Open Access Article
Open Access ArticleEnzyme cascade reactions encapsulated in a liposome compartment with size-limited molecular transport
Shiwei Zhanga,
Peng Lin a,
Futa Komatsubaraa,
Eiji Nakataa and
Takashi Morii
a,
Futa Komatsubaraa,
Eiji Nakataa and
Takashi Morii *ab
*ab
aInstitute of Advanced Energy, Kyoto University, Uji, Kyoto 611-0011, Japan
bDepartment of Health and Nutrition, Kyoto Koka Women's University, Ukyo-ku, Kyoto 615-0882, Japan. E-mail: morii.takashi.68s@st.kyoto-u.ac.jp
First published on 2nd January 2026
Abstract
We designed a liposome-coated DNA origami compartment to encapsulate a defined number of cascade enzymes with retained cofactors. Confinement enhanced cascade reaction efficiency particularly at low cofactor levels. Efficiency gains plateaued at higher NADH concentrations, indicating that intermediate diffusion and enzyme spatial organization influence cascade performance in confined environments.
Cellular metabolic pathways face many challenges in organizing individual enzymatic reactions, including slow turnover, loss of volatile intermediates, formation of toxic by-products, and competition for metabolites.1 Cells overcome these limitations by compartmentalizing biomacromolecules, including enzymes, within organelles such as the nucleus, endoplasmic reticulum, and Golgi complex. These compartments create distinct microenvironments with specific pH values, ionic strengths, and polarities, thereby facilitating cascade reactions.2 Organelle membranes act as selective barriers, while confined spaces such as carboxysomes and lysosomes3 are thought to enhance enzyme activity by increasing the local concentration of substrates and intermediates, thereby promoting collision frequency in diffusion-driven processes.4 In addition, the specific microenvironment and the spatial organization of multienzyme complexes enable substrate channelling, allowing direct metabolite transfer between enzymes, minimizing diffusion into the cytoplasm, reducing cross-talk with other pathways, and ultimately enhancing both efficiency and selectivity.
Synthetic compartments offer powerful platforms for mimicking such enzymatic organization by encapsulating defined metabolic pathways.5 However, quantitatively evaluating compartmentalized enzyme cascade reactions requires compartments of controlled size and composition, as well as the precise number and arrangement of enzymes. Previous studies have examined proximity effects on substrate transfer7 using enzymes immobilized on the DNA scaffold.6 Nevertheless, it remains challenging to directly verifying how compartmentalization and spatial organization contribute to the efficiency of synthetic metabolic pathways, largely due to the difficulty of assembling a defined number of cascade enzymes within compartments of uniform size.
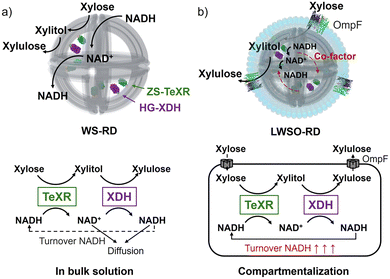
To address this challenge, we previously developed a transporting liposomal compartment with an internal DNA origami wireframe skeleton (WS), equipped with the bacterial membrane transporter, OmpF, which selectively permits passage of molecules smaller than 600 Da.8 In the present study, we employed this platform to construct a two-step xylose cascade composed of Talaromyces emersonii xylose reductase (TeXR),9 which reduces xylose to xylitol using NADH, and xylitol dehydrogenase (XDH),10 which oxidizes xylitol to xylulose using NAD+. In this system, the substrate xylose, the intermediate xylitol, and the product xylulose would freely diffuse through OmpF, while the essential cofactors NADH and NAD+ remain encapsulated (Fig. 1).
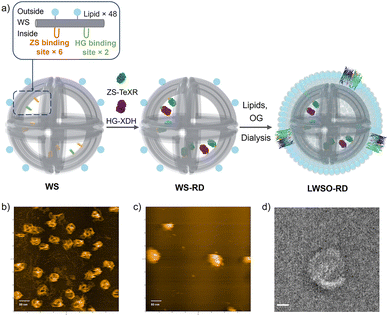
In earlier work, we assembled a defined number of cascade enzymes on DNA scaffolds by fusing Pichia stipitis XR11 to the modular adaptor ZF-SNAP (ZS),12 which consists of a monomeric zinc finger protein (zif268)13 linked to a SNAP tag protein,14 enabling XR-XDH cascade reaction on 2D or 3D DNA scaffolds.12 However, ZS-XR exhibited marked loss of activity under the liposome formation condition. To overcome this limitation, we replaced XR with the thermostable TeXR to construct ZS-TeXR (Fig. S1). In parallel, XDH was fused to HG, a modular adaptor composed of a dimeric leucine zipper protein (GCN4)15 and a HALO tag protein16 to construct HG-XDH. This strategy enabled stable encapsulation of a defined number of enzymes, allowing us to evaluate cascade performance within a synthetic transporter-equipped compartment.
ZS-TeXR specifically bound to the zif268 recognition sequence and reacted with the SNAP-tag substrate benzylguanine (BG) embedded within the zif268 DNA sequence. Unlike ZS-XR, the activity of ZS-TeXR remained stable for 48 hours (Fig. S2), although its reaction rate was significantly lower than that of ZS-XR in the presence of 500 µM NADH. Kinetic analysis revealed that ZS-TeXR exhibited a Km for xylose comparable to ZS-XR17 (Fig. S3), while the Km for NADH (3.15 mM) was markedly higher than that of ZS-XR (0.15 mM). Although ZS-TeXR exhibited a higher reaction rate with NADPH than with NADH due to its lower Km for NADPH than for NADH (Fig. S4), NAD+ was much preferred to NADP+ as a cofactor for XDH (Fig. S5). Therefore, NADH was used as the initial cofactor in the ZS-TeXR/HG-XDH cascade reaction.
Based on these parameters, the DNA origami skeleton WS was designed with six binding sites for ZS-TeXR and two for HG-XDH (Fig. 2a, b and Fig. S6). DNA strands were modified with BG for ZS-TeXR (Fig. S6) and with chlorohexane (CH) for HG-XDH (Fig. S6). After incubation of WS with ZS-TeXR and/or HG-XDH at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio, gel filtration removed unbound proteins and yielded purified WS assembled with both ZS-TeXR and HG-XDH (WS-RD). Enzyme loading was quantified by SDS-PAGE because an AFM imaging was limited by the 3D shape of WS (Fig. 2c and Fig. S7). In the individually assembled system, there are 5 ± 0.2 ZS-TeXR monomers and 1.9 ± 0.3 HG-XDH dimers per WS, WS-R and WS-D, respectively (Fig. S8 and S9). In the co-assembled system, 5.1 ± 0.1 ZS-TeXR monomers and 1.9 ± 0.2 HG-XDH dimers were loaded onto each WS-RD (Fig. S10). No significant difference was observed in the enzyme loading numbers between the individually assembled (WS-R: 5 ± 0.1; WS-D: 2 ± 0.1) and co-assembled systems, indicating that the simultaneous immobilization of ZS-TeXR and HG-XDH did not interfere with their binding efficiency or the structural integrity of the WS scaffold.
2 molar ratio, gel filtration removed unbound proteins and yielded purified WS assembled with both ZS-TeXR and HG-XDH (WS-RD). Enzyme loading was quantified by SDS-PAGE because an AFM imaging was limited by the 3D shape of WS (Fig. 2c and Fig. S7). In the individually assembled system, there are 5 ± 0.2 ZS-TeXR monomers and 1.9 ± 0.3 HG-XDH dimers per WS, WS-R and WS-D, respectively (Fig. S8 and S9). In the co-assembled system, 5.1 ± 0.1 ZS-TeXR monomers and 1.9 ± 0.2 HG-XDH dimers were loaded onto each WS-RD (Fig. S10). No significant difference was observed in the enzyme loading numbers between the individually assembled (WS-R: 5 ± 0.1; WS-D: 2 ± 0.1) and co-assembled systems, indicating that the simultaneous immobilization of ZS-TeXR and HG-XDH did not interfere with their binding efficiency or the structural integrity of the WS scaffold.
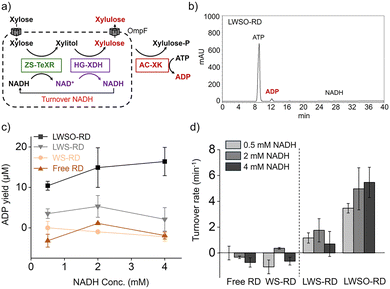
In this cascade (Fig. 1), the efficiency can be directly estimated by quantifying xylulose production by XDH. To quantify xylulose, we employed a coupled reaction with xylulose kinase (XK), which converts xylulose to xylulose-5-phosphate by consuming ATP.17 The amount of ADP yielded from this reaction is the same as the amount of xylulose (Fig. S11a). Verification experiments confirmed that ADP yield correlated closely with NADH production by HG-XDH, with >93% conversion efficiency (Fig. S11). Thus, XK provided a reliable readout of cascade efficiency.
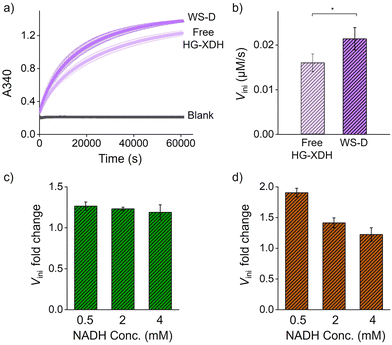
We next evaluated the cascade efficiency of free enzymes and enzymes assembled on WS by monitoring xylulose production (Fig. 1a). Comparison of free and scaffolded enzymes showed modest but reproducible activity enhancement. The initial reaction rate (Vini) of HG-XDH (10 nM) on WS-D (5 nM) was approximately 1.3-fold higher than that of free HG-XDH (10 nM) (Fig. 3a and b). Similarly, ZS-TeXR (25 nM) on WS-R (5 nM) displayed a 1.2-fold increase in Vini over free ZS-TeXR at different NADH concentrations ranging from 500 µM to 4 mM (Fig. 3c and Fig. S13–S15). When both enzymes were co-assembled (WS-RD), cascade activity increased by 1.5-fold relative to free enzymes (Fig. 3d and Fig. S16–S18). These results are consistent with our previous findings that the enzyme catalyzing hydrophilic substrates exhibit higher reaction rates on DNA scaffolds.18 The cascade reaction efficiency was particularly enhanced at low cofactor levels (Fig. 3d).
To assess the effect of compartmentalization, WS-RD was encapsulated within liposomes containing NADH. The liposome-coated WS-RD (LWS-RD) was purified by density-gradient ultracentrifugation (Fig. S19).8 The LWS-RD fractions were then incubated with OmpF to form LWSO-RD and analyzed by TEM. TEM images confirmed liposome diameter of 77.8 ± 10.4 nm (n = 172), consistent with the designed diameter of 75 nm (Fig. 2d and Fig. S20).8 Using our previously established protocol,8 we quantified the encapsulation yields for LWS, LWS-RD, and LWSO-RD as 83 ± 4%, 79 ± 5%, and 78 ± 4%, respectively (Fig. S21), The high and similar encapsulation efficiencies across all constructs, regardless of the presence or absence of anchored proteins, demonstrate that our method is robust and broadly applicable.
Three types of liposomes (LWS-RD and LWSO-RD) were prepared, each encapsulating different concentrations of NADH (500 µM, 2 mM, and 4 mM). Initial internal NADH concentrations of 500 µM, 2 mM, and 4 mM in LWS-RD (5 nM) or LWSO-RD (5 nM) correspond to effective concentration of 0.35, 1.4, and 2.7 µM NADH, respectively, in 100 µL of the cascade reaction mixture (for the details of effective NADH concentration, see “estimation of the effective NADH concentration in the bulk solution” in Methods). Successful internalization of NADH into the liposomes was confirmed by variations in peak intensity that correlated with the initial NADH concentrations (see Fig. S22 and Table S4).
Cascade reactions of free ZS-TeXR and HG-XDH (free RD), WS-RD, LWS-RD, and LWSO-RD (Fig. 1b) were compared under equivalent enzyme and NADH conditions (Fig. 4a). All reaction systems contained an equivalent number of moles of ZS-TeXR and HG-XDH. The initial NADH concentrations were equivalent to those of the liposome-encapsulated NADH.
Each reaction was analyzed by HPLC to quantify the ADP produced at steady state after 10 hours (Fig. 4b and Fig. S23–S25). Monitoring of NADH absorbance and ADP production revealed that only LWSO-RD supported efficient cascade activity (Fig. 4c). At a low internal concentration of NADH (500 µM), the ADP yield was 10 to 15 µM for LWSO-RD. No significant activity was detected for free RD or WS-RD (Fig. 4d). The lack of activity for LWS-RD indicates that OmpF effectively introduces xylose into the LWSO-RD compartment. These results suggest that the cascade reaction is significantly more efficient under compartmentalization than in a bulk solution when the effective concentration of the cofactor NADH is low. This can be attributed to the microenvironment within the liposomes, where local enzyme and NADH concentrations are significantly higher. This accelerates the cascade reaction compared to the free enzyme system.
However, increasing the internal NADH concentration to 2 or 4 mM did not improve the ADP yield in LWSO-RD further (Fig. 4c and Fig. S26). The same trend was observed in the non-liposomal WS-RD reaction (Fig. 3d). Since the Michaelis–Menten kinetics of the ZS-TeXR reaction with 4 mM NADH showed no inhibition at this concentration (Fig. S3), the plateau observed in the efficiency of the cascade reaction could not be explained by ZS-TeXR inhibition at higher internal NADH concentrations. In LWSO-RD, OmpF permits bidirectional flux of xylose, xylitol, and xylulose, though not as freely as in WS-RD. Therefore, intermediates formed in LWSO-RD are prone to escape unless they are transferred directly from ZS-TeXR to HG-XDH. Consequently, the turnover rate of the second step, catalyzed by HG-XDH, remained relatively unchanged. This resulted in little variation in cascade efficiency across different initial NADH concentrations (Fig. 4d). Our results suggest that tuning the stoichiometry and/or spacing of ZS-TeXR and HG-XDH would further improve the efficiency of the cascade reaction in LWSO-RD.
In summary, defined numbers of xylose cascade enzymes, ZS-TeXR and HG-XDH, were successfully assembled on a DNA origami scaffold and encapsulated within liposomes. Incorporation of the size-selective membrane transporter protein OmpF enabled internal retention of essential cofactors while allowing free diffusion of substrate and intermediate. The compartmentalized system exhibited markedly enhanced cascade efficiency compared to non-compartmentalized enzymes, particularly at low NADH concentrations. The observed plateau in efficiency at higher NADH levels suggests that enzyme stoichiometry and spacing critically influence the performance of the cascade, even within confined compartments. It should be noted that there are potential limitations to our enzyme anchoring strategy. Although the modular adaptor provides covalent stability and a high assembly yield, it may restrict the conformational flexibility of enzymes tethered to the DNA scaffold in some cases. This could lead to suboptimal orientations and contribute to the activity plateau observed at high enzyme densities. This effect is likely enzyme-dependent, as the catalytic performance of ZS-TeXR was more constrained than that of HG-XDH, possibly due to the structural sensitivity of TeXR itself. The current arrangement of enzymes is not a fixed limitation, but rather a deliberate design choice. Both stoichiometry and inter-enzyme spacing can be easily modified by changing the number and locations of binding sites through staple strand redesign. This inherent programmability highlights the adaptability of our platform for systematically investigating how spatial organization influences cascade reactions. Future studies manipulating inter-enzyme spacing and stoichiometry in our compartmentalized systems will further illuminate the design principles of efficient enzymatic cascades. These studies will also advance applications in synthetic biology and biocatalysis.
This work was supported by JSPS KAKENHI, Grant Numbers 23H02083 and 23K26776 (T. M.), 24K17787 (P. L.), 24H01129 and 24K01629 (E. N.), Japan. The TEM measurements in this study were supported by the Kyoto University Nano Technology Hub in the “Nanotechnology Platform Project,” sponsored by MEXT, Japan. The Analysis and Development System for Advanced Materials (ADAM) at the Research Institute for Sustainable Humanosphere (RISH) of Kyoto University also supported this study.
Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cc06049d.Notes and references
- (a) A. H. Chen and P. A. Silver, Trends Cell Biol., 2012, 22, 662–670 CrossRef CAS PubMed; (b) H. Eichelmann, E. Talts, V. Oja, E. Padu and A. Laisk, J. Exp. Bot, 2009, 60, 4077–4088 CrossRef CAS PubMed; (c) K. Jørgensen, A. V. Rasmussen, M. Morant, A. H. Nielsen, N. Bjarnholt, M. Zagrobelny, S. Bak and B. L. Møller, Curr. Opin. Plant Biol., 2005, 8, 280–291 CrossRef; (d) B. A. Manjasetty, J. Powlowski and A. Vrielink, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 6992–6997 CrossRef CAS; (e) Y. Liu, L. L. Beer and W. B. Whitman, Trends Microbiol., 2012, 20, 251–258 CrossRef CAS PubMed.
- (a) S. Hurtley, Science, 2009, 326, 1205 CrossRef CAS PubMed; (b) C. A. Kerfeld, M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby and T. O. Yeates, Science, 2005, 309, 936–938 CrossRef CAS PubMed; (c) I. A. Berlatzky, A. Rouvinski and S. Ben-Yehuda, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14136–14140 CrossRef CAS; (d) A. Küchler, M. Yoshimoto, S. Luginbühl, F. Mavelli and P. Walde, Nat. Nanotechnol., 2016, 11, 409–420 CrossRef.
- B. H. Lipshutz, N. A. Isley, J. C. Fennewald and E. D. Slack, Angew. Chem., Int. Ed., 2013, 52, 10952–10958 CrossRef CAS PubMed.
- (a) Y. A. Takagi, D. H. Nguyen, T. B. Wexler and A. D. Goldman, J. Mol. Evol., 2020, 88, 598–617 CrossRef CAS PubMed; (b) S. Koga, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2011, 3, 720–724 CrossRef CAS PubMed; (c) J. P. Schrum, T. F. Zhu and J. W. Szostak, Cold Spring Harb. Perspect. Biol., 2010, 2, a002212 Search PubMed.
- (a) O. Staufer, M. Schröter, I. Platzman and J. P. Spatz, Small, 2020, 16, 1906424 CrossRef CAS PubMed; (b) C. D. Reinkemeier, G. E. Girona and E. A. Lemke, Science, 2019, 363, eaaw2644 CrossRef CAS; (c) E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC; (d) B. C. Buddingh and J. C. M. Van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef CAS; (e) S. Kumar, M. Karmacharya, I. J. Michael, Y. Choi, J. Kim, I. Kim and Y. K. Cho, Nat. Catal., 2021, 4, 763–774 CrossRef CAS.
- J. Fu, M. Liu, Y. Liu, N. W. Woodbury and H. Yan, J. Am. Chem. Soc., 2012, 134, 2023 Search PubMed.
- (a) Z. G. Wang, O. I. Wilner and I. Willner, Nano Lett., 2009, 9, 4098–4102 CrossRef CAS; (b) L. Sun, Y. Gao, Y. Xu, J. Chao, H. Liu, L. Wang, D. Li and C. Fan, J. Am. Chem. Soc., 2017, 139, 17525–17532 CrossRef CAS; (c) G. Ke, M. Liu, S. Jiang, X. Qi, Y. R. Yang, S. Wooten, F. Zhang, Z. Zhu, Y. Liu, C. J. Yang and H. Yan, Angew. Chem., 2016, 128, 7609–7612 CrossRef; (d) V. Linko, M. Eerikäinen and M. A. Kostiainen, Chem. Commun., 2015, 51, 5351–5354 RSC.
- (a) S. Zhang, E. Nakata, P. Lin and T. Morii, Chem. – Eur. J., 2023, 29, e202302093 CrossRef CAS PubMed; (b) S. Zhang, P. Lin, F. Komatsubara, E. Nakata and T. Morii, ChemBioChem, 2025, 26, e202401041 CrossRef CAS.
- S. Fernandes, M. G. Tuohy and P. G. Murray, J. Biosci., 2009, 34, 881–890 CrossRef CAS.
- S. Watanabe, T. Kodaki and K. Makino, J. Biol. Chem., 2005, 280, 10340–10349 CrossRef CAS.
- S. Watanabe, A. Abu Saleh, S. P. Pack, N. Annaluru, T. Kodaki and K. Makino, Microbiology, 2007, 153, 3044–3054 CrossRef CAS PubMed.
- (a) T. A. Ngo, E. Nakata, M. Saimura and T. Morii, J. Am. Chem. Soc., 2016, 138, 3012–3021 CrossRef CAS; (b) P. Lin, H. Dinh, Y. Morita, E. Nakata and T. Morii, Adv. Funct. Mater., 2023, 33, 2215023 CrossRef CAS.
- (a) N. P. Pavletich and C. O. Pabo, Science, 1991, 252, 809 CrossRef CAS; (b) E. Nakata, F. F. Liew, C. Uwatoko, S. Kiyonaka, Y. Mori, Y. Katsuda, M. Endo, H. Sugiyama and T. Morii, Angew. Chem., Int. Ed., 2012, 51, 2421–2424 CrossRef CAS.
- (a) A. Keppler, S. Gendreizig, T. Gronemeyer, H. Pick, H. Vogel and K. Johnsson, Nat. Biotechnol., 2003, 21, 86 CrossRef CAS; (b) E. Nakata, H. Dinh, T. A. Ngo, M. Saimura and T. Morii, Chem. Commun., 2014, 51, 1016–1019 RSC; (c) T. M. Nguyen, E. Nakata, M. Saimura, H. Dinh and T. Morii, J. Am. Chem. Soc., 2017, 139, 8487–8496 CrossRef CAS; (d) T. M. Nguyen, E. Nakata, Z. Zhang, M. Saimura, H. Dinh and T. Morii, Chem. Sci., 2019, 10, 9315–9325 RSC; (e) Z. Zhang, E. Nakata, H. Dinh, M. Saimura, A. Rajendran, K. Matsuda and T. Morii, Chem. – Eur. J., 2021, 27, 18118–18128 CrossRef CAS PubMed.
- T. E. Ellenberger, C. J. Brandl, K. Struhl and S. C. Harrison, Cell, 1992, 71, 1223–1237 CrossRef CAS.
- C. G. England, H. Luo and W. Cai, Bioconjugate Chem., 2015, 26, 975–986 CrossRef CAS.
- P. Lin, H. Dinh, E. Nakata and T. Morii, Chem. Commun., 2021, 57, 11197–11200 RSC.
- P. Lin, T. Hayashi, H. Dinh, E. Nakata, M. Kinoshita and T. Morii, ACS Appl. Mater. Interfaces, 2025, 17, 15775–15792 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |