Open Access Article
Open Access ArticleCold argon plasmas for non-enzymatic digestion of macromolecules
Daniel McGill *ab,
Daniel Simon
*ab,
Daniel Simon bc,
Siva Ramadurai
bc,
Siva Ramadurai a,
Felicia Green
a,
Felicia Green a and
Zoltan Takats
a and
Zoltan Takats *abc
*abc
aThe Rosalind Franklin Institute, Building R113 Rutherford Appleton Laboratory, Harwell Campus, Didcot, Oxfordshire OX11 0QX, UK. E-mail: daniel.mcgill@rfi.ac.uk
bDepartment of Metabolism, Digestion, and Reproduction, Imperial College London, Hammersmith Campus, Du Cane Road, London, W12 0NN, UK. E-mail: z.takats@imperial.ac.uk
cInstitut für Funktionelle Genomik, University Regensburg, 9 Am Biopark, Regensburg 93053, Germany
First published on 2nd December 2025
Abstract
We have developed surface treatment techniques utilising radiofrequency current cold argon plasmas (RF-CAP) for non-enzymatic digestion of macromolecules, exemplified here with peptides and proteins of varying sizes. These plasmas are able to rapidly digest condensed-phase macromolecules with minimal sample preparation, producing predictable fragments visible by LC-MS analysis.
The relative size and complexity of macromolecules such as proteins, oligosaccharides, and oligoconjugates pose a challenge for unique identification by mass spectrometry.1–3 Two broad analytical approaches (colloquially, ‘top-down’ and ‘bottom-up’) are commonly used for identification and quantitation. Using proteins as an exemplar, top-down mass spectrometry-based proteomics aims to analyse native analytes whole, including both the parent mass and its constituent fragments, alongside any post-translational modifications (PTMs).4,5 By contrast, bottom-up proteomics typically utilises chemical6 or enzymatic7,8 digestions (typically trypsin, but also elastase, pepsin, etc) to break bonds within the molecule, rendering smaller peptides available for MS analysis. However, sample preparation required for bottom-up analysis is also non-trivial, requiring significant time, expense, and experience.
Non-enzymatic, non-liquid, and/or reagentless digestion and degradation approaches to macromolecular analysis can be utilised to increase analytical throughput. Current non-enzymatic degradation approaches include thermal decomposition/digestion (TDD)9 and irradiation with continuous wave lasers;10 other approaches include the use of nonthermal plasmas (also known as cold plasmas), which currently see use in the emerging field of plasma medicine. Cold plasma methods incorporate techniques such as argon plasma coagulation (APC), already commonplace in electrosurgery, with particular use in endoscopy.11,12 APC utilises a radiofrequency alternating current (through either monopolar or bipolar electrodes) to generate gas discharges, which in turn generate desirable thermal effects in tissue – these thermal effects can be used for haemostasis, devitalisation of tumours, and tissue reduction.13 One understudied side-product of nonthermal argon plasmas is the generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS), which can be utilised for reactions with various subtrates14 – generating these RONS particles using cold argon plasmas typically also produces metastable noble gas atoms and free elections, which can be exploited for use as a method for in situ digestion of biological macromolecules.
To generate radiofrequency cold argon plasma (RF-CAP) we utilised a custom handpiece (Ambimass, HU) with inlets for gas flow and current, which was in turn connected to a Valleylab Forcetriad electrosurgical unit (Medtronic, US) and an in-house argon supply at 0.1 bar. Below the corona pin, glass slides with substrates to be examined were placed on top of a grounding pad, acting as an insulator. With the handpiece activated a dielectric barrier discharge (DBD) was generated, forming a plasma of around 30 mm diameter (Fig. 1b). The standard was exposed for between 5 and 60 seconds, then dissolved in 50% acetonitrile or 0.1% formic acid and injected by direct infusion or LC-MS, respectively.
After exposure of leucine enkephalin (YGGFL) and bradykinin (RPPGFSPFR) peptide standards to RF-CAP for between 5 and 60 seconds, digestion products visible by direct infusion were annotated – these included ‘classical’ abc–xyz peptide fragments (particularly ‘c-‘, ‘z-‘, and ‘(z + 1)-type’ fragments, which we describe in this paper using ‘all-explicit’ notation as [cm + (n + 1)H]n+, [zm + nH]n+, and [zm + (n + 1)H]n+ respectively15), as well as neutral loss products such as [M–CO2 + H]+ (SI1.1, 1.2, and 1.3). The presence of ‘c-‘ and ‘z-type’ fragments suggests a mechanism reliant on free electron- or radical-mediated reactions, as in-spec fragmentation methods such as electron transfer dissociation (ETD) and electron capture dissociation (ECD) often produce these fragments through charge-remote fragmentation characteristic of odd-electron reactions.16,17
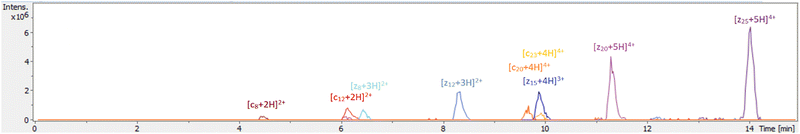
Applying RF-CAP on the timescales used for smaller peptides (5–60 seconds) on larger macromolecules did not produce easily identifiable peaks; additionally, direct infusion approaches suffered from significant ion suppression and loss of information from generated analytes. Consequently, for digestion of proteins, the plasma was applied with pulses of approximately 100 µs at a rate of 1 Hz (10% duty cycle) for 30 seconds, with the plasma-treated substrate then dissolved and injected by LC-MS onto a reversed-phase C18 column (such as for haemoglobin – see SI2) – this workflow afforded, among other analytes, a range of ‘c-‘, ‘z-‘, and ‘(z + 1)-type’ fragments ([cm + (n + 1)H]n+, [zm + nH]n+, and [zm + (n + 1)H]n+, respectively) (Fig. 2). These terminal ions were found to be replicable on other spectrometers across different timescales using the same LC-MS method (SI1.4). One such ion generated from ubiquitin, annotated as [c5 + 2H]+, was isolated and fragmented by CID, affording a series of ‘b-type’ ions corroborating the annotation (SI1.5).
By building an intensity curve across six concentrations, recording abundances of four product peaks using three biological replicates, we were able to determine that the workflow would reliably produce these products, among others; additionally, we found that the limit of detection for these products was reached when the concentration of ubiquitin was approximately 195 nanomolar (195 fmol µL−1) when using this workflow (SI1.6).
Digestion of ubiquitin and haemoglobin using the RF-CAP setup produced not only ‘c’, ‘z’, and ‘(z + 1)’ fragments (including from both haemoglobin subunits – SI2), but was noted to additionally produce a significant number of ‘a’, ‘b’, ‘x’, and ‘y’ ions ([am + (n − 1)H]n+, [bm + (n − 1)H]n+, [xm + (n − 1)H]n+, and [ym + (n + 1)H]n+, respectively – Fig. 3 and SI2). For ubiquitin and haemoglobin treated by RF-CAP, the sequence coverage was 100%; by comparison, while the sequence coverage for ubiquitin was 100% by ECD, it represented only 70% for haemoglobin subunit beta. The simultaneous fragmentation of both subunits with greater sequence coverage proves more flexible and powerful than ECD.
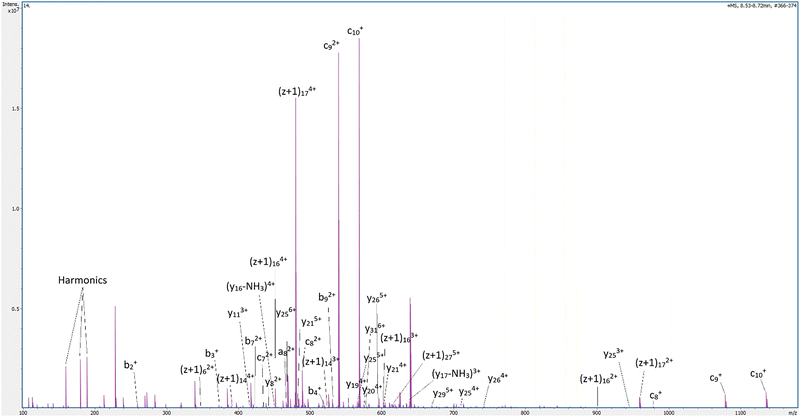 | ||
| Fig. 3 Annotated LC-MS spectrum (retention time 8.53–8.72 minutes) of ubiquitin after exposure to 1 Hz pulsed RF-CAP for 30 seconds showing multiple abc–xyz fragments, as well as several unknowns. | ||
Beyond the ‘abc–xyz’ terminal ions annotated, a great number of additional peaks not corresponding to these products (or a single neutral loss) were also present in plasma-exposed standards but not easily annotated, such as several multiply charged analytes across the 800–850 m/z range (SI1.7). Some of these peaks are believed to be internal ion products – which we here describe as ‘by’-type ions – stemming from serial amide cleavages (or N–Cα bond breaking and net loss of –NH followed by amide bond cleavage, or vice versa), as peaks corresponding to [H2N-RLIF-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]+, [H2N-QRLIF-C
O]+, [H2N-QRLIF-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]+, [H2N-QRLIFA-C
O]+, [H2N-QRLIFA-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]+, [H2N-QRLIFAGKQ-C
O]+, [H2N-QRLIFAGKQ-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]2+, and even [H2N-QRLIFAGKQLEDGRTLSDYNIQK-C
O]2+, and even [H2N-QRLIFAGKQLEDGRTLSDYNIQK-C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]5+ could be putatively annotated in ubiquitin plasma-treated samples (Table 1). These products are notable for a mass difference of +2.016 Da compared to typically calculated internal ions, possibly suggesting the presence of a –C
O]5+ could be putatively annotated in ubiquitin plasma-treated samples (Table 1). These products are notable for a mass difference of +2.016 Da compared to typically calculated internal ions, possibly suggesting the presence of a –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O terminus rather than the –C
O terminus rather than the –C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O+ characteristic of typical ‘b-type’ ions. We intend to develop tools to further examine the data with the hope of discovering additional exotic internal ion fragments.
O+ characteristic of typical ‘b-type’ ions. We intend to develop tools to further examine the data with the hope of discovering additional exotic internal ion fragments.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) annotated in ubiquitin LC-MS spectrum. Further internal ion annotations for ubiquitin can be found in SI2
O) annotated in ubiquitin LC-MS spectrum. Further internal ion annotations for ubiquitin can be found in SI2
| b–y internal ion | Formula | m/z | Error (ppm) |
|---|---|---|---|
| [RL]+/[LR]+ | C12H25N5O2 | 272.208 | −2.66 |
| [RLI]+ | C18H36N6O3Hf | 385.292 | −0.86 |
| [RLIF]+ | C27H45N7O4 | 532.361 | −0.69 |
| [QRLIF]+ | C32H53N9O6 | 660.419 | −0.03 |
| [QRLIFA]+ | C35H58N10O7 | 731.456 | −0.25 |
| [QRLIFAG]+ | C37H61N11O8 | 788.478 | −0.94 |
| [QRLIFAGK]2+ | C43H73N13O9 | 458.789 | −2.55 |
| [QRLIFAGKQ]2+/[QQRLIFAGK]2+ | C48H81N15O11 | 522.818 | −1.45 |
| [QRLIFAGKQL]2+ | C54H92N16O12 | 579.361 | −1.48 |
| [QRLIFAGKQLE]2+ | C59H99N17O15 | 643.882 | −1.57 |
| [QRLIFAGKQLED]2+ | C63H104N18O18 | 701.395 | −1.57 |
| [QRLIFAGKQLEDG]2+ | C65H107N19O19 | 729.906 | −1.3 |
Finally, we considered the application of RF-CAP to proteins with PTMs, specifically of phosphorylated proteins. Typically, characterisation of the phosphate groups is hindered by their lability; CID MS/MS of phosphopeptides (such as those produced by tryptic digestion) is known to cause neutral losses of HPO3, generating dominant fragments and hindering localisation of the PTM. Here, beta-casein – a model protein whose most common isoforms contain phosphorylated serine groups at positions 15, 17, 18, 19, and 35 – was exposed to RF-CAP for 30 seconds at a 10% duty cycle. Subsequent LC-MS analysis revealed [c7–16 + H]+ products with an intact phosphate group on Ser15 (SI1.8); higher-order products like [c17 + H]+ were not observed, likely due to the increased negativity from the phosphate groups conferred to the digest products rendering them not visible in positive mode, as has been previously reported in a study applying ECD to beta-casein.18
In conclusion, we have developed a novel method using a cold argon plasma for off-line digestion of proteins and peptides, which produces terminal and internal fragments from condensed-phase samples comparable to those produced by odd-electron fragmentation techniques such as ECD. This was demonstrated using a range of proteins and peptides, including ubiquitin, haemoglobin, and beta-casein; in the latter example, we demonstrated that the Ser15 phosphate group is kept intact during plasma digestion. The key advantage of this approach over comparable methods is a substantial reduction in sample preparation time and effort and the total elimination of reagents, including any use of enzymes; we intend in future to demonstrate the preservation of additional PTMs with this workflow. Furthermore, the technique may be used as an alternative to direct-infusion ECD for identification of proteins, with hyphenated liquid chromatography not only allowing for simultaneous characterisation of different subunits revealing significantly more sequence ions than with ECD – sometimes resulting in greater sequence coverage, as in the case of haemoglobin – as well as additional product ions not yet elucidated, including in complex mixtures.
Initially, this method could be used to develop new databases of reporter ions for targeted analysis (where generated peak lists are provided for an arbitrary number of target proteins, either puriss or in complex mixtures) – however, we intend to continue this research by fully elucidating the mechanisms of action, enabling rapid prediction of digestion products and, consequently, enabling total structural elucidation of proteins – including PTMs – from their plasma-exposed fragments (including any modifications), as well as the high-throughput imaging of proteins and other macromolecules in condensed-phase samples.
The authors acknowledge The Rosalind Franklin Institute, funded by the UK Research and Innovation, Engineering and Physical Sciences Research Council. Thank you to Logan Mackay for assistance with the Bruker solariX FT-ICR.
Conflicts of interest
DM, DS, and ZT are co-inventors on a patent application related to the findings of this study (publication number WO/2024/256813).Data availability
Data for this article is available at SciDB: https://doi.org/10.57760/sciencedb.29139.The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: (1) experimental and data; (2) annotated peak lists for Haemoglobin, Beta-casein, Ubiquitin, and Angiotensin I. See DOI: https://doi.org/10.1039/d5cc06333g.
Notes and references
- E. J. Dupree, M. Jayathirtha, H. Yorkey, M. Mihasan, B. A. Petre and C. C. Darie, A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field, Proteomes, 2020, 8(3), 1–26, DOI:10.3390/PROTEOMES8030014.
- S. Cristoni and L. R. Bernardi, Development of New Methodologies for the Mass Spectrometry Study of Bioorganic Macromolecules, Mass Spectrom. Rev., 2003, 22(6), 369–406, DOI:10.1002/mas.10062.
- L. Konermann, S. Vahidi and M. A. Sowole, Mass Spectrometry Methods for Studying Structure and Dynamics of Biological Macromolecules, Anal. Chem., 2014, 86(1), 213–232, DOI:10.1021/ac4039306.
- J. A. Melby, D. S. Roberts, E. J. Larson, K. A. Brown, E. F. Bayne, S. Jin and Y. Ge, Novel Strategies to Address the Challenges in Top-Down Proteomics, J. Am. Soc. Mass Spectrom., 2021, 32(6), 1278, DOI:10.1021/JASMS.1C00099.
- A. D. Catherman, O. S. Skinner and N. L. Kelleher, Top Down Proteomics: Facts and Perspectives, Biochem. Biophys. Res. Commun., 2014, 445(4), 683, DOI:10.1016/J.BBRC.2014.02.041.
- C. Harkin, K. W. Smith, F. L. Cruickshank, C. Logan Mackay, B. Flinders, R. M. A. Heeren, T. Moore, S. Brockbank and D. F. Cobice, On-Tissue Chemical Derivatization in Mass Spectrometry Imaging, Mass Spectrom. Rev., 2022, 41(5), 662–694, DOI:10.1002/MAS.21680.
- B. Cillero-Pastor and R. M. A. Heeren, Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging for Peptide and Protein Analyses: A Critical Review of on-Tissue Digestion, J. Proteome Res., 2014, 13(2), 325–335, DOI:10.1021/pr400743a.
- R. M. Miller and L. M. Smith, Overview and Considerations in Bottom-up Proteomics, Analyst, 2023, 148(3), 475–486, 10.1039/D2AN01246D.
- F. Basile, S. Zhang, S. K. Kandar and L. Lu, Mass Spectrometry Characterization of the Thermal Decomposition/Digestion (TDD) at Cysteine in Peptides and Proteins in the Condensed Phase, J. Am. Soc. Mass Spectrom., 2011, 22(11), 1926–1940, DOI:10.1007/s13361-011-0222-9.
- R. Zhou and F. Basile, Plasmonic Thermal Decomposition/Digestion of Proteins: A Rapid On-Surface Protein Digestion Technique for Mass Spectrometry Imaging, Anal. Chem., 2017, 89(17), 8704–8712, DOI:10.1021/acs.analchem.7b00430.
- G. Farin and K. E. Grund, Technology of Argon Plasma Coagulation with Particular Regard to Endoscopic Applications, Endosc. Surg. Allied Technol., 1994, 2(1), 71–77 CAS.
- H. Manner, Argon Plasma Coagulation Therapy, Curr. Opin. Gastroenterol., 2008, 24(5), 612–616, DOI:10.1097/MOG.0B013E32830BF825.
- J. W. Birk and A. Kaur, Argon Plasma Coagulation, GMS Krankenhaushygiene Interdisziplinar, 2008, 3 (1), 91–99 DOI:10.1007/978-1-59745-044-7_6.
- P. Lamichhane, T. R. Acharya, N. Kaushik, L. N. Nguyen, J. S. Lim, V. Hessel, N. K. Kaushik and E. H. Choi, Non-Thermal Argon Plasma Jets of Various Lengths for Selective Reactive Oxygen and Nitrogen Species Production, J. Environ. Chem. Eng., 2022, 10(3), 107782, DOI:10.1016/J.JECE.2022.107782.
- I. K. Chu, C.-K. Siu, J. K.-C. Lau, W. K. Tang, X. Mu, C. K. Lai, X. Guo, X. Wang, N. Li, Y. Xia, X. Kong, H. B. Oh, V. Ryzhov, F. Tureček, A. C. Hopkinson and K. W. M. Siu, Proposed Nomenclature for Peptide Ion Fragmentation, Int. J. Mass Spectrom., 2015, 390, 24–27, DOI:10.1016/j.ijms.2015.07.021.
- H. J. Cooper, K. Håkansson and A. G. Marshall, The Role of Electron Capture Dissociation in Biomolecular Analysis, Mass Spectrom. Rev., 2005, 24(2), 201–222, DOI:10.1002/mas.20014.
- B. Paizs and S. Suhai, Fragmentation Pathways of Protonated Peptides, Mass Spectrom. Rev., 2005, 24(4), 508–548, DOI:10.1002/mas.20024.
- B. Chen, X. Guo, T. Tucholski, Z. Lin, S. McIlwain and Y. Ge, The Impact of Phosphorylation on Electron Capture Dissociation of Proteins: A Top-down Perspective, J. Am. Soc. Mass Spectrom., 2017, 28(9), 1805–1814, DOI:10.1007/s13361-017-1710-3.
| This journal is © The Royal Society of Chemistry 2026 |