Open Access Article
Open Access ArticleBackbone engineering in the hydrophobic core of villin headpiece
Yuhan
Lin
,
Ryley M.
David
,
Dyllan M.
Amin
,
Shane W. J.
Osborne
and
W. Seth
Horne
 *
*
Department of Chemistry, University of Pittsburgh, Pittsburgh, PA 15211, USA. E-mail: horne@pitt.edu
First published on 17th December 2025
Abstract
Changing the backbone connectivity of proteins can impart useful new traits while maintaining essential structural and functional features. In design of artificial proteomimetic agents, backbone modification is usually isolated to sites that are solvent-exposed in the folded state, as similar changes at buried residues can alter the fold. Recent work has shown that core backbone modification without structural perturbation is possible; however, the modifications in that study were consistently destabilizing and made in a prototype of exceptionally high conformational stability. Here, we report efforts to broaden the scope and improve the efficacy of core backbone engineering by applying it to the C-terminal subdomain of villin headpiece. A series of variants are prepared in which different artificial residue types are incorporated at core positions throughout the sequence, including a crucial aromatic triad. Impacts on folding energetics are quantified by biophysical methods, and high-resolution structures of several variants determined by NMR. We go on to construct a variant with ∼40% of its core modified that adopts a fold identical to the prototype while showing enhanced thermodynamic stability.
Introduction
Engineering the chemical composition of peptides and proteins can yield biomimetic macromolecules with intricate folded structures and potent biological functions.1–4 Expanding beyond ribosomally encoded amino acids as the monomer pool for construction of protein mimetics has been used to hone structure–activity relationships as well as facilitate biomedical applications.5,6 One chemical change commonly applied in such work is the incorporation of amino acids with modified backbones.7–9 Examples include alteration of stereochemistry (i.e., D-α-residues), addition of new substituents (e.g., N-Me-α-, N-amino-α-, Cα-Me-α-residues), and backbone elongation (e.g., β-residues). Compared to side-chain substitution, altering backbone connectivity can lead to large effects on folded structure, folding energetics, and biological properties from relatively small chemical changes.10While modification of backbone composition has proven utility in construction of artificial peptide and protein mimetics, moving from an isolated secondary structure (e.g., a helical peptide) to a higher order tertiary fold (e.g., a small protein) as the target for mimicry poses challenges.1 In tertiary structures, local conformational preferences must be considered alongside a complex network of weak, long-range interactions critical to the fold. Related to this challenge, backbone modification in the hydrophobic core of sequences with tertiary folding patterns has been shown to lead to a variety of unexpected effects.11–13
Seeking to broaden the scope of strategies for protein mimetic design to encompass reliable modification within the hydrophobic core, we recently reported efforts to engineer the backbone of the G-related albumin-binding module from bacterial protein PAB (1PRB).14 In that work, we showed judicious backbone alteration in the core could be structurally tolerated but was consistently destabilizing. While promising, these results left open questions. Would the design strategies prove viable in a protein domain with a more delicate fold than 1PRB, which has a thermal unfolding temperature ∼90 °C? Further, would expanding the pool of artificial monomers used in the construction of core-modified protein mimetics enable the creation of variants with enhanced conformational stability relative to the prototype? Here, we report efforts to address these questions through the application of backbone modification at core positions of the C-terminal subdomain of villin headpiece (HP35, Fig. 1).15–17
HP35 is one of the smallest domains that folds autonomously and enjoys a rich history as a model for fundamental studies of protein folding.18–29 In the realm of protein mimetics, backbone modification in HP35 has been made at solvent-exposed sites, and the structural and energetic effects of β-residue and Cα-Me-α-residue incorporation described by Gellman30 and our lab,31 respectively. Backbone modification to the HP35 core has not been reported; however, Berlicki described a computationally designed de novo protein mimetic that contained several carbocyclic β-residues in its core and was intended to mimic HP35.13 Interestingly, this sequence was observed to form a domain-swapped dimer fold that deviated significantly from the design.13 In the present study, we set out to systematically examine the impacts of altered backbone composition in the hydrophobic core of HP35 on folded structure and stability of the protein. Our results show artificial residues can be accommodated at several core positions in this system—individually or in tandem—without altering folded structure and that some modifications enhance folded stability relative to the canonical backbone.
Results and discussion
Design of core-modified HP35 variants
The hydrophobic core of HP35 (Fig. 1(A) and (B)) is primarily composed of three phenylalanine side chains (F6, F10, F17), which are located within the first and second helices and form an aromatic cluster that is critical to stabilizing the tertiary fold of the domain.32 Solvent-accessible surface area (SASA) analysis shows these three residues are each >90% buried. In addition to the phenylalanine triad, M12 in the loop between helix 1 and 2, L20 in the loop between helix 2 and 3, and L28 in helix 3 are partially buried (74–81%). Finally, polar residues Q25 and K29 in helix 3 occupy hydrophobic core-flanking positions and are also partially buried (67–85%). Based on the above analysis, we selected five sites from the core of HP35 as targets for backbone modification (Fig. 1(A)): the key aromatic cluster F6, F10 and F17, buried aliphatic L28, and interfacial charged K29.Three different artificial residue types were employed in design of core-modified HP35 variants: β3-residues, chiral Cα-Me-α-residues, and the cyclic β-residue trans-2-aminocyclopentane carboxylic acid (ACPC) (Fig. 1(C)). All three of these classes are known to be well accommodated in solvent-exposed helical contexts in a variety of heterogeneous-backbone tertiary structure mimetics, including HP35.30,31 Relative to a canonical α-residue, a methylene group is inserted between the carbonyl and Cα in a β3 monomer, which adds a rotatable bond and increases backbone flexibility. In a Cα-Me-α residue, the Hα is replaced by a methyl group, which restricts backbone conformational freedom. In the cyclic β-residue ACPC, the cyclopentane ring constrains the central torsion, which increases rigidity relative to the β3 residue at the expense of a lost side chain. Collectively, the application of these three monomer types allows for a comparative analysis of the importance of rigidity and side chain functionality when modifying the backbone at buried core positions.
For HP35 variants modified at core position L28 and core-flanking position K29, the canonical α-residue was replaced by a β3-residue or ACPC. As a control to understand context-dependent effects of these modifications at a comparable non-core site, we incorporated the same monomer types at neighboring solvent-exposed residue K30. Given the crucial role of side-chain packing of the phenylalanine triad in HP35, backbone substitutions at those positions were limited to β3 and Cα-Me-α monomers that maintain the side chain.
The above design considerations yielded a set of 12 single-site backbone substitution variants of HP35 for analysis. As data for the β3K30 variant have been reported,30 the remaining 11 peptides were prepared by Fmoc solid-phase methods. All syntheses were completed without major issues and yielded good crude purities (Fig. S1). Peptides were purified by preparative reverse phase high-performance liquid chromatography (HPLC). The identity of isolated products was confirmed by electrospray ionization mass spectrometry (ESI-MS) and purity assessed by analytical HPLC (Fig. S2–S13) prior to biophysical and structural characterization. To avoid complications from oxidation during storage and handling, M12 was replaced by norleucine in the HP35 prototype and all variants.
β-Residue substitution at a core, core-flanking, and solvent-exposed site
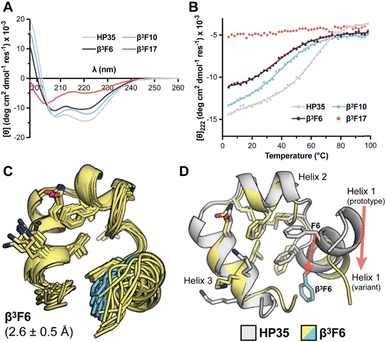
We first examined folding behavior of the HP35 variants modified in helix 3, comparing effects of β3 or ACPC substitution at three neighboring sites: core L28, core-flanking K29, and solvent-exposed K30. Results for the variants were benchmarked against our recently reported biophysical data set for prototype HP35.31 Circular dichroism (CD) scans at 20 °C on samples consisting of 50 µM peptide in 50 mM phosphate pH 7 show minima at ∼208 and ∼222 nm consistent with the predominantly helical fold of the prototype; however, the magnitude of the CD peaks in β3L28 and β3K29 is slightly attenuated (Fig. 2(A)).Thermal unfolding experiments monitoring CD signal at 222 nm as a function of temperature exhibit cooperative unfolding transitions in all cases (Fig. 2(B)), with temperature midpoint (Tm) values ranging from 47–69 °C (Table 1). Backbone modification at core position L28 results in a consistent reduction in the thermal stability of the fold (ΔTm −19 °C for β3L28 and −14 °C for ACPC28). At core-flanking position K29, variant β3K29 is comparably destabilized (ΔTm −15 °C), while ACPC29 shows a small increase in thermal stability relative to prototype (ΔTm +3 °C). For solvent-exposed position 30, ACPC incorporation is slightly destabilizing (ΔTm of −2 °C), and prior results indicate β3 substitution is moderately destabilizing (ΔTm of −12 °C).30
 | ||
| Fig. 2 CD scans at 20 °C (A) and thermal melts (B) for HP35 and indicated variants. Conditions: 50 µM peptide in 50 mM phosphate buffer, pH 7. | ||
| T m (°C) | ΔTm (°C) | (kcal mol−1) | (kcal mol−1) | |
|---|---|---|---|---|
a Temperature midpoint (Tm) of the thermal unfolding transition and change in Tm relative to HP35 (ΔTm) determined by variable temperature CD (conditions: 50 µM peptide in 50 mM phosphate buffer, pH 7). Folding free energy (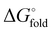 ) and change in folding free energy relative to HP35 ( ) and change in folding free energy relative to HP35 (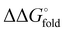 ) determined via chemical denaturation with guanidinium chloride monitored by tryptophan fluorescence (conditions: 10 µM peptide in 50 mM phosphate buffer, pH 7, 25 °C). Uncertainties for Tm and ) determined via chemical denaturation with guanidinium chloride monitored by tryptophan fluorescence (conditions: 10 µM peptide in 50 mM phosphate buffer, pH 7, 25 °C). Uncertainties for Tm and 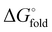 are standard errors from the fits and uncertainties for ΔTm and are standard errors from the fits and uncertainties for ΔTm and  determined by error propagation.
b Data sourced from published report;30 the sequence (M12/H27) and buffer composition (acetate pH 5) differed slightly from the present work, so the ΔTm value is reported relative to the prototype Tm from that study. determined by error propagation.
b Data sourced from published report;30 the sequence (M12/H27) and buffer composition (acetate pH 5) differed slightly from the present work, so the ΔTm value is reported relative to the prototype Tm from that study.
|
||||
| HP35 | 65.6 ± 0.7 | — | −2.14 ± 0.12 | 0 |
| β3L28 | 47.0 ± 2.6 | −19 ± 3 | — | — |
| ACPC28 | 51.2 ± 1.3 | −14 ± 1 | — | — |
| β3K29 | 50.6 ± 1.7 | −15 ± 2 | — | — |
| ACPC29 | 68.7 ± 0.7 | +3 ± 1 | −2.38 ± 0.15 | −0.2 ± 0.2 |
| β3K30 | 57.2 ± 1.7b | −12 ± 2b | — | — |
| ACPC30 | 63.3 ± 0.8 | −2 ± 1 | — | — |
| β3F6 | 36.3 ± 3.5 | −29 ± 4 | — | — |
| β3F10 | 47.3 ± 2.5 | −18 ± 3 | — | — |
| β3F17 | — | — | — | — |
| αMeF6 | 71.5 ± 0.7 | +6 ± 1 | −3.63 ± 0.26 | −1.5 ± 0.3 |
| αMeF10 | 45.3 ± 5.0 | −20 ± 5 | — | — |
| αMeF17 | 57.4 ± 1.3 | −8 ± 1 | −0.90 ± 0.21 | +1.2 ± 0.2 |
| αMeF6/αMeF17/ACPC29 | — | — | −3.00 ± 0.24 | −0.9 ± 0.3 |
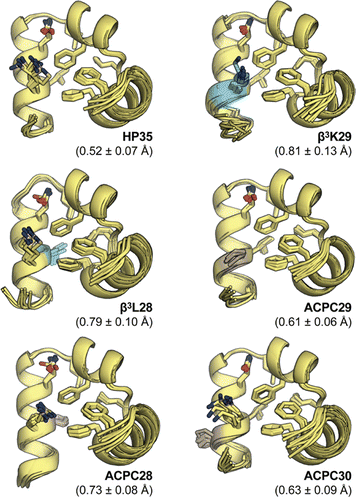
To determine the effects of backbone modification at core residues on the folded structure of HP35, we subjected the helix-3 variants to analysis by NMR spectroscopy. 1-Dimensional 1H and 2-dimesional 1H/1H COSY, TOCSY, and NOESY spectra were recorded for β3L28, β3K29, ACPC28, ACPC29, and ACPC30 at 1 mM peptide in 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 H2O/D2O at pH 5 (Fig. S15–S20). Assignment of proton resonances followed by simulated annealing with restraints derived from the NMR measurements yielded a 10-model structure ensemble for each peptide (Fig. 3 and Tables S1–S5).
1 H2O/D2O at pH 5 (Fig. S15–S20). Assignment of proton resonances followed by simulated annealing with restraints derived from the NMR measurements yielded a 10-model structure ensemble for each peptide (Fig. 3 and Tables S1–S5).
All the helix-3 variants adopt tertiary folds close to that of HP35 (0.5–0.8 Å backbone rmsd to the X-ray structure). Further support for this finding comes from observed backbone Hα chemical shifts throughout the domain, which are also very similar to the prototype (Fig. S27). Inspection of hydrophobic core packing shows that modification at L28 does not perturb the key aromatic triad; however, the artificial residues pack less efficiently (94%, 89%, and 70% burial for the α, ACPC, and β3 residue at position 28, respectively) (Fig. S28). This reduction in packing efficiency may give rise to the thermal stability loss observed by CD. At core-flanking position 29, both the β3K side chain as well as the ACPC ring engage core residues F10 and F17 in a manner similar to K29 in HP35.
From a structural perspective, all the modifications examined in helix 3 of HP35 are well tolerated. Even when backbone composition is altered at buried aliphatic L28, the variant folded structure is indistinguishable from the prototype. With respect to stability, effects of backbone modification vary with monomer type and sequence context. Results for the β3 variants point to a similar impact on thermal stability from substitution at the core, core-flanking, or solvent-exposed site. This suggests the thermal stability loss from β3 substitution arises primarily from properties inherent to the monomer, such as enhanced flexibility. In contrast, effects of ACPC substitution are context dependent—destabilizing at core site L28 but stabilizing at neighboring core-flanking K29. This finding indicates long-range interactions involving the ACPC side chain may influence fold stability in HP35. Prior studies have shown that methionine or norleucine substitution at K29 stabilizes HP35 due to elimination of unfavorable electrostatic interactions.33 Incorporation of ACPC at this position may provide a similar benefit. Correlations between the side chain of K29 and the aromatic rings of F10 and F17 are observed in the NOESY spectrum of HP35, and similar interactions are seen for the ACPC residue in variant ACPC29 (Fig. S29). These results support the hypothesis that long-range interactions involving residue 29 are important to the folded stability of HP35.
β-Residue substitution in the HP35 core aromatic triad
Given β-residue incorporation was viable at aliphatic core position L28 in HP35, we hypothesized it might also be tolerated in the crucial aromatic core cluster of F6, F10, and F17. To test this hypothesis, each of these residues was individually replaced by a β3 analogue. CD scans and thermal melts suggest that α → β3 substitution in the aromatic triad is consistently disruptive, though the degree depends on the site modified (Fig. 4(A), (B) and Table 1). β3F6 and β3F10 show evidence for a predominantly helical tertiary fold but are both significantly destabilized relative to the prototype (ΔTm = −30 °C and −20 °C, respectively). Backbone alteration in β3F17 leads to CD spectral properties characteristic of a random coil and complete loss of a cooperative thermal folding transition, suggesting this variant does not adopt an ordered fold.Comparison of the NMR spectra of β3F6 and β3F10 to that of HP35 shows reduced peak dispersion and increased peak broadening (Fig. S21 and S22). This effect suggests exchange among multiple ordered conformational states on an intermediate time scale.34,35 Peak overlap and broadening in β3F10 precluded resonance assignment; however, we were able to generate the NMR structure for β3F6 (Fig. 4(C) and Table S6). β3F6 adopts a predominantly helical fold; however, tertiary packing is altered relative to prototype HP35. A shift in the β3F side chain away from the core is accompanied by an increase in local conformational heterogeneity in helix 1 and a change in the position of helix 1 relative to the remainder of the protein (Fig. 4(D)). Additional experimental support for a greater degree of disorder in this variant comes from the Hα chemical shifts, which show a lower magnitude deviation from random coil values throughout the domain than in any other folded variant (Fig. S27). The structural rearrangement of helices is accompanied by a change in composition of the hydrophobic core. Residues β3F6, F10, and K29 become more exposed in the variant, while residues T13, K24, and K32 become more buried and form new long-range contacts.
Overall, these findings show that β3 residue incorporation in the core aromatic triad of HP35 is significantly disruptive to both folded structure and folded stability. The differences among the three β3F variants reinforce the idea that the position of modification is crucial. Among the three isomers (β3F6, β3F10, β3F17), which differ only in the placement of a single methylene group in the backbone, one adopts an ordered but non-native fold, one a partially ordered molten globule, and the other a disordered random coil.
Cα-Me-α-residue substitution in the HP35 core aromatic triad
Given β3 substitution at core aromatic positions in HP35 was not tolerated, we wondered whether an alternate monomer might be better accommodated at these sites. Thus, we examined a corresponding set of variants in which each of the key phenylalanine residues was individually replaced by a chiral Cα-Me-α analogue bearing the same side chain. Given chiral Cα-methyl residues are strong helix inducers,31,36–42 we were interested to see how side chain retaining α → Cα-Me substitution would compare to α → β3 replacement when targeted to critical buried core positions in HP35.CD spectra of variants αMeF6 and αMeF17 are similar to HP35 (Fig. 5(A)), while that of αMeF10 suggests significantly reduced helicity. Variable-temperature CD measurements exhibit cooperative folding transitions; however, the folded baseline for αMeF10 is poorly defined (Fig. 5(B)). Tm values range from 45–72 °C (Table 1), and relative thermal stabilities follow the trend αMeF6 > HP35 > αMeF17 > αMeF10. Folded structures of αMeF6 and αMeF17 determined by NMR show tertiary folds close to HP35 (Fig. 5(C) and Tables S7, S8). Packing of the aromatic triad in these variants is unchanged compared to the prototype, including at the artificial Cα-Me-α residues. The α-methyl group at each of these sites is buried in the core, in close contact with other side chains. In αMeF6, the organization of the remainder of the core is indistinguishable from HP35. In the case of αMeF17, the side chains of norleucine 12 and L20 shift slightly to accommodate the new methyl group. While subtle, this change may give rise to the observed decrease in thermal stability. NMR spectra of αMeF10 show significant peak overlap compared to the other αMeF variants (Fig. S23–S25), preventing resonance assignment. Taken with a low intensity CD signature and minimal thermal unfolding cooperativity, the small chemical shift dispersion suggests this variant does not adopt an ordered tertiary fold. The origin of the destabilization of αMeF10 relative to the other two αMeF variants is unclear.
Generation of an HP35 variant with a backbone modification throughout the core
Given that several individual core sites in HP35 proved amenable to backbone modification, we next sought to establish whether multiple core residues could be replaced in tandem. Thus, we synthesized and characterized a triple substitution variant αMeF6/αMeF17/ACPC29. With one modification in each of the three helices, this analogue has a core that is 38% artificial in backbone composition. As detailed above, each of the corresponding individual substitutions combined in this sequence are innocuous with respect to the folded structure and two are thermally stabilizing.CD scans suggest that the triple variant adopts a fold very similar to HP35 (Fig. 6(A)). Thermal melts reveal an exceptionally high thermal stability, with the almost complete absence of a defined unfolding transition (Fig. 6(B)). To obtain quantitative insights into thermodynamic effects of the altered backbone composition in this analogue, we compared the free energy of folding (ΔG°) for it, the prototype and each corresponding single-site variant (i.e., αMeF6, αMeF17, and ACPC29) through chemical denaturation with guanidinium chloride monitored by tryptophan fluorescence (Fig. 6(C)). All five sequences showed a cooperative unfolding transition. Folding free energies (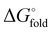 ) obtained from the fits follow a similar trend as Tm (Table 1). Relative to HP35, single-site variant αMeF6 is significantly stabilized (ΔΔG° −1.5 kcal mol−1), ACPC29 is comparable in stability (ΔΔG° −0.2 kcal mol−1), and αMeF17 is slightly destabilized (ΔΔG° +1.2 kcal mol−1). The triple mutant is also more thermodynamically stable than the prototype (ΔΔG° −0.9 kcal mol−1), and more stable than all but one of the single-site variants. NMR analysis for the αMeF6/αMeF17/ACPC29 triple variant (Fig. S26 and Table S9) shows that, despite substantial fraction of the core being backbone modified, it adopts a very similar fold to HP35 (Fig. 6(D)).
) obtained from the fits follow a similar trend as Tm (Table 1). Relative to HP35, single-site variant αMeF6 is significantly stabilized (ΔΔG° −1.5 kcal mol−1), ACPC29 is comparable in stability (ΔΔG° −0.2 kcal mol−1), and αMeF17 is slightly destabilized (ΔΔG° +1.2 kcal mol−1). The triple mutant is also more thermodynamically stable than the prototype (ΔΔG° −0.9 kcal mol−1), and more stable than all but one of the single-site variants. NMR analysis for the αMeF6/αMeF17/ACPC29 triple variant (Fig. S26 and Table S9) shows that, despite substantial fraction of the core being backbone modified, it adopts a very similar fold to HP35 (Fig. 6(D)).
Conclusions
In summary, we have reported here a systematic examination of the impacts of backbone modification in the hydrophobic core of the C-terminal subdomain of the villin headpiece on the folded structure and conformational stability of the protein. We find that backbone alteration through substitution of α-residues with β3, ACPC, or Cα-Me-α analogues can be made at core or core-flanking positions without perturbing the fold of the protein. The sequence context for a given backbone modification is an important factor in determining its exact effect; however, even residues with side chains that engage in crucial tertiary packing interactions in the hydrophobic core can be replaced. Many of the single-site variants are destabilized relative to the prototype, but some show improved conformational stability. Combining three well tolerated modifications into a single construct yields a variant with 38% of its core residues modified, a fold virtually identical to the prototype, and significantly enhanced thermal stability and folding free energy.Compared to prior work on the application of backbone modification at core positions in villin and other domains, the fact that many of the variants adopt folds very similar to prototype is noteworthy. Further, the enhanced stability seen for several of the variants is significant. Backbone modification in tertiary structure contexts is often destabilizing—even when made at solvent-exposed sites. Examples of tertiary structure mimetics with multiple backbone modifications and enhanced conformational stability relative to a prototype protein are rare.43–46 Achieving this outcome when the modification sites are in the core of the domain is noteworthy, considering the unique composition of the hydrophobic core, the small size, and the moderate unfolding free energy of HP35. Collectively, the present results demonstrate that the entirety of a sequence—both buried core residues and solvent-exposed sites—should be considered fair game in the design of heterogeneous-backbone protein mimetics in the context of α-helical secondary structure.
A significant challenge in development of protein mimetics based on artificial backbones is the incorporation of non-proteogenic amino acids. Although methods for ribosomal synthesis with non-canonical backbones continue to advance,47 chemical synthesis remains the method of choice for larger sequences with multiple modifications. Reliance on total chemical synthesis poses limitations to the size of entities that can be produced and, thus, the complexity of folds that can be mimicked. Methods such as native chemical ligation and semi-synthesis can expand the chain lengths accessible;48 however, spontaneous refolding of large synthetic constructs absent the typical cellular milieu can prove difficult.49 The above technical challenges notwithstanding, studies on the folding behavior of artificial protein-like backbones—such as the present work—show what is possible in these entities and provides insights into their design. Improved understanding of these fundamental issues along with continued advances in methods for construction of protein-like artificial macromolecules will help to realize the full potential of proteomimetics as a concept.1
Conflicts of interest
There are no conflicts to declare.Abbreviations
| 1PRB | G-Related albumin-binding module from bacterial protein PAB |
| ACPC | trans-2-Aminocyclopentane carboxylic acid |
| BMRB | Biological magnetic resonance bank |
| CD | Circular dichroism |
| COSY | Correlated spectroscopy |
| ESI-MS | Electrospray ionization mass spectrometry |
| Gnd | Guanidinium chloride |
| HP35 | Villin headpiece |
| HPLC | High-performance liquid chromatography |
| NMR | Nuclear magnetic resonance |
| NOESY | Nuclear Overhauser effect spectroscopy |
| PDB | Protein data bank |
| rmsd | Root mean square deviation |
| SASA | Solvent-accessible surface area |
| TOCSY | Total correlation spectroscopy |
Data availability
Coordinates and experimental data for newly reported NMR structures are deposited in the PDB (9YLZ, 9YM0, 9YM1, 9YM2, 9YM3, 9YM4, 9YM5, 9YM6, 9YM7) and BMRB (31270, 31271, 31272, 31273, 31274, 31275, 31276, 31277, 31278). Other data are available from the corresponding author upon request.Additional data supporting the findings of this study can be found in the SI for the article. Supplementary information: Fig. S1–S29, Tables S1–S9, materials and methods. See DOI: https://doi.org/10.1039/d5cb00269a.
Acknowledgements
Funding for this work was provided by a grant from the National Institutes of Health (R35GM149220 to W. S. H.). R. M. D. was supported by the National Science Foundation (REU 2244200).Notes and references
- W. S. Horne and T. N. Grossmann, Nat. Chem., 2020, 12, 331–337 CrossRef CAS PubMed.
- C. S. Swenson, G. Mandava, D. M. Thomas and R. E. Moellering, Chem. Rev., 2024, 124, 13020–13093 CrossRef CAS PubMed.
- S. H. Hong, T. Nguyen, J. F. Ongkingco, A. Nazzaro and P. S. Arora, Chem. Rev., 2025, 125, 6819–6869 CrossRef CAS PubMed.
- L. Lombardi, V. D. Genio, F. Albericio and D. R. Williams, Chem. Rev., 2025, 125, 7099–7166 CrossRef CAS PubMed.
- E. Lenci and A. Trabocchi, Chem. Soc. Rev., 2020, 49, 3262–3277 RSC.
- J. L. Hickey, D. Sindhikara, S. L. Zultanski and D. M. Schultz, ACS Med. Chem. Lett., 2023, 14, 557–565 CrossRef CAS PubMed.
- Z. E. Reinert and W. S. Horne, Org. Biomol. Chem., 2014, 12, 8796–8802 RSC.
- K. L. George and W. S. Horne, Acc. Chem. Res., 2018, 51, 1220–1228 CrossRef CAS PubMed.
- P. Sang and J. Cai, Chem. Soc. Rev., 2023, 52, 4843–4877 RSC.
- M. M. Müller, Biochemistry, 2018, 57, 177–185 CrossRef PubMed.
- W. S. Horne, J. L. Price and S. H. Gellman, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9151–9156 CrossRef CAS PubMed.
- M. W. Giuliano, W. S. Horne and S. H. Gellman, J. Am. Chem. Soc., 2009, 131, 9860–9861 CrossRef CAS PubMed.
- M. Bejger, P. Fortuna, M. Drewniak-Świtalska, J. Plewka, W. Rypniewski and Ł. Berlicki, Chem. Commun., 2021, 57, 6015–6018 RSC.
- Y. Lin and W. S. Horne, Chem. – Eur. J., 2024, 30, e202401890 CrossRef CAS PubMed.
- J. C. McKnight, D. S. Doering, P. T. Matsudaira and P. S. Kim, J. Mol. Biol., 1996, 260, 126–134 CrossRef PubMed.
- C. J. McKnight, P. T. Matsudaira and P. S. Kim, Nat. Struct. Biol., 1997, 4, 180–184 CrossRef CAS PubMed.
- D. E. Mortenson, K. A. Satyshur, I. A. Guzei, K. T. Forest and S. H. Gellman, J. Am. Chem. Soc., 2012, 134, 2473–2476 CrossRef CAS PubMed.
- Y. Duan and P. A. Kollman, Science, 1998, 282, 740–744 CrossRef CAS PubMed.
- C. D. Snow, H. Nguyen, V. S. Pande and M. Gruebele, Nature, 2002, 420, 102–106 CrossRef CAS PubMed.
- J. Kubelka, W. A. Eaton and J. Hofrichter, J. Mol. Biol., 2003, 329, 625–630 CrossRef CAS PubMed.
- M. Wang, Y. Tang, S. Sato, L. Vugmeyster, C. J. McKnight and D. P. Raleigh, J. Am. Chem. Soc., 2003, 125, 6032–6033 CrossRef CAS PubMed.
- A. Fernández, M.-y Shen, A. Colubri, T. R. Sosnick, R. S. Berry and K. F. Freed, Biochemistry, 2003, 42, 664–671 CrossRef PubMed.
- S. H. Brewer, D. M. Vu, Y. Tang, Y. Li, S. Franzen, D. P. Raleigh and R. B. Dyer, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 16662–16667 CrossRef CAS PubMed.
- H. Lei, C. Wu, H. Liu and Y. Duan, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 4925–4930 CrossRef CAS PubMed.
- D. L. Ensign, P. M. Kasson and V. S. Pande, J. Mol. Biol., 2007, 374, 806–816 CrossRef CAS PubMed.
- D. E. Shaw, P. Maragakis, K. Lindorff-Larsen, S. Piana, R. O. Dror, M. P. Eastwood, J. A. Bank, J. M. Jumper, J. K. Salmon, Y. Shan and W. Wriggers, Science, 2010, 330, 341–346 CrossRef CAS PubMed.
- K.-N. Hu, W.-M. Yau and R. Tycko, J. Am. Chem. Soc., 2010, 132, 24–25 CrossRef CAS PubMed.
- J. K. Chung, M. C. Thielges and M. D. Fayer, J. Am. Chem. Soc., 2012, 134, 12118–12124 CrossRef CAS PubMed.
- G. Žoldák, J. Stigler, B. Pelz, H. Li and M. Rief, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18156–18161 CrossRef PubMed.
- D. F. Kreitler, D. E. Mortenson, K. T. Forest and S. H. Gellman, J. Am. Chem. Soc., 2016, 138, 6498–6505 CrossRef CAS PubMed.
- T. W. Harmon, Y. Lin, R. T. Sutton, S. W. J. Osborne and W. S. Horne, ChemBioChem, 2025, 26, e202401022 CrossRef CAS PubMed.
- B. S. Frank, D. Vardar, D. A. Buckley and C. J. McKnight, Protein Sci., 2002, 11, 680–687 CrossRef CAS PubMed.
- J. Kubelka, T. K. Chiu, D. R. Davies, W. A. Eaton and J. Hofrichter, J. Mol. Biol., 2006, 359, 546–553 CrossRef CAS PubMed.
- H. J. Dyson and P. E. Wright, Methods Enzymol., 2001, 339, 258–270 CAS.
- A. K. Mittermaier and L. E. Kay, Trends Biochem. Sci., 2009, 34, 601–611 CrossRef CAS PubMed.
- C. Zhang, W. Miller, K. J. Valenzano and D. J. Kyle, J. Med. Chem., 2002, 45, 5280–5286 CrossRef CAS PubMed.
- N. A. Tavenor, Z. E. Reinert, G. A. Lengyel, B. D. Griffith and W. S. Horne, Chem. Commun., 2016, 52, 3789–3792 RSC.
- R. Islam, D. O. Sviridov, S. K. Drake, J. Tunyi, G. Abdoulaeva, L. A. Freeman, R. W. Pastor and A. T. Remaley, Biochem. Biophys. Res. Commun., 2020, 526, 349–354 CrossRef CAS PubMed.
- V. Bauer, B. Schmidtgall, G. Gógl, J. Dolenc, J. Osz, Y. Nominé, C. Kostmann, A. Cousido-Siah, A. Mitschler, N. Rochel, G. Travé, B. Kieffer and V. Torbeev, Chem. Sci., 2021, 12, 1080–1089 RSC.
- R. A. D. Bathgate, P. Praveen, A. Sethi, W. I. Furuya, R. R. Dhingra, M. Kocan, Q. Ou, A. L. Valkovic, I. Gil-Miravet, M. Navarro-Sánchez, F. E. Olucha-Bordonau, A. L. Gundlach, K. J. Rosengren, P. R. Gooley, M. Dutschmann and M. A. Hossain, J. Am. Chem. Soc., 2023, 145, 20242–20247 CrossRef CAS PubMed.
- A. Chandramohan, H. Josien, T. Y. Yuen, R. Duggal, D. Spiegelberg, L. Yan, Y.-C. A. Juang, L. Ge, P. G. Aronica, H. Y. K. Kaan, Y. H. Lim, A. Peier, B. Sherborne, J. Hochman, S. Lin, K. Biswas, M. Nestor, C. S. Verma, D. P. Lane, T. K. Sawyer, R. Garbaccio, B. Henry, S. Kannan, C. J. Brown, C. W. Johannes and A. W. Partridge, Nat. Commun., 2024, 15, 489 CrossRef CAS PubMed.
- T. W. Harmon, J. Song, A. J. Gulewicz, Y. P. Di and W. S. Horne, ChemBioChem, 2025, 26, e202400951 CrossRef CAS PubMed.
- K. L. George and W. S. Horne, J. Am. Chem. Soc., 2017, 139, 7931–7938 CrossRef CAS PubMed.
- S. D. Melton, E. A. E. Brackhahn, S. J. Orlin, P. Jin and D. M. Chenoweth, Chem. Sci., 2020, 11, 10638–10646 RSC.
- J. R. Santhouse, J. M. G. Leung, L. T. Chong and W. S. Horne, Chem. Sci., 2022, 13, 11798–11806 RSC.
- M. M. Wright, B. H. Rajewski, T. A. Gerrein, Z. Xu, L. J. Smith, W. S. Horne and J. R. Del Valle, Commun. Chem., 2025, 8, 76 CrossRef CAS PubMed.
- M. Sigal, S. Matsumoto, A. Beattie, T. Katoh and H. Suga, Chem. Rev., 2024, 124, 6444–6500 CrossRef CAS PubMed.
- V. Agouridas, O. El Mahdi, V. Diemer, M. Cargoët, J.-C. M. Monbaliu and O. Melnyk, Chem. Rev., 2019, 119, 7328–7443 CrossRef CAS PubMed.
- A. J. Callahan, A. Rondon, R. M. Reja, L. L. Salazar, S. Gandhesiri, J. Rodriguez, A. Loas and B. L. Pentelute, J. Am. Chem. Soc., 2024, 146, 28696–28706 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |