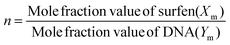Insights on the binding and selectivity of surfen towards different DNA topologies
Laxmi
Kashyap
,
Kritika
Varshney
and
Manoj
Munde
 *
*
School of Physical Sciences, Jawaharlal Nehru University, New Delhi-110067, India. E-mail: mundemanoj@gmail.com
First published on 28th November 2025
Abstract
Surfen is widely recognized for its anticoagulant properties in clinical applications. Recent interest, however, has shifted toward exploring its potential interactions with DNA, which may pave the way for novel therapeutic strategies, particularly in areas where regulation of DNA structure and function is critical. In view of this, we have investigated surfen's interaction with various DNA topologies, namely double stranded DNA (dsDNA), antiparallel G-quadruplex (AP GQ-DNA), hybrid G-quadruplex (HB GQ-DNA) and triplex DNA (tDNA) through a combination of experimental approaches and molecular dynamics simulations. This integrated strategy provided a comprehensive understanding of surfen's binding preferences and its structural impact on different DNA topologies. Experimental data, including conformational and binding analysis, demonstrated that surfen stabilizes dsDNA more effectively than G-quadruplex and triplex forms through the groove binding mode. Fluorescence intensity, lifetime changes, and competitive drug displacement data support the minor groove binding mode for surfen with dsDNA. These findings are supported by molecular dynamics (MD) simulations, which revealed a higher number of stable hydrogen bonds and a notable decrease in the radius of gyration (Rg) for dsDNA upon surfen binding, indicating compaction and structural stabilization. In contrast, surfen binding to AP and HB G-quadruplexes led to increased RMSD values and fluctuating hydrogen bond profiles, suggesting more transient and less stable interactions. Notably, HB GQ-DNA exhibited a dynamic structural transition during the simulation, implying a possible conversion to a more stable anti-parallel form upon extended drug interaction, also supported by circular dichroism (CD) study. The docking study emphasizes the critical role of DNA groove architecture, surfen shape, and functional group positioning in molecular recognition. These combined results highlight surfen's structural preference for duplex DNA and its potential to selectively stabilize specific DNA conformations, with implications for therapeutic targeting of genomic regions with distinct structural motifs.
1. Introduction
Since the discovery of the structure of DNA, it has become a key target for a wide range of therapeutically important small molecules, including anticancer drugs, antibiotics, and other pharmacological agents. These compounds exert their effects not only by binding directly to DNA but also by interacting with DNA-associated proteins or interfering with DNA-related biological processes. Thus, the targeting of biologically relevant forms of DNA such as double stranded DNA (dsDNA), G-quadruplex DNA (GQ-DNA), and triplex DNA (tDNA) by DNA-binding agents is an exciting area of study due to the unique structural characteristics and the functional roles that these DNA forms play in the genome.1 These forms of DNA offer distinct opportunities for targeted therapeutic intervention and gene regulation. Recently, the ENCODE project2 has shed light on the critical role of functional control sequences in DNA for gene regulation. In this direction, duplex DNA binding small molecules have been identified as having great therapeutic potential.3–5 Further, GQ-DNA binding agents such as bisquinolinium salts, acridine derivatives, and porphyrins have also been identified as potential anticancer agents.6–9 Similarly, triplex-forming oligonucleotides (TFOs) have garnered a lot of attention due to their potential to bind to double-stranded DNA and inhibit transcription.9 Notably, compounds like acridines, benzopyridoindoles, and benzopyridoquinoxalines have been reported as effective triplex-binding agents.10,11However, a critical challenge in designing DNA binding small molecules is ensuring that the active compounds exert their biological effects only on the desired DNA sequence. They should not bind indiscriminately to other sequences within the genome in the same concentration range, as this could lead to off-target effects that may be harmful. Therefore, in targeting specific DNA sequences, the challenge remains in designing ligands that can stabilize one type of DNA with high selectivity and specificity over other diverse DNA forms. Interestingly, quadruplex, duplex and triplex structures of DNA differ completely from each other in terms of contrasting geometries and functional groups, and these differences can be exploited to design newer compounds with better selectivity.
Evidence of selective targeting, such as the ability to target G-quadruplex DNA,12 and the success of minor groove-binding agents in clinical settings,13 clearly supports the possibility of utilizing DNA-targeting compounds for therapeutic development. While agents that meet these criteria do exist, they are rare. The difficulty lies in the fact that there are no established rules for designing such selective DNA-targeting agents. On the other hand, understanding the finer details of DNA structure might help researchers design more effective molecules that can specifically target certain DNA sequences while avoiding non-specific binding to others. Emerging research on the local microstructural states of DNA could provide insights into how selective DNA-targeting agents work.14,15
In view of this, we have explored the small molecule, bis-2-methyl-4-amino-quinolyl-6-carbamide, called surfen (Fig. 1), for its binding ability and selectivity towards a variety of DNA structures. Surfen was initially used as an excipient in depot insulin formulations in 1938.16 It has been shown to have mild heparin-neutralizing effects in rat oral feeding experiments. Surfen is thought to potentially disrupt heparin's capacity to bind and activate antithrombin.17 Subsequently, surfen was also shown to have trypanocidal properties.18 Biophysical studies of surfen with various glycoaminoglycans have been reported previously.19 To our knowledge, detailed information on its binding mode and selectivity towards a variety of DNA structures is not available in the literature. Understanding how surfen interacts with DNA could open new avenues for expanding its therapeutic applications, particularly in contexts where modulation of DNA structure and function is crucial. Therefore, the objective of this paper is to gain molecular insights into how surfen affects the G-quadruplex (hybrid and antiparallel) DNA, dsDNA and tDNA through the use of fluorescence spectroscopy, time-resolved photoluminescence, and circular dichroism (CD) and molecular dynamics (MD) studies.
2. Materials and methods
2.1. DNAs, buffers and reagents
Our study focuses on 21-mer human telomeric DNA oligonucleotide (5′GGGTTAGGGTTAGGGTTAGGG-3′), as well as triplex sequences: 5′-GAG AGG AGA GAG AAG AGG AAG-3′, 5′-CTC TCC TCTCTC TTC TCC TTC-3′, and 5′-CTC TCC TCT CTC TTC TCC TTC-3′, all of HPLC grade. Furthermore, oligomer sequences such as AT-DNA d(5′-GCAAATTTGC-3′) GC-DNA d(5′-GCGCGCGCGC-3′) were also used for the selected study. All DNA sequences were purchased from Integrated DNA Technologies (IDT) and used without further purification. DNA from the calf thymus (ctDNA) was purchased from Sigma-Aldrich Corporation. The molar extinction coefficient used for ctDNA is 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1. After verifying the drug's purity through impurity emission testing in the targeted wavelength range using an extinction coefficient of 74
200 M−1 cm−1. After verifying the drug's purity through impurity emission testing in the targeted wavelength range using an extinction coefficient of 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 cm−1,19 we procured surfen hydrate (98% pure) from Sigma and used it in our investigation. For the formation of G quadruplex DNA, the strands were individually dissolved in 10 mM HEPES, 1 mM ethylenediaminetetraacetic acid, and 100 mM KCl or NaCl at pH 7.4 to create oligonucleotide solutions. The telomeric strands were first annealed for five minutes at 90 °C, gradually cooled down to room temperature and then they were stored for forty-eight hours at 4 °C. Before each experiment, CD was used to confirm the formation of hybrid GQ-DNA topology under K+ ions (Fig. 1A) and the antiparallel basket-type GQ-DNA topology under Na+ ions (Fig. 1). For the formation of triplex DNA, equimolar amounts of the relevant strands were combined with a phosphate buffer (10 mM PBS, 100 mM NaNO3 at pH 7.4) to create triplex DNAs.1 The mixture was heated to 90 °C and then gradually cooled to room temperature. Before every experiment, CD was used to confirm the desired structure of the DNA.
400 M−1 cm−1,19 we procured surfen hydrate (98% pure) from Sigma and used it in our investigation. For the formation of G quadruplex DNA, the strands were individually dissolved in 10 mM HEPES, 1 mM ethylenediaminetetraacetic acid, and 100 mM KCl or NaCl at pH 7.4 to create oligonucleotide solutions. The telomeric strands were first annealed for five minutes at 90 °C, gradually cooled down to room temperature and then they were stored for forty-eight hours at 4 °C. Before each experiment, CD was used to confirm the formation of hybrid GQ-DNA topology under K+ ions (Fig. 1A) and the antiparallel basket-type GQ-DNA topology under Na+ ions (Fig. 1). For the formation of triplex DNA, equimolar amounts of the relevant strands were combined with a phosphate buffer (10 mM PBS, 100 mM NaNO3 at pH 7.4) to create triplex DNAs.1 The mixture was heated to 90 °C and then gradually cooled to room temperature. Before every experiment, CD was used to confirm the desired structure of the DNA.
2.2. UV-vis and fluorescence
We recorded the steady-state absorption spectra to calculate the concentration of the sample utilizing an absorption spectrophotometer (Cary 100 UV-vis). Fluorescence emission spectra were recorded using a Cary Eclipse fluorescence spectrometer to evaluate the interaction of surfen (1 µM) during the titration of GQ-DNA (with Na+ and K+ ions present), tDNA, and ctDNA. In every experiment, rectangular quartz cells with a path length of 1 cm were used. Scatchard plots were used for analysing the dissociation constant (Kd), especially in receptor–ligand interactions.The non-linear Scatchard plot can be expressed as:
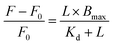 | (1) |
 . Nonlinear regression methods can provide more accurate estimates of the dissociation constant (Kd) and maximum binding capacity (Bmax) by directly fitting the original data to a hyperbolic model.
. Nonlinear regression methods can provide more accurate estimates of the dissociation constant (Kd) and maximum binding capacity (Bmax) by directly fitting the original data to a hyperbolic model.
2.3. CD spectroscopic studies
We recorded CD spectra using a Chirascan Applied Photo Physics CD spectrometer that was attached to a thermostable cell holder using 10 mM HEPES with a salt concentration of 100 mM for all the DNAs in the absence and presence of surfen. At 20 °C, a cuvette with a 0.1 cm path length was employed. After performing baseline correction with the blank buffer, the spectra were recorded in the wavelength range of 220–320 nm, obtained by averaging five scans at a speed of 75 nm min−1. The measurements were conducted with a concentration of 10 µM for the GQDNA (in Na+ or K+) and tDNA, and 100 µM for ctDNA.2.4. Time-resolved fluorescence estimation
A time-resolved photoluminescence spectrofluorometer (Horiba) was used to acquire time-resolved fluorescence decay data. With a wavelength of 350 nm and a pulse width of 64.2 ps, a picosecond pulsed diode laser was used to excite the samples, which included surfen (3 µM) and its 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with GQ-DNA (surfen-GQ DNA). The emission was detected at wavelengths between 400 and 600 nm to perform time-resolved fluorescence measurements. A 4096-channel analyser with a time resolution of 24 ps per channel was used to record all decay traces. The peak counts were approximately 7000. Fitting χ2 values between 1 and 1.3 were obtained by analysing the data using a reconvolution method that included the instrument response function (IRF). The data were fitted to the following multiexponential fitting for lifetime measurements:
1 complex with GQ-DNA (surfen-GQ DNA). The emission was detected at wavelengths between 400 and 600 nm to perform time-resolved fluorescence measurements. A 4096-channel analyser with a time resolution of 24 ps per channel was used to record all decay traces. The peak counts were approximately 7000. Fitting χ2 values between 1 and 1.3 were obtained by analysing the data using a reconvolution method that included the instrument response function (IRF). The data were fitted to the following multiexponential fitting for lifetime measurements:| A + B1 × exp(−i/T1) + B2 × exp(−i/T2) + B3 × exp(−i/T3) |
2.5. Fluorescence displacement assay (FDA)
The fluorescence displacement assay (FDA) was carried out using a spectrophotometer (Cary Eclipse). A quartz cuvette with a 1 cm path length and a scanning speed of 60 nm per minute was used for the experiments, which were conducted at room temperature. The surfen was titrated into the ligand–DNA complex to investigate ligand displacement by surfen from the complex. In this study, TMPyP4 was used as a ligand (dye), which exhibited emission at 432 nm, with the emission spectrum recorded across the 600–800 nm wavelength range. For the GQ-DNA experiments, a buffer of 10 mM HEPES along with 100 mM NaCl/KCl was used. In ctDNA experiments, a buffer of 10 mM PBS along with 100 mM NaCl was used, while for the experiments of triplex DNA, 10 mM PBS and 100 mM NaNO3 were used. All experiments were conducted at room temperature.2.6. Molecular docking
To determine the precise binding mode of surfen to double stranded DNA (PDB ID: 1BNA),20 and hybrid (PDB ID: 2JSM)21 and antiparallel (PDB ID: 143D)22 G-quadruplex DNAs, blind docking was performed using surfen (PubChem CID-71166) to the above mentioned DNA structures using AutoDock 4.0. During the docking process, surfen and G-quadruplexes were given the Gasteiger–Marsili atomic charges and Kollman united atom partial charges, respectively. Grid maps covering the entire DNA molecule were created with the resolution set to 0.375 Å. With a maximum generation count of 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, an initial population of 150 individuals, 20 runs per docking operation, and a maximum of 2
000, an initial population of 150 individuals, 20 runs per docking operation, and a maximum of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 energy evaluations, the Lamarckian genetic algorithm was employed. For crossover and mutation, the default values were set to 0.8 and 0.02 respectively. The same cluster was assigned to results with a root-mean-square deviation (RMSD) of 2.0 Å. Apart from RMSD clustering, AutoDock was used to evaluate the ligand's binding free energies in each active site, and the lowest-energy conformations were chosen from the largest cluster. Groove binding, end stacking, and other binding types were categorized. For G4, an even number of user-specified grid points = 42 x-points, 36 y-points, 46 z-points. For dsDNA, an even number of user-specified grid points = 56 x-points, 72 y-points, 120 z-points.
000 energy evaluations, the Lamarckian genetic algorithm was employed. For crossover and mutation, the default values were set to 0.8 and 0.02 respectively. The same cluster was assigned to results with a root-mean-square deviation (RMSD) of 2.0 Å. Apart from RMSD clustering, AutoDock was used to evaluate the ligand's binding free energies in each active site, and the lowest-energy conformations were chosen from the largest cluster. Groove binding, end stacking, and other binding types were categorized. For G4, an even number of user-specified grid points = 42 x-points, 36 y-points, 46 z-points. For dsDNA, an even number of user-specified grid points = 56 x-points, 72 y-points, 120 z-points.
2.7. All-atom molecule dynamics simulation
Gromacs package 2018.1 was used for MD studies.23 All-atom molecular dynamics (AA-MD) was initiated by employing the Parmbsc1 forcefield24 for DNA and the TIP3P model for water. ctDNA, hybrid and anti-parallel DNA were taken. Ligand parameters were generated using the acepype server (https://www.bio2byte.be/acpype/). To achieve this, the DNA or the complex was placed within a triclinic box, ensuring a minimum distance of 1 nm between the DNA and the nearest box face. Subsequently, the systems were solvated and ions were introduced to neutralise the system while attaining a NaCl concentration of 100 mM for anti-parallel and ctDNA. While for the hybrid 100 mM KCl was added to mimic the experimental studies.The LINCS algorithm was used to constrain all bond lengths along with the hydrogen bonds. Minimization was performed exploiting the steepest descent method for 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps set as the upper limit. The system was further equilibrated under the isochoric–isothermal (NVT) ensemble for 200 ps with a 2 fs time step using a modified Berendsen thermostat for maintaining a temperature of 300 K. Furthermore, the system was equilibrated under isothermal–isobaric (NPT) conditions for 800 ns using a modified Berendsen thermostat and Parrinello–Rahman barostat to stabilize both temperature and pressure at 300 K and 1 bar, respectively. Until this point the polymorphs of DNA and surfen were position restrained using a spring constant of 1 kcal mol−1 Å−2. For neighbour searching, the Verlet cutoff scheme was utilized. The smooth particle mesh Ewald summation method (PME) along with the cubic interpolation method and a Fourier grid spacing of 16 Å was used for long range electrostatic determination. All nonbonded interactions were evaluated using a cutoff of 1 nm and were gpu accelerated. Then, the position restraints were removed and the system was further equilibrated for 2 ns. A production run was performed for 150 ns. Velocity snapshots were generated using the render option in Visual Molecular Dynamics (VMD),57 and finally, Origin Pro 2025 was utilized for graph generation.
000 steps set as the upper limit. The system was further equilibrated under the isochoric–isothermal (NVT) ensemble for 200 ps with a 2 fs time step using a modified Berendsen thermostat for maintaining a temperature of 300 K. Furthermore, the system was equilibrated under isothermal–isobaric (NPT) conditions for 800 ns using a modified Berendsen thermostat and Parrinello–Rahman barostat to stabilize both temperature and pressure at 300 K and 1 bar, respectively. Until this point the polymorphs of DNA and surfen were position restrained using a spring constant of 1 kcal mol−1 Å−2. For neighbour searching, the Verlet cutoff scheme was utilized. The smooth particle mesh Ewald summation method (PME) along with the cubic interpolation method and a Fourier grid spacing of 16 Å was used for long range electrostatic determination. All nonbonded interactions were evaluated using a cutoff of 1 nm and were gpu accelerated. Then, the position restraints were removed and the system was further equilibrated for 2 ns. A production run was performed for 150 ns. Velocity snapshots were generated using the render option in Visual Molecular Dynamics (VMD),57 and finally, Origin Pro 2025 was utilized for graph generation.
3. Results and discussion
3.1. DNA conformational changes using CD spectroscopy
To gain deeper insights into the configuration and topology behaviour of DNAs in the presence of surfen, we have performed CD experiments. As shown in Fig. 2A, GQ-DNA in the presence of sodium (Na+) exhibits positive peaks near 295 nm and 245 nm, along with a negative band at 265 nm, indicating the formation of antiparallel GQ-DNA (AP GQ-DNA).25,26 Upon addition of surfen, there is a progressive decrease in these bands, indicating the binding of surfen to AP GQ-DNA; however the overall configuration remains unchanged (Fig. 2A), implying that AP GQ-DNA preserves its original structure upon surfen binding. Groove binding has been shown to induce changes in magnitude of 290, 265 and 243 nm bands, whereas end stacking does not show any significant change. Therefore, external groove binding may occur in the present investigations.27 The CD spectrum of GQ-DNA in K+ exhibits a broad positive peak at 290 nm, a smaller peak at 273 nm, and a negative band in the 234–240 nm region, indicating the formation of hybrid G-quadruplex DNA (HB GQ-DNA), as shown in Fig. 2B. The gradual addition of surfen to mixed hybrid GQ DNA led to a decrease in these bands and the appearance of a negative peak around 275 nm and 250 nm, which suggests a likely switch in configuration from the hybrid to an antiparallel one. Similar results have been observed for other ligands in the past.28,29Next, the CD spectrum of duplex DNA (ctDNA) shows a positive band in the 273–280 nm range and a negative band at 245 nm, confirming its right-handed helicity. The intensities of the positive and negative bands of ctDNA increase with increasing surfen concentration (Fig. 2C), suggesting that there are significant interactions between surfen and ctDNA. Generally, this type of binding is shown by minor groove binders confirming that surfen binds in the minor groove.30–32 In Fig. 2D, the negative peak at approximately 247 nm and the positive peak at approximately 277 nm increased due to base stacking, accompanying a negative peak at approximately 225 nm, indicating the formation of triplex DNA.33 Upon addition of surfen, there is a continuous reduction in CD signal intensity suggesting that the drug binding may lead to aggregation of DNA.
3.2. Binding studies using fluorescence emission spectroscopy
Fluorescence emission studies were performed to investigate the binding interaction of surfen with GQ-DNA, ctDNA and tDNA. In this instance, the surfen excitation wavelength at 340 nm was used to excite the sample, and the emission spectra were collected in the range of 400 to 600 nm. The resultant spectra and corresponding binding plots are shown in Fig. 3. As illustrated in Fig. 3A and B, the fluorescence intensity of surfen increased as AP GQ-DNA and HB GQ-DNA were gradually added. This increase in fluorescence is due to the transfer of surfen from the aqueous environment to the hydrophobic environment within DNA.34 This transfer reduces solvent exposure, leading to enhanced fluorescence intensity. AP GQ-DNA shows a ten times stronger binding to surfen (2.1 × 106 M−1) compared to HB GQ-DNA (2.0 × 105 M−1). This moderate binding affinity was supported by the CD melting study (Fig. S1), in which surfen induced only a 2–3 °C thermal stabilization of the AP and HB GQ-DNA structures.Next, adding ctDNA to surfen (Fig. 3C) results in a sharp increase in the intensity, indicating the formation of a strong surfen–ctDNA complex (8.3 × 108 M−1). In contrast, there is a very weak binding between tDNA and surfen as shown by its weaker intensity change in Fig. 3D. Thus, the binding constant values indicate that the order of binding affinity of surfen for different DNA sequences is ctDNA > AP GQ-DNA > HB GQ-DNA > tDNA (Table 1).
| Complex | K a/M−1 |
|---|---|
| Surfen–ctDNA | 8.3 × 108 |
| Surfen–GQ DNA(AP) | 2.1 × 106 |
| Surfen–GQ DNA(HB) | 2.0 × 105 |
| Surfen–tDNA | 4.9 × 104 |
Next, in order to explore the binding specificity of surfen within duplex sequences, we performed a selective fluorescence binding experiment with AT- and GC-rich duplex DNAs (Fig. S3). The results reveal that surfen exhibits much more enhanced fluorescence intensity in the presence of AT-rich DNA compared to GC-rich DNA. This preferential binding can be attributed to structural and electronic features of the AT-rich regions.35,36 The minor groove of AT-rich DNA is considered as narrower and deeper, providing a more favorable binding pocket for small cationic ligands and groove binders.3–5,35,36 It is well established that the absence of the exocyclic amino group at the C2 position of guanine in the AT base pairs reduces steric hindrance and enhances hydrophobic and van der Waals interactions between the ligand and the groove.36 In contrast, GC-rich regions have a wider minor groove and higher base stacking energy, which generally hinders the accommodation of such small molecules.36
3.3. Binding stoichiometry using fluorescence-based continuous variation experiment (Job plot)
Job plots serve as a valuable tool for analysing binding stoichiometries in molecular systems. In fluorescence spectroscopy, the binding stoichiometry of the surfen–DNA complex was determined using the continuous variation method, also known as Job's plot.37 Variations in the molar ratio of surfen to DNA were achieved while keeping the total molar concentration constant. Emission intensities at 488 nm were plotted against mole fraction, as depicted in Fig. 4. The least-squares fitted lines for DNA and surfen intersected at a mole fraction of ∼0.5. The Job's plot yielded a binding stoichiometry ratio, n of ∼1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1,
1,which corresponds to the binding of one surfen molecule per one DNA strand of AP and HB GQ-DNA (Fig. 4A and B). In the case of ctDNA (Fig. 4C), the stoichiometry of 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 means one surfen molecule binding per 10 base pairs. As reported in the literature, employing a fluorescence Job's plot, the interaction of Hoechst 33258 with calf thymus DNA revealed a binding ratio of one dye molecule per 10 bp strand of DNA.38 Similarly, previous studies have shown that most groove-binding molecules tend to interact with DNA in a ratio of approximately one ligand per DNA strand of 10 base pairs.31,39,40 Thus, we used this estimate to calculate the stoichiometry in the Job's plot in Fig. 4. As far as triplex DNA is concerned, due to weak binding of surfen, the stoichiometry value may have error on the higher side (Fig. 4D).
1 means one surfen molecule binding per 10 base pairs. As reported in the literature, employing a fluorescence Job's plot, the interaction of Hoechst 33258 with calf thymus DNA revealed a binding ratio of one dye molecule per 10 bp strand of DNA.38 Similarly, previous studies have shown that most groove-binding molecules tend to interact with DNA in a ratio of approximately one ligand per DNA strand of 10 base pairs.31,39,40 Thus, we used this estimate to calculate the stoichiometry in the Job's plot in Fig. 4. As far as triplex DNA is concerned, due to weak binding of surfen, the stoichiometry value may have error on the higher side (Fig. 4D).
3.4. Time-resolved studies
To elucidate deeper insights into the binding phenomenon, we further performed time-resolved studies. Measuring the fluorescence lifetime is an effective method for studying the excited-state environment of fluorophores as well as to understand the binding interactions between DNA and ligand. Surfen displays a single exponential decay characterised by a lifetime of approximately 1.45 ns as shown in Fig. 5. It is observed that upon adding DNA to surfen, there is an increase in the lifetime decay profile (Fig. 5), indicating that surfen's environment changes, leading to the creation of a drug–DNA complex. We have not performed an experiment for triplex DNA as its binding with surfen was not significant. | ||
Fig. 5 (A) Surfen (3 µM) fluorescence lifetime decay profiles in (A) AP GQ-DNA, (B) HB GQ-DNA and (C) ctDNA. Experiments were done at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DNA 1 DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) drug ratio in buffer with pH 7.0. drug ratio in buffer with pH 7.0. | ||
As shown in Fig. 5, the fluorescence lifetime of surfen increases significantly in the presence of duplex DNA (4.23 ns) compared to AP GQ-DNA (1.96 ns) and HB GQ-DNA (2.19 ns). This suggests that the surfen chromophores are more effectively shielded from solvent interactions when bound to duplex DNA. The increased fluorescence lifetime upon binding to both duplex and GQ-DNA indicates that DNA protects surfen from solvent quenching, thereby promoting its interaction with DNA. The lifetime enhancement reflects the quenching mechanism of surfen in the presence of DNA (Fig. 5 and Table 2), and the data imply that quenching occurs via excited-state collisions or diffusion. Therefore, the quenching mechanism is consistent with a dynamic (collisional) quenching process.41,42
| Complex | Average lifetime (τa) (ns) | χ 2 |
|---|---|---|
| Surfen | 1.459 | 1.13357 |
| Surfen–GQ(AP) | 2.195 | 1.050276 |
| Surfen–GQ(HB) | 1.968 | 1.031221 |
| Surfen–Duplex DNA | 4.235 | 1.23653 |
3.5. Fluorescent displacement assay
To investigate the mode of drug–DNA interactions, a variety of well-characterized DNA-binding fluorescent dyes are frequently employed.43,44 These dyes have well-established binding modes, making them valuable tools in competitive displacement assays. In such assays, the ability of a small molecule to displace a pre-bound dye from DNA suggests that the molecule interacts with DNA in a similar binding mode as the displaced dye. The resulting changes in fluorescence provide a straightforward and reliable means to interpret the nature of the small molecule's interaction with DNA.Here, fluorescent displacement assays were conducted to evaluate the ligand binding mode of surfen with GQ-DNA (AP and HB) and ctDNA. In the case of GQ-DNA, a mixture of DNA and TMPyP4 was titrated with surfen. TMPyP4 exhibits emission peaks at 661 nm and 713 nm upon excitation of 432 nm. The short bands of permitted π−π* are responsible for the two peaks, which are the Q(0,0) and Q(1,0) band37,45 electronic transitions (Fig. 6). Surfen emission does not interfere in this range. Here, the addition of GQ-DNA (AP and HB) to TMPyP4 results in a decrease in fluorescence intensity, which suggests π–π* stacking interactions between TMPyP4 and GQ-DNA (in both KCl and NaCl). This supports earlier studies involving crystallography and NMR that have demonstrated that TMPyP4 binds to G4-DNA in end-stacking mode with G4 tetrads.23 Here, if addition of surfen to the TMPyP4-G4 complex causes quenching of the fluorescence intensity, it means that surfen displaces TMPyP4 and end-stacks with the G-quadruplex DNA. However, upon gradually adding surfen to TMPyP4-GQ DNA (AP and HB), there is no measurable change in the fluorescence intensity, indicating that there is no displacement of TMPyP4 from the TMPyP4-G4 complexes. This clearly rules out end stacking interaction between surfen and GQ-DNA (in Na+ and K+).
Furthermore, the intercalation of small molecules between the tetrads of GQ-DNA is generally considered difficult, as GQ structures are extremely stable and rigid. Each G-tetrad is stabilized by eight hydrogen bonds significantly more than the two or three hydrogen bonds found in AT or GC base pairs, respectively. Consequently, any distortion of the G4 structure would entail a high energetic cost.46 Therefore, based on this and the CD results, binding of surfen within the grooves of the GQ-DNA would represent the most probable mode of interaction, which would also be energetically more favourable.47 Several peptide–hemicyanine conjugates have been proposed to be specific for binding to quadruplex grooves and loops without significant π stacking with DNA bases.48
Next, in order to test the binding mode of surfen in duplex DNA, DAPI as a minor groove agent was used.43,44 The DAPI–ctDNA complex results in an increase in the fluorescence intensity as shown in Fig. 6C, indicating that DAPI binds in the minor groove of DNA. However, when surfen was added to the DAPI–ctDNA complex, there is a decrease in fluorescence intensity, indicating that surfen successfully displaces DAPI from the minor groove of ctDNA.
3.6. Molecular docking
In order to obtain a clearer picture of the DNA binding mode and atomic-level interactions of surfen with DNA, a molecular docking study was performed using the AutoDock 4.0 software suite. This approach enabled the prediction of the most favourable binding orientations, identification of key interacting bases, and analysis of non-covalent interactions between the ligand and the DNA. Docking results (Fig. 7A) revealed that surfen preferentially binds within the groove region of the AP GQ-DNA, adopting a single dominant binding conformation associated with the lowest binding free energy among all the tested structures (Table 3). This favourable interaction is stabilized by four interactions between surfen and the DNA (Fig. 7D); the –NH2 group of the aminoquinoline ring forms a hydrogen bond with the exocyclic amino group of G20, specifically targeting the nitrogen attached to the C2 carbon of the guanine base. Two –NH groups of the aminoquinoline ring engage in hydrogen bonding with the phosphate backbone (the OP2 atom is a non-bridging oxygen of the phosphate group connected to G16). An additional hydrogen bond is observed between the same –NH2 group with the phosphate backbone (the OP2 atom connected to thymine T17). The shallower groove of the AP GQ-DNA facilitates optimal molecular recognition that contributes to the stability of the surfen–DNA complex.| S. no. | DNA | Sequence | Binding energy (kcal mol−1) |
|---|---|---|---|
| 1 | AP GQ-DNA | A1G2G3G4T5T6A7G8G9G10T11T12A13G14G15G16T17T18A19G20G21G22 | −7.92 |
| 2 | HB GQ-DNA | T1A2G3G4G5T6T7A8G9G10G11T12T13A14G15G16G17T18T19A20G21G22G23 | −7.39 |
| 3 | dsDNA | C1G2C3G4A5A6T7T8C9G10C11G12G24C23G22C21T20T19A18A17G16C15G14C13 | −11.70 |
In contrast, the docking analysis of HB GQ-DNA (Fig. 7B and E) reveals that surfen binds in the groove with only two hydrogen bonds involving G11. A first hydrogen bond involves the –NH2 group of surfen and the exocyclic amino nitrogen of G11 (attached to the C2 carbon of guanine) and the second one with the phosphate backbone (between the –NH group of the aminoquinoline ring of surfen and the oxygen atom bonded to the 3′ carbon of the sugar moiety connected to G11). The HB GQ-DNA structure features a narrower and wider groove, which results in a reduced binding affinity for surfen. The larger and less compact groove architecture fails to accommodate the ligand as effectively, leading to suboptimal interactions and higher binding free energies (Table S1).
Docking analysis of surfen with dsDNA (Fig. 7C) reveals the conformation with the lowest binding free energy (Table 3) and the largest cluster population both correspond to surfen occupying the minor groove, in alignment with experimental observations. Structural evaluation of the surfen–dsDNA complex indicates that surfen's molecular curvature is well-suited for the minor groove. Both amidinium groups engage in hydrogen bonding with the base pair edges at the floor of the groove, ensuring a snug fit and optimal interaction (Fig. 7F). The –NH2 group of surfen forms a hydrogen bond with T19 (carbonyl oxygen at C2 position of thymine, chain B). Another –NH2 group of the aminoquinoline ring interacts with C9 (carbonyl oxygen double-bonded to the C2 carbon of cytosine, chain A). Two distinct –NH groups of surfen interact through a bifurcated H-bond with T8 (carbonyl oxygen at C2 of thymine, chain A). The higher number of interactions observed between surfen and dsDNA contributes to a stronger binding affinity compared to its interactions with other DNA types.
3.7. MD studies
The molecular dynamics (MD) study (Fig. 8) aimed to compare the relative structural stability of dsDNA, AP GQ-DNA, and HB GQ-DNA, both in their free forms and when complexed with surfen by evaluating key structural parameters. RMSD is a fundamental metric for assessing the structural integrity and stability of biomolecules over time and it is used as the starting point.49 The RMSD of the AP GQ-DNA exhibits fluctuations before it stabilizes after approximately 100 ns (Fig. 8B). This behaviour suggests that the AP GQ-DNA samples multiple conformational sub-states during the timescale, reflecting inherent dynamic flexibility rather than complete unfolding. Similar slow equilibration and dynamic sampling have been reported in the literature50 for G-quadruplexes of this topology. In contrast, the RMSD of the surfen-bound AP GQ-DNA complex stabilizes much earlier, plateauing around 20 ns (Fig. 8B), suggesting increased stability of AP GQ-DNA upon surfen binding.In the case of the surfen-bound HB GQ-DNA complex, frequent RMSD fluctuations are observed compared to its drug-free form, persisting until approximately 50 ns before stabilizing (Fig. 8C). This suggests that surfen induces conformational rearrangements in the HB G-quadruplex structure (discussed later). In the case of native dsDNA (Fig. 8A), RMSD fluctuations were observed, which may primarily arise from terminal fraying events, where the ends of the DNA transiently open and close dynamically during the simulation.51 However, surfen-bound dsDNA achieves stability rapidly and exhibits minimal RMSD fluctuations compared to the free dsDNA, indicating significantly enhanced structural stability. This stabilization likely arises from specific, sustained interactions within the minor groove, as supported by docking studies.
The radius of gyration (Rg) is a key indicator of the compactness of DNA structures.52 The abrupt increase in Rg observed around 120 ns for AP native DNA is indicative of a significant conformational event, such as partial unwinding or expansion of the structure (Fig. 8E). This is consistent with unfolding or rearrangement of the G-quadruplex loops or G-quartets. However, in the presence of surfen, Rg remained stable at around 1.06 nm to 1.05 nm. Similarly, in the case of HB GQ-DNA, there is an insignificant increase in Rg from 1.07 nm to 1.11 nm (Fig. 8F). However, for the surfen complex, there is sudden increase in Rg around 50 ns, indicating a drug induced conformational change. For dsDNA, the average Rg decreased significantly from 1.36 nm to 1.2 nm upon surfen binding (Fig. 8D). This substantial reduction suggests strong compaction and stabilization of the dsDNA, highlighting surfen's structural preference for and stronger interaction with duplex over G-quadruplex DNA.
Intermolecular hydrogen bonds formed between a drug and DNA play a crucial role in maintaining the three-dimensional structure and stability of the resulting complex. Studying these hydrogen bonds provides valuable insights into the intermolecular stability of the drug–DNA complex, including how the drug recognizes and interacts with DNA, and the specificity of that interaction.30,31,53,54 Hydrogen bond analysis of the surfen–dsDNA complex reveals the highest average number of hydrogen bonds (Fig. 8G), with these bonds remaining consistent throughout the simulation. As supported by docking results, the observed shorter bond lengths (2.06, 1.94, 2.2 and 1.9 Å; Table S1) also suggest stronger interaction in the case of dsDNA. This observation supports experimental findings indicating that surfen most effectively stabilizes dsDNA. In contrast, complexes of surfen with AP and HB GQ-DNA (Fig. 8H and I) structures exhibit fewer average hydrogen bonds below 3.0 Å (three in AP and 2 in HB; Table S1) and more frequent fluctuations to zero or near-zero levels (Fig. 8H and I). These results suggest that surfen's interaction with GQ-DNA structures is less stable and more transient compared to its interaction with dsDNA.
Furthermore, we have quantitatively assessed the contribution of electrostatic and van der Waals interaction energies of surfen binding with different DNA topologies as shown in Fig. S2. As shown in the Fig. S2A and B, dsDNA presents the most favorable environment for surfen binding, evidenced by strong and sustained electrostatic and van der Waals interactions attributable to its well-organized minor groove. The AP GQ-DNA (Fig. S2C and D) showed less favorable electrostatic and van der Waals energies compared to dsDNA. Although surfen still interacts with the AP quadruplex form, the interaction is weaker and more transient, as reflected by higher fluctuations and less negative energy values. The HB GQ-DNA (Fig. S2E and F) also displays weaker and variable interaction energies due to its flexible and poorly defined grooves.
3.8. Conformational switch in HB GQ-DNA
Detailed analysis of the RMSD plot of the HB GQ-DNA in the presence of surfen (Fig. 9A) reveals initial fluctuations, followed by a sudden increase in RMSD around 60 ns, after which the system re-equilibrates. Based on the fluctuation of RMSD values, the process can be divided into different stages for trajectory analysis to evaluate the dynamic behavior. Structural snapshots taken from UCSF ChimeraX at various time points provide deeper insight into the DNA's flexibility and conformational dynamics during surfen binding. During the initial 60 ns, hybrid GQ-DNA, characterized by its combination of parallel and anti-parallel strands and flexible loops connecting guanine tetrads, maintains its native topology with wider and shallower grooves. However, between 60 and 150 ns, the simulations show a marked structural opening, particularly in the major loop region. The observation aligns with the radius of gyration results as evidenced by an increase in the radius of gyration in Fig. 8F. This opening is accompanied by increased conformational flexibility, allowing the guanine tetrads and connecting loops to rearrange. The mechanism underlying this transition involves the destabilization of the hybrid structure's loops. As the loops collapse and the strands realign, the structure becomes more compact and the grooves will narrow-up. Similar results were reported earlier for GQ-DNA in the presence of clinical drug sorafenib.55 These computational findings are consistent with experimental observations. In contrast, the RMSD plot of anti-parallel GQ-DNA upon surfen binding (Fig. 9B) indicates that the DNA undergoes initial fluctuations until 20 ns and thereafter remains stable through 150 ns, indicating that the overall conformation of the anti-parallel DNA remains consistent throughout the period. | ||
| Fig. 9 Snapshots from UCF Chimera-X of (A) HB GQ-DNA and (B) AP GQ-DNA at specific time points (0 ns, 10 ns, 100 ns and 150 ns) to illustrate the structural changes. | ||
3.9. Conformation of surfen
The central structural feature of surfen consists of two identical quinoline units (Fig. 1). Each quinoline ring is a bicyclic aromatic heterocycle containing a nitrogen atom in its structure. These rings are substituted with a methyl group at the 2-position and an amino group at the 4-position. The two 4-aminoquinoline moieties are linked through a urea bridge at the 6-position, forming the characteristic bis-quinoline framework. This urea linkage plays a critical role in surfen's ability to bind its molecular targets. Here, MD simulation studies have shown that the rotational flexibility around the urea bond allows aminoquinoline rings to adopt multiple conformations, thereby influencing surfen's interaction with DNA.The presence of the aromatic quinoline rings and the basic amino groups makes surfen a cationic molecule, especially when protonated. During the analysis of how surfen undergoes conformational change upon binding with dsDNA, we observed that surfen rotates (Fig. 10) to align its H-bond donating groups such as –NH or –NH2 with H-bond acceptor groups of the DNA minor groove of dsDNA (Fig. 10B). This alignment also helps in optimizing the van der Waals and electrostatic interactions, strengthening the overall binding affinity. In the previous study, amines, and the urea linker region were shown to play essential roles in the interaction of surfen with heparin sulphate.56 In the case of dsDNA, surfen undergoes a conformational change and takes a shape complementary to the size of the groove. In fact, surfen's molecular twist helps both amidinium groups engage in hydrogen bonding with the base pair edges at the floor of the groove, ensuring a snug fit and optimal interaction (Fig. 7). In the AP-bound conformation (Fig. 10D), surfen exhibits a higher degree of twist between its quinoline rings compared to the HB-bound GQ-DNA (Fig. 10C). Generally, the degree of twisting is determined by a complex interplay of the ligand properties and the GQ-DNA's dynamic structural features. The antiparallel topology's alternating strand orientation often creates wider, more accessible grooves compared to the more rigid parallel core of hybrid structures.57 Thus, it seems, that between AP and HB GQ-DNA, surfen prefers binding in the less constrained groove regions of an AP GQ-DNA so that it can experience more freedom to twist than getting trapped in the tighter, more rigid stacked-tetrad region of a HB GQ-DNA. Overall, in the case of dsDNA, a higher number of interactions were observed between surfen and dsDNA resulting in a stronger binding affinity compared to its interactions with other DNA types.
 | ||
| Fig. 10 Conformation of surfen in free and DNA-bound forms: (A) free surfen, (B) dsDNA-bound, (C) HB-bound and (D) AP-bound. | ||
Thus, MD simulations demonstrate that surfen preferentially binds and stabilizes duplex DNA compared to G-quadruplex structures. The urea-linked bis-quinoline framework of surfen allows conformational flexibility, enabling optimal alignment within the dsDNA groove to enhance H-bond, electrostatic and van der Waals contacts. Notably, surfen induced a conformational switch in HB GQ-DNA, while AP GQ-DNA retained its topology. It is true that 150 ns sampling may be insufficient to fully capture all slow conformational transitions in DNA, especially for complex G-quadruplex structures known to undergo slow folding/unfolding dynamics on micro- to millisecond timescales. In this context, longer simulations would be beneficial for more definitive conclusions on rare transitions. Nonetheless, our combined experimental and computational data provide meaningful insights into surfen's differential effects on DNA topologies within the accessible timescale.
4. Conclusion
This study employed a combination of biophysical techniques and computational methods to investigate the binding mode and selectivity of surfen toward various DNA topologies, including dsDNA, tDNA, and GQ-DNA. The results reveal that surfen exhibits preferential binding to duplex DNA over both GQ-DNA and tDNA. Notably, surfen shows high specificity for DNA groove structures, particularly favoring the deeper and more defined minor groove of duplex DNA. In contrast, its affinity for GQ-DNA is significantly lower, likely due to the poorly defined groove architecture characteristic of G-quadruplex structures. For triplex DNA, the presence of a third strand induces structural perturbations that may reduce its stability and compatibility with surfen binding. The selective recognition of duplex DNA by surfen can be attributed to the unique structural features of the minor groove in dsDNA, as well as the complementary shape and functional groups of surfen that facilitate groove fitting and hydrogen bonding. These findings highlight the importance of groove topology in small molecule–DNA interactions and support the rational design of DNA-targeting ligands that exploit structural complementarity for sequence- and topology-specific recognition.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: A table demonstrating the interactions between DNA bases and surfen. See DOI: https://doi.org/10.1039/d5cp02724a.Acknowledgements
This work was supported by grants from SERB (CRG/2023/007393). LK acknowledges fellowship support by UGC. This work was also supported by ‘JNU-PAIR Network on Science for Sustainable Future’ (ANRF/PAIR/2025/000029/PAIR). We also thank AIRF for the instrument facility. Furthermore, we acknowledge the SATHI Facility, IIT Delhi, Sonipat Campus for Time Resolved Fluorescence.References
- L. H. Hurley and F. L. Boyd, Trends Pharmacol. Sci., 1988, 9, 402–407 CrossRef CAS PubMed.
- E. P. Consortium, Nature, 2012, 489, 57–74 CrossRef.
- A. Paul, J. R. Terrell, A. A. Farahat, E. N. Ogbonna, A. Kumar, D. W. Boykin, S. Neidle and W. D. Wilson, ACS Chem. Biol., 2025, 20, 489–506 CrossRef CAS PubMed.
- A. Paul, R. Nanjunda and W. D. Wilson, Curr. Protoc., 2023, 3, e729 CrossRef CAS.
- P. Guo, A. A. Farahat, A. Paul, D. W. Boykin and W. D. Wilson, Chem. Sci., 2021, 12, 15849–15861 RSC.
- S. Neidle, Molecules, 2024, 29, 505 CrossRef.
- R. C. Monsen, J. O. Trent and J. B. Chaires, Acc. Chem. Res., 2022, 55, 3242–3252 CrossRef CAS PubMed.
- S. Roy, P. Pramanik and S. Bhattacharya, Biochimie, 2025, 234, 120–145 CrossRef CAS PubMed.
- E. A. Owens, H. T. Huynh, E. M. Stroeva, A. Barman, K. Ziabrev, A. Paul, S. V. Nguyen, M. Laramie, D. Hamelberg, M. W. Germann, W. D. Wilson and M. Henary, Bioconjugate Chem., 2019, 30, 2647–2663 CrossRef CAS.
- S. A. Cassidy, L. Strekowski, W. D. Wilson and K. R. Fox, Biochemistry, 1994, 33, 15338–15347 CrossRef CAS.
- J. L. Mergny, G. Duval-Valentin, C. H. Nguyen, L. Perrouault, B. Faucon, M. Rougée, T. Montenay-Garestier, E. Bisagni and C. Hélène, Science, 1992, 256, 1681–1684 CrossRef CAS PubMed.
- E. Y. Lam, D. Beraldi, D. Tannahill and S. Balasubramanian, Nat. Commun., 2013, 4, 1796 CrossRef PubMed.
- A. Vidal, C. Muñoz, M. J. Guillén, J. Moretó, S. Puertas, M. Martínez-Iniesta, A. Figueras, L. Padullés, F. J. García-Rodriguez, M. Berdiel-Acer, M. A. Pujana, R. Salazar, M. Gil-Martin, L. Martí, J. Ponce, D. G. Molleví, G. Capella, E. Condom, F. Viñals, D. Huertas, C. Cuevas, M. Esteller, P. Avilés and A. Villanueva, Clin. Cancer Res., 2012, 18, 5399–5411 CrossRef CAS PubMed.
- S. P. Hancock, T. Ghane, D. Cascio, R. Rohs, R. Di Felice and R. C. Johnson, Nucleic Acids Res., 2013, 41, 6750–6760 CrossRef CAS PubMed.
- R. Rohs, S. M. West, A. Sosinsky, P. Liu, R. S. Mann and B. Honig, Nature, 2009, 461, 1248–1253 CrossRef CAS PubMed.
- F. Umber, F. Stoerring and W. Foellmer, Klin. Wochenschr., 1938, 17, 443–446 CrossRef.
- D. T. Hunter and J. M. Hill, Nature, 1961, 191, 1378–1379 CrossRef CAS PubMed.
- F. C. Goble, J. Pharmacol. Exp. Ther., 1950, 98, 49–61 CrossRef CAS.
- M. Schuksz, M. M. Fuster, J. R. Brown, B. E. Crawford, D. P. Ditto, R. Lawrence, C. A. Glass, L. Wang, Y. Tor and J. D. Esko, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 13075–13080 CrossRef CAS.
- H. R. Drew, R. M. Wing, T. Takano, C. Broka, S. Tanaka, K. Itakura and R. E. Dickerson, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 2179–2183 CrossRef CAS.
- A. T. Phan, V. Kuryavyi, K. N. Luu and D. J. Patel, Nucleic Acids Res., 2007, 35, 6517–6525 CrossRef CAS.
- Y. Wang and D. J. Patel, Structure, 1993, 1, 263–282 CrossRef CAS PubMed.
- D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef CAS PubMed.
- I. Ivani, P. D. Dans, A. Noy, A. Pérez, I. Faustino, A. Hospital, J. Walther, P. Andrio, R. Goñi, A. Balaceanu, G. Portella, F. Battistini, J. L. Gelpí, C. González, M. Vendruscolo, C. A. Laughton, S. A. Harris, D. A. Case and M. Orozco, Nat. Methods, 2016, 13, 55–58 CrossRef CAS.
- M. Vorlíčková, I. Kejnovská, J. Sagi, D. Renčiuk, K. Bednářová, J. Motlová and J. Kypr, Methods, 2012, 57, 64–75 CrossRef PubMed.
- K. N. Luu, A. T. Phan, V. Kuryavyi, L. Lacroix and D. J. Patel, J. Am. Chem. Soc., 2006, 128, 9963–9970 CrossRef CAS PubMed.
- E. W. White, F. Tanious, M. A. Ismail, A. P. Reszka, S. Neidle, D. W. Boykin and W. D. Wilson, Biophys. Chem., 2007, 126, 140–153 CrossRef CAS.
- S. A. Ohnmacht, E. Varavipour, R. Nanjunda, I. Pazitna, G. Di Vita, M. Gunaratnam, A. Kumar, M. A. Ismail, D. W. Boykin, W. D. Wilson and S. Neidle, Chem. Commun., 2014, 50, 960–963 RSC.
- T. Biver, Molecules, 2022, 27, 4165 CrossRef PubMed.
- M. Munde, M. A. Ismail, R. Arafa, P. Peixoto, C. J. Collar, Y. Liu, L. Hu, M.-H. David-Cordonnier, A. Lansiaux, C. Bailly, D. W. Boykin and W. D. Wilson, J. Am. Chem. Soc., 2007, 129, 13732–13743 CrossRef CAS PubMed.
- M. Munde, M. Lee, S. Neidle, R. Arafa, D. W. Boykin, Y. Liu, C. Bailly and W. D. Wilson, J. Am. Chem. Soc., 2007, 129, 5688–5698 CrossRef CAS PubMed.
- M. M. Munde, A. Kumar, R. Nhili, S. Depauw, M.-H. David-Cordonnier, D. Boykin and D. Wilson, Biophys. J., 2011, 100, 234–235 CrossRef.
- N. Lohani and M. R. Rajeswari, RSC Adv., 2016, 6, 39903–39917 RSC.
- M. Sirajuddin, S. Ali and A. Badshah, J. Photochem. Photobiol., B, 2013, 124, 1–19 CrossRef CAS PubMed.
- N. Narayanaswamy, S. Das, P. K. Samanta, K. Banu, G. P. Sharma, N. Mondal, S. K. Dhar, S. K. Pati and T. Govindaraju, Nucleic Acids Res., 2015, 43, 8651–8663 CrossRef CAS PubMed.
- B. Wang, J. Tan and L. Zhu, Colloids Surf., B, 2010, 79, 1–4 CrossRef CAS PubMed.
- S. Luikham, A. Mavani, D. Sinha and J. Bhattacharyya, J. Phys. Chem. B, 2023, 127, 4966–4978 CrossRef CAS PubMed.
- F. G. Loontiens, P. Regenfuss, A. Zechel, L. Dumortier and R. M. Clegg, Biochemistry, 1990, 29, 9029–9039 CrossRef CAS PubMed.
- A. Paul, R. Nanjunda and W. D. Wilson, Curr. Protoc., 2023, 3, e729 CrossRef CAS.
- M. Munde, A. Kumar, P. Peixoto, S. Depauw, M. A. Ismail, A. A. Farahat, A. Paul, M. V. Say, M.-H. David-Cordonnier, D. W. Boykin and W. D. Wilson, Biochemistry, 2014, 53, 1218–1227 CrossRef CAS.
- P. Mandal, D. Sahoo, S. Saha and J. Chowdhury, J. Phys. Chem. B, 2018, 122, 10279–10290 CrossRef CAS PubMed.
- A. Ghosh, M. Trajkovski, M. P. Teulade-Fichou, V. Gabelica and J. Plavec, Angew. Chem., Int. Ed., 2022, 61, e202207384 Search PubMed.
- S. Gupta, S. Aggarwal and M. Munde, ACS Omega, 2023, 8, 4554–4565 CrossRef CAS PubMed.
- S. Gupta, N. Tiwari and M. Munde, Sci. Rep., 2019, 9, 5891 CrossRef PubMed.
- C. Wei, G. Jia, J. Zhou, G. Han and C. Li, Phys. Chem. Chem. Phys., 2009, 11, 4025–4032 RSC.
- H. Han, D. R. Langley, A. Rangan and L. H. Hurley, J. Am. Chem. Soc., 2001, 123, 8902–8913 CrossRef CAS PubMed.
- T. M. Ou, Y. J. Lu, J. H. Tan, Z. S. Huang, K. Y. Wong and L. Q. Gu, ChemMedChem, 2008, 3, 690–713 CrossRef CAS PubMed.
- J. A. Schouten, S. Ladame, S. J. Mason, M. A. Cooper and S. Balasubramanian, J. Am. Chem. Soc., 2003, 125, 5594–5595 CrossRef CAS.
- Y. Wang, Q. C. Zheng, C. P. Kong, Y. Tian, J. Zhan, J. L. Zhang and H. X. Zhang, Mol. BioSyst., 2015, 11, 252–261 RSC.
- B. Islam, P. Stadlbauer, A. Gil-Ley, G. Pérez-Hernández, S. Haider, S. Neidle, G. Bussi, P. Banas, M. Otyepka and J. Sponer, J. Chem. Theory Comput., 2017, 13, 2458–2480 CrossRef CAS PubMed.
- M. Zgarbová, M. Otyepka, J. Šponer, F. Lankaš and P. Jurečka, J. Chem. Theory Comput., 2014, 10, 3177–3189 CrossRef.
- F. M. Aldakheel, S. A. Alduraywish and K. H. Dabwan, Sci. Rep., 2025, 15, 12764 CrossRef CAS PubMed.
- D. Herschlag and M. M. Pinney, Biochemistry, 2018, 57, 3338–3352 CrossRef CAS PubMed.
- M. Munde, A. Kumar, R. Nhili, S. Depauw, M.-H. David-Cordonnier, M. A. Ismail, C. E. Stephens, A. A. Farahat, A. Batista-Parra, D. W. Boykin and W. D. Wilson, J. Mol. Biol., 2010, 402, 847–864 CrossRef CAS PubMed.
- W. Wu, X. Hu, Z. Zeng, D. Wu and H. Li, J. Phys. Chem. B, 2023, 127, 874–883 CrossRef CAS.
- R. J. Weiss, P. L. Gordts, D. Le, D. Xu, J. D. Esko and Y. Tor, Chem. Sci., 2015, 6, 5984–5993 RSC.
- M. Farag and L. Mouawad, Nucleic Acids Res., 2024, 52, 3522–3546 CrossRef CAS.
| This journal is © the Owner Societies 2026 |