 Open Access Article
Open Access ArticleAQbD-driven UHPLC method for simultaneous quantification of budesonide, glycopyrronium bromide, and salbutamol sulphate: a unified approach for inhalation product and bioanalytical applications
Alessio Gaggero a,
Dalibor Jeremicb,
Anna Fedorkoa and
Jesús Alberto Afonso Urich
a,
Dalibor Jeremicb,
Anna Fedorkoa and
Jesús Alberto Afonso Urich *ac
*ac
aResearch Center Pharmaceutical Engineering GmbH, Inffeldgasse 13, 8010 Graz, Austria. E-mail: jesus.afonso@rcpe.at; afonsourich@student.tugraz.at
bDepartment of Health Studies – Biomedical Science, FH JOANNEUM, Alte Poststrasse 149, 8020 Graz, Austria
cInstitute of Process and Particle Engineering, Graz University of Technology, Inffeldgasse 13, 8010 Graz, Austria
First published on 15th December 2025
Abstract
Inhalation therapies often combine budesonide, glycopyrronium bromide, and salbutamol sulphate, necessitating analytical methods capable of their simultaneous quantification. Conventional UHPLC methods typically address these active pharmaceutical ingredients (APIs) individually, leading to inefficiencies in development, validation, and regulatory alignment. Analytical Quality by Design (AQbD) has transformed pharmaceutical analytical method development through systematic risk assessment and structured optimization, but its application in the bioanalytical domain remains limited. The challenge addressed in this study is the creation of a single robust UHPLC method that meets both pharmaceutical and bioanalytical requirements within regulatory frameworks. A unified UHPLC method was developed using AQbD principles, employing Design of Experiments to identify and optimize critical method parameters. The optimized method achieved baseline separation of all three APIs on a YMC UltraHT Hydrosphere C18 (2.1 × 100 mm; 2.0 μm) column under gradient elution with methanol and 0.1% formic acid in 10 mM ammonium formate. Robustness was established through a defined Method Operable Design Region. The method was first validated under ICH Q2 (R2) for pharmaceutical applications, confirming accuracy, precision, and sensitivity. Subsequently, it was extended to the bioanalytical domain and validated under ICH M10 guidelines in simulated lung fluid, demonstrating reproducibility in complex matrices. This dual validation highlights the method's versatility and regulatory robustness, underscoring AQbD's ability to unify pharmaceutical and bioanalytical method development into a single lifecycle appropriate platform. This study demonstrates the first AQbD-driven UHPLC method validated under both ICH Q2 (R2) and ICH M10, bridging pharmaceutical and bioanalytical applications. Extending AQbD principles into bioanalysis provides a regulatory-relevant framework that enhances robustness, lifecycle flexibility, and compliance. The work establishes a unified strategy for inhalation therapies and beyond, supporting broader adoption of science- and risk-based analytical development.
1. Introduction
Chronic obstructive pulmonary disease (COPD) and asthma are two of the most prevalent and debilitating respiratory disorders globally, affecting hundreds of millions of individuals and contributing significantly to healthcare burdens.1,2 COPD ranks as the fourth leading cause of death worldwide, accounting for approximately 3.5 million deaths in 2021,3 while asthma impacted over 262 million people and was responsible for 455![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2019.4 Although both conditions involve airflow obstruction and inflammation, they differ in pathophysiology: COPD is defined by persistent and progressive airflow limitation driven by chronic inflammation and alveolar destruction, whereas asthma is characterized by variable airflow limitation, airway hyperresponsiveness, and typically reversible bronchoconstriction triggered by allergens or environmental stimuli.5
000 deaths in 2019.4 Although both conditions involve airflow obstruction and inflammation, they differ in pathophysiology: COPD is defined by persistent and progressive airflow limitation driven by chronic inflammation and alveolar destruction, whereas asthma is characterized by variable airflow limitation, airway hyperresponsiveness, and typically reversible bronchoconstriction triggered by allergens or environmental stimuli.5
To manage these complex diseases, especially in moderate to severe stages, current therapeutic strategies often rely on combination therapies that target multiple underlying mechanisms. Among the most widely used agents are short-acting β2-agonists (SABAs) like salbutamol sulphate (SBS), which provide rapid bronchodilation;6 long-acting muscarinic antagonists (LAMAs) such as glycopyrronium bromide (GB), which reduce cholinergic-mediated bronchoconstriction;7 and inhaled corticosteroids (ICS) like budesonide (BUD), which attenuate airway inflammation.8 These three APIs are frequently co-formulated in fixed-dose combinations (e.g., ICS-LAMA, ICS-SABA), offering a synergistic approach to bronchodilation and inflammation control.9,10 Their combination is particularly relevant in treating asthma-COPD overlap (ACO), a phenotype associated with frequent exacerbations and poor treatment response when managed with monotherapy alone.11,12 The molecular structures and corresponding pKa values of the APIs are depicted in Fig. 1.
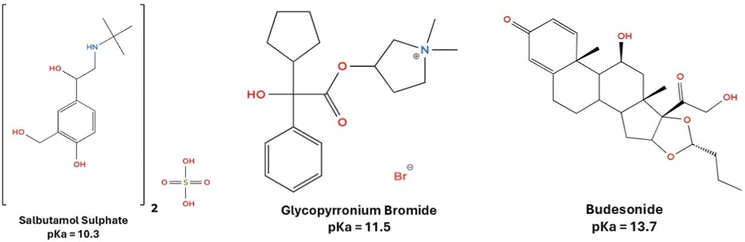 | ||
| Fig. 1 Chemical structures of the selected APIs with the relative pKa (strongest acid).13–15 | ||
Inhalation-based delivery of these agents ensures targeted action and reduced systemic side effects but also presents analytical challenges. Ensuring precise quantification and consistent performance of combination inhalation products requires highly sensitive, selective, and robust analytical methodologies.16 This is particularly important given the low doses involved and the need to differentiate among multiple APIs with diverse chemical properties.17 Furthermore, the inhalation products’ performance is nowadays carried out according to the United States Pharmacopeia (USP) chapter 〈601〉, where typically a cascade impactor is recommended for the testing.18 These cascade impactors (e.g., Next Generation Impactor – NGI) artificially simulate the lung surface, and the powder's particle size distribution can be measured. Usually, an analytical methodology is developed and dedicated to powder recovery in the different stages of the impactor.19
To address these needs, we developed a novel ultra-high-performance liquid chromatography (UHPLC) method to quantify BUD, GB, and SBS simultaneously. The technique was designed and optimized using AQbD principles, a modern approach that integrates risk assessment, critical method attributes (CMAs) identification, and design of experiments (DoE) to build quality into the method from its inception.20 The method was validated following ICH Q2 (R2) guidelines,21 ensuring suitability for quality control of pharmaceutical inhalation products.
Importantly, this work extends the AQbD paradigm to the bioanalytical field, an area where conventional method development practices, typically linear and empirical, still dominate. Bioanalytical methods, governed by ICH M1022 and other regulatory guidelines,23 are essential for quantifying drug levels in biological matrices for pharmacokinetic, toxicokinetic, and bioequivalence studies.24 However, these methods often suffer from matrix effects, high variability, and limited flexibility due to their empirical design.25,26
Applying AQbD to bioanalytical method development represents a transformative shift. It enables systematic understanding of critical method parameters, such as extraction efficiency, matrix interference, and ion suppression. While using DoE and risk assessment tools (e.g., Ishikawa diagrams, Failure Mode and Effect Analysis), AQbD facilitates the creation of robust methods27,28 that perform reliably across variable biological conditions and over extended study durations.
In this study, we also developed and validated a parallel bioanalytical method for quantifying the same three APIs in a lung fluid scenario. The method was optimized and validated under ICH M10,22 demonstrating regulatory compliance and suitability for clinical applications. Implementing AQbD across pharmaceutical and bioanalytical workflows underscores its versatility and the value of a unified, science-based development framework.
This work serves as a first step toward broader integration of AQbD into bioanalytical science, demonstrating its potential to improve method robustness, streamline development timelines, and elevate the reliability of pharmacokinetic assessments. By bridging pharmaceutical and bioanalytical method development through a single, scientifically justified platform, we offer a pathway for more unified, lifecycle-based analytical strategies in drug development.
2. Materials and methods
2.1. Reagents and consumables
The ammonium formate used for the mobile phase was acquired from Sigma-Aldrich GmbH (Vienna, Austria). The Methanol (HPLC gradient) used for the mobile phases and diluent was purchased from M&B Stricker Laborfachhandel GbR (Bernried am Starnberger See, Germany). The purified water for all analyses was obtained from Triton UV purification equipment from Neptec (Elbtal, Germany). The formic acid (≥98%) used for pH modifications was purchased from Carl Roth (Karlsruhe, Germany). Additional solvents used for forced degradation studies, such as 30% w/v hydrogen peroxide, sodium hydroxide solution, and hydrochloric acid, were acquired from Sigma-Aldrich GmbH (Vienna, Austria). The chloroform for the simulated lung fluid (SLF) preparation was acquired from Sigma-Aldrich GmbH (Vienna, Austria). The buffers used for the Phosphate Buffer Saline (PBS) preparation were purchased from VWR International GmbH (Vienna, Austria). All sample solutions were filtered before being injected into the chromatographic systems using PTFE syringe filters (0.45 μm) from YETI Merz Brothers GmbH (Haid, Austria).2.2. Standards, samples, and excipients
The standards for quantification of Budesonide (Pharmaceutical Secondary Standard), glycopyrronium bromide (European Pharmacopeia Reference Standard), and Salbutamol Sulphate (Albuterol Sulfate, Pharmaceutical Secondary Standard) were acquired from Sigma-Aldrich GmbH (Vienna, Austria). The identical standards batches were employed for sample preparation and method validation. Dipalmitoylphosphatidylcholine (DPPC) and dipalmitoylphosphatidylglycerol (DPPG), included in the SLF preparation, were obtained from Avanti Polar Solutions, Inc. (Alabama, USA). A complete protocol for the SLF preparation is provided in the SI (chapter 1.1).2.3. Equipment and software
A Reversed-Phase UHPLC H-Class from Waters Corp. (Milford, MA, USA) coupled with a Photo-Diode Array Detector (PDA) and a QDa single-quadrupole Mass Detector (QDa) was used to acquire chromatographic data. The Empower 3 (v.3.8.0) software from Waters Corp. (Milford, MA, USA) was used to control the HPLC and acquire method development and validation data. The screening DoE was performed using the Acquity HSS T3 (2.1 × 100 mm; 1.8 μm) column from Waters Corp. (Milford, MA, USA), the Triart C18 (2.1 × 100 mm; 1.9 μm), the Triart Phenyl (2.1 × 100 mm; 1.9 μm), and the UltraHT Hydrosphere C18 (2.1 × 100 mm; 2.0 μm) columns provided by YMC Europe GmbH (Dinslaken, Germany). The robustness DoE included a Kinetex C18 (2.1 × 100 mm; 1.7 μm) from Phenomenex. The DoEs’ conceptualization and statistical analysis were performed on Design Expert v.13 (Stat-Ease Inc., Minneapolis, MN, USA). The regression analysis was performed on OriginPro 2023b v.10.0.5.157 (OriginLab Corp., Northampton, MA, USA). The Centrivap centrifugal concentrator used for SLF preparation was obtained from LabConco Corporation (Kansas City, MO, USA). The FiveEasy™ FP20 pH meter used for mobile phase preparation was acquired from Mettler Toledo GmbH (Columbus, OH, USA).2.4. Analytical method validation by ICH Q2 (R2)
Limit of detection (LOD) and quantification (LOQ) were calculated using the signal-to-noise ratio approach. The signal-to-noise ratio is determined by comparing measured signals from samples with known low analyte concentrations with those of blank injections. A signal-to-noise ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was considered acceptable for estimating LOD; for LOQ, a ratio of at least 10
1 was considered acceptable for estimating LOD; for LOQ, a ratio of at least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was acceptable.21
1 was acceptable.21
2.5. Analytical method validation by ICH M10
Accuracy and precision were determined by analysing the QCs within each run (within-run) and in two different runs (between-run). Within-run accuracy and precision were evaluated by analysing five replicates at each QC concentration level. Between-run accuracy and precision were assessed by examining each QC concentration level in three analytical runs over two days (n = 15 total). The accuracy (RE%) at each concentration level should be within ±15% of the nominal concentration, except at the LLOQ, where it should be within ±20%.22 The precision (RSD%) of the concentrations determined at each level should not exceed 15%, except at the LLOQ, which should not exceed 20%.22
During this stage, one analytical run used for accuracy and precision was injected again on a different day to prove reinjection reproducibility, following the same acceptance criteria.22
3. Results and discussion
3.1. Method development by AQbD principles
The AQbD workflow consists of multiple steps that align with the most recent version of the USP 〈1220〉 chapter and ICH Q14 revisions, including an extended review of the analytical procedure development and lifecycle.31,32 Following this rigorous process ensures that the developed analytical methodology fits the intended scope by producing robust and reliable results.27,33 An overview of the steps involved in the AQbD stream is provided in Fig. 2.The initial stage involves defining the Analytical Target Profile (ATP), which outlines the intended performance criteria that the analytical method is expected to achieve. The developed method should detect the three selected APIs with desirable chromatography standards, including adequate peak resolution, a fundamental property to ensure specificity and peak purity, without sacrificing the fast analysis setup provided by UHPLC. The solubility pattern of the APIs presents another challenge in terms of retention: SBS and GB are very hydrophilic, while BUD is strongly hydrophobic due to the steroid groups (Fig. 1). In that context, the choice of mobile phase, gradient, and column is pivotal to secure a correct elution of the analytes.34 Sensitivity-wise, the method should reach interesting LOD and LOQ values, laying a solid basis for future tuning via MS. Inhalation performance testing often requires further dilution (e.g., via NGI) to recover a quantity of sample to test afterward via HPLC, making reaching low sensitivity standards a fundamental feature for this method. Therefore, in a later stage, the technique was asked to robustly quantify the analytes in the range of a few ng ml−1, especially given the bioavailability of these drugs once they reach the lungs and then the bloodstream.35–38 This study aimed to develop a robust method with trustworthy performance across a broad range of applications, ensuring its suitability for translation into an industrial setting, where consistency and routine operation still play an essential role. The ATP with the performance characteristics and its criteria is shown in Table 1.
Regarding the detection setup, the PDA mode simultaneously captured the full UV spectrum during each injection, with validation data evaluated at a selected wavelength of 225 nm. A baseline filter was applied to minimize futile noise. Continuing with the AQbD process, the identification of critical method parameters (CMPs) focused on factors such as column selection, column temperature, flow rate, injection volume, organic modifier, and mobile phase buffer. Each factor was optimized to ensure compatibility with potential MS-based method development. A Quality Risk Management (QRM) tool like the Ishikawa fishbone diagram was employed to assess the intrinsic relationship between the CMPs and CMAs (Fig. 3).
To complement the Ishikawa diagram, a semi-quantitative risk matrix was constructed to prioritize method parameters based on their potential influence on CMAs; it is included in the SI (section 2.1.; Table S2). Due to the inherent complexity of developing a method with broad applicability, an initial DoE was conducted to screen and evaluate key chromatographic characteristics and identify significant factors that influence method performance. Moreover, the DoE serves as a preliminary screening tool to establish a baseline for the method in terms of both detection and retention properties.42 A set of four different C18 columns was explored to achieve optimal retention, given the diverse separation behavior of the chosen substances, including an HSS T3 from Waters and a Triart Phenyl from YMC, which are standard options for retaining more hydrophilic APIs and can take up to a 100% aqueous mobile phase composition. Three MS-compatible buffers were examined to provide the best setup for sensitivity purposes, providing a broader scenario of retention compatibility with our APIs. The tested gradient started with a 95% aqueous buffer phase and proceeded towards the opposite side of the organic phase, to shape a scouting gradient, an ideal choice when the retention behavior of the analytes remains unknown.43 The gradient speed was kept linear. Table 2 provides a complete overview of the chosen factors and levels of the development DoE.
| Analytical column | Organic modifier (%) | Mobile phase buffer | Flow (mL min−1) | Injection volume (μL) | Column temperature (°C) |
|---|---|---|---|---|---|
| Column letters corresponding to (A) Acquity HSS T3 (2.1 × 100 mm; 1.8 μm); (B) Triart C18 (2.1 × 100 mm; 1.9 μm); (C) Triart Phenyl (2.1 × 100 mm; 1.9 μm); (D) UltraHT Hydrosphere C18 (2.1 × 100 mm; 2.0 μm). | |||||
| (A) | Acetonitrile methanol | 10 mM ammonium formate | 0.3–0.5 | 1–10 | 30–60 |
| (B) | 10 mM ammonium Acetate | ||||
| (C) | 0.1% formic acid | ||||
| (D) | |||||
3.2. Statistical evaluation of the development DoE
A response surface methodology (RSM) approach was employed to systematically optimize the HPLC method following Quality by Design principles. An I-optimal design for quadratic effects was implemented, comprising 50 experimental runs to ensure adequate model fitting and prediction capability. Due to the practical constraints of changing column temperature and organic modifier during the experimental sequence, these factors were designated as “hard-to-change” parameters, necessitating a split-plot design structure. The experimental design was organized into seven whole plots with two levels of organic modifier, where column temperature settings were nested within each organic modifier level. The split-plot arrangement resulted in acetonitrile and methanol being tested as separate whole plots with varying column temperature profiles. The chromatographic performance was evaluated through four critical response variables for the three APIs: USP signal-to-noise (USP s/n), k′, peak tailing, and USP resolution, currently listed in Table 1. The optimization strategy prioritized compliance with the selected ATP (Table 1) which is based on USP pharmacopeial requirements for k′, tailing, and resolution, while USP s/n served as a secondary objective for method robustness enhancement.40Model predictions were validated through 13 independent confirmation runs using the optimized conditions, with injection volume adjusted to 5 μL to fulfil a satisfactory chromatographic performance aligned with our ATP (Table 1). Statistical evaluation compared predicted mean values against experimental results using 95% prediction intervals (PI), with results summarized in the SI (chapter 2.2.1). The optimization results derived from the development DoE, bridged with the robustness results from method validation, helped us establish the Method Operable Design Region (MODR). MODR is a multidimensional space that represents a scientific warrant that the method perform robustly and efficiently.
The I-optimal response surface design effectively identified optimal chromatographic conditions (flow rate 0.478 mL min−1, column temperature 30 °C, injection volume 5 μL, YMC UltraHT Hydrosphere C18 column, ammonium formate buffer, methanol organic modifier), yielding an overall desirability of 0.689. Factor effect analysis revealed column type and mobile phase buffer composition as the most critical parameters influencing separation performance, with the YMC Ultra HT Hydrosphere C18 column providing superior chromatographic characteristics across the investigated design space.
Model validation through confirmation runs demonstrated that while not all predicted values fell within 95% prediction intervals, all experimental results remained well within the ATP frame (Table 1). This outcome highlights the practical success of optimization despite minor prediction deviations, confirming that the DoE approach delivered a specification-compliant analytical method.
3.3. Analytical method definition
As a result of the DoE evaluation, the final conditions of the analytical method were assessed (Table 3) and the same parameters were utilized for the validation according to ICH Q2 (R2) and M10 guidelines. An exemplary chromatogram obtained with this validated methodology is given in Fig. 5, where complete separation could be achieved in 9 minutes.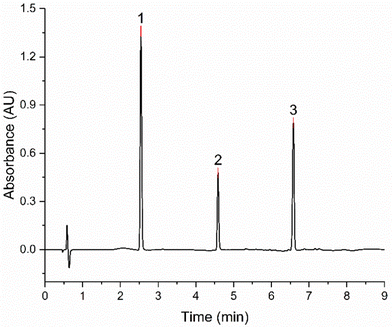 | ||
| Fig. 5 Chromatogram of a 250 μg mL−1 injection of the three APIs mixture obtained with the final method conditions. (1) Salbutamol sulphate, (2) glycopyrronium bromide, (3) budesonide. | ||
| Validation conditions | |
|---|---|
| Column | YMC UltraHT hydrosphere C18 |
| Column temperature | 30 °C |
| Flow rate | 0.478 mL min−1 |
| Gradient | %B: 0.0 min 5%, 0.5 min 5%, 6.0 min 95%, 6.5 min 95%, 9.0 min 5% |
| Injection volume | 5 μL |
| Mobile phase buffer (A) | 10 mM ammonium formate |
| Organic modifier (B) | Methanol |
| Sample diluent | Methanol/water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) 50) |
| Wavelength | 225 nm |
3.4. Analytical method validation results ICH Q2 (R2)
 | ||
| Fig. 6 Chromatograms of specificity studies. (A) Blank solution injection; (B) mobile phase injection; (C) SLF injection. | ||
The forced degradation studies and purity plots analysis confirmed the absence of interference under distinct stressing conditions, and the results highlighting the samples’ purity are presented in the SI (chapter 2.3.1).
The LOD and LOQ were assessed by estimating the USP s/n ratio of prepared solutions at the lowest calibration point (100 µg mL−1) against a blank injection in the interested region of the analyte. The results are summarized in the SI (chapter 2.3.2).
| API | Percent of target (%) | Recovery (%) | Mean (%) | RSD (%) |
|---|---|---|---|---|
| Salbutamol sulphate | 70% | 99.1 | 99.3 | 0.2 |
| 99.3 | ||||
| 99.5 | ||||
| 100% | 102.0 | 102.3 | 0.8 | |
| 101.7 | ||||
| 103.2 | ||||
| 130% | 102.4 | 102.8 | 0.8 | |
| 102.2 | ||||
| 103.7 | ||||
| 70% | 101.0 | 99.9 | 1.2 | |
| 98.6 | ||||
| 100.1 | ||||
| Glycopyrronium bromide | 100% | 100.6 | 100.4 | 0.4 |
| 100.7 | ||||
| 99.9 | ||||
| 130% | 100.6 | 100.4 | 0.3 | |
| 100.6 | ||||
| 100.2 | ||||
| 70% | 99.7 | 99.9 | 1.2 | |
| 99.7 | ||||
| 99.9 | ||||
| Budesonide | 100% | 99.9 | 100.4 | 0.4 |
| 99.7 | ||||
| 99.9 | ||||
| 130% | 100.4 | 100.2 | 0.2 | |
| 100.2 | ||||
| 100.0 | ||||
The developed method exhibited high precision and reproducibility, with RSD below 2% for all measurements (Table 5). A Student's t-test revealed no statistically significant difference between the two analysts’ results (p > 0.05), indicating insufficient evidence to conclude that the datasets differ.45
| API | Parameter | Analyst I | Analyst II |
|---|---|---|---|
| Salbutamol sulphate | Mean | 101.4 | 98.3 |
| SD | 1.4 | 1.7 | |
| RSD | 1.4 | 1.8 | |
| SEM | 0.6 | 0.8 | |
| n | 6 | 6 | |
| p-Value | 1.0 | ||
| Glycopyrronium bromide | Mean | 99.7 | 99.3 |
| SD | 0.5 | 1.6 | |
| RSD | 0.5 | 1.6 | |
| SEM | 0.2 | 0.6 | |
| n | 6 | 6 | |
| p-Value | 0.5 | ||
| Budesonide | Mean | 100.1 | 101.4 |
| SD | 1.8 | 2.0 | |
| RSD | 1.8 | 2.0 | |
| SEM | 0.7 | 0.7 | |
| n | 6 | 6 | |
| p-Value | 0.3 | ||
Three factors were systematically varied within realistic operational ranges: sample diluent composition (A: 40–60% methanol in sample preparation), formic acid concentration in mobile phase (B: 0.05–0.15%), and column variability (C: two different C18 stationary phases). The column factor included the optimized YMC UltraHT Hydrosphere C18 column alongside a Phenomenex Kinetex C18 column. The same response variables used in the optimization study were monitored: USP s/n, k′, peak tailing, and USP resolution for each API, with the same limits described in the ATP (Table 1) serving as acceptance criteria for the evaluation. The robustness assessment demonstrated that the optimized method maintained acceptable performance across all tested conditions, without violating the selected specifications throughout the 30 experimental runs. This outcome provides strong evidence for method robustness under the investigated parameter variations. The data is shown in the SI (chapter 2.3.3, Table S8).
Statistical modeling revealed varying degrees of complexity across the different API responses, as summarized in the SI (chapter 2.3.3, Tables S9–S19).
SBS responses presented the most challenging modeling scenarios:
• USP s/n ratio (adjusted R2 = 0.53): required complex higher-order terms including A2B and A2C interactions to achieve an acceptable model fit. Despite including these quadratic interaction terms, the lack-of-fit test remained marginally significant (p = 0.074). The large difference between adjusted R2 (0.53) and predicted R2 (0.33) indicated potential issues with model adequacy. However, attempts at model reduction consistently resulted in a significant lack of fit, necessitating the retention of the complex model structure.
• Capacity factor (adjusted R2 = 0.62): modeling required inclusion of the B2 term for adequate fit, despite the main factor B (formic acid concentration) showing no significant effect (p = 0.663). This counterintuitive result highlights the complex response surface behavior.
• Tailing factor (adjusted R2 = 0.93): achieved excellent model fit but required a complex nine-term model including multiple quadratic and interaction terms (A, C, AC, BC, A2, B2, A2C, B2C).
GB and BUD responses demonstrated much simpler and more interpretable patterns:
• For GB, the column was the dominant significant factor across all responses: k′ (adjusted R2 = 0.97), USP s/n (adjusted R2 = 0.49), tailing (adjusted R2 = 0.14), and resolution (adjusted R2 = 0.44).
• For BUD, column type similarly dominated: USP s/n (adjusted R2 = 0.96), k′ (adjusted R2 = 0.20), and tailing (adjusted R2 = 0.82). The exception was resolution (adjusted R2 = 0.61), where diluent composition (A, p = 0.044) and the AC interaction (p = 0.049) were also statistically significant alongside column type.
Technical investigation suggested that sample diluent composition (factor A) may contribute to baseline variability, potentially affecting USP s/n ratios and peak tailing. This observation provides one possible explanation for the complex modeling behavior observed, particularly with SBS responses. All experimental results remained within the specification limits across the entire investigated parameter space, demonstrating maintenance of acceptable analytical performance under the tested robustness conditions.
The subsequent robustness evaluation across 30 experimental runs confirmed method stability under realistic operational variations. Despite encountering complex statistical modeling challenges, particularly for Salbutamol responses requiring higher-order polynomial terms, the fundamental robustness criterion was consistently met: all analytical results remained within USP pharmacopeial limits across the entire investigated parameter space.
The statistical complexity observed, especially the requirement for quadratic interaction terms in Salbutamol models and the counterintuitive significance of B2 terms without corresponding main effects, likely reflects the influence of sample diluent composition on chromatographic baseline behavior. These findings underscore the multifactorial nature of chromatographic systems and the value of comprehensive experimental design in revealing such interactions.
The successful demonstration of method robustness across different C18 stationary phases (YMC Hydrosphere and Kinetex) addresses regulatory requirements for method transferability and provides confidence for implementation across multiple laboratory environments. The consistent performance of glycopyrronium and budesonide responses, contrasted with the more complex behavior of Salbutamol, suggests compound-specific susceptibility to analytical conditions that should be considered during method transfer protocols.
This study demonstrates both the power and limitations of statistical modeling in analytical method development. The varying model adequacy across robustness responses (e.g., R2-adj ranging from 0.14 to 0.97) reflects the different sensitivity levels of individual analytical parameters to experimental variability. While ideally robustness studies would show no significant effects, the detection of significant but poorly predictable effects (e.g., glycopyrronium tailing, R2-adj = 0.14) indicates inherent analytical variability that, while statistically detectable, remains within acceptable specification limits.
While complex response surfaces may challenge conventional modeling approaches, the ultimate measure of method suitability remains compliance with analytical specifications. The QbD approach successfully guided method development decisions despite statistical modeling complexities, emphasizing the importance of focusing on practical performance criteria alongside statistical rigor.
The comprehensive experimental design provided valuable insights into factor interactions that would be difficult to detect through traditional one-factor-at-a-time approaches, justifying the investment in systematic DoE methodology for critical analytical method development projects.
3.5. Analytical method validation results ICH M10
Specificity was proven by injecting a 1 µg mL−1 solution prepared in working diluent (methanol/water) and recording the signal at the same SIR where the analytes were processed, as per selectivity. These injections were compared to the analytes’ response at the LLOQ in SLF diluent. The chromatograms confirmed identical retention times, while the MS scan verified the compounds’ identities (Fig. S11).
Regarding the matrix effect, no significant interference was observed when different lots of prepared SLF were used, defining a stable analysis regardless of the sample composition. The quantification showed no evidence of matrix effects, ion suppression, or ion enhancement, as the accuracy and precision fell within the target of ±15% of nominal concentration and RSD% (Table S21).
| API | Calibration level (ng mL−1) | Mean back-calculated (ng mL−1) | Accuracy (RE, %) | Precision (RSD, %) |
|---|---|---|---|---|
| Salbutamol sulphate | 1.0 | 1.1 | +10.7 | 3.0 |
| 2.5 | 2.5 | −1.6 | 1.2 | |
| 5.0 | 5.0 | −0.5 | 1.4 | |
| 10.0 | 10.1 | +1.4 | 2.5 | |
| 15.0 | 14.9 | −1.0 | 0.7 | |
| 20.0 | 20.3 | +1.3 | 0.9 | |
| Glycopyrronium bromide | 1.0 | 1.0 | +0.7 | 10.9 |
| 2.5 | 2.5 | −2.1 | 6.7 | |
| 5.0 | 5.4 | +8.3 | 8.7 | |
| 10.0 | 10.3 | +3.4 | 5.0 | |
| 15.0 | 16.1 | +7.3 | 4.0 | |
| 20.0 | 21.5 | +7.6 | 5.4 | |
| Budesonide | 1.0 | 1.1 | +8.0 | 11.0 |
| 2.5 | 2.7 | +6.5 | 6.6 | |
| 5.0 | 5.0 | +0.8 | 4.8 | |
| 10.0 | 10.4 | +3.6 | 2.4 | |
| 15.0 | 16.0 | +6.9 | 0.2 | |
| 20.0 | 20.3 | +1.6 | 0.8 |
| API | Spiked concentration (ng ml−1) | Intra-day (n = 5) | Inter-day (n = 10) | ||||
|---|---|---|---|---|---|---|---|
| mean (ng ml−1) | Accuracy (RE, %) | Precision (RSD, %) | Mean (ng ml−1) | Accuracy (RE, %) | Precision (RSD, %) | ||
| RE, relative error; RSD, relative standard deviation. | |||||||
| Salbutamol sulphate | 1.0 | 1.0 | −0.3 | 12.8 | 1.0 | 3.9 | 4.8 |
| 3.0 | 3.2 | +6.3 | 8.3 | 3.2 | 5.4 | 11.0 | |
| 10.0 | 10.4 | +4.2 | 10.7 | 9.8 | −1.9 | 2.1 | |
| 15.0 | 15.8 | +5.3 | 8.7 | 14.7 | −2.1 | 5.1 | |
| Glycopyrronium bromide | 1.0 | 1.2 | +8.1 | 7.4 | 1.0 | −5.3 | 10.5 |
| 3.0 | 3.3 | +5.3 | 10.2 | 3.3 | 9.6 | 7.5 | |
| 10.0 | 11.3 | +9.1 | 11.7 | 10.1 | 0.5 | 6.9 | |
| 15.0 | 16.9 | +7.8 | 11.4 | 15.7 | 4.2 | 5.4 | |
| Budesonide | 1.0 | 1.1 | +9.9 | 11.5 | 1.1 | 9.1 | 7.3 |
| 3.0 | 2.7 | −12.4 | 6.3 | 3.2 | 4.8 | 7.9 | |
| 10.0 | 9.8 | −1.8 | 6.0 | 9.8 | −2.2 | 7.1 | |
| 15.0 | 15.4 | +2.8 | 5.0 | 16.0 | 6.1 | 5.3 | |
4. Conclusions
A unified UHPLC method was successfully developed and validated for the simultaneous quantification of budesonide, glycopyrronium bromide, and salbutamol sulphate, key APIs in inhalation therapy for asthma, COPD, and ACO. Using AQbD principles, the method was optimized with a strong understanding of critical parameters and validated according to ICH Q2 (R2) for pharmaceutical use. The approach was further extended to the bioanalytical field, with successful adaptation and validation in human plasma following ICH M10 guidelines. This demonstrates the feasibility and value of applying AQbD to bioanalysis, enhancing method robustness, flexibility, and regulatory confidence. By integrating AQbD into both pharmaceutical and bioanalytical workflows, this work supports more efficient, consistent, and science-based analytical development across the drug lifecycle.Author contributions
Alessio Gaggero: methodology, data curation, investigation, visualization, formal analysis, writing – original draft preparation. Dalibor Jeremic: formal analysis, investigation, writing – original draft preparation. Anna Fedorko: investigation, formal analysis, methodology, writing – reviewing and editing. Jesús Alberto Afonso Urich: conceptualization, visualization, investigation, supervision, project administration, resources, writing – original draft preparation.Conflicts of interest
The authors declare no conflict of interest.Data availability
All data and materials are present in the manuscript and supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5an01086a.Acknowledgements
This research was funded within the framework of COMET - Competence Centers for Excellent Technologies by BMK, BMAW, Land Steiermark, and SFG. The COMET program is managed by the FFG.References
- M. Hardin, E. K. Silverman, R. G. Barr, N. N. Hansel, J. D. Schroeder, B. J. Make, J. D. Crapo and C. P. Hersh, The clinical features of the overlap between COPD and asthma for the COPDGene Investigators, Respir. Res., 2011, 12, 1–8 CrossRef https://respiratory-research.biomedcentral.com/track/pdf/10.1186/1465-9921-12-127?site=respiratory-research.biomedcentral.com.
- D. J. Maselli, M. Hardin, S. A. Christenson, N. A. Hanania, C. P. Hersh, S. G. Adams, A. Anzueto, J. I. Peters, M. L. K. Han and F. J. Martinez, Clinical Approach to the Therapy of Asthma-COPD Overlap, Chest, 2019, 155, 168–177, DOI:10.1016/j.chest.2018.07.028.
- World Health Organization (WHO), Chronic obstructive pulmonary disease (COPD), 2024. https://www.who.int/news-room/fact-sheets/detail/chronic-obstructive-pulmonary-disease-(copd) (accessed February 24, 2025).
- C. Abbafati, K. M. Abbas and M. Abbasi, et al., Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019, Lancet, 2020, 396, 1204–1222, DOI:10.1016/S0140-6736(20)30925-9.
- N. J. Gross and P. J. Barnes, New therapies for asthma and chronic obstructive pulmonary disease, Am. J. Respir. Crit. Care Med., 2017, 195, 159–166, DOI:10.1164/rccm.201610-2074PP.
- D. Nikjoo, I. van der Zwaan, J. Rudén and G. Frenning, Engineered microparticles of hyaluronic acid hydrogel for controlled pulmonary release of salbutamol sulphate, Int. J. Pharm., 2023, 643, 1–10, DOI:10.1016/j.ijpharm.2023.123225.
- H. Arievich, T. Overend, D. Renard, M. Gibbs, V. Alagappan, M. Looby and D. Banerji, A novel model-based approach for dose determination of glycopyrronium bromide in COPD, BMC Pulm. Med., 2012, 12, 74, DOI:10.1186/1471-2466-12-74.
- D. J. S. Singh, J. J. Parmar, D. D. Hegde, A. A. Lohade, P. Soni, A. Samad and M. D. Menon, Development and evaluation of dry powder inhalation system of terbutaline sulphate for better management of asthma, Int. J. Adv. Pharm. Sci., 2010, 1, 133–141, DOI:10.5138/ijaps.2010.0976.1055.01015.
- X. Tu, C. Donovan, R. Y. Kim, P. A. B. Wark, J. C. Horvat and P. M. Hansbro, Asthma-COPD overlap: Current understanding and the utility of experimental models, Eur. Respir. Rev., 2021, 30, 159, DOI:10.1183/16000617.0185-2019.
- M. Olszanecka-Glinianowicz, J. Chudek and A. Almgren-Rachtan, Therapeutic preferences and factors determining the use of inhaled corticosteroids with long-acting β2-agonists in patients with asthma and chronic obstructive pulmonary disease, Postep. Dermatologii i Alergol., 2021, 38, 752–760, DOI:10.5114/ada.2020.99945.
- B. G. Cosio, J. B. Soriano, J. L. López-Campos, M. Calle-Rubio, J. J. Soler-Cataluna, J. P. De-Torres, J. M. Marín, C. Martínez-Gonzalez, P. De Lucas, I. Mir, G. Peces-Barba, N. Feu-Collado, I. Solanes, I. Alfageme and C. Casanova, Defining the asthma-COPD overlap syndrome in a COPD Cohort, Chest, 2016, 149, 45–52, DOI:10.1378/chest.15-1055.
- M. J. Parnham, V. Norris, J. A. Kricker, T. Gudjonsson and C. P. Page, Prospects for macrolide therapy of asthma and COPD, Adv. Pharmacol., 2023, 98, 83–110, DOI:10.1016/bs.apha.2023.03.002.
- Drug Bank, Albuterol Sulfate Drug Entry. https://go.drugbank.com/salts/DBSALT000257 (accessed August 27, 2025).
- Drug Bank, Glycopyrronium Drug Entry. https://go.drugbank.com/drugs/DB00986 (accessed August 27, 2025).
- Drug Bank, Budesonide Drug Entry. https://go.drugbank.com/drugs/DB01222 (accessed August 27, 2025).
- T. Verch, C. Campa, C. C. Chéry, R. Frenkel, T. Graul, N. Jaya, B. Nakhle, J. Springall, J. Starkey, J. Wypych and T. Ranheim, Analytical Quality by Design, Life Cycle Management, and Method Control, AAPS J., 2022, 24, 34, DOI:10.1208/s12248-022-00685-2.
- J. Zheng, Formulation and Analytical Development for Low–Dose Oral Drug Products, John Wiley & Sons Inc., 2009. DOI:10.1002/9780470386361.
- United States Pharmacopeia (USP), 〈601〉 Inhalation and Nasal Drug Products: Aerosols, Sprays, and Powders—Performance Quality Tests, 2024. DOI:10.31003/USPNF_M99360_06_01(accessed May 13, 2025).
- H. Yoshida, A. Kuwana, H. Shibata, K. Izutsu and Y. Goda, Comparison of Aerodynamic Particle Size Distribution Between a Next Generation Impactor and a Cascade Impactor at a Range of Flow Rates, AAPS PharmSciTech, 2017, 18, 646–653, DOI:10.1208/s12249-016-0544-9.
- S. Beg and M. Rahman, Analytical quality by design for liquid chromatographic method development, in: Handb. Anal. Qual. by Des, Elsevier, 2021, pp. 87–97. DOI:10.1016/B978-0-12-820332-3.00010-8.
- International Council for Harmonisation, Guideline Q2 (R2) Validation of Analytical Procedures, 2022, pp. 1–24. https://database.ich.org/sites/default/files/ICH_Q2%28R2%29_Guideline_2023_1130.pdf (accessed August 27, 2025) Search PubMed.
- International Council of Harmonization, ICH guideline M10 on bioanalytical method validation and study sample analysis, 2022. https://database.ich.org/sites/default/files/M10_Guideline_Step4_2022_0524.pdf (accessed May 8, 2025) Search PubMed.
- C. for B.E. and R. U.S. Food and Drug Administration, Center for Drug Evaluation and Research, Bioanalytical Method Validation - Guidance for Industry, 2018. https://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/default.htmand/ or https://www.fda.gov/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/default.htm (accessed May 8, 2025) Search PubMed.
- M. Gu, A. Gehman, B. Nifong, A. P. Mayer, V. Li, M. Birchler, K. Wang and H. Tang, From Guidelines to Implementation: A Case Study on Applying ICH M10 for Bioanalytical Assay Cross-Validation, AAPS J., 2025, 27, 1–10, DOI:10.1208/s12248-025-01038-5.
- J. P. Danaceau and M. E. Trudeau, A Simple, Broadly Applicable Automated Bioanalytical Sample Preparation Strategy for LC-MS Quantification of Apixaban: Evaluation of Common Bioanalytical Extraction Techniques, 2023. https://www.waters.com/content/dam/waters/en/app-notes/2023/720007946/720007946-en.pdf (accessed August 27, 2025) Search PubMed.
- M. L. Williams, A. A. Olomukoro, R. V. Emmons, N. H. Godage and E. Gionfriddo, Matrix effects demystified: Strategies for resolving challenges in analytical separations of complex samples, J. Sep. Sci., 2023, 46, 41–74, DOI:10.1002/jssc.202300571.
- A. Gaggero, V. Marko, D. Jeremic, C. Tetyczka, P. Caisse and J. A. Afonso Urich, AQbD-Based UPLC-ELSD Method for Quantifying Medium Chain Triglycerides in Labrafac™ WL 1349 for Nanoemulsion Applications, Molecules, 2025, 30, 1–19, DOI:10.3390/molecules30030486.
- J. A. Afonso Urich, V. Marko, K. Boehm, R. A. Lara Garcia, A. Fedorko, S. Salar-Behzadi and D. Jeremic, Stability-Indicating UPLC-PDA-QDa Methodology for Carvedilol and Felodipine in Fixed-Dose Combinations Using AQbD Principles, Sci. Pharm., 2024, 92, 22, DOI:10.3390/scipharm92020022.
- S. Venkataraman and M. Manasa, Forced degradation studies: Regulatory guidance, characterization of drugs, and their degradation products - A review, Drug Invent. Today, 2018, 10, 2 Search PubMed , accessed August 27, 2025: https://www.scribd.com/document/407201667/DIT-article-pdf.
- O.O.R.Q. Food and Drug Administration Affairs, ORA Lab Manual Vol. II - Methods, Method Verification and Validation (ORA- LAB.5.4.5), Food Drug Adm. Off. Regul. Aff. Qual., 2023, 1–34. https://www.fda.gov/science-research/field-science-and-laboratories/field-science-laboratory-manual (accessed August 27, 2025).
- International Council for Harmonisation, Guideline Q14, Analytical Procedure Development, 2023, pp. 1–36. https://database.ich.org/sites/default/files/ICH_Q14_Guideline_2023_1116_1.pdf (accessed August 27, 2025) Search PubMed.
- USP-NF 〈1220〉 Analytical Procedure Life Cycle, (n.d.). https://online.uspnf.com/uspnf/document/1_GUID-35D7E47E-65E5-49B7-B4CC-4D96FA230821_2_en-US?source=Activity (accessed March 27, 2025).
- L. Volta e Sousa, R. Gonçalves, J. C. Menezes and A. Ramos, Analytical Method Lifecycle Management in Pharmaceutical Industry: a Review, AAPS PharmSciTech, 2021, 22, 128, DOI:10.1208/s12249-021-01960-9.
- A. Gaggero, D. Jeremic, R. Sattler, A. Paudel and J. A. Afonso Urich, Evaluation of Analytical Quality by Design Approaches on the Development and Optimization of Size Exclusion Chromatography Analytical Procedures for Bovine Serum Albumin, J. Sep. Sci., 2025, 48, e70168, DOI:10.1002/jssc.70168.
- B. J. Lipworth and D. J. Clark, Comparative lung delivery of salbutamol given via Turbuhaler and Diskus dry powder inhaler devices, Eur. J. Clin. Pharmacol., 1997, 53, 47–49, DOI:10.1007/s002280050335.
- AstraZeneca Canada Inc., Pulmicort Turbuhaler® 200 mcg Product Monograph, 2017, pp. 1–22. https://www.astrazeneca.ca/content/dam/az-ca/downloads/productinformation/pulmicort-turbuhaler-product-monograph-en.pdf (accessed August 27, 2025) Search PubMed.
- AstraZeneca, Product Monograph Pulmicort Nebuamp, 2017, pp. 1–24. https://www.astrazeneca.ca/content/dam/az-ca/downloads/productinformation/pulmicort-nebuamp-product-monograph-en.pdf (accessed August 27, 2025) Search PubMed.
- C. Bartels, M. Looby, R. Sechaud and G. Kaiser, Determination of the pharmacokinetics of glycopyrronium in the lung using a population pharmacokinetic modelling approach, Br. J. Clin. Pharmacol., 2013, 76, 868–879, DOI:10.1111/bcp.12118.
- J. A. Afonso Urich, V. Marko, A. Fedorko and D. Jeremic, AQbD-based development of a stability-indicating UHPLC-PDA-QDa method for triptorelin in parenteral formulations, Anal. Methods, 2025, 6459–6469, 10.1039/d5ay00919g.
- United States Pharmacopeia (USP), USP-NF 〈621〉 Chromatography. https://online.uspnf.com/uspnf/document/1_GUID-6C3DF8B8-D12E-4253-A0E7-6855670CDB7B_8_en-US?source=Activity (accessed March 27, 2025).
- United States Pharmacopeia (USP), USP43-NF38, <2> Oral Drug Prod. Qual. Tests (n.d.). https://doi.usp.org/USPNF/USPNF_M3211_06_01.html (accessed August 27, 2025).
- S. Focaroli, P. T. Mah, J. E. Hastedt, I. Gitlin, S. Oscarson, J. V. Fahy and A. M. Healy, A Design of Experiment (DoE) approach to optimise spray drying process conditions for the production of trehalose/leucine formulations with application in pulmonary delivery, Int. J. Pharm., 2019, 562, 228–240, DOI:10.1016/j.ijpharm.2019.03.004.
- A. Alcázar, J. M. Jurado and A. G. González, Gradient scouting in reversed-phase HPLC revisited, J. Chem. Educ., 2011, 88, 74–76, DOI:10.1021/ed100506c.
- S. S. Shapiro and M. B. Wilk, An Analysis of Variance Test for Normality (Complete Samples), Biometrika, 1965, 52, 591, DOI:10.2307/2333709.
- P. Mishra, U. Singh, C. Pandey, P. Mishra and G. Pandey, Application of student’s t-test, analysis of variance, and covariance, Ann. Cardiac Anaesth., 2019, 22, 407, DOI:10.4103/aca.ACA_94_19.
- S. Belouafa, F. Habti, S. Benhar, B. Belafkih, S. Tayane, S. Hamdouch, A. Bennamara and A. Abourriche, Statistical tools and approaches to validate analytical methods: Methodology and practical examples, Int. J. Metrol. Qual. Eng., 2017, 8, 9, DOI:10.1051/ijmqe/2016030.
- T. Tome, N. Žigart, Z. Časar and A. Obreza, Development and Optimization of Liquid Chromatography Analytical Methods by Using AQbD Principles: Overview and Recent Advances, Org. Process Res. Dev., 2019, 23, 1784–1802, DOI:10.1021/acs.oprd.9b00238.
| This journal is © The Royal Society of Chemistry 2026 |



