calculations for peptides in aqueous solution is critically dependent on the proper treatment of solvent system. For terminal residues, particularly the charged aspartic acid and lysine residues, implicit solvation models may inadequately capture the specific hydrogen bonding networks and electrostatic interactions with water molecules. Recent computational studies have demonstrated that explicit solvation models are essential for accurate description of peptide-solvent interactions, particularly for systems involving strong hydrogen bonding and charged residues.58,59 The central leucine residue, being surrounded by neighbouring residues and possessing a hydrophobic side chain, experiences less solvent interaction, making implicit solvation models more suitable for this residue.
In conclusion, we have successfully determined scaling factors for NMR chemical shift calculations in aqueous environments, demonstrating their reliability and accuracy for both small molecule and peptide systems. This systematic validation provides a robust framework for future applications in predicting NMR parameters of biomolecules in their aqueous environment.
The Boltzmann-weighted averaging of 15N chemical shifts from 12 representative conformers yielded good agreement with experimental HSQC measurements, with deviations of 2.5–5.6 ppm for most residues and an improved accuracy (1.1 ppm) for leucine. These results validate the ensemble approach for capturing the dynamic nature of intrinsically disordered proteins and demonstrate that experimental chemical shifts represent population-weighted averages over accessible conformational states.
The computational protocol developed in this work provides a reliable foundation for future investigations of larger and more flexible systems, bridging the gap between theoretical predictions and experimental NMR observations. This methodology offers valuable insights into how conformational dynamics modulate 15N chemical shifts in biomolecules for which conformational flexibility is central to biological function.
Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cp03354c.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (no. RS-2025-005194288, NRF-2022R1I1A2073122, and RS-2024-00459815).Notes and references
- H. Le and E. Oldfield, J. Phys. Chem., 1996, 100, 16423–16428 CrossRef CAS
.
-
J. Cavanagh, W. J. Fairbrother, A. G. Palmer, M. Rance and N. J. Skelton, in Protein NMR Spectroscopy, ed. J. Cavanagh, W. J. Fairbrother, A. G. Palmer, M. Rance and N. J. Skelton, Academic Press, Burlington, 2nd edn, 2007, ch. 10, pp. 781–817 Search PubMed
.
-
R. Gupta and M. Lechner, Chemical shifts and coupling constants for Fluorine-19 and Nitrogen-15, Springer Berlin, Heidelberg, 1998 Search PubMed
.
- Q. Gao, S. Yokojima, T. Kohno, T. Ishida, D. G. Fedorov, K. Kitaura, M. Fujihira and S. Nakamura, Chem. Phys. Lett., 2007, 445, 331–339 CrossRef CAS
.
- X. Yi, L. Zhang, R. A. Friesner and A. McDermott, J. Phys. Chem. Lett., 2024, 15, 2270–2278 CrossRef CAS PubMed
.
- Q. N. N. Nguyen, J. Schwochert, D. J. Tantillo and R. S. Lokey, Phys. Chem. Chem. Phys., 2018, 20, 14003–14012 RSC
.
- J. Li, J. Liang, Z. Wang, A. L. Ptaszek, X. Liu, B. Ganoe, M. Head-Gordon and T. Head-Gordon, J. Chem. Theory Comput., 2024, 20, 2152–2166 CrossRef PubMed
.
- S. Liu, J. Li, K. C. Bennett, B. Ganoe, T. Stauch, M. Head-Gordon, A. Hexemer, D. Ushizima and T. Head-Gordon, J. Phys. Chem. Lett., 2019, 10, 4558–4565 CrossRef CAS
.
- W. Gerrard, L. A. Bratholm, M. J. Packer, A. J. Mulholland, D. R. Glowacki and C. P. Butts, Chem. Sci., 2020, 11, 508–515 RSC
.
- C. Yiu, B. Honoré, W. Gerrard, J. Napolitano-Farina, D. Russell, I. M. L. Trist, R. Dooley and C. P. Butts, Chem. Sci., 2025, 16, 8377–8382 RSC
.
- V. A. Semenov, D. O. Samultsev and L. B. Krivdin, J. Phys. Chem. A, 2019, 123, 8417–8426 CrossRef CAS PubMed
.
- P. Gao, X. Wang and H. Yu, Adv. Theory Simul., 2019, 2, 1800148 CrossRef
.
- D. Xin, C. A. Sader, U. Fischer, K. Wagner, P.-J. Jones, M. Xing, K. R. Fandrick and N. C. Gonnella, Org. Biomol. Chem., 2017, 15, 928–936 RSC
.
- S. E. Soss, P. F. Flynn, R. J. Iuliucci, R. P. Young, L. J. Mueller, J. Hartman, G. J. O. Beran and J. K. Harper, ChemPhysChem, 2017, 18, 2225–2232 CrossRef CAS PubMed
.
- J. D. Hartman and G. J. O. Beran, Solid State Nucl. Magn. Reson., 2018, 96, 10–18 CrossRef CAS
.
-
E. Delaforge, T. N. Cordeiro, P. Bernadó and N. Sibille, in Modern Magnetic Resonance, ed. G. A. Webb, Springer International Publishing, Cham, 2018, pp. 381–399 DOI:10.1007/978-3-319-28388-3_52
.
- E. Benassi, J. Comput. Chem., 2022, 43, 170–183 CrossRef CAS PubMed
.
- Y. Sugita and Y. Okamoto, Chem. Phys. Lett., 1999, 314, 141–151 CrossRef CAS
.
- H. J. C. Berendsen, D. van der Spoel and R. van Drunen, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS
.
- E. Lindahl, B. Hess and D. van der Spoel, Mol. Model. Annu., 2001, 7, 306–317 CrossRef CAS
.
- D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef CAS PubMed
.
- B. Hess, C. Kutzner, D. van der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS
.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess and E. Lindahl, Bioinformatics, 2013, 29, 845–854 CrossRef CAS
.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef
.
-
M. Abraham, A. Alekseenko, V. Basov, C. Bergh, E. Briand, A. Brown, M. Doijade, G. Fiorin, S. Fleischmann, S. Gorelov, G. Gouaillardet, A. Gray, M. E. Irrgang, F. Jalalypour, J. Jordan, C. Kutzner, J. A. Lemkul, M. Lundborg, P. Merz, V. Miletic, D. Morozov, J. Nabet, S. Pall, A. Pasquadibisceglie, M. Pellegrino, H. Santuz, R. Schulz, T. Shugaeva, A. Shvetsov, A. Villa, S. Wingbermuehle, B. Hess and E. Lindahl, Zenodo, 2024 Search PubMed
.
-
S. Páll, M. J. Abraham, C. Kutzner, B. Hess and E. Lindahl, in Solving Software Challenges for Exascale, ed. S. Markidis and E. Laure, Springer International Publishing, Cham, 2015, pp. 3–27 Search PubMed
.
- V. Hornak, R. Abel, A. Okur, B. Strockbine, A. Roitberg and C. Simmerling, Proteins: Struct., Funct., Bioinform., 2006, 65, 712–725 CrossRef CAS
.
- H. J. Berendsen, J.-R. Grigera and T. P. Straatsma, J. Phys. Chem., 1987, 91, 6269–6271 CrossRef CAS
.
- X. Daura, K. Gademann, B. Jaun, D. Seebach, W. F. van Gunsteren and A. E. Mark, Angew. Chem., Int. Ed., 1999, 38, 236–240 CrossRef CAS
.
- A. D. Becke, J. Chem. Phys., 1986, 84, 4524–4529 CrossRef CAS
.
- A. D. Becke, Phys. Rev. A: At., Mol., Opt. Phys., 1988, 38, 3098–3100 CrossRef CAS
.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed
.
- W. J. Hehre, R. Ditchfield and J. A. Pople, J. Chem. Phys., 1972, 56, 2257–2261 CrossRef CAS
.
- P. C. Hariharan and J. A. Pople, Theor. Chim. Acta, 1973, 28, 213–222 CrossRef CAS
.
- M. M. Francl, W. J. Pietro, W. J. Hehre, J. S. Binkley, M. S. Gordon, D. J. DeFrees and J. A. Pople, J. Chem. Phys., 1982, 77, 3654–3665 CrossRef CAS
.
-
M. J. Frisch, et al., Gaussian 16, Revision A.03, Gaussian, Inc., Wallingford CT, 2016 Search PubMed
.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS
.
- R. C. Binning Jr and L. A. Curtiss, J. Comput. Chem., 1990, 11, 1206–1216 CrossRef
.
- J.-P. Blaudeau, M. P. McGrath, L. A. Curtiss and L. Radom, J. Chem. Phys., 1997, 107, 5016–5021 CrossRef CAS
.
- A. D. McLean and G. S. Chandler, J. Chem. Phys., 1980, 72, 5639–5648 CrossRef CAS
.
- K. Raghavachari and G. W. Trucks, J. Chem. Phys., 1989, 91, 1062–1065 CrossRef
.
- A. J. H. Wachters, J. Chem. Phys., 1970, 52, 1033–1036 CrossRef CAS
.
- P. J. Hay, J. Chem. Phys., 1977, 66, 4377–4384 CrossRef CAS
.
- R. Krishnan, J. S. Binkley, R. Seeger and J. A. Pople, J. Chem. Phys., 1980, 72, 650–654 CrossRef CAS
.
- M. P. McGrath and L. Radom, J. Chem. Phys., 1991, 94, 511–516 CrossRef CAS
.
- L. A. Curtiss, M. P. McGrath, J. P. Blaudeau, N. E. Davis, R. C. Binning, Jr. and L. Radom, J. Chem. Phys., 1995, 103, 6104–6113 CrossRef CAS
.
- T. M. Henderson, A. F. Izmaylov, G. Scalmani and G. E. Scuseria, J. Chem. Phys., 2009, 131, 044108 CrossRef PubMed
.
- O. A. Vydrov and G. E. Scuseria, J. Chem. Phys., 2006, 125, 234109 CrossRef PubMed
.
- O. A. Vydrov, G. E. Scuseria and J. P. Perdew, J. Chem. Phys., 2007, 126, 154109 CrossRef PubMed
.
- O. A. Vydrov, J. Heyd, A. V. Krukau and G. E. Scuseria, J. Chem. Phys., 2006, 125, 074106 CrossRef
.
- R. Ditchfield, W. J. Hehre and J. A. Pople, J. Chem. Phys., 1971, 54, 724–728 CrossRef CAS
.
- P. C. Hariharan and J. A. Pople, Mol. Phys., 1974, 27, 209–214 CrossRef CAS
.
- M. S. Gordon, Chem. Phys. Lett., 1980, 76, 163–168 CrossRef CAS
.
- V. A. Rassolov, J. A. Pople, M. A. Ratner and T. L. Windus, J. Chem. Phys., 1998, 109, 1223–1229 CrossRef CAS
.
- V. A. Rassolov, M. A. Ratner, J. A. Pople, P. C. Redfern and L. A. Curtiss, J. Comput. Chem., 2001, 22, 976–984 CrossRef CAS
.
- J. C. Facelli, Prog. Nucl. Magn. Reson. Spectrosc., 2011, 58, 176–201 CrossRef CAS
.
- A. S. Larsen, L. A. Bratholm, A. S. Christensen, M. Channir and J. H. Jensen, PeerJ, 2015, 3, e1344 CrossRef PubMed
.
- T. Zhu, J. Z. H. Zhang and X. He, J. Chem. Theory Comput., 2013, 9, 2104–2114 CrossRef CAS
.
- K. Scholten and C. Merten, Phys. Chem. Chem. Phys., 2022, 24, 3611–3617 RSC
.
| This journal is © the Owner Societies 2026 |