Open Access Article
Open Access ArticleSustainable reservoir computing with liquid egg albumen
Raphael
Fortulan
 *ac,
Noushin
Raeisi Kheirabadi
*ac,
Noushin
Raeisi Kheirabadi
 a,
Davin
Browner
a,
Davin
Browner
 a,
Alessandro
Chiolerio
a,
Alessandro
Chiolerio
 ab and
Andrew
Adamatzky
ab and
Andrew
Adamatzky
 a
a
aUnconventional Computing Laboratory, UWE, Bristol, UK. E-mail: raphael.vicentefortulan@uwe.ac.uk; noushin.raeisikheirabadi@uwe.ac.uk; davin.browner@uwe.ac.uk; andrew.adamatzky@uwe.ac.uk
bBioinspired Soft Robotics, Istituto Italiano di Tecnologia, Via Morego 30, 16163, Genova, Italy. E-mail: alessandro.chiolerio@iit.it
cSchool of Computing and Engineering, University of Huddersfield, Huddersfield, UK. E-mail: r.vicentefortulan@hud.ac.uk
First published on 7th March 2025
Abstract
While physical reservoir computing offers a promising approach for efficient information processing, identifying suitable substrates remains challenging. Here, we demonstrated that colloidal albumen proteins could function as an effective physical reservoir for classifying multivariate datasets and electrocardiogram (ECG) signals. We exploited the nonlinear dynamics of protein macromolecules and ions in the albumen to perform high-dimensional mappings of input data. Our albumen-based reservoir achieved classification accuracy comparable to conventional machine learning methods on benchmark datasets while consuming over 5000 times less energy during training. Notably, the reservoir exhibited short-term plasticity analogous to biological synapses, with conductance spikes and fading memory. This bio-inspired computing paradigm not only offered a sustainable alternative to traditional architectures but also provided insights into the information-processing capabilities of biological systems. Our findings opened new avenues for low-power, environmentally friendly computing solutions with potential applications in real-time health monitoring and edge computing.
Introduction
Edible electronics represent a promising innovation in the biomedical, pharmaceutical, and food industries. These ingestible devices, made from biocompatible and eco-friendly materials, offer real-time, non-invasive monitoring, particularly useful for gastrointestinal diagnostics. Their ability to degrade naturally makes them both cost-effective and sustainable, with potential applications extending to food quality monitoring and anti-counterfeiting.1–4This shift towards biologically based technology parallels advancements in unconventional computing, which explores alternatives to traditional silicon-based computers. By leveraging biological, chemical, and physical processes, unconventional computing enables novel computational approaches, such as chemical and slime mold computing.5 These systems exhibit adaptability, parallelism, and energy efficiency, addressing the limitations of classical computing and opening new possibilities in AI, optimization, and sustainable technologies.6
A key application of unconventional computing is reservoir computing (RC), a machine learning (ML) framework designed to process temporal and sequential data. By using highly nonlinear systems such as spintronic oscillators, photonic systems, and soft robotics, RC systems perform high-dimensional mappings and complex transformations, making them ideal for tasks requiring real-time, adaptive computation.7–12
Among the unconventional substrates explored for RC, liquid cybernetic systems have shown remarkable promise. Colloidal suspensions, such as zinc oxide (ZnO)13 and magnetite (Fe3O4) ferrofluid,14 demonstrate unique information processing capabilities through their dynamic and self-organizing properties. These systems can encode and process information via complex electrohydrodynamic and magnetohydrodynamic interactions, respectively. Experimental studies have illustrated their potential as neuromorphic processors, capable of synaptic plasticity-like learning and pattern recognition tasks, thus broadening the scope of physical reservoirs in unconventional computing.15,16
This paper explores the use of a biological physical reservoir based on colloidal albumen proteins derived from eggs for classification tasks. Egg albumen, or egg white, has drawn considerable attention in recent years due to its low cost, high transparency, and ease of extraction. Its electrical properties have been studied since the early 1900s,17 with later research confirming its excellent dielectric properties.18–20 Studies have shown that thermally treated albumen can serve as an effective dielectric layer in organic field-effect transistors,21 and modified albumen has been used to create reliable and efficient memory cells.22 Heat-denatured albumen has demonstrated stable switching endurance, high resistance ratios, and long retention times, making it suitable for memory storage applications.23,24
In contrast, this work utilizes egg albumen in its colloidal form. The choice to use colloidal albumen derives from its unique ability to exploit colloidal particles under external fields. Colloids interact naturally through van der Waals forces and electrostatic repulsion, but under an external field, they can follow alternative thermodynamic pathways, forming metastable suprastructures.25–27 This property is critical for RC as it allows the system to leverage nonlinear behavior, global connectivity, and memory effects for efficient information processing.28,29 The nonlinear interactions within the colloidal suspension, influenced by ion drift and local electric fields, enhance the system's capacity to map complex time-varying signals.
Our system leverages the nonlinear dynamics of protein macromolecules and ions in liquid albumen to create high-dimensional nonlinear mappings of input data, converting the results into conductance values that can be used as inputs for linear classifiers to perform classification tasks. We demonstrate that this bio-inspired reservoir can classify multivariate datasets, and, following previous studies,30,31 we also used it to classify electrocardiogram (ECG) signals with accuracy comparable to conventional ML models. Lastly, we discuss the potential of this technology for low-power, sustainable computing solutions with applications in real-time health monitoring and edge computing.
Results and discussion
Fig. 1(a) illustrates the current–voltage (I–V) characteristics of the albumen colloid under cyclic DC voltage sweeps from 0 V → 5 V → 0 V → −5 V at a scan rate of 1 V s−1. The measured I–V curves exhibit a hysteresis loop, which is indicative of a circuit containing a memory element.16,32,33 The changes in the hysteresis loops shown in Fig. 1(b) at scan rates of 1 V s−1, 2 V s−1, and 4 V s−1 for 100 cycles demonstrate a dependence between frequency and the current/voltage characteristics of the albumen-based reservoir, similar to what is seen in memristive-like materials.34–36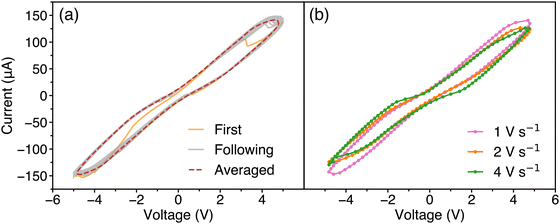 | ||
| Fig. 1 Current–voltage characteristics of the reservoir for (a) 100 sweeps at a scan rate of 1 V s−1 and (b) averaged 100 sweeps at scan rates of 1 V s−1, 2 V s−1, and 4 V s−1. | ||
According to the USDA (United States Department of Agriculture), raw egg whites are primarily composed of water (87.57%) and proteins (10.9%), with the remainder consisting of carbohydrates (0.73%), lipids (0.17%), and minerals.37 The principal protein molecules present in egg white are ovalbumin (54%), ovotransferrin (12%), ovomucoid (11%), lysozyme (3.5%), and ovomucin (3.5%).38 Without denaturation, these protein macromolecules are folded and linked to mineral ions such as iron (Fe), potassium (K), and sodium (Na),39,40 as illustrated in Fig. 2(a). The presence of a high concentration of ions bound to folded proteins creates a liquid ionic conductor, which enables the memory and resistivity switching behavior of the reservoir.
 | ||
| Fig. 2 (a) Idealization of the biological reservoir. The presence of a high concentration of ions bound to folded proteins in raw egg white creates an ionic fluid ideal for memory and resistivity switching behavior. (b) Neuromorphic behavior of the reservoir exhibits short term plasticity due to −5 V, 10 ms pulses. Illustration of the biological synapse was adapted from ref. 41. (c) Illustration of the fading memory of the reservoir. | ||
When a negative voltage sweep is applied to the reservoir, electrons are released from the redox sites, as oxidized Fe ions are preferentially reduced due to their similar work functions to the Cu electrode.23 This process creates conductive paths that decrease resistivity. Conversely, when a sufficiently strong positive bias is applied, electrons are injected into the redox sites, causing ion movement that breaks these conductive paths and increases resistivity.
At the positively charged anode, copper metal undergoes oxidation, losing electrons to form copper ions (Cu2+). These copper ions then migrate into the liquid, where they encounter hydroxide ions (OH−) drawn towards the anode. The copper and hydroxide ions react, forming copper(II) hydroxide (Cu(OH)2). Conversely, at the negatively charged cathode, copper ions in the electrolyte gain electrons and are reduced back to copper metal.
The presence of memory in the system is likely attributable to the metallization of the electrodes, which results in diminishing conductance increases as voltage sweeps or pulses are applied, as illustrated in Fig. 2(b). Additionally, ion-binding proteins,42 in particular the copper ion binding ovalbumin protein,43 create traps for charges and ions (see Fig. 2(a)), further contributing to the memory effect.
These electrical characteristics of the reservoir bear a notable resemblance to the properties of synapses and synaptic plasticity observed in neurons. Specifically, the occurrence of conductance (G) spikes in the materials is analogous to the spikes associated with short-term plasticity in neurons.44,45 This similarity suggests potential parallels between the behavior of our albumen-based system and neuronal information processing mechanisms.
An important property exhibited by the biological reservoir is the capacity of forgetting, which plays a key role in the brain's biological system for managing memories.46–48 This characteristic is also essential for RC, with fading memory being a crucial component.49 In the case of our albumen-based reservoir, we have verified that the device demonstrates this property, as it can recover its conductance state after some time has elapsed, as shown in Fig. 2(c).
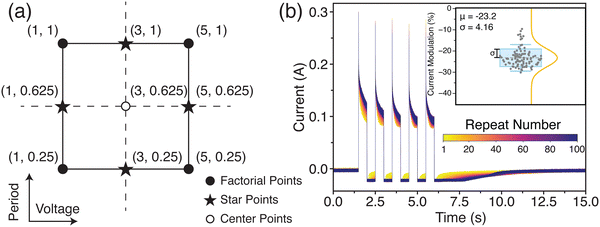
Due to the memfractor-like nature of colloidal reservoirs,50 both the frequency/period and voltage magnitude are possible factors that can influence the response of the reservoir and change important variables such as the memory and conductance modulation of the reservoir. To investigate this, a design of experiments (DoE) statistical approach was used to investigate the effects of these factors on the response of the system. Here, we employed a face-centered central composite design as illustrated in Fig. 3(a). We applied a set of five pulses to the material. The high and low levels for the voltage magnitude and pulse period were set to −5 V and −1 V, and 1 s and 250 ms, respectively. A total of five center points were included, with the total number of repetitions set to five, resulting in 65 experiments.51 The output variables were defined as the percentage change in conductance and the relaxation time of the conductance after all pulses had been applied to the reservoir.
The two-way ANOVA analysis for the conductance modulation is shown in Table 1 and indicates that both main factors, voltage (F(1,64) = 13.47, p < 0.001) and period (F(1,64) = 13.42, p < 0.001), had statistically significant effects on the percentage conductance modulation at α = 0.05. The interaction effect between voltage and period was not statistically significant (F(1, 64) = 3.96, p = 0.051).
| Source | DF | F-value | p-value |
|---|---|---|---|
| Voltage | 1 | 13.47 | 0.001 |
| Period | 1 | 13.42 | 0.001 |
| 2-way interaction: voltage × period | 1 | 3.96 | 0.051 |
On the other hand, the two-way ANOVA analysis for the relaxation time, presented in Table 2, revealed that the period (F(1,64) = 32.43, p < 0.001) had a statistically significant effect on the system's memory time constant at α = 0.05. Neither the voltage (F(1,64) = 2.44, p = 0.124) nor the voltage-period interaction (F(1,64) = 1.53, p = 0.220) showed statistically significant effects.
| Source | DF | F-value | p-value |
|---|---|---|---|
| Voltage | 1 | 2.44 | 0.124 |
| Period | 1 | 62.43 | 0.000 |
| 2-way interaction: voltage × period | 1 | 1.53 | 0.220 |
These results show that it is possible to control the system's memory by adjusting the frequency of the applied pulses. Additionally, the change in the conductance of the material can be modulated by varying either the frequency or the applied voltage. The voltage-period interaction can never be exploited to program the device output, and this reveals to us that the independent variable magnetic flux (1 Wb = 1 V × 1 s) does not play any role, as it usually happens in memristors.
The repeatability of the system was tested by applying a train of five 5 V pulses 100 times and measuring the percentage change in the system's current. The 100 repetitions are shown in Fig. 3(b), where it can be seen that the system's response maintains a very similar behavior. The inset in Fig. 3(b) shows the raincloud plot of the measured percentage change in current. As observed, most of the measured values fall within 10% of the average value, indicating a repeatable response.
Reservoir performance – Iris
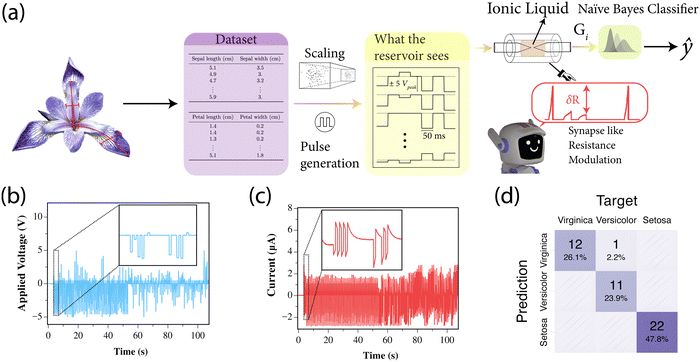
We tested the capabilities of the proposed architecture to classify multivariate datasets using the traditional Iris dataset.52 The dataset consists of physical measurements of Iris flowers (see the Methods section for more details).In Fig. 4(a), the architecture used for the classification is presented. Each feature is individually scaled to a range of ±1 and then mapped to a voltage pulse. Specifically, a feature value of +1 corresponds to a 5 V, 50 ms pulse, while a value of −1 corresponds to a −5 V, 50 ms pulse, with intermediate values mapped linearly within this range. All four features for each example are sequentially applied, and the final conductance value is then measured using a 0.5 V, 50 ms pulse. The conductance of the liquid albumen is then reset by waiting 1 s and the system is ready for the next example. The final classification is then performed using a Naïve Bayes classifier.
As discussed previously, the device exhibits synaptic-like behavior. When we apply short-interval voltage pulses into the liquid albumen, the conductance increases, mimicking memory retention, and the conductance change is correlated with the voltage pulse magnitude. However, if no voltage is applied, the conductance gradually resets, effectively “forgetting” the previous input. By encoding each example in the dataset as a conductance value in the material, we can classify multivariate datasets using a single-port device.
The applied voltage applied to the reservoir and the measured current during the training phase are shown in Fig. 4(b) and (c). The insets in both figures display a two-second snapshot of measurements, illustrating the highly nonlinear behavior of the reservoir. The confusion matrix shown in Fig. 4(d) demonstrates the effectiveness of the classifier, with an overall accuracy of approximately 98% and a nearly diagonal matrix with only one misclassified example. The advantage of using this simple dataset is to provide a quick benchmark for the classification capabilities of the reservoir, and the performance observed here is similar to that of other physically based reservoirs.53
Reservoir performance – physionet
The proposed albumen-based reservoir is also capable of encoding and processing time series data. For this, the input signal u(t), which represents the heart's electrical activity, was first encoded using Mel-frequency cepstral coefficients (MFCCs).54,55 MFCCs are widely employed as a preprocessing layer in RC architectures56,57 since they can extract useful features from time series and improve classification accuracy.58 The resulting Mel-frequency cepstrum's four coefficients for each signal were binarized using a threshold algorithm, creating a binary map which could then be applied to the reservoir following a similar methodology previously validated with the MNIST dataset.59,60As illustrated in Fig. 5, the 4-bit binary map was applied sequentially row-wise, with a logical “1” translated to a 5 V, 250 μs pulse and a logical “0” translated to no voltage being applied. After the pulse train was applied, the conductance, Gi, was measured using a 0.5 V, 250 μs pulse. The reservoir was then left to rest for 1 ms so the conductance was reset and a new set of cepstral coefficients could be applied.
After the full binary map was applied, a conductance vector G was obtained for each example and classified using a support vector machine (SVM) layer.
Given the inherent characteristics of the ECG dataset, which exhibits a highly skewed distribution favoring normal cardiac rhythms—an expected outcome due to the predominance of healthy individuals—utilizing accuracy as the sole metric for classifier evaluation is inadequate. Instead, we employed the F1-score, interpreted as the harmonic mean of precision and recall. Precision quantifies the proportion of true positive results (instances where the model correctly identifies a class) among all instances predicted as positive by the model. In contrast, recall measures the proportion of true positive results among all actual positive instances in the dataset. The F1-score is defined as
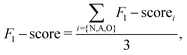 | (1) |
 | (2) |
The PhysioNet ECG dataset presents significantly greater classification challenges compared to the comparatively simpler Iris dataset.61 To ensure a fair comparison, we trained a convolutional neural network (CNN) on the same dataset, applying identical preprocessing steps. The CNN architecture comprised three convolutional layers with batch normalization and ReLU activations, followed by max-pooling, global average pooling, and dense layers with dropout, culminating in a softmax-based classification layer.
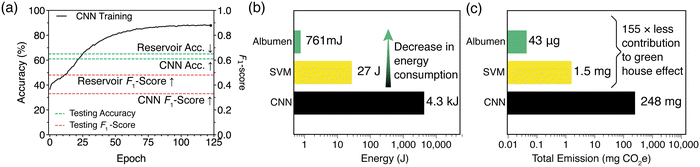
Fig. 6(a) compares the performance metrics of our albumen-based reservoir with the CNN. As shown, the reservoir outperforms the CNN in both accuracy and F1-score on the test dataset, achieving 65% accuracy and an F1-score of 0.48, compared to the CNN's 61% accuracy and 0.32 F1-score. Although the performance is reasonable, it is still below that of more sophisticated approaches such as those using XGBoost (F1-score of 0.83).62 However, the most significant advantage of the biological reservoir lies in its energy consumption and environmental impact.
Fig. 6(b) shows that the albumen-based reservoir is over 5000 times more energy-efficient during the training phase compared to the CNN. However, this efficiency is partially offset by the energy consumption of the SVM layer, which uses 35 times more energy than the reservoir. From a practical standpoint, this increased computational efficiency reduces the system's contribution to global warming. Fig. 6(c) shows the equivalent CO2 emissions, calculated using the UK 2023 energy conversion factors,63 for the training phase of both architectures. These results reveal that bio-based reservoirs have a much smaller environmental footprint, making them a promising green alternative to conventional computing systems.64
These results are promising, as historically, cardiac conditions were diagnosed solely based on physicians' interpretation of ECG features. Although modern techniques now enable the use of raw ECG data in conjunction with machine learning and deep learning,62,65 detecting these conditions remains challenging due to their episodic and non-regular nature.
From a healthcare perspective, detecting atrial fibrillation (AF) is particularly important, as it affects approximately 2% of the general population.66 AF is associated with significant mortality and morbidity, increasing the risk of death, stroke, hospitalization, heart failure, and coronary artery disease. Its incidence rate rises gradually from 0.9 per 1000 person-years at ages 40–49 to 17.7 per 1000 person-years in individuals aged 70 and older.67
A simple, low-cost, and effective system for AF detection is therefore of great interest. The results presented here demonstrate that our bio-based system can perform this classification with a fraction of the energy cost, offering a sustainable and environmentally friendly solution. Interestingly, it also highlights the potential of using biological substrates to process biological signals.
Conclusion
In conclusion, this study demonstrated the potential of a biological physical reservoir using colloidal albumen proteins for classification tasks. By harnessing the unique electrical properties of colloidal particles under external fields, we developed a system exhibiting key features essential for RC: nonlinearity and memory effect. This bio-based approach, combined with a simple classifier, achieved competitive performance on multivariate datasets and in ECG signal classification.While the more complex PhysioNet dataset was more challenging, the biologically inspired reservoir showed significant advantages in energy efficiency and environmental sustainability compared to traditional models such as CNNs. The low power consumption of this system translated into a reduced carbon footprint, positioning bio-based computing systems as promising green alternatives to conventional architectures. Additionally, the reservoir's ability to perform memory-related tasks mimicked biological synaptic behavior, offering the potential for developing neuromorphic systems.
Methods
Two datasets were employed to evaluate the physical reservoir. The first dataset, the Iris dataset,52 comprises four features (petal and sepal length and diameter) of three Iris species: Iris setosa, Iris virginica, and Iris versicolor. This dataset contains 50 examples for each class, with two classes being linearly separable and one not.The second dataset, from the PhysioNet 2017 Challenge,61,68 is accessible at https://physionet.org/content/challenge-2017/1.0.0/. It consists of ECG signals sampled at 300 Hz, classified by experts into four categories: normal (N), atrial fibrillation (A)†, other rhythm (O), and noisy recording (∼). The dataset contains 8422 measurements of varying durations, averaging 9000 data points.
To ensure consistency, we implemented preprocessing steps for the PhysioNet dataset: arrays with fewer than 9000 points were discarded, and longer arrays were truncated. Noisy recordings were also excluded. Due to the imbalanced nature of the dataset, with A, N, and O categories containing 738, 5050, and 2456 examples, respectively, we performed data augmentation for the underrepresented classes using ADASYN.70 Specifically, the data for categories A and O were replicated seven and two times, respectively. This augmentation was crucial to mitigate classifier bias, as the model without it would likely default to predicting N.
Both datasets were divided into training and testing sets using a 66%/33% split. We evaluated the results using Python 3.11.5 on an ARM-based CPU running macOS. The output layer was trained using scikit-learn 1.5.2, while the convolutional neural network (CNN) was trained using TensorFlow 2.12.0. Energy consumption during training was measured using the built-in powermetrics utility on macOS.
For the experiments, signals were applied to the colloid using a pulse generator (Sanworks, Pulse Pal v2). Voltage and current measurements were obtained with a PC-based oscilloscope (Pico Technology, PicoScope 5442D) coupled with a shunt and amplifier (LowPowerLab, CurrentRanger). The reservoir was constructed using two copper electrodes (Thermo Scientific Chemicals, copper wire, ∅0.5 mm, annealed, 99.9% purity), 10 μl of albumen extracted from hen eggs, and 20 μl micropipettes (intraMARK, BLAUBRAND®). The physical realization of the albumen-based reservoir is shown in Fig. 7(a), and the overall architecture for RC is depicted in Fig. 7(b). The energy consumption of the physical reservoir was estimated from voltage and current measurements.
Data availability
Data sets generated during the current study are available from the corresponding author on reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work received support from the European Innovation Council and SMEs Executive Agency (EISMEA) under grant agreement No. 964388.References
- W. Xu, H. Yang, W. Zeng, T. Houghton, X. Wang, R. Murthy, H. Kim, Y. Lin, M. Mignolet and H. Duan, et al. , Adv. Mater. Technol., 2017, 2, 1700181 CrossRef.
- A. S. Sharova, F. Melloni, G. Lanzani, C. J. Bettinger and M. Caironi, Adv. Mater. Technol., 2021, 6, 2000757 CrossRef.
- Y. Wu, D. Ye, Y. Shan, S. He, Z. Su, J. Liang, J. Zheng, Z. Yang, H. Yang and W. Xu, et al. , Adv. Mater. Technol., 2020, 5, 2000100 CrossRef.
- A. S. Sharova, F. Modena, A. Luzio, F. Melloni, P. Cataldi, F. Viola, L. Lamanna, N. F. Zorn, M. Sassi and C. Ronchi, et al. , Nanoscale, 2023, 15, 10808–10819 RSC.
- S. Harding, J. Koutnik, J. Schmidhuber and A. Adamatzky, Inspired by Nature: Essays Presented to Julian F. Miller on the Occasion of his 60th Birthday, 2018, 323–337 Search PubMed.
- A. Adamatzky, Handbook of Unconventional Computing: Volume 2: Implementations, World Scientific, 2021, vol. 2 Search PubMed.
- D. Verstraeten, B. Schrauwen, M. d’Haene and D. Stroobandt, Neural Networks, 2007, 20, 391–403 CrossRef CAS PubMed.
- O. Obst and J. Boedecker, Guided self-organization: Inception, Springer, 2014, pp. 319–340 Search PubMed.
- M. Dale, J. F. Miller and S. Stepney, Advances in Unconventional Computing: Volume 1: Theory, 2017, 533–571 Search PubMed.
- Z. Konkoli, S. Nichele, M. Dale and S. Stepney, Comput. Matter., 2018, 269–293 Search PubMed.
- V. Athanasiou and Z. Konkoli, Int. J. Parallel, Emergent Distrib. Syst., 2018, 33, 367–386 CrossRef.
- M. Dale, J. F. Miller, S. Stepney and M. A. Trefzer, Proc. R. Soc. A, 2019, 475, 20180723 CrossRef PubMed.
- N. Raeisi Kheirabadi, A. Chiolerio and A. Adamatzky, Bionanoscience, 2024, 1–9 Search PubMed.
- M. Crepaldi, C. Mohan, E. Garofalo, A. Adamatzky, K. Szaciłski and A. Chiolerio, Adv. Mater., 2023, 35, 2211406 CrossRef CAS.
- A. Chiolerio, Adv. Intell. Syst., 2020, 2, 2000120 CrossRef.
- N. R. Kheirabadi, A. Chiolerio, N. Phillips and A. Adamatzky, Neurocomputing, 2023, 557, 126710 CrossRef.
- E. F. Northrup, J. Franklin Inst., 1913, 175, 413–419 CrossRef CAS.
- J. Wang, J. Tang, Y. Wang and B. Swanson, LWT – Food Sci. Technol., 2009, 42, 1204–1212 CrossRef CAS.
- L. Ragni, A. Al-Shami, G. Mikhaylenko and J. Tang, J. Food Eng., 2007, 82, 450–459 CrossRef.
- N. R. Kheirabadi, F. Karimzadeh, M. H. Enayati and E. N. Kalali, Adv. Electron. Mater., 2023, 9, 2200839 CrossRef CAS.
- M. Ma, X. Xu, L. Shi and L. Li, RSC Adv., 2014, 4, 58720–58723 RSC.
- G. Zhou, Z. Ren, L. Wang, B. Sun, S. Duan and Q. Song, Mater. Horiz., 2019, 6, 1877–1882 RSC.
- Y.-C. Chen, H.-C. Yu, C.-Y. Huang, W.-L. Chung, S.-L. Wu and Y.-K. Su, Sci. Rep., 2015, 5, 10022 CrossRef CAS PubMed.
- X. He, J. Zhang, W. Wang, W. Xuan, X. Wang, Q. Zhang, C. G. Smith and J. Luo, ACS Appl. Mater. Interfaces, 2016, 8, 10954–10960 CrossRef CAS PubMed.
- J. A. Ferrar and M. J. Solomon, Soft Matter, 2015, 11, 3599–3611 RSC.
- J. Zhang, J. Yang, Y. Zhang and M. A. Bevan, Sci. Adv., 2020, 6, eabd6716 CrossRef PubMed.
- M. T. Sullivan, K. Zhao, A. D. Hollingsworth, R. H. Austin, W. B. Russel and P. M. Chaikin, Phys. Rev. Lett., 2006, 96, 015703 CrossRef CAS PubMed.
- W. Poon, Science, 2004, 304, 830–831 CrossRef CAS PubMed.
- J. N. Israelachvili, Intermolecular and Surface Forces, Elsevier Science, 2011 Search PubMed.
- S. Jiang, J. Sun, M. Pei, L. Peng, Q. Dai, C. Wu, J. Gu, Y. Yang, J. Su, D. Gu, H. Zhang, H. Guo and Y. Li, J. Phys. Chem. Lett., 2024, 15, 8501–8509 CrossRef CAS PubMed.
- M. Cucchi, C. Gruener, L. Petrauskas, P. Steiner, H. Tseng, A. Fischer, B. Penkovsky, C. Matthus, P. Birkholz, H. Kleemann and K. Leo, Sci. Adv., 2021, 7, eabh0693 CrossRef PubMed.
- M. Gur, F. Akar, K. Orman, Y. Babacan, A. Yesil and F. Gul, Circuits, Syst., Sig. Proc., 2023, 42, 6481–6493 CrossRef.
- M. E. Fouda, A. S. Elwakil and A. G. Radwan, Microelectron. J., 2015, 46, 834–838 CrossRef.
- L. O. Chua and S. M. Kang, Proc. IEEE, 1976, 64, 209–223 Search PubMed.
- L. Chua, IEEE Trans. Circuit Theory, 1971, 18, 507–519 Search PubMed.
- T. D. Dongale, K. P. Patil, P. K. Gaikwad and R. K. Kamat, Mater. Sci. Semicond. Proc., 2015, 38, 228–233 CrossRef CAS.
- USDA, Egg, White Only, Raw, United States Department of Agriculture, Economic Research Service Survey (FNDDS) 1100199, 2020.
- E. D. N. S. Abeyrathne, H. Y. Lee and D. U. Ahn, Poult. Sci., 2013, 92, 3292–3299 CrossRef CAS PubMed.
- L. A. Julien, F. Baron, S. Bonnassie, F. Nau, C. Guérin, S. Jan and S. C. Andrews, Biometals, 2019, 32, 453–467 CrossRef CAS PubMed.
- J. Kovacs-Nolan, M. Phillips and Y. Mine, J. Agric. Food Chem., 2005, 53, 8421–8431 CrossRef CAS PubMed.
- J. G. Betts, K. A. Young, J. A. Wise, E. Johnson, B. Poe, D. H. Kruse, O. Korol, J. E. Johnson, M. Womble and P. DeSaix, Anatomy and Physiology, OpenStax, Houston, Texas, 2013 Search PubMed.
- M. J. Puglisi and M. L. Fernandez, Nutrients, 2022, 14, 2904 CrossRef CAS PubMed.
- H. J. McGee, S. R. Long and W. R. Briggs, Nature, 1984, 308, 667–668 CrossRef CAS.
- R. S. Zucker and W. G. Regehr, Annu. Rev. Physiol., 2002, 64, 355–405 CrossRef CAS PubMed.
- A. Citri and R. C. Malenka, Neuropsychopharmacology, 2008, 33, 18–41 CrossRef.
- R. L. Davis and Y. Zhong, Neuron, 2017, 95, 490–503 CrossRef CAS.
- B. A. Richards and P. W. Frankland, Neuron, 2017, 94, 1071–1084 CrossRef CAS.
- M. Mayford, S. A. Siegelbaum and E. R. Kandel, Cold Spring Harbor Perspect. Biol., 2012, 4, a005751 Search PubMed.
- M. Cucchi, S. Abreu, G. Ciccone, D. Brunner and H. Kleemann, Neuromorph. Comput. Eng., 2022, 2, 032002 CrossRef.
- R. Fortulan, N. R. Kheirabadi, N. Raeisi-Kheirabadi, A. Nezamzadeh-Ejhieh, A. Chiolerio and A. Adamatzky, Phys. Rev. E, 2024, 110, 034607 CrossRef CAS PubMed.
- D. C. Montgomery, Design and Analysis of Experiments, 10th edn, EMEA edn, Wiley, Hoboken, NJ, 2020 Search PubMed.
- R. A. Fisher, Iris Species, https://www.kaggle.com/datasets/uciml/iris, 2016 Search PubMed.
- Z. Ma, J. Ge and S. Pan, Adv. Electron. Mater., 2024, 2400469 CrossRef.
- Md Sahidullah and G. Saha, Speech Commun., 2012, 54, 543–565 CrossRef.
- Z. K. Abdul and A. K. Al-Talabani, IEEE Access, 2022, 10, 122136–122158 Search PubMed.
- H. Tan and S. van Dijken, Nat. Commun., 2023, 14, 2169 CrossRef CAS PubMed.
- E. Picco, A. Lupo and S. Massar, Deep Photonic Reservoir Computer for Speech Recognition, 2023.
- D. Mitrovic, M. Zeppelzauer and C. Breiteneder, Advances in Computers, Elsevier, 2010, vol. 78 of Advances in Computers: Improving the Web, pp. 71–150 Search PubMed.
- M. Lian, C. Gao, Z. Lin, L. Shan, C. Chen, Y. Zou, E. Cheng, C. Liu, T. Guo, W. Chen and H. Chen, Light: Sci. Appl., 2024, 13, 179 CrossRef CAS PubMed.
- Y. Yamazaki and K. Kinoshita, Adv. Sci., 2024, 11, 2304804 CrossRef CAS PubMed.
- G. D. Clifford, C. Liu, B. Moody, H. L. Li-wei, I. Silva, Q. Li, A. E. Johnson and R. G. Mark, Comput. Cardiol., 2017, 1–4 Search PubMed.
- T. Teijeiro, C. A. Garcia, D. Castro and P. Félix, Arrhythmia classification from the abductive interpretation of short single-lead ECG records, 2017 Computing in Cardiology (CinC), Rennes, France, 2017, pp. 1–4 DOI:10.22489/CinC.2017.166-054.
- T. C. Trust, Energy And Carbon Conversions 2023 Update, CTV085, 2024.
- S. I. Zandalinas, F. B. Fritschi and R. Mittler, Trends Plant Sci., 2021, 26, 588–599 CrossRef CAS PubMed.
- S. Hong, M. Wu, Y. Zhou, Q. Wang, J. Shang, H. Li and J. Xie, ENCASE: An ENsemble ClASsifiEr for ECG classification using expert features and deep neural networks, 2017 Computing in Cardiology (CinC), Rennes, France, 2017, pp. 1–4 DOI:10.22489/CinC.2017.178-245.
- R. C. Davis, F. R. Hobbs, J. E. Kenkre, A. K. Roalfe, R. Iles, G. Y. Lip and M. K. Davies, EP Eur., 2012, 14, 1553–1559 Search PubMed.
- B. Morseth, B. Geelhoed, A. Linneberg, L. Johansson, K. Kuulasmaa, V. Salomaa, L. Iacoviello, S. Costanzo, S. Söderberg, T. J. Niiranen, J. K. K. Vishram-Nielsen, I. Njølstad, T. Wilsgaard, E. B. Mathiesen, M.-L. Løchen, T. Zeller, S. Blankenberg, F. M. Ojeda and R. B. Schnabel, Open Heart, 2021, 8, e001624 CrossRef PubMed.
- A. L. Goldberger, L. A. Amaral, L. Glass, J. M. Hausdorff, P. C. Ivanov, R. G. Mark, J. E. Mietus, G. B. Moody, C.-K. Peng and H. E. Stanley, Circulation, 2000, 101, e215–e220 CAS.
- B. J. J. M. Brundel, X. Ai, M. T. Hills, M. F. Kuipers, G. Y. H. Lip and N. M. S. de Groot, Nat. Rev. Dis. Primers, 2022, 8, 1–23 CrossRef.
- H. He, Y. Bai, E. A. Garcia and S. Li, IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), 2008, pp. 1322–1328.
Footnote |
| † Atrial fibrillation is an irregular heartbeat where the heart's upper chambers (atria) beat out of coordination with the lower chambers (ventricles).69 |
| This journal is © The Royal Society of Chemistry 2025 |