Boosting living Bacillus spore identification: Kolmogorov–Arnold network-guided convolutional neural network combined with laser tweezers Raman spectroscopy
Yifan
Sun†
a,
Xiao
Peng†
a,
Fusheng
Du
b,
Lin
He
b,
Yuan
Lu
*c,
Yufeng
Yuan
 *b and
Junle
Qu
*b and
Junle
Qu
 a
a
aState Key Laboratory of Radio Frequency Heterogeneous Integration (Shenzhen University), College of Physics and Optoelectronic Engineering, Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province, Shenzhen University, Shenzhen, Guangdong 518060, China
bSchool of Electronic Engineering and Intelligentization, Dongguan University of Technology, Dongguan, Guangdong 523808, China. E-mail: yufengyuan@dgut.edu.cn
cThe Sixth People's Hospital of Shenzhen University, Shenzhen University, Shenzhen, Guangdong 518060, China. E-mail: chfsums@163.com
First published on 14th November 2025
Abstract
As primary carriers of foodborne and zoonotic diseases, Bacillus spores can pose a serious threat to food microbiology and human disease. Thus, the precise identification of Bacillus spores is of great significance for ensuring food safety and human health. Herein, this study proposed a living Bacillus spore identification platform: an adaptive Kolmogorov-Arnold network (KAN)-guided convolutional neural network (CNN) configuration combined with laser tweezers Raman spectroscopy (LTRS). To address the small size of the original single-cell Raman spectral datasets, Gaussian noise-based spectra augmentation was employed to significantly enlarge and enrich it. When the adaptive KAN was introduced into the dense layer of CNN, the prediction accuracy of five Bacillus spore species was as high as 97.80% ± 1.79%. Moreover, the KAN-guided CNN configuration has strong robustness and generalization ability, providing a prediction accuracy of 96% for an independent spectral dataset. To figure out the classification contribution of each Raman band, a blocking individual Raman band method was proposed. The Raman band located at 1655 cm−1, belonging to the amide I vibration of protein, was determined as the dominant contributor, surpassing two Raman bands belonging to Ca-DPA at 1576 cm−1 and 1449 cm−1. It can be foreseen that the KAN-guided CNN configuration combined with LTRS shows great promise for determining microbial identity, especially for unculturable microorganisms.
1. Introduction
Bacterial survival in various extreme environments is highly dependent on well-known adaptive strategies, enabling them to transiently orchestrate gene expression.1 Among these adaptive strategies, sporulation is a paradigm of evolutionary ingenuity,2 in which vegetative bacterial cells experience a typical developmental process and change into metabolically inert spores. It is well-known that the typical bacterial spore structure can be widely found in various ecological sites, such as soil matrices, aquatic ecosystems, and atmospheric aerosols.3 Moreover, dormant bacterial spores can tolerate various harsh external stresses, such as radiation,4 heat,5 and long-term drought.6 Unfortunately, the resultant Bacillus spores can pose a serious threat for both food microbiology and human disease. For example, as primary carriers of foodborne and zoonotic diseases, Bacillus spores are the main culprit of multiple diseases, such as botulism, anthrax, and tetanus, seriously threatening the global public health security.7 Therefore, developing a Bacillus spore identification platform that integrates high sensitivity with specificity is an imperative need.Generally, conventional approaches that are employed for spore identification heavily rely on phenotypic characterization. Standard protocols involve culturing spores on solid agar media, followed by both morphological and physiological analysis guided by skilled staff.8,9 In addition, molecular techniques including species-specific genetic probes and sequence-based assays have enhanced the discrimination accuracy of Bacillus spores.10,11 Moreover, both biosensors12 and ratiometric fluorescence nanoprobes13 have been proposed, utilizing engineered molecular recognition for targeted detection. Although the reliability of these mature methods have been verified, they still have inherent limitations. For example, phenotypic characterization requires a long incubation period, rendering it unfriendly for non-culturable spore species. In addition, biochemical assays usually require destructive sampling, making it unsuitable for living cell characterization. Moreover, both biosensor and nanoprobe designs require intricate preparation processes. Finally, it is almost impossible to achieve single-cell resolution through conventional platforms. Therefore, it is of great significance to develop a non-invasive, rapid, and high-precision characterization strategy at a single-cell level.
Confocal Raman spectroscopy has been considered as a promising solution for revealing rich information about the biological composition in living cells through identifying molecular fingerprint vibrations. However, confocal Raman spectroscopy usually suffers from low signal-to-noise ratios (SNRs) of an individual cell. Fortunately, this drawback can be addressed by well-known laser tweezers Raman spectroscopy (LTRS), which integrates an optical trapping technology with confocal Raman spectroscopy. To overcome the signal fluctuation of Raman scattering caused by Brownian motion, the optical trapping technology can efficiently capture and stabilize individual microparticles, resulting in high-quality Raman scattering signals.14,15 To date, LTRS has been widely employed in revealing biomolecular information in bacteria, spores,16 organelles,17 and environmental pollutant microplastics.18 Although the high SNR Raman scattering of a single cell is possible, it remains to be improved in directly distinguishing similar species, due to the spectral similarity originating from the overlapped biomolecular composition. Moreover, raw Raman spectra usually contain background noise, requiring a data processing algorithm to extract discriminative spectral features.
Conventional chemometric approaches, including hierarchical cluster analysis (HCA), linear discriminant analysis (LDA), and principal component analysis (PCA), were firstly proposed to deal with the species classification of Raman spectra, depending on the statistical models to reduce the data dimensionality.19,20 However, they cannot efficiently process high-dimensional datasets. Afterwards, various early machine learning algorithms such as support vector machines (SVM) and Random Forest were further developed by guiding computers to learn from large spectra datasets, significantly enhancing the analytical efficiency.19,21,22 However, they also possess respective drawbacks. For example, SVM is proficient in binary classification tasks rather than in multiclass tasks, and Random Forest may neglect critical spectral features in the ensemble learning process. Based on these observations, both conventional chemometrics and early machine learning approaches cannot satisfy the specific demand for accurately classifying similar species.
In recent years, deep learning, an advanced branch of machine learning, has brought significant advancements in Raman spectral analysis. Among various networks, CNN is the most widely employed to hierarchically extract spectra features from massive datasets, enabling it to automatically identify discriminative patterns. To date, the excellent identification capability of the CNN model has facilitated the precise prediction of marine bacteria and pathogens,23,24 gradually extending to perform cancer diagnostics.25–27 However, single CNN exhibits limited feature extraction capability when processing highly similar datasets. The reason is that the multilayer perceptron (MLP) widely employed in the dense layer of the CNN model has a low capability of feature extraction only via fixed activation functions. Fortunately, an adaptive network called KAN shows strong learning capability through the use of learnable activation functions. Therefore, a co-functioned configuration incorporating KAN with CNN has shown enhanced flexibility in capturing nonlinear data patterns.28,29
Inspired by these observations, we proposed a novel strategy to precisely identify living Bacillus spores using a single-cell platform LTRS combining with KAN-guided CNN architecture. Firstly, the original single-cell Raman spectra from five Bacillus spore species were collected by our home-built LTRS platform. To overcome the limit of the small size of the original single-cell Raman spectra datasets, the Gaussian noise-based data augmentation approach was employed to significantly enlarge the size of the original single-cell Raman spectra datasets. Afterwards, the classification performance of the KAN-guided CNN architecture was optimized by tuning the CNN iteration. Finally, through the introduction of a blocking band algorithm, the classification weight from each Raman band was systematically evaluated. The main innovations of our proposed method can be summarized in three aspects: (1) Our home-built LTRS platform can achieve non-invasive and high-quality Raman scattering signals from Bacillus spores in a single-cell level; (2) A novel deep learning architecture integrating CNN with KAN was developed to achieve Bacillus spore identification with high prediction accuracy, providing better performance than conventional machine learning models; (3) Beyond achieving high prediction accuracy, the interpretable blocking band algorithm was introduced to explain the classification process of the model and provide biologically interpretable insights.
2. Experimental section
2.1. Bacteria strains and spore preparation
In our study, five Bacillus strains were employed to cultivate mature spores: Bacillus marisflavi (MCCC1K02430), Bacillus aryabhattai (MCCC1K02966), Bacillus subtilis (CICC63501), Bacillus pumilus (CICC22276), and Bacillus cereus (CICC22369). Both Bacillus marisflavi and Bacillus aryabhattai strains were gifted by the Institute of Microbiology, Chinese Academy of Sciences (Beijing, China), and the other three strains were bought from the China Center of Industrial Culture Collection (CICC).To reduce the yielded spore heterogeneity in the same Bacillus strain, a well-known approach named Percoll discontinuous density gradient centrifugation was employed to synchronize the vegetative Bacillus cells. Afterwards, agar plates containing nutrient broth were employed to cultivate vegetative Bacillus cells. It is worth noting that there were two different nutrient broths involved in the spore culture process. In addition, the nutrient broth (CM1168; Thermo Scientific, UK) was employed to incubate Bacillus subtilis, Bacillus pumilus, and Bacillus cereus seed cells, respectively. Both Bacillus marisflavi and Bacillus aryabhattai seed cells were cultured in marine nutrient broth 2216 (Becton Dickinson, USA). To enhance the spore yield, 10 μL MnSO4 solution (50 g L−1, ≥99.5% purity; Tianjin Kermel Chemical Reagent Co., China) was injected into 100 mL nutrient broth supplemented with 1.5% agar, due to the supplementation of Mn2+ significantly improving the sporulation yield in the Bacillus species.30 Prior to cell inoculation, the agar plates were sterilized by autoclave. Then, the vegetative seed cells from five Bacillus strains were coated onto five sterilized agar plates, and all plates were held into a culture incubator at 37 °C. After a culture period of 120 hours, many Bacillus colonies had formed on the agar plates. To obtain synchronized spores, a single Bacillus colony of each Bacillus strain was selected and further purified by high-speed centrifugation, which was performed at 5000 rpm for 25 minutes. Next, the spore precipitate was washed by high-speed centrifugation three times. Finally, the yielded mature spores were conserved and sealed into sterile ultrapure water at a temperature of 4 °C.
2.2. Laser tweezers Raman spectroscopy
To acquire the single-cell Raman spectra of Bacillus spores, a single-beam LTRS setup was constructed, as shown in Fig. 1. Unlike commercial LTRS systems equipped with two laser beams (optical trapping beam, Raman scattering excitation beam), our home-built LTRS system was constructed based on a single laser beam, which has dual functions of both optical trapping and Raman scattering excitation. In brief, a 532 nm laser beam (solid state, continuous wave) was incident with an inverted microscope (Nikon Eclipse Ti2). With the help of an oil immersion objective (100×, numerical aperture = 1.40), a stable optical trapping well was generated in the focal plane, enabling it to capture individual cells in the liquid physiological environment. Meanwhile, the Raman scattering signals of trapped cells were excited by the same 532 nm laser beam. Finally, the Raman scattering signals were collected and transported into a high-resolution spectrometer (Horiba LabRAM HR Evolution). The spectral resolution in our LTRS system is approximately 4 cm−1. To guarantee both spectra quality and reproducibility, the LTRS platform was calibrated by capturing 2 µm polystyrene spheres, which have three well-known Raman bands at 620, 1001 and 1602 cm−1.16In addition, the laser spot focused on the spore sample had a diameter of 1.5 μm, and the laser power on the spore sample was measured to be 3.8 mW. Our previous study has shown that a laser power of 3.8 mW has little to no damage on the Raman spectra of Bacillus spores.31 For each species of Bacillus spore, approximately 150 single-cell Raman spectra of trapped spores were measured, and the acquisition time for each spore was 10 s. Furthermore, for each species of Bacillus spore, they were collected from two independent experiment runs of spore culture including 100 (first run) and 50 (second run), respectively. The background Raman spectra were also measured, facilitating the extraction of the original Raman spectra of the trapped spores.
2.3. Spectra augmentation
After removing the background Raman spectra, the original Raman spectra of the trapped spores were further preprocessed by baseline correction, Savitzky–Golay smoothing, and min-max normalization. In detail, the baseline correction was performed by the well-known Adaptive Iteratively Reweighted Penalized Least Squares (airPLS) algorithm, and the critical parameters in Savitzky–Golay smoothing were fixed in a window length of 5 and a polynomial order of 2. In addition, the min-max normalization was performed by scaling the intensities of each Raman spectrum into the range from 0 to 1. Considering that CNN is a heavily data-hungry configuration, it must be optimized and trained on massive data. In theory, the classification performance of a CNN improves with the amount of data it is trained on. In contrast, small sample sizes are likely to significantly increase the risk of overfitting. In our single-cell Raman spectra datasets, there were a total of 753 spectra from five Bacillus species. To ensure the diversity of datasets, the spectra were obtained from two independent experiment runs of spore culture. Obviously, the number of Raman spectra from each Bacillus species was small, which was insufficient for training a CNN model. Therefore, it is important to enlarge the spectral data size via a reliable approach.It has been reported that data augmentation strategies such as Gaussian noise injection, spectra shifting, and spectra combination, were proposed to enlarge the size of datasets.32,33 It can be found that an obvious advantage of spectral augmentation is to significantly reduce the time required for measuring the spectra.
In our study, a controlled Gaussian noise injection strategy was employed to augment both the size and diversity of the single-cell Raman spectra datasets. In brief, the Gaussian noise was extracted by estimating the standard deviation at each wavenumber point across the original Raman spectra. This process was defined by the equation: y′(v) = y(v) + ε, where ε ∼ N(0(α·σ(ν))2). Here, ε represents the Gaussian noise term extracted from a normal distribution with zero mean and standard deviation α·σ(ν). The parameter α denotes the noise intensity level, and σ(ν) stands for the standard deviation of spectral intensity at wavenumber ν computed across the entire training dataset.
Afterwards, the simulated Gaussian noise was injected into the original spectra data. Therefore, the new single-cell Raman spectra dataset was composed of two parts: the original spectra and the newly generated simulated Raman spectra. In detail, 10% of standard deviation was injected into the original spectra data to form the new single-cell Raman spectra datasets. With the help of various degrees of spectral augmentation, the obtained single-cell Raman spectra datasets are presented in Table S1. For each augmentation operation, in addition to the original single-cell Raman spectra from each Bacillus species, there was another simulated Raman spectra.
2.4. KAN-guided CNN configuration
To match with the one-dimensional single-cell Raman spectra of the trapped spores, the one-dimensional CNN framework was selected to reveal the local spectral features through multiple convolution operations. The KAN-guided CNN architecture was created by embedding a KAN function into the dense layer of CNN, as shown in Fig. 2. In the CNN configuration, there is an input layer, two convolutional layers, two max pooling layers, a flatten layer, a dense layer and an output layer. First, the input layer serves as the initial layer, which obtains the input Raman spectra datasets. Second, the role of the convolution layer is to extract the features of the input Raman spectral data. For the two convolution layers with a stride of 1, the kernel size is fixed at 1 × 3, and the number of convolution kernels is 16 and 64, respectively. Third, the max pooling layer is employed to reduce the dimension of output spectra feature vectors and store the critical spectra feature information. For the two pooling layers with a stride of 2, the number of input channels is 16, and 64, respectively. After the pooling layer, the introduction of flatten layer is to reshape the spectra feature map into a one-dimensional vector array. Subsequently, a dense layer, also called a fully connected layer, is employed to linearly transform the spectral feature vector through a weight matrix. Finally, the SoftMax function is the activation function of the output layer, providing a probability distribution of the input validation Raman spectra datasets belonging to a specific Bacillus species.It is worth noting that, in the dense layer of the CNN model, the MLP layer is usually introduced. As shown in Fig. S1, the MLP consists of interconnected neurons, where each neuron can process the input through learnable weights that can determine the influence of the specific input on the output. Moreover, these weights in combination with fixed activation functions can transform the input into a temporary output, and the final output highly depends on the CNN training. During the CNN training, the MLP can enhance the identification ability via iteratively optimizing weights. However, the MLP layer also has inherent limitations. For example, the high dependency on both fixed activation functions and learnable weights restricts the CNN model to match complex data sequences, resulting in low feature discrimination.34
To overcome the drawbacks of MLP, the KAN algorithm was proposed to replace MLP by introducing a paradigm shift, as illustrated in Fig. 2. In the KAN model, each input neuron is required to pass through adaptive functions, and the outputs are performed through a sum operation rather than linear weight multiplication in MLP, as shown in Fig. S1. The structure development enables KAN to represent a multivariate continuous function with superior flexibility, especially for complex spectral patterns. In brief, the calculation formulation of KAN is defined as:
 | (1) |
ϕ(x) = wbb(x) + ws![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) spline(x) spline(x) | (2) |
2.5. Conventional machine learning models
To evaluate the classification performance of the KAN-guided CNN platform, four conventional machine learning algorithms (including Random Forest, XGBoost, SVM, and K-Nearest Neighbors (KNN)) were employed to process the same Raman spectra datasets as our proposed KAN-guided CNN platform. To eliminate the ordering bias, the single-cell Raman spectra of Bacillus spores were initially randomized. Afterwards, a grouped cross-validation strategy was employed to separate the single-cell Raman spectra datasets into three parts: training (70%), testing (20%), and validating (10%) subdatasets. To guarantee the general robustness, at least 10 independent operations were performed for each conventional machine learning model.2.6. Classification contribution of Raman bands
To systematically evaluate the effect of specific spectral regions on the classification performance of the KAN-guided CNN platform, the whole Raman spectra from 400 to 1800 cm−1 were initially divided into seven sub-intervals with an identical range of 200 cm−1. Afterwards, a defined method called blocking single Raman band was proposed. In this study, all the intensity values within a blocked band were set to be zero, effectively removing the spectra information in this region. Then, these modified Raman spectra were input to the KAN-guided CNN platform for training and classification. The relative contribution C of each blocked band was calculated as: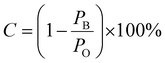 , where PO denotes the classification accuracy using the whole original spectra dataset, and PB represents the classification accuracy obtained by blocking the target band. To comprehensively evaluate all the typical Raman bands, a sliding window with an identical range of 200 cm−1 was gradually moved across all the intervals. Finally, the relative contribution for each Raman band was averaged by running 10 independent operations to mitigate the stochastic variations.
, where PO denotes the classification accuracy using the whole original spectra dataset, and PB represents the classification accuracy obtained by blocking the target band. To comprehensively evaluate all the typical Raman bands, a sliding window with an identical range of 200 cm−1 was gradually moved across all the intervals. Finally, the relative contribution for each Raman band was averaged by running 10 independent operations to mitigate the stochastic variations.
3. Results and discussion
3.1. Single-cell Raman spectra of Bacillus spores
With the help of our home-built LTRS system, the normalized single-cell Raman spectra originating from five Bacillus spore species, including Bacillus marisflavi (MCCC1K02430), Bacillus aryabhattai (MCCC1K02966), Bacillus subtilis (CICC63501), Bacillus pumilus (CICC22276), and Bacillus cereus (CICC22369), were obtained, as shown in Fig. 3. It can be found that, the single-cell Raman spectra can reveal the signatures of both calcium dipicolinic acid (Ca-DPA) and proteins in living Bacillus spores, as shown in Table 1. Moreover, the typical biomarker Ca-DPA is the main component, whose characteristic Raman bands are located at 660, 826, 1017, 1397, 1449, and 1576 cm−1, respectively.35 In fact, the spectral heterogeneity in both intra-species and inter-species comparison was observed in the original single-cell Raman spectra of five Bacillus species. For the trapped spores originating from an identical single clonal colony, the Raman intensity of four typical Raman bands (1017, 1397, 1449, and 1576 cm−1) were obviously heterogeneous, as depicted in Fig. S2(A–E). Except for the spectral heterogeneity of the intra-species, the spectral heterogeneity of the inter-species was also found in Fig. S2(A–F). To further reduce the effect of intra-species spectral variability on the CNN model classification, spectral normalization was employed to deal with the original single-cell Raman spectra of five Bacillus spores, largely weakening the heterogeneity in the relative peak intensity. Although the spectral normalization was performed, there was slight spectral heterogeneity in an identical clonal colony due to the phenotypic heterogeneity of Bacillus spores. It is worth noting that this slight spectral heterogeneity is not an obstacle for CNN training iteration. Conversely, it stands for spectral data diversity, which is advantageous for enhancing the robustness of the CNN model. Although the spectral heterogeneity exists in five Bacillus species, it is still impossible to differentiate the Bacillus species through directly extracting the spectral differences. We also confirmed that the laser irradiation in our LTRS hardly generated phototoxicity on living Bacillus spores in our previous work.| Raman band (cm−1) | Assignment | Vibrational mode |
|---|---|---|
| 660 | Ca-DPA | Bending vibration of C–C in pyridine ring |
| 826 | Ca-DPA | Out-of-plane deformation of C–H |
| 1017 | Ca-DPA | Symmetric stretching of pyridine ring |
| 1250 | Amide III | C–N stretching and N–H bending |
| 1397 | Ca-DPA | Symmetric stretching of O–C–O |
| 1449 | Lipids and Ca-DPA | Symmetric bending of C–H in pyridine ring |
| 1576 | Ca-DPA | Asymmetric stretching of O–C–O |
| 1657 | Amide I | Amide I vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
3.2. Bacillus spore identification via KAN-guided CNN model
For the 753 original single-cell Raman spectra from five Bacillus species, a single CNN model was firstly employed to classify the Bacillus species. Similar to conventional machine learning methods, a grouped cross-validation mechanism was also introduced to divide the original single-cell Raman spectral datasets into three sub-datasets: training (70%), testing (20%), and validating (10%) groups. Significantly, the training group was derived from one independent cultured spore batch, while the validating and testing groups were shared with a distant batch. As shown in Fig. S3(A), the spectral dataset was partitioned into 30 splits, with each split containing two distant batches. One entire batch containing seven folds (70%) was used for training. From the other batch, one fold (10%) was designated as the testing dataset, and two folds (20%) were randomly selected from the remaining six folds as the validating dataset. Additionally, two batches were employed as training in sequence. There is a significant advantage that the testing datasets are identical across every 15 splits, but both the training and validating datasets are entirely different. Both Table 2 and Fig. S3(B) show that with the assistance of the grouped cross-validation mechanism, the single CNN model can provide an average prediction accuracy of 89.20% ± 4.21% by running 10 independent training operations. It can be found that the prediction accuracy provided by the single CNN model cannot meet the demand for identifying the Bacillus species with high sensitivity. The main reason is that the number of original single-cell Raman spectra is limited. However, CNN requires massive spectral datasets to be trained.| Data volume | CNN | KAN-guided CNN |
|---|---|---|
| Without data augmentation | 89.20% ± 4.21% | 91.80% ± 3.94% |
| With data augmentation by 200 | 91.60% ± 5.37% | 93.70% ± 3.38% |
| With data augmentation by 300 | 95.87% ± 1.07% | 97.80% ± 1.79% |
| With data augmentation by 400 | 96.10% ± 1.34% | 97.91% ± 1.92% |
| With data augmentation by 500 | 96.52% ± 1.73% | 98.15% ± 2.21% |
To address the spectral data scarcity, Gaussian noise-based spectra augmentation was employed to enlarge the size of the single-cell Raman spectra datasets by extracting the standard deviation of the original Raman spectra dataset. Through injecting a standard deviation of 10% into the original Raman spectra, newly simulated single-cell Raman spectra were formed. It is worth noting that the single-cell Raman spectral datasets employed for CNN training contain two parts: original and simulated Raman spectral datasets, which significantly enhanced the spectra diversity. More cautiously, it is essential to prevent the simulated Raman spectra generated from the validation or testing sets being mixed into the training set, otherwise the risk of data leakage will increase. Therefore, the simulated Raman spectra from the training set should only be employed for the training process.
To validate the reliability of newly simulated single-cell Raman spectra, the comparison between the original and newly simulated Raman spectra datasets was plotted, as shown in Fig. 4 and Fig. S4.
It can be found that the global distribution of newly simulated Raman spectra is well aligned with the original Raman spectra, confirming that the Gaussian noise-based spectra augmentation was effective without distorting critical spectral features. Here, the extraction of 10% noise intensity was driven by calculating the relationships between the spectral intensity and perturbation magnitude. Gaussian noise was assumed to introduce these deviations proportional to the original signal intensity. It means that high-intensity Raman bands are likely to experience larger spectral variations. However, these Raman bands inherently exhibit stronger baseline signals, making them more resilient to noise introduction. Conversely, low-intensity Raman bands holding subtle discriminative features are highly sensitive to the perturbations of spectral signals, and even minor noise could cover these tiny features. In this study, the effects of different levels of Gaussian noise were also studied, as shown in Table S3. When the Gaussian noise level was determined to be 10%, the yielded prediction accuracy was largest. Therefore, the standard deviation of 10% was determined to balance the spectra augmentation outcome and feature preservation.
Next, the effect on the prediction accuracy by integrating a KAN layer into CNN was systematically studied, as shown in Table 2. Compared with the average prediction accuracy of 89.20% ± 4.21% provided by the single CNN, the KAN-guided CNN platform can achieve a higher prediction accuracy of 91.80% ± 3.94% due to the introduction of the KAN layer. More importantly, with further assistance of the Gaussian noise-based augmentation technique, the prediction accuracy enhancement of the KAN-guided CNN platform was faster than the single CNN model. Amazingly, when the newly simulated Raman spectral datasets were further augmented by 300, the prediction accuracy of the KAN-guided CNN platform was as high as 97.80% ± 1.79%, indicating that it can almost perfectly identify five Bacillus species. Therefore, the high prediction accuracy of the KAN-guided CNN platform was attributed to the synergistic mechanism. Firstly, the Gaussian noise-based augmentation enriched the spectral diversity, enabling the CNN model to generalize the subtle interspecies variations. Secondly, the adaptive activation functions of the KAN layer can efficiently capture the nonlinear relationships in spectral datasets, overcoming the limitation of the MLP layer in the dense layer. To further demonstrate the performance of the Gaussian noise-based spectra augmentation, comparative studies were performed by introducing three augmentation strategies including baseline variations, Poisson noise and GAN spectra, as shown in Table S4. It can be seen that they are comparable. However, the baseline variation method may distort the significant Raman spectra. For the Poisson noise, it is essential to dynamically adjust the parameter (scaling factor) to guarantee that the noise is effective for each wavelength. Finally, the GAN spectra augmentation acquires complicated calculation loads, making it difficult to achieve the astringency of the GAN model.36
The effects of different spline order were studied to determine the optimal parameter for KAN, as shown in Table S5. When the spline order of KAN was determined to be 10, the yielded prediction accuracy was the highest. Then, the iteration dynamics of the KAN-guided CNN platform were monitored using two loss functions and Adam optimization algorithm with these parameters (β1 = 0.9, β2 = 0.999, and learning rate = 0.001), and the spline order (the parameter of KAN) was 10. Next, the KAN-guided CNN platform was further optimized by varying the number of epochs from 10 to 500, as shown in Fig. S5. When the number of iteration epochs was smaller than 300, both the training and validation function curves significantly fluctuated, indicating that the KAN-guided CNN platform was underfitting. However, when the number of iteration epochs was larger than 300, both the training and validation function curves exhibited almost stable states, as shown in Fig. 5(A) and Fig. S5(D–F). Moreover, the optimized accuracy from both training and validation datasets were approximately 0.98. More importantly, the KAN-guided CNN platform demonstrated exceptional robustness, achieving an ultra-high prediction accuracy of 97.80% ± 1.79% for all five Bacillus species in at least 10 independent operations, as shown in Table S6. Interestingly, Fig. 5(B) shows that for a single independent operation, the prediction accuracies on both original and simulated spectra are almost the same, indicating that the simulated Raman spectra were well aligned with the experimental Raman spectra. In addition, the misjudged Raman spectra was extracted, as shown in Fig. S7. It clearly shows that the Raman bands at 1657 cm−1 of the misjudged Raman spectra (B. 2430) are highly similar with those of B. pumilus, causing the misjudgment of the KAN-guided CNN platform. Moreover, the yielded receiver operating characteristics (ROC) curves are shown in Fig. 5(C). The per-class precision, recall and F1 scores are also given in Table S7. It can be found that the true positive rates for the five Bacillus species are approximately equal to 1 and the F1 scores are larger than 90%, indicating that our proposed KAN-guided CNN platform has high specificity.
To further verify the excellent robustness and generalization of the KAN-guided CNN platform, a newly constructed single-cell Raman spectral dataset containing 250 spectra from newly cultivated Bacillus spores was collected and introduced to work as an independent validation dataset. Particularly, the dataset was derived from completely new cultured strains in separate batches on different dates, distinct from those used for model training. Through running 10-time independent operation, the optimal KAN-guided CNN platform can provide a prediction accuracy of 96.00% ± 0.63%, as shown in Fig. 5(D) and Table S8. It can be found that our proposed KAN-guided CNN model has excellent generalization capacity.
To show the significant superiority of the KAN-guided CNN platform, comparative studies were performed by introducing four conventional machine learning models and two advanced deep learning models, as shown in Fig. S6 and Table 3. For four conventional machine learning models, Random Forest can achieve the highest accuracy of 88.89% ± 1.09%, followed by SVM (88.59% ± 1.06%), XGBoost (88.22% ± 1.99%), and K-nearest neighbors (84.89% ± 2.57%), respectively. For the two advanced deep learning models, the prediction accuracy of ResNet was approximately 96.83% ± 1.25%, and the prediction accuracy of Transformer was 96.40% ± 1.78%. It can be found that the yielded prediction accuracy of the two advanced deep learning models is a little higher than that for the single CNN model, but lower than the KAN-guided CNN model. It can be concluded that our proposed KAN-guided CNN platform can consistently provide ideal prediction accuracy.
| Classifier | Prediction accuracy |
|---|---|
| KAN-guided CNN | 97.80% ± 1.79% |
| ResNet | 96.83% ± 1.25% |
| Transformer | 96.40% ± 1.78% |
| Single CNN | 95.87% ± 1.07% |
| Random Forest | 88.89% ± 1.09% |
| SVM | 88.59% ± 1.06% |
| XGBoost | 88.22% ± 1.99% |
| KNN | 84.89% ± 2.57% |
3.3. Precise classification contribution based on blocking a single Raman band
To systematically verify the classification contribution of an individual Raman band, the spectral range from 400 to 1800 cm−1 was initially divided into seven contiguous parts with an interval of 200 cm−1. As shown in Fig. 6, the prediction accuracy of each interval was quantified via training the KAN-guided CNN platform with the individual interval of 200 cm−1. For example, the interval of 1000–1200 cm−1 showed the highest prediction accuracy of 94.96%, while the interval of 1400–1600 cm−1 exhibited a prediction accuracy of 86.58%. In the two intervals with high prediction accuracy, there are the main Raman bands at 1017, 1449, and 1576 cm−1. In contrast, the two intervals of 600–800 cm−1 and 1200–1400 cm−1 showed moderate prediction accuracies of <80%. There was one detectable Raman band in each interval. However, for the interval of 400–600 cm−1, the detectable Raman peaks were quite weak, exhibiting poor prediction accuracy of 67.33%. These observations indicated that the presence of Raman spectral bands is a critical determinant of the prediction accuracy. Here, we assumed that bacteria classification, particularly for Bacillus species, highly relies on the Raman spectral bands due to their discriminative spectral features. To validate the relationship between the Raman band and classification performance, 12 spectral intervals were redefined and each spectral interval covered no more than one unique Raman band. By successively blocking an individual Raman band, the modified Raman spectral datasets continued to be trained and predicted by our proposed KAN-guided CNN platform. Then, the relative classification contribution of each Raman band could be quantified.As shown in Table 4 and Fig. 6, the relative weights of band-specific contributions were quantitatively determined. Most prominently, the Raman band at 1657 cm−1 was determined as the highest contributor (3.26%), surpassing the second highest contributor at 1576 cm−1 and the third highest contributor at 1449 cm−1, with registered contributions of 3.04% and 2.89%, respectively. In addition, the two intervals containing Raman bands at 1397 and 1017 cm−1 exhibited moderate contributions. Finally, the Raman bands at 660, 826 and 1250 cm−1 showed negligible effects on the classification contribution. In general, the spectral intervals with higher signal intensities were generally correlated with greater contributions. However, the correlation may deviate when the spectral variability outweighs the intensity. For example, the Raman band at 1655 cm−1 with modest signal intensity demonstrated high classification contribution. In fact, the variability in interspecies reflects the significant differences observed in the Raman spectra from various Bacillus species, as shown in Fig. 3 and Fig. S2. Thus, the classification efficacy is determined not only by peak intensity, but also by the synergistic interplay of both spectral feature and inter-species difference.
| Blocked Raman band (cm−1) | Prediction accuracy | Classification contribution |
|---|---|---|
| 649–687 containing 660 | 98.33% | 1.67% |
| 798–849 containing 826 | 97.33% | 2.67% |
| 982–1062 containing 1017 | 97.25% | 2.75% |
| 1062–1357 containing 1250 | 99.08% | 0.92% |
| 1358–1421 containing 1397 | 97.18% | 2.82% |
| 1421–1515 containing 1449 | 97.11% | 2.89% |
| 1515–1625 containing 1576 | 96.96% | 3.04% |
| 1625–1716 containing 1657 | 96.74% | 3.26% |
4. Conclusion
In this study, we proposed a novel Bacillus spore identification strategy through integrating the LTRS technology with the KAN-guided CNN platform at a single-cell resolution level. To overcome the data scarcity of the original Raman spectra datasets, the controlled Gaussian noise augmentation method was employed to significantly enlarge the size of the Raman spectral datasets and enrich the diversity of spectral datasets though simulating new batches of Raman spectral datasets. By replacing the commonly employed MPL with adaptive KAN, the KAN-guided CNN platform can provide a prediction accuracy of 97.80% ± 1.79% for five Bacillus species, exceeding that of a single CNN model and conventional machine learning methods such as Random Forest, SVM, XGBoost, and KNN. Systematic band-blocking analysis revealed that the Raman band at 1655 cm−1, belonging to the amide I vibration of protein, was determined as the dominant contributor, surpassing the Raman bands of Ca-DPA at 1576 cm−1 and 1449 cm−1. It can be expected that the KAN-guided CNN Raman platform can overcome the limitations of spectral specificity and high-throughput identification, while retaining spore viability, with potential adaptability extending to unculturable microorganisms in food safety, biomedicine diagnostics, and environmental monitoring. By synergizing advanced single-cell Raman spectroscopy with adaptive deep learning algorithms, the platform can reveal the microbial identity and dynamics in physiological states, further advancing single-cell microbiology changing from morphology characterization to intracellular biomolecule insight.Author contributions
Y. F. S. designed the KAN-guided CNN model, drew the graphics and wrote the original manuscript. X. P. revised the graphics and reviewed the original manuscript. F. S. D. measured the single-cell Raman spectra of Bacillus spores. L. H. optimized the LTRS setup. Y. L. supervised the project and reviewed and edited the original manuscript. Y. F. Y. conceived the study, supervised the project, and reviewed and revised original manuscript. J. L. Q. discussed and analyzed the KAN-guided CNN model. Y. F. S. and X. P. contributed equally to this work. All the authors have approved the final version of the manuscript.Conflicts of interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Data availability
Data will be made available from the corresponding author (yufengyuan@dgut.edu.cn) on request. Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5an00834d.Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (62075137), the Guangdong Basic and Applied Basic Research Foundation (2023A1515140161), the Dongguan Science and Technology of Social Development Program (20231800936312), the Sanming Project of Medicine in Shenzhen (No. SZSM202103014), and the high-level talent program of Dongguan University of Technology (No. 221110080).References
- C. Bang, T. Dagan, P. Deines, N. Dubilier, W. J. Duschl, S. Fraune, U. Hentschel, H. Hirt, N. Hülter, T. Lachnit, D. Picazo, L. Pita, C. Pogoreutz, N. Rädecker, M. M. Saad, R. A. Schmitz, H. Schulenburg, C. R. Voolstra, N. Weiland-Bräuer, M. Ziegler and T. C. G. Bosch, Zoology, 2018, 127, 1–19 CrossRef PubMed
.
- N. Koopman, L. Remijas, J. Seppen, P. Setlow and S. Brul, Int. J. Mol. Sci., 2022, 23, 3405 CrossRef CAS PubMed
.
- X. Xu and Á. T. Kovács, Environ. Microbiol., 2024, 26, e16593 CrossRef CAS PubMed
.
- P. Setlow, J. Appl. Microbiol., 2006, 101, 514–525 CrossRef CAS PubMed
.
-
I. Mandic-Mulec, P. Stefanic and J. D. van Elsas, The bacterial spore: From molecules to systems, ASM Press, 2016, pp. 59–85 Search PubMed
.
- S. Vardharajula, S. Zulfikar Ali, M. Grover, G. Reddy and V. Bandi, J. Plant Interact., 2011, 6, 1–14 CrossRef CAS
.
- P. Setlow and G. Christie, Microbiol. Mol. Biol. Rev., 2023, 87, e00080-22 CrossRef
.
- P. Foegeding and M. Fulp, J. Appl. Bacteriol., 1988, 65, 249–259 CrossRef
.
- M. L. Stecchini, M. Spaziani, M. Del Torre and S. Pacor, J. Appl. Microbiol., 2009, 106, 1838–1848 CrossRef CAS PubMed
.
- D. Xu and J. C. Cote, Int. J. Syst. Evol. Microbiol., 2003, 53, 695–704 CrossRef CAS PubMed
.
- C. Ash, J. A. E. Farrow, S. Wallbanks and M. D. Collins, Lett. Appl. Microbiol., 1991, 13, 202–206 CrossRef CAS
.
- R. Tyskiewicz, M. Fedorowicz, A. Nakonieczna, P. Zielinska, M. Kwiatek and L. Mizak, Anal. Biochem., 2023, 675, 115215 CrossRef CAS
.
- X. Zhang, Q. Wang, X. Sun, M. Asif, A. Aziz, Y. Zhang, C. Dong, R. Wang and S. Shuang, Chem. Eng. J., 2024, 490, 151582 CrossRef CAS
.
- P. C. Ashok and K. Dholakia, Curr. Opin. Biotechnol, 2012, 23, 16–21 CrossRef CAS PubMed
.
- J. W. Chan, J. Biophotonics, 2013, 6, 36–48 CrossRef CAS PubMed
.
- L. Kong, P. Zhang, G. Wang, J. Yu, P. Setlow and Y. Q. Li, Nat. Protoc., 2011, 6, 625–639 CrossRef CAS PubMed
.
- N. Mithun, S. Shastry, G. Mohan, J. Lukose, M. V. Matham and S. Chidangil, Sci. Rep., 2025, 15, 6049 CrossRef CAS
.
- R. Gillibert, A. Magazzù, A. Callegari, D. Bronte-Ciriza, A. Foti, M. G. Donato, O. M. Maragò, G. Volpe, M. Lamy de La Chapelle, F. Lagarde and P. G. Gucciardi, Environ. Sci.: Nano, 2022, 9, 145–161 RSC
.
- Y. Qi, D. Hu, Y. Jiang, Z. Wu, M. Zheng, E. X. Chen, Y. Liang, M. A. Sadi, K. Zhang and Y. P. Chen, Adv. Opt. Mater., 2023, 11, 2203104 CrossRef CAS
.
- Y. Li, W. Huang, J. Pan, Q. Ye, S. Lin, S. Feng, S. Xie, H. Zeng and R. Chen, Mol. Clin. Oncol., 2015, 3, 375–380 CrossRef PubMed
.
- S. Khan, R. Ullah, S. Shahzad, S. Javaid and A. Khan, Optik, 2018, 157, 565–570 CrossRef CAS
.
- D. P. Dos Santos, M. M. Sena, M. R. Almeida, I. O. Mazali, A. C. Olivieri and J. E. L. Villa, Anal. Bioanal. Chem., 2023, 415, 3945–3966 CrossRef CAS PubMed
.
- L. Wang, J. W. Tang, F. Li, M. Usman, C. Y. Wu, Q. H. Liu, H. Q. Kang, W. Liu and B. Gu, Microbiol. Spectrum, 2022, 10, e02580-22 CrossRef
.
- Y. Liu, J. Xu, Y. Tao, T. Fang, W. Du and A. Ye, Analyst, 2020, 145, 3297–3305 RSC
.
- L. Huang, H. Sun, L. Sun, K. Shi, Y. Chen, X. Ren, Y. Ge, D. Jiang, X. Liu, W. Knoll, Q. Zhang and Y. Wang, Nat. Commun., 2023, 14, 48 CrossRef CAS PubMed
.
- Z. Cao, X. Pan, H. Yu, S. Hua, D. Wang, D. Z. Chen, M. Zhou and J. Wu, BME Front., 2022, 2022, 9872028 CrossRef PubMed
.
- D. Ma, L. Shang, J. Tang, Y. Bao, J. Fu and J. Yin, Spectrochim. Acta, Part A, 2021, 256, 119732 CrossRef CAS PubMed
.
- Z. Pei, Z. Zhang, J. Chen, W. Liu, B. Chen, Y. Huang, H. Yang and Y. Lu, Electronics, 2025, 14, 414 CrossRef
.
- M. A. A. Al-Qaness and S. Ni, IEEE J. Biomed. Health Inf., 2025, 29, 188–197 Search PubMed
.
- A. C. Granger, E. K. Gaidamakova, V. Y. Matrosova, M. J. Daly and P. Setlow, Appl. Environ. Microbiol., 2011, 77, 32–40 CrossRef CAS
.
- F. Du, L. He, X. Lu, Y.-q. Li and Y. Yuan, Spectrochim. Acta, Part A, 2023, 289, 122216 CrossRef CAS
.
- J. Zhao, H. Lui, S. Kalia, T. K. Lee and H. Zeng, Front. Oncol., 2024, 14, 1320220 CrossRef
.
- A. R. Flanagan, D. Dalal and F. G. Glavin, Chem. Rev., 2025, 125, 6130–6155 CrossRef CAS
.
- Y. LeCun, Y. Bengio and G. Hinton, Nature, 2015, 521, 436–444 CrossRef CAS PubMed
.
- P. Zhang, L. Kong, G. Wang, P. Setlow and Y. Q. Li, Appl. Environ. Microbiol., 2011, 77, 4754–4769 CrossRef CAS PubMed
.
- A. R. Flanagan, D. Dalal and F. G. Glavin, Chem. Rev., 2025, 125, 6130–6155 CrossRef CAS
.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2026 |






