Open Access Article
Open Access ArticleGamma-FIT-PNAs as sensitive RNA probes
Manoj Kumar Gupta,
Salam Maree and
Eylon Yavin *
*
Institute for Drug Research, School of Pharmacy, The Hebrew University of Jerusalem, Hadassah Ein-Kerem, Jerusalem 91120, Israel. E-mail: manojkumar.gupta@mail.huji.ac.il; eylony@ekmd.huji.ac.il
First published on 20th January 2026
Abstract
A variety of oligonucleotide-based probes have been developed for specific and selective sensing of RNA and DNA. Among these, FIT-PNAs (forced intercalation-peptide nucleic acids) and FIT probes (DNA- and RNA-based sensors) have been studied for a variety of RNA biomarkers in cell culture and tissues, and in vivo. FIT-PNAs and FIT probes are RNA/DNA sensors that exhibit fluorescence upon sequence-specific RNA/DNA hybridization. Several synthetic approaches have been successfully applied to increase the brightness and selectivity of these molecules, including the introduction of cyclopentane (cp) modified PNA monomers (cpPNA) as well as locked nucleic acids (LNAs—for FIT probes). In this report, we have explored the biophysical properties of FIT-PNAs that are modified with gamma-L-serine PNAs (γPNAs). We found that introducing a single γ-PNA flanking the fluorophore (BisQ) in the FIT-PNA sequence is sufficient to achieve a 46-fold increase in fluorescence for the PNA:RNA duplex, similarly to cpPNA. Interestingly, when two γ-PNAs flank BisQ on both sides, a significant increase in RNA affinity is observed (over an 8 °C increase in melting temperature, Tm). Altogether, γ-PNAs are a beneficial chemical modification that leads to brighter FIT-PNAs with improved binding affinities to targeted RNA.
Introduction
Peptide nucleic acids (PNAs), discovered over three decades ago, are synthetic nucleic acid analogs obtained by replacing the ribose sugar and phosphate groups of the DNA/RNA backbone with a pseudopeptide backbone.1,2 PNAs contain repeating units of uncharged N-(2-aminoethyl) glycine (aeg) units as a backbone. Nucleobases such as adenine, thymine, cytosine, and guanine (A/T/C/G) are attached via a carboxy-methylene linker to the secondary amine of the PNA backbone (Fig. 1a). The non-natural backbone of PNA contributes to its unique enzymatic stability while maintaining resistance to both proteases/peptidases and nucleases.3 Furthermore, PNAs can bind to the target DNA or RNA with high affinity and specificity via Watson–Crick-Franklin base pairing even in the presence of low salt concentrations. Due to these properties, PNAs have been extensively used to target both single-stranded and duplex DNA and RNA for gene regulation, and other diverse biomedical applications like RNA biosensing,4,5 polymerase chain reaction (PCR),6 and genomic barcoding.7 However, classical PNAs are limited by numerous challenges such as poor aqueous solubility,8 aggregation, non-specific interactions with macromolecules, low cellular uptake, and fast in vivo elimination half-life.9 Hence, efforts have been made to overcome these challenges for PNAs through chemical modifications to the PNA backbone.10 Several appended ring systems on the PNA backbone have shown favorable binding and cellular uptake. Examples include cyclopentane- (cp)11 and THF-modified backbones (thyclotides),12 as well as cis-cyclohexyl13 and pyrrolidinyl backbones.14–16 In addition, since PNAs are synthesized by solid-phase peptide chemistry, a variety of cell-penetrating peptides have been conjugated to these oligomers for improving solubility and cellular uptake.17,18 Some notable examples include Tat,19 NLS,20,21 and poly-L-lysine,22,23 as well as bacteria-targeting peptides.24 For the same reason, further chemical modifications on the PNA backbone have been made at the α-, β- or γ-position, resulting in numerous novel PNAs, some with higher specificity to duplex DNA16,25–27 and duplex RNA.18,28–32 Chemical diversity at the gamma position is further exemplified by the characteristics of these modifications that include a negative charge,33,34 a positive charge,35–38 and hydrophilic39 and hydrophobic residues.40–42 Notably, γ-MP (mini-PEG) PNA25 and γ-L-serine PNA43 (Fig. 1b) have been extensively studied in a variety of applications, such as antisense44–47 and antigene25,48,49 therapeutics as well as biosensing.50–55FIT-PNAs (forced intercalation-peptide nucleic acids)56–64 are a type of PNA-based fluorescent probes in which a dye molecule (e.g. thiazole orange, a.k.a. the surrogate base) is built directly into the PNA strand (instead of a nucleobase). When the probe binds its complementary DNA or RNA target, the dye intercalates between the bases, leading to a strong increase in fluorescence. Forced-intercalation (FIT) probes have since been extended to 2′-O-methyl RNA and DNA backbones.65–71 Incorporation of a locked nucleic acid (LNA)—a sugar-modified, conformationally restricted nucleotide—flanking the FIT surrogate base quinoline blue (QB) or bis-quinoline (BisQ) in PNA, markedly enhances probe brightness.67 More recently, we demonstrated that placing a cyclopentane-modified monomer—cpT (Fig. 1c) or cpC—adjacent to the BisQ-monomer (Fig. 1d), in the FIT-PNA oligomer, particularly at the 3′ position, further increases -the brightness and quantum yield.72 These optimized probes were successfully applied for the detection of the highly expressed long non-coding RNA FLJ22447 in ovarian cancer cells.73
It has been reported that the γ-(L)-serine-modified PNA has been shown to increase binding affinity to complementary DNA/RNA, presumably by a preformed alpha-helical fold of the single-stranded PNA.43 As such, we hypothesized that both the rigidity of the γ-PNA monomer and its higher affinity to complementary RNA, as corroborated by melting temperature (Tm) analysis,74 would result in brighter FIT-PNA probes as RNA sensors. This report examines the impact of chemical modification at the gamma position in FIT-PNA-based probes, focusing on biophysical properties with complementary RNA, including thermal melting, fluorescence emission, mismatch discrimination, and overall cellular uptake in ovarian cancer cells (OVCA433).
Results and discussion
Design and synthesis of gamma-L-serine FIT-PNA oligomers
In this study, we have designed and synthesized a series of γ-L-serine-modified FIT-PNAs based on an 11-mer model system (Table 1). Four D-lysine residues were attached at the C-terminus to afford water solubility and cellular uptake of these molecules.57,61,73 In our previous studies, the incorporation of a cyclopentane (cp) moiety adjacent to BisQ (the fluorophore, a.k.a. the surrogate base) revealed that the position of the modification/s affects the (enhanced) fluorescence emission upon binding to complementary RNA, likely due to increased structural stability and rigidity of the formed duplex.72,73 In those studies, cpT and cpC bases were introduced at either side of BisQ. In general, the introduction of cpT at the 3′ position to BisQ was effective in increasing quantum yields and brightness (BR) compared to the unmodified FIT-PNA. The addition of cpC or cpT at the 5′ position to BisQ had a very modest effect on the observed photophysical parameters.72 Accordingly, in the present study, we employed these γ-modifications, which are known to improve duplex stability and increase water solubility.43| S. no. | ID | FIT-PNA (C–N terminus) | Cal. M.W. | Obs. M.W. |
|---|---|---|---|---|
| 4KD: 4-(D)-lysine, with the underline denoting the gamma-(L)-serine modification and cpT indicating the cyclopentane T-modification in the FIT-PNA oligomer. | ||||
| 1 | aeg-BisQ | 4KD ATA CAT BisQ CA AC | 3619.8 [M + H]+ | 3619.9 |
| 2 | aeg-cpT-BisQ | 4KD ATA CA cpT BisQ CA AC | 3657.7 [M]+ | 3657.7 |
| 3 | (γ-Ser)1-BisQ | 4KD ATA CA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) BisQ CA AC BisQ CA AC |
3649.8 [M + H]+ | 3650.0 |
| 4 | (γ-Ser)2-BisQ | 4KD ATA CA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) BisQ BisQ ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) A AC A AC |
3679.8 [M + H]+ | 3680.6 |
| 5 | (γ-Ser)5-BisQ | 4KD A![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) A A ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) A A![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) BisQ C BisQ C![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) A A![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) |
3768.9 [M]+ | 3766.4 |
| 6 | (γ-Ser)10-BisQ | 4KD ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) BisQ BisQ ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) |
3919.0 [M]+ | 3918.4 |
We therefore introduced the γ-modified PNA monomers at different positions to examine the effect of the number and position of these modifications (Table 1). FIT-PNA 2 contained a cpT PNA modification adjacent to BisQ and served as a control. For direct comparison, this cpT was substituted with a single γ-modification in FIT-PNA 3. The BisQ unit was flanked by two γ-modifications introduced by FIT-PNA 4, whereas FIT-PNA 5 had five γ-modifications (50% γ-modified) but had only one γ-modified monomer flanking BisQ. The FIT-PNA 6 oligomer was entirely γ-modified (100%, Scheme 1). All six FIT-PNA oligomers were successfully synthesized using a manual solid-phase method, purified via RP-HPLC (Fig. S1–S10), and characterized by MALDI-TOF MS (Table 1 and Fig. S11–S15). Their thermal stabilities were subsequently assessed in the presence of the complementary synthetic 11-mer RNA sequence (Table S1).
Thermal melting stability and brightness of RNA:gamma-FIT-PNAs
It has been known that, regardless of the overall charge of the PNAs (unmodified, negative or positive), they form duplexes with complementary DNA or RNA strands in 10 mM PBS buffer across a wide range of NaCl concentrations.34 In the current study, 1× PBS buffer (10 mM phosphate, 138 mM NaCl and 2.7 mM KCl) was chosen as it balances the maintenance of physiological ionic strength with compatibility for biophysical measurements and cell culture studies. The thermal stability of FIT-PNAs (1–6) to complementary RNA was assessed by increasing the temperature from 20 to 90 °C at a rate of 1 °C per minute, monitoring the absorbance at 260 nm (Fig. S16). Brightness (BR) was calculated from the duplex extinction coefficient maximum (εmax = 586 nm), which is derived from the linearly fitted UV-dilution experiments at λmax = 586 nm (Fig. S17) multiplied by the corresponding quantum yield value. The Tm and BR values of the unmodified FIT-PNA vs. gamma-FIT PNA:RNA duplexes are summarized in Table 2. The average thermal melting (Tm) stability of the FIT-PNA:RNA duplex was measured by multiple thermal melting experiments, with standard deviations summarized in Table S2 (SI). In this study, we have observed that the FIT-PNA 1 (unmodified) duplex with complementary RNA exhibits a melting temperature (Tm) of 43.0 °C (Fig. S16a) and a brightness (BR) of 12. In contrast, FIT-PNA 2 (with cpT) exhibits a Tm of 45.2 °C (Fig. S16b), which is stabilized by +2.2 °C. FIT-PNA 3, which has a single γ-T PNA monomer at the same position as cpT, demonstrated thermal stability comparable to that of the unmodified FIT-PNA 1:RNA duplex (Fig. S16c), and its brightness (in the duplex form) increased by 1.5-fold (BR = 18). FIT-PNA 4 exhibited a thermal stability of +2.7 °C (Fig. S16d) and increased brightness by about 2.1-fold (BR = 24.6). This improvement may be attributed to the symmetrical alignment of the nucleobase pairing in the duplex due to the γ-modification on both sides of the BisQ unit. This advantage is further demonstrated by the installation of the γ-modification far from BisQ in FIT-PNA 5. In this case, adding five γ-modifications but having only one monomer flanking BisQ results in a thermal stability of +1.6 °C, which is nearly identical to that of the unmodified duplex (Fig. S16e), with an increased brightness by 1.9-fold (BR = 23.3). Under identical conditions, the thermal stability of the fully γ-modified FIT-PNA 6 duplex (Fig. S16f) exhibits a +8.0 °C increase in stability and enhanced brightness by 2.3-fold (BR = 27.6), highlighting the stabilization effect for a fully modified γ-L-serine FIT-PNA. It is interesting to note that, in comparison to a single cpT modification on the cpFIT-PNA, adding two flanking cpPNA monomers around BisQ led to either no increase in Tm or an increase of only 1 °C.72 In the case of the γ-modifications, having two flanking γ-PNA monomers (FIT-PNA 4) had a more stabilizing effect (ΔTm = +2.7 °C).| S. no. | RNA duplex | λmax, abs (nm) | ε586 (mM−1 cm−1) | ϕ | BR (mM−1 cm−1) | I/I0 | Tm (°C) | ΔTm (°C) |
|---|---|---|---|---|---|---|---|---|
| I/I0 = fluorescence ratios for FIT-PNA:RNA/FIT-PNA (n = 3). | ||||||||
| 1 | aeg-BisQ-PNA | 586 | 59.8 | 0.20 | 12.0 | 24 | 43.0 ± 0.6 | — |
| 2 | aeg-cpT-BisQ-PNA | 586 | 78.4 | 0.32 | 25.1 | 39 | 45.2 ± 1.0 | +2.2 |
| 3 | (γ-ser)1-BisQ-PNA | 586 | 48.7 | 0.37 | 18.0 | 46 | 44.0 ± 0.4 | +1.0 |
| 4 | (γ-ser)2-BisQ-PNA | 586 | 72.3 | 0.34 | 24.6 | 41 | 45.7 ± 0.9 | +2.7 |
| 5 | (γ-ser)5-BisQ-PNA | 586 | 63.0 | 0.37 | 23.3 | 40 | 44.6 ± 0.3 | +1.6 |
| 6 | (γ-ser)10-BisQ-PNA | 586 | 83.5 | 0.33 | 27.6 | 41 | 51.0 ± 0.4 | +8.0 |
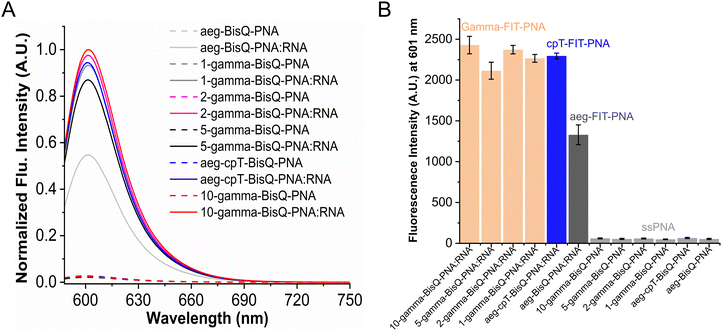
Fluorescence emission of FIT-PNA:RNA duplexes
Our previous report demonstrated that the introduction of a cyclopentane unit adjacent to the BisQ moiety enhanced fluorescence emission relative to the unmodified FIT-PNA.72 The present investigation compares fluorescence emission between cyclopentane- and γ-L-serine-modified derivatives at identical positions and further investigates the impact of γ-modification at different positions (Table 2, Fig. 2, and Fig. S18). The fluorescence emission of the unmodified aeg-FIT PNA 1 demonstrated a 24-fold increase in fluorescence when hybridized to complementary RNA (I/I0 = 24). The addition of a cyclopentane unit adjacent to BisQ in FIT-PNA 2 led to a 39-fold increase in fluorescence (I/I0 = 39), yielding a ca. 1.6-fold increase in intensity compared to the unmodified duplex. Substituting cyclopentane with a γ-modification at the same position in FIT-PNA 3 resulted in a 46-fold enhancement (I/I0 = 46) compared to the single strand and a 1.9-fold increase relative to the unmodified duplex. All other γ-modified FIT-PNAs (FIT-PNAs 4-6) led to similar I/I0 values (Table 2 and Fig. 2).In addition to cpPNA monomers, a well-studied FIT probe is based on DNA or 2′-OMe RNA sequences with either TO69 or QB67 as the surrogate base, which are modified with LNA to increase the brightness of these RNA/DNA sensors. In most cases, introducing an LNA-T modified base flanking QB (BisQ) results in a 3-fold increase in I/I0 and a ca. 2-fold increase in quantum yields.69 These effects are greater than those in cp- and gamma-modified FIT-PNAs but are, nonetheless, comparable. With TO,67 a similar trend is observed. Introducing a single LNA-T or LNA-A monomer flanking TO results in a ca. 2-fold increase in quantum yields. Moreover, a heavily modified LNA FIT probe (8 LNAs out of the 16 bases/50%) resulted in a greater increase in quantum yield (ca. 2.7-fold vs. unmodified FIT probe). This was not observed in our case (50% gamma-modified PNA, FIT-PNA 5), as we did not observe a change in quantum yields compared to the single γ-modified gamma FIT-PNA (FIT-PNA 3). This may be related to the fact that we are looking at a much shorter sequence (11-mer).
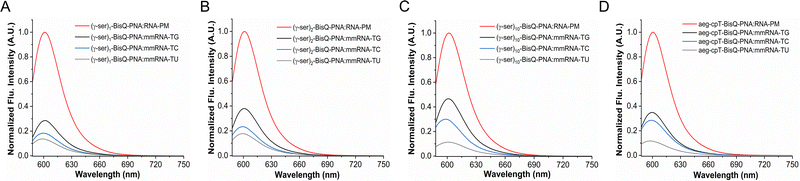
The binding selectivity of FIT-PNAs for RNA was further elucidated using mismatched RNA. We conducted mismatch selectivity based on fluorescence readout for single γ-modified FIT-PNA 3, double γ-modified FIT-PNA 4, and fully γ-modified FIT-PNA 6, and compared these with cpFIT-PNA 2 (Fig. 3). As anticipated, this experiment revealed a decrease in fluorescence emission intensity attributable to mismatch discrimination between perfectly matched and all types of single-mismatch FIT-PNA:RNA duplexes (TG, TC, and TU). In summary, γ-modified FIT-PNAs demonstrated mismatch discrimination comparable to that of cp-modified FIT-PNA 2.
Cellular uptake of FIT-PNAs (1–6) in OVCA433 cells
To gain a better understanding of the impact of gamma modifications on cellular uptake, we conducted a comparative cellular uptake study. The measurement of the cellular uptake of FIT-PNA (1–6) oligomers into OVCA433 cells was conducted using fluorescence-activated cell sorting (FACS) as illustrated in Fig. 4 and Fig. S19, S20. These FIT-PNAs target many RNA sequences in the cell milieu (being short 11-mers). As such, we assume that fluorescence in cells is primarily a feature of cellular uptake and, to a lesser extent, RNA targeting/binding. We compared fluorescence in cells for unmodified FIT-PNA 1, single modified cpFIT-PNA 2, and gamma modified FIT-PNAs that have only one gamma modification flanking BisQ (FIT-PNA 3 and FIT-PNA-5; Fig. 4a and b). In addition, we compared FIT-PNAs with the 2-gamma modification affixed to both sides of BisQ (FIT-PNAs 4 and 6; Fig. 4c and d). It is assumed that FIT-PNAs with comparable affinities to their complementary RNA would produce comparable fluorescence readouts when subjected to FACS analysis. Therefore, variations in fluorescence readouts between FIT-PNAs with comparable Tm values may indicate variations in cellular uptake. Fig. 4a and b present the outcomes for a series of FIT-PNAs with comparable Tms. We observed that the uptake levels of FIT-PNA 2 (with a cpPNA) were comparable to those of FIT-PNA 1 (unmodified), whereas FIT-PNAs 3 and 5 (with 1 and 5 γ-modifications, respectively) showed a slight decrease in fluorescence (Fig. 4a and b). The two other gamma FIT-PNAs, FIT-PNA-4 and FIT-PNA-6, exhibit Tm values of 45.7 °C and 51.0 °C (respectively), while differing in the number of γ-substitutions (2 vs. 10). Our observations indicate that fully γ-modified FIT-PNA 6 exhibits a larger fluorescence signal than that of doubly γ-modified FIT-PNA 4, as shown by the histogram plot and mean fluorescence intensity in the FACS results (Fig. 4c and d). This may be related to the enhanced structural rigidity of the FIT-PNA that may promote more effective interactions with cellular components and/or cell membranes. It is also interesting to note that the FACS profile for γ-modified FIT-PNAs flanking both sides of BisQ (FIT-PNA 4 and FIT-PNA 6; Fig. 4d) is narrower than all other profiles (Fig. 4b). Altogether, it can be speculated that a fully γ-modified FIT-PNA may offer improved RNA sensitivity, binding affinity, and cellular uptake. However, for the latter, further studies on longer γ-modified FIT-PNAs are needed to confirm this hypothesis—a direction we are currently pursuing.Conclusions
In summary, we have systematically examined the effect of γ-L-serine-modified FIT-PNA on the photophysical and hybridization properties with synthetic RNA as well as the cell permeability of these FIT-PNAs by FACS analysis (using the ovarian cancer cell line OVCA433).Incorporation of γ-PNA monomers adjacent to BisQ in the 2-gamma-FIT-PNA resulted in an increase in duplex stability (ΔTm ≈ +2.8 °C) and a significant increase in brightness (BR = 24.6) and fluorescence turn-on (I/I0 = 41) compared to the unmodified duplex (aeg-FIT-PNA, BR = 12 and I/I0 = 24.0). Interestingly, the 5-gamma-BisQ-PNA:RNA duplex exhibits a thermal stability of 44.2 °C and a BR of 23.3, although its stability is comparable to that of the unmodified aeg-PNA duplex (Tm = 43.9 °C). This similarity in thermal stability may be attributed to the gamma-modification positioned away from BisQ in FIT-PNA design. In addition, we observed a trend for all gamma-modified FIT-PNA, namely, higher thermal stability coinciding with greater brightness, with the fully γ-modified FIT-PNA (FIT-PNA 6) leading the series with ΔTm = +7.5 °C and BR = 27.6 (Table 2).
The mismatch selectivity of FIT-PNA:RNA to a single nucleobase mismatch adjacent to BisQ was evaluated using 1, 2, and 10-gamma-BisQ-PNA:RNA duplexes by testing all possible mismatches (TG, TC, and TU). These were compared to our previously reported mono-substituted cp-FIT-PNA:RNA duplex.72 It was observed that all gamma-FIT-PNAs demonstrate comparable discrimination to that of the cpFIT-PNA 2:RNA duplex (Fig. 3).
According to FACS results (Fig. 4), following cellular uptake of γ-modified FIT-PNAs in OVCA433 cells, we observed similar fluorescence intensities for all probes. Thus, a clearer understanding of the alignment of the gamma modification and the arrangement of BisQ in the helix of modified gamma-FIT-PNA:RNA duplexes may be achieved through future studies using single-crystal structures or by exploring longer FIT-PNA sequences.
These results indicate that the γ-modification serves as a structural motif for designing next-generation RNA-sensing probes with improved brightness and thermal stability. By using a peptide conjugation strategy combined with targeted γ-FIT-PNAs to specific RNA biomarkers, we may enhance the diagnostic potential of such RNA sensors.
Experimental procedure
Solid-phase synthesis of FIT-PNA oligomers
FIT-PNA oligomers (1–6) were synthesized using manual Fmoc solid-phase synthesis on a Nova-Syn TGA resin (0.25 mmol gm−1), in accordance with the standard synthesis protocol and a resin cleavage method.75 This was followed by purification using RP-HPLC and characterization via MALDI-TOF MS (for further details, see the experimental section).Tm analysis
The melting temperatures (Tm) of the FIT-PNA/RNA duplexes were determined using the sigmoidal UV melting curves obtained using an Evolution One Plus UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, USA). FIT-PNAs and their complementary RNAs were prepared in 1× PBS buffer (pH 7.0) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, achieving a final duplex concentration of 2.0 µM. Before analysis, the samples were heated from 20 °C to 90 °C at a rate of 5 °C min−1 and subsequently cooled to the initial temperature at a rate of 2 °C min−1. The absorbance change was observed at 260 nm while increasing the temperature to 90 °C at a rate of 1 °C min−1, and the Tm data were analyzed using Origin 2024 software.
1 ratio, achieving a final duplex concentration of 2.0 µM. Before analysis, the samples were heated from 20 °C to 90 °C at a rate of 5 °C min−1 and subsequently cooled to the initial temperature at a rate of 2 °C min−1. The absorbance change was observed at 260 nm while increasing the temperature to 90 °C at a rate of 1 °C min−1, and the Tm data were analyzed using Origin 2024 software.
Fluorescence quantum yield determination
We used a reported procedure to calculate the fluorescence quantum yield of FIT-PNA:RNA duplexes.76 The reference dye used for the BisQ-FIT-PNA:RNA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5) duplex was cresyl violet, which was chosen based on the absorbance range of the FIT-PNA duplex. To verify that the spectral shape was consistent, serial dilutions of FIT-PNA:RNA duplexes were prepared to ensure optical density (OD) values below 0.1 at the absorbance maximum (Fig. S17). The reference dye solutions and the FIT-PNA duplexes were both kept at ODs below 0.1 at their respective highest wavelengths (λmax = 586 nm for FIT-PNA duplexes and 592 nm for cresyl violet) to determine the cut-point/excitation wavelength. Cresyl violet solutions and FIT-PNA duplexes were excited at 580 nm and fluorescence emission spectra were recorded between 588 to 750 nm. Using the equation reported in the literature,72 the relative fluorescence quantum yields of the FIT-PNA:RNA duplexes were determined from the integrated emission intensities, absorbance at excitation wavelength, and solvents’ refractive indexes with respect to the known dye.
1.5) duplex was cresyl violet, which was chosen based on the absorbance range of the FIT-PNA duplex. To verify that the spectral shape was consistent, serial dilutions of FIT-PNA:RNA duplexes were prepared to ensure optical density (OD) values below 0.1 at the absorbance maximum (Fig. S17). The reference dye solutions and the FIT-PNA duplexes were both kept at ODs below 0.1 at their respective highest wavelengths (λmax = 586 nm for FIT-PNA duplexes and 592 nm for cresyl violet) to determine the cut-point/excitation wavelength. Cresyl violet solutions and FIT-PNA duplexes were excited at 580 nm and fluorescence emission spectra were recorded between 588 to 750 nm. Using the equation reported in the literature,72 the relative fluorescence quantum yields of the FIT-PNA:RNA duplexes were determined from the integrated emission intensities, absorbance at excitation wavelength, and solvents’ refractive indexes with respect to the known dye.
Fluorescence measurements
Fluorescence spectra of FIT-PNAs with and without synthetic RNA were recorded using a Jasco FP-6500 spectrofluorometer (Jasco Inc., Co., Ltd, Tokyo, Japan) with a bandwidth of 2.5 nm and a response time of 0.2 s. For FIT-PNAs, excitation was determined at 580 nm, and the emission spectra were recorded from 588 to 750 nm. Solutions of FIT-PNAs and synthetic RNA in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 ratio were prepared in 1× PBS. The hybridization of FIT-PNAs with synthetic RNA was conducted by annealing at 65 °C for 5 minutes, followed by incubation at 37 °C for 2–3 hours. The used RNA sequence was 5′-UAUGUAUGUUG-3′. RNAs showing mismatches were as follows: TU mm: 5′-UAUGU
1.5 ratio were prepared in 1× PBS. The hybridization of FIT-PNAs with synthetic RNA was conducted by annealing at 65 °C for 5 minutes, followed by incubation at 37 °C for 2–3 hours. The used RNA sequence was 5′-UAUGUAUGUUG-3′. RNAs showing mismatches were as follows: TU mm: 5′-UAUGU![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) UGUUG-3′; TC mm: 5′-UAUGU
UGUUG-3′; TC mm: 5′-UAUGU![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) UGUUG-3′; and TG mm: 5′-UAUGU
UGUUG-3′; and TG mm: 5′-UAUGU![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) UGUUG-3′.
UGUUG-3′.
Cell lines and culture conditions
The ovarian cancer cell line OVCA433 was purchased from the American Type Culture Collection (ATCC) and maintained according to the guidelines provided by the repository. OVCA433 cells were maintained in EMEM (ThermoFisher Scientific, Waltham, USA) augmented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA). The cell culture was maintained at 37 °C with 5% CO2.Flow cytometry analysis
FACS investigation of FIT-PNA intake was conducted by culturing OVCA433 (150 × 103) to around 80% confluency in 6-well plates. Following two washes with 1× PBS, the cells were treated with 5 µM of FIT-PNA oligomer for 5 h at 37 °C. Following extensive washing, the cells were collected with 0.25% Trypsin-EDTA, centrifuged at 1500 rpm, and resuspended in 300 µL of 1× PBS, which was subsequently filtered and analyzed using a Fortessa FACS analyzer (Core Research Facilities, The Hebrew University of Jerusalem, Jerusalem, Israel). The cells were gated based on the mCherry positivity observed in those treated with FIT-PNAs relative to their untreated controls. A minimum of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events, defined by gating on forward and side scatter plots, were obtained for each sample. The findings were analyzed using FlowJo v10.10 software, and the histogram was shown on a logarithmic scale.
000 events, defined by gating on forward and side scatter plots, were obtained for each sample. The findings were analyzed using FlowJo v10.10 software, and the histogram was shown on a logarithmic scale.
Statistical analysis for FACS analysis
FACS data from experiments are presented as mean ± SD. Two independent experiments were performed per assay, each with three technical replicates. Statistical significance was determined using a one-way ANOVA test with P < 0.001 considered extremely significant (***), P < 0.01 highly significant (**), and P < 0.05 statistically significant (*).Author contributions
E. Y. and M. K. G. planned the experiments and wrote the paper. M. K. G. conducted all chemical syntheses and biophysical studies. S. M. conducted the cell culture experiment (FACS). All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflicts of interest.Data availability
The data supporting the findings of this study are available within the article and its supplementary information (SI). Supplementary information: materials and methods and supporting figures (HPLC and MALDI-TOF MS of FIT-PNAs, melting profiles, UV-Vis spectra, and FACS data). See DOI: https://doi.org/10.1039/d5cb00292c.Acknowledgements
This research was supported by the Len & Susan Mark Initiative for Ovarian and Uterine/MMMT Cancers-Phase III grant from the Israel Cancer Research Fund (grant no. 21-305-MI) and by the Israel Science Foundation (grant no. 572/21). We thank Dr Daniel H. Appella and Dr Hongchao Zheng for providing the cpT-PNA monomer.Notes and references
- M. Egholm, O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden and P. E. Nielsen, Nature, 1993, 365, 566–568 Search PubMed.
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497–1500 CAS.
- V. V. Demidov, V. N. Potaman, M. D. Frankkamenetskii, M. Egholm, O. Buchard, S. H. Sonnichsen and P. E. Nielsen, Biochem. Pharmacol., 1994, 48, 1310–1313 CAS.
- R. D’Agata, M. C. Giuffrida and G. Spoto, Molecules, 2017, 22(11), 1951 Search PubMed.
- F. Hovelmann and O. Seitz, Acc. Chem. Res., 2016, 49, 714–723 CrossRef PubMed.
- G. Silvia, F. Barbara, T. Maurizio, C. Cristina, R. Gabriella, S. Luca, S. Maddalena, P. Federica, F. Augusto, F. Maurizio and C. Laura, Haematologica, 2008, 93, 610–614 Search PubMed.
- A. Singer, S. Rapireddy, D. H. Ly and A. Meller, Nano Lett., 2012, 12, 1722–1728 CrossRef CAS PubMed.
- E. E. Watson, Org. Biomol. Chem., 2025, 23, 9797–9814 Search PubMed.
- B. M. McMahon, D. Mays, J. Lipsky, J. A. Stewart, A. Fauq and E. Richelson, Antisense Nucleic Acid Drug Dev., 2002, 12, 65–70 Search PubMed.
- C. Suparpprom and T. Vilaivan, RSC Chem. Biol., 2022, 3, 648–697 Search PubMed.
- H. C. Zheng, I. Botos, V. Clausse, H. Nikolayevskiy, E. E. Rastede, M. F. Fouz, S. J. Mazur and D. H. Appella, Nucleic Acids Res., 2021, 49, 713–725 Search PubMed.
- H. C. Zheng, V. Clausse, H. Amarasekara, S. J. Mazur, I. Botos and D. H. Appella, JACS AU, 2023, 3, 1952–1964 CrossRef CAS PubMed.
- T. Govindaraju, V. Madhuri, V. A. Kumar and K. N. Ganesh, J. Org. Chem., 2006, 71, 14–21 CrossRef CAS PubMed.
- B. Ditmangklo, W. Sittiwong, T. Boddaert, T. Vilaivan and D. J. Aitken, Biopolymers, 2021, 112, e23459 Search PubMed.
- C. Vilaivan, C. Srisuwannaket, C. Ananthanawat, C. Suparpprom, J. Kawakami, Y. Yamaguchi, Y. Tanaka and T. Vilaivan, Artif. DNA PNA XNA, 2011, 2, 50–59 Search PubMed.
- T. Vilaivan, Acc. Chem. Res., 2015, 48, 1645–1656 CrossRef CAS PubMed.
- J. J. Turner, G. D. Ivanova, B. Verbeure, D. Williams, A. A. Arzumanov, S. Abes, B. Lebleu and M. J. Gait, Nucleic Acids Res., 2005, 33, 6837–6849 CrossRef CAS.
- N. Brodyagin, M. Katkevics, V. Kotikam, C. A. Ryan and E. Rozners, Beilstein J. Org. Chem., 2021, 17, 1641–1688 CrossRef CAS PubMed.
- Z. Zeng, S. Han, W. Hong, Y. Lang, F. Li, Y. Liu, Z. Li, Y. Wu, W. Li, X. Zhang and Z. Cao, Mol. Ther. – Nucleic Acids, 2016, 5, e295 CrossRef CAS PubMed.
- G. Cutrona, E. M. Carpaneto, M. Ulivi, S. Roncella, O. Landt, M. Ferrarini and L. C. Boffa, Nat. Biotechnol., 2000, 18, 300–303 CrossRef CAS.
- A. L. Scardovi, D. Bartolucci, L. Montemurro, S. Bortolotti, S. Angelucci, C. Amadesi, G. Nieddu, S. Oosterholt, L. Cerisoli, O. Della Pasqua, P. Hrelia and R. Tonelli, Nucleic Acid Ther., 2024, 34, 173–187 Search PubMed.
- J. Hu and D. R. Corey, Biochemistry, 2007, 46, 7581–7589 CrossRef CAS PubMed.
- J. Hu, M. Matsui, K. T. Gagnon, J. C. Schwartz, S. Gabillet, K. Arar, J. Wu, I. Bezprozvanny and D. R. Corey, Nat. Biotechnol., 2009, 27, 478–484 CrossRef CAS PubMed.
- U. Tsylents, I. Siekierska and J. Trylska, Eur. Biophys. J., 2023, 52, 533–544 CrossRef CAS.
- R. Bahal, B. Sahu, S. Rapireddy, C. M. Lee and D. H. Ly, ChemBioChem, 2012, 13, 56–60 CrossRef CAS PubMed.
- R. G. Emehiser, K. Dhuri, C. Shepard, S. Karmakar, R. Bahal and P. J. Hrdlicka, Org. Biomol. Chem., 2022, 20, 8714–8724 RSC.
- R. G. Emehiser, E. Hall, D. C. Guenther, S. Karmakar and P. J. Hrdlicka, Org. Biomol. Chem., 2020, 18, 56–65 RSC.
- E. Rozners, in Chemical Biology of Nucleic Acids: Fundamentals and Clinical Applications ed. V. A. Erdmann, W. T. Markiewicz and J. Barciszewski, 2014, pp. 167–180 DOI:10.1007/978-3-642-54452-1_10.
- C. A. Ryan, V. Baskevics, M. Katkevics and E. Rozners, Chem. Commun., 2022, 58, 7148–7151 RSC.
- T. X. Han, Y. Sato, E. Rozners and S. Nishizawa, Chem. Commun., 2025, 61, 11778–11781 RSC.
- M. Katkevics, J. A. Mackay and E. Rozners, Chem. Commun., 2024, 60, 1999–2008 RSC.
- X. Zhan, L. P. Deng and G. Chen, Biopolymers, 2022, 113, e23476 CrossRef CAS PubMed.
- C. Avitabile, L. Moggio, G. Malgieri, D. Capasso, S. Di Gaetano, M. Saviano, C. Pedone and A. Romanelli, PLoS One, 2012, 7, e35774 CrossRef CAS PubMed.
- N. Tilani, S. De Costa and J. M. Heemstra, PLoS One, 2013, 8, e58670 Search PubMed.
- E. A. Englund and D. H. Appella, Angew. Chem., Int. Ed., 2007, 46, 1414–1418 Search PubMed.
- D. R. Jain, L. Anandi, M. Lahiri and K. N. Ganesh, J. Org. Chem., 2014, 79, 9567–9577 CrossRef CAS PubMed.
- R. Mitra and K. N. Ganesh, J. Org. Chem., 2012, 77, 5696–5704 CrossRef CAS PubMed.
- V. Tähtinen, A. Verhassel, J. Tuomela and P. Virta, ChemBioChem, 2019, 20, 3041–3051 CrossRef PubMed.
- K. T. Kim, S. Angerani and N. Winssinger, Chem. Sci., 2021, 12, 8218–8223 RSC.
- S. Ellipilli and K. N. Ganesh, J. Org. Chem., 2015, 80, 9185–9191 CrossRef CAS PubMed.
- S. Rapireddy, R. Bahal and D. H. Ly, Biochemistry, 2011, 50, 3913–3918 Search PubMed.
- S. Rapireddy, G. He, S. Roy, B. A. Armitage and D. H. Ly, J. Am. Chem. Soc., 2007, 129, 15596–15600 Search PubMed.
- A. Dragulescu-Andrasi, S. Rapireddy, B. M. Frezza, C. Gayathri, R. R. Gil and D. H. Ly, J. Am. Chem. Soc., 2006, 128, 10258–10267 CrossRef CAS PubMed.
- R. R. Gaddam, K. Dhuri, Y. R. Kim, J. S. Jacobs, V. Kumar, Q. X. Li, K. Irani, R. Bahal and A. Vikram, J. Med. Chem., 2022, 65, 3332–3342 CrossRef CAS PubMed.
- A. R. Kaplan, H. Pham, Y. F. Liu, S. Oyaghire, R. Bahal, D. M. Engelman and P. M. Glazer, Mol. Cancer Res., 2020, 18, 873–882 Search PubMed.
- S. Sarkar and B. A. Armitage, ACS Infect. Dis., 2021, 7, 1445–1456 CrossRef CAS PubMed.
- Y. Z. Wang, S. Malik, H. W. Suh, Y. Xiao, Y. X. Deng, R. Fan, A. Huttner, R. S. Bindra, V. Singh, W. M. Saltzman and R. Bahal, Sci. Adv., 2023, 9, eabq7459 Search PubMed.
- R. Bahal, E. Quijano, N. A. McNeer, Y. Liu, D. C. Bhunia, F. Lopez-Giraldez, R. J. Fields, W. M. Saltzman, D. H. Ly and P. M. Glazer, Curr. Gene Ther., 2014, 14, 331–342 Search PubMed.
- A. S. Ricciardi, R. Bahal, J. S. Farrelly, E. Quijano, A. H. Bianchi, V. L. Luks, R. Putman, F. López-Giráldez, S. Coşkun, E. Song, Y. Liu, W.-C. Hsieh, D. H. Ly, D. H. Stitelman, P. M. Glazer and W. M. Saltzman, Nat. Commun., 2018, 9, 2481 CrossRef PubMed.
- B. Dong, K. X. Nie, H. H. Shi, L. M. Chao, M. Y. Ma, F. X. Gao, B. Liang, W. Chen, M. Q. Long and Z. C. Liu, Biosens. Bioelect., 2019, 136, 1–7 Search PubMed.
- J. M. Goldman, S. Kim, S. Narburgh, B. A. Armitage and J. W. Schneider, Biopolymers, 2022, 113, e23479 CrossRef CAS PubMed.
- J. Nölling, S. Rapireddy, J. I. Amburg, E. M. Crawford, R. A. Prakash, A. R. Rabson, Y. W. Tang and A. Singer, mBio, 2016, 7, e00345 CrossRef PubMed.
- A. Orenstein, A. S. Berlyoung, E. E. Rastede, H. H. Pham, E. Fouquerel, C. T. Murphy, B. J. Leibowitz, J. Yu, T. Srivastava, B. A. Armitage and P. L. Opresko, Molecules, 2017, 22, 2117 CrossRef PubMed.
- H. H. Pham, C. T. Murphy, G. Sureshkumar, D. H. Ly, P. L. Opresko and B. A. Armitage, Org. Biomol. Chem., 2014, 12, 7345–7354 RSC.
- K. K. Sadhu and N. Winssinger, Chem. – Eur. J., 2013, 19, 8182–8189 CrossRef CAS.
- G.-m Fang, J. Chamiolo, S. Kankowski, F. Hövelmann, D. Friedrich, A. Löwer, J. C. Meier and O. Seitz, Chem, Sci., 2018, 9, 4794–4800 Search PubMed.
- D. Hashoul, R. Shapira, M. Falchenko, O. Tepper, V. Paviov, A. Nissan and E. Yavin, Biosens. Bioelectron., 2019, 137, 271–278 CrossRef CAS PubMed.
- Y. Kam, A. Rubinstein, S. Naik, I. Djavsarov, D. Halle, I. Ariel, A. O. Gure, A. Stojadinovic, H. Pan, V. Tsivin, A. Nissan and E. Yavin, Cancer Lett., 2014, 352, 90–96 CrossRef CAS PubMed.
- Y. Kam, A. Rubinstein, A. Nissan, D. Halle and E. Yavin, Mol. Pharm., 2012, 9, 685–693 CrossRef CAS PubMed.
- O. Koehler, D. V. Jarikote and O. Seitz, ChemBioChem, 2005, 6, 69–77 CrossRef CAS PubMed.
- N. Kolevzon, D. Hashoul, S. Naik, A. Rubinstein and E. Yavin, Chem. Commun., 2016, 52, 2405–2407 RSC.
- S. Kummer, A. Knoll, E. Socher, L. Bethge, A. Herrmann and O. Seitz, Angew. Chem., Int. Ed., 2011, 50, 1931–1934 CrossRef CAS PubMed.
- S. Kummer, A. Knoll, E. Socher, L. Bethge, A. Herrmann and O. Seitz, Bioconjugate Chem., 2012, 23, 2051–2060 CrossRef CAS PubMed.
- N. Loibl, C. Arenz and O. Seitz, ChemBioChem, 2020, 21, 2527–2532 CrossRef CAS PubMed.
- J. Chamiolo, I. Gaspar, A. Ephrussi and O. Seitz, in RNA DETECTION: Methods and Protocols, ed. I. Gaspar, 2018, vol. 1649, pp. 273–287 Search PubMed.
- I. Haralampiev, M. Schade, J. Chamiolo, F. Jolmes, S. Prisner, P. T. Witkowski, M. Behrent, F. Hövelmann, T. Wolff, O. Seitz and A. Herrmann, ChemBioChem, 2017, 18, 1589–1592 CrossRef CAS PubMed.
- F. Hoevelmann, I. Gaspar, J. Chamiolo, M. Kasper, J. Steffen, A. Ephrussi and O. Seitz, Chem. Sci., 2016, 7, 128–135 RSC.
- F. Hoevelmann, I. Gaspar, A. Ephrussi and O. Seitz, J. Am. Chem. Soc., 2013, 135, 19025–19032 CrossRef CAS PubMed.
- F. Hoevelmann, I. Gaspar, S. Loibl, E. A. Ermilov, B. Roeder, J. Wengel, A. Ephrussi and O. Seitz, Angew. Chem., Int. Ed., 2014, 53, 11370–11375 CrossRef CAS PubMed.
- A. Homer, A. Knoll, U. Gruber and O. Seitz, Chem. Sci., 2025, 16, 846–853 RSC.
- S. Schöllkopf, A. Knoll, A. Homer and O. Seitz, Chem. Sci., 2023, 14, 4166–4173 RSC.
- O. Tepper, H. C. Zheng, D. H. Appella, E. Yavin, Chem. Commun., 2021, 57, 540–543; Erratum: 2023, 59, 11593 Search PubMed.
- S. T. Mannully, R. Mahajna, H. Nazzal, S. Maree, H. Zheng, D. H. Appella, R. Reich and E. Yavin, Biomolecules, 2024, 14, 609 CrossRef CAS PubMed.
- B. Sahu, I. Sacui, S. Rapireddy, K. J. Zanotti, R. Bahal, B. A. Armitage and D. H. Ly, J. Org. Chem., 2011, 76, 5614–5627 Search PubMed.
- H. Nazzal and E. Yavin, Methods Mol. Biol., 2025, 2934, 189–207 CrossRef CAS PubMed.
- C. Würth, M. Grabolle, J. Pauli, M. Spieles and U. Resch-Genger, Nat. Protoc., 2013, 8, 1535–1550 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |