Open Access Article
Open Access ArticleDevelopment of a glycine-modified iron oxide nanoparticle-electrochemical biosensor for specific detection of Klebsiella pneumoniae DNA†
Rutuja Prashant
Gambhir
a,
Somnath
Kundale
b,
Sohel B.
Shaikh
a,
Amol S.
Vedpathak
 c,
Rajanish K.
Kamat
de,
Tukaram D.
Dongale
c,
Rajanish K.
Kamat
de,
Tukaram D.
Dongale
 b,
Yogendra Kumar
Mishra
b,
Yogendra Kumar
Mishra
 f,
Ravindra N.
Bulakhe
*g,
Ji Man
Kim
f,
Ravindra N.
Bulakhe
*g,
Ji Man
Kim
 *g and
Arpita Pandey
Tiwari
*g and
Arpita Pandey
Tiwari
 *ah
*ah
aDepartment of Medical Biotechnology and Stem Cells and Regenerative Medicine, Centre for Interdisciplinary Research, D. Y. Patil Education Society (Deemed to be University), Kolhapur, Maharashtra 416006, India. E-mail: arpitaptiwari@gmail.com
bSchool of Nanoscience and Biotechnology, Shivaji University, Kolhapur, Maharashtra 416004, India
cSymbiosis Centre for Nanoscience and Nanotechnology (SCNN), Symbiosis International (Deemed University), Lavale, Pune, 412115, India
dDepartment of Electronics, Shivaji University, Kolhapur 416004, India
eDr Homi Bhabha State University, 15, Madam Cama Road, Mumbai 400032, India
fSmart Materials, Nano SYD, Mads Clausen Institute, University of Southern Denmark, Alsion 2, Sønderborg 6400, Denmark
gDepartment of Chemistry, Sungkyunkwan University, Suwon, 16419, Republic of Korea. E-mail: bulakhe@skku.edu; jimankim@skku.edu
hGlobal Innovative Center of Advanced Nanomaterials, College of Engineering, Science and Environment, University of Newcastle, Callaghan, NSW 2308, Australia
First published on 13th June 2025
Abstract
This study presents a simple electrochemical DNA biosensor to accurately detect Klebsiella pneumoniae DNA. The biosensor uses glycine functionalized iron oxide nanoparticles (glycine@Fe3O4) to capture Klebsiella pneumoniae DNA. The as-synthesized nanoparticles were characterized using X-ray diffraction, X-ray photoelectron spectroscopy, Raman spectroscopy, and transmission electron microscopy, and created a sensitive electrode surface that produced strong electrochemical signals when DNA attached. Electrochemical techniques, including cyclic voltammetry and square wave voltammetry, were used to develop a biosensor for detecting Klebsiella pneumoniae DNA. The biosensor showed limits of detection of 3.27 nM in the 30–90 nM range and 3.94 nM in the 120–270 nM range, with limits of quantification of 3.90 nM and 11.95 nM, respectively. The sensitivity, determined from calibration curves, was 0.1009 μA nM−1 for the low range and 0.0838 μA nM−1 for the high range. The biosensor demonstrated high sensitivity and selectivity, effectively distinguishing the analyte from interferents like albumin and folic acid. Molecular docking showed strong DNA binding (−6 kcal mol−1). Lab tests confirmed detection of Klebsiella pneumoniae antimicrobial resistance genes (SHV, TEM, CTX-M, OXA-1) via PCR-based gel electrophoresis. This easy-to-use non-enzymatic biosensor offers fast, accurate, rapid, sensitive, and specific Klebsiella pneumoniae DNA detection, valuable for point-of-care diagnostics and antimicrobial resistance monitoring.
1. Introduction
Hospital-borne infections, particularly those caused by Klebsiella pneumoniae (K. pneumoniae), are a significant threat within healthcare settings. K. pneumoniae is a Gram-negative bacterium known for its high virulence and antimicrobial resistance, making it a leading cause of severe infections in these environments. Traditional methods of detecting K. pneumoniae rely on lengthy culturing processes and the use of specialized equipment. These methods are time-consuming, which highlights the urgent need for faster and more sensitive detection techniques. Electrochemical DNA sensing has emerged as a promising solution for rapid and specific pathogen detection.1–5 The primary objective of this study was to present an electrochemical DNA sensing platform specifically designed for the detection of K. pneumoniae DNA. This was achieved by using glycine-functionalized magnetic nanoparticles (glycine@Fe3O4). Magnetic nanoparticles enhance the sensitivity of the biosensor by improving the capture and separation of target DNA, thereby easing more efficient detection.6–8 The sensing process involves two key steps: capturing the target DNA and amplifying the signal, which results in increased sensitivity and reduced assay time.9–11 This approach is particularly important since K. pneumoniae DNA is often present in extremely low quantities in clinical samples, presenting significant analytical challenges.12,13A major concern with K. pneumoniae infections is the presence of genes associated with multidrug resistance, such as Sulphydryl Variable (SHV), Temoniera (TEM), Cefotaxime-Munich (CTX-M), Oxacillinase (OXA), and CMY AmpC β-lactamase (CMY). The detection of these resistance genes is key for understanding and controlling the spread of resistant strains. The developed electrochemical DNA sensing platform offers a label-free detection method, eliminating the need for additional probe DNA or hybridizing enzymes, which simplifies the detection process.
To assess the performance of this sensor, clinical samples were analysed and the results compared to traditional techniques. The electrochemical DNA sensing platform demonstrated significant promise for rapid diagnosis, surveillance, and monitoring of K. pneumoniae infections in healthcare settings. By enabling quicker detection, it can facilitate the timely administration of appropriate antibiotics, thereby reducing the risk of severe infections and improving patient outcomes.
Electrochemical techniques, such as cyclic voltammetry (CV), square wave voltammetry (SWV) and electrochemical impedance spectroscopy (EIS), are commonly used for DNA sensing. Among these, EIS is particularly effective for analysing bio-recognition processes at the electrode surface, such as nucleic acid interactions.14–16 EIS-based biosensors are known for their low cost, quick reaction times, and potential for miniaturization, making them ideal for real-time, label-free DNA detection with minimal sample preparation.17–21
In conclusion, this study advances the field of electrochemical DNA sensing by introducing a non-enzymatic adsorption mechanism. As summarized in Table 1, our findings exhibit high specificity and competitive sensitivity for DNA detection (0–300 nM) using CV, EIS, and SWV techniques, while utilizing fewer chemical components compared to other electrochemical biosensors reported between 2020 and 2025. The platform developed in this research provides a valuable tool for detecting and monitoring K. pneumoniae infections and holds promise for future applications in detecting other pathogenic bacteria as well.
| No. | Detection range | Detection molecule | Probe used | Specificity | Electrochemical methods | Electrolyte used | Number of chemicals used | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | 10–500 nM | Cancer biomarkers | Yes, aptamer-based | High | Cyclic voltammetry (CV), EIS | PBS (pH 7.4) | 5 | 22 |
| 2 | 5–200 nM | Glycoproteins | No | Moderate | Differential pulse voltammetry (DPV) | 0.1 M PBS | 4 | 23 |
| 3 | 1–100 nM | DNA | Yes, DNA probe | High | Square wave voltammetry (SWV) | 1 M PBS | 6 | 24 |
| 4 | 50 nM–1 μM | Proteins | No | Moderate | Linear sweep voltammetry (LSV) | 0.5 M NaCl | 3 | 25 |
| 5 | 2–50 nM | RNA | Yes, RNA probe | High | CV, EIS | 0.1 M PBS | 7 | 26 |
| 6 | 10–100 μM | Glucose | No | Low | Amperometry | 0.1 M KCl | 2 | 27 |
| 7 | 1–10 μM | Heavy metals | No | Moderate | DPV | 0.1 M HCl | 4 | 28 |
| 8 | 5–500 nM | miRNA | Yes, miRNA probe | High | CV, SWV | 1 M PBS | 6 | 29 |
| 9 | 10 nM–1 μM | Pathogens | Yes, antibody-based | High | EIS | 0.1 M PBS | 5 | 30 |
| 10 | 100 nM–10 μM | Enzymes | No | Low | CV | 0.1 M NaOH | 3 | 31 |
| 11 | 1–100 nM | DNA | Yes, aptamer-based | High | DPV, EIS | 0.5 M PBS | 8 | 32 |
| 12 | 10–100 μM | Environmental toxins | No | Moderate | Amperometry | 0.1 M NaCl | 4 | 33 |
| 13 | 5–200 nM | COVID-19 antigens | Yes, antibody-based | High | CV, DPV | 1 M PBS | 6 | 34 |
| 14 | 50 nM–1 μM | Biomolecules | No | Moderate | SWV | 0.1 M PBS | 5 | 35 |
| 15 | 1–10 μM | Enzymes | No | Low | CV | 0.1 M NaOH | 3 | 36 |
| 16 | 0 – 300 nm | DNA | No | High | CV, EIS, SWV | 1 M PBS | 4 | Our findings |
2. Materials and methods
Analytical grade (AR grade 99.9%) phosphate buffer saline solution (PBS), sodium hydroxide (NaOH), glycine (C2H5NO2), ferric (FeCl2·4H2O), and ferrous (FeCl3·6H2O) chloride were purchased from Himedia Pvt. Ltd. They were used directly without additional purification after purchase. Double distilled water (DDW) was incorporated for subsequent reactions. An electrochemical workstation (Biologic VSP) was used for all electrochemical measurements. Scanning electron microscopy (SEM, JSM-7900F, JEOL, Japan), X-ray photoelectron spectroscopy (XPS, K alpha plus), transmission electron microscopy (TEM, JEM-2010, JEOL, Japan. 200 kV), and Raman spectroscopy (Model: Flex G, Tokyo Instruments) were performed in this work.2.1. Synthesis of glycine@Fe3O4 nanoparticles
The glycine@Fe3O4 nanoparticles were synthesized with a co-precipitation method as reported earlier.37 The resulting magnetic nanoparticles were characterized using various analytical techniques such as X-ray diffraction (XRD), scanning electron microscopy (SEM), transmission electron microscopy (TEM), X-ray photoelectron spectroscopy (XPS), and Raman spectroscopy to determine their size, crystalline structure, and morphology.2.2. Glycine@Fe3O4–DNA conjugation
K. pneumoniae DNA containing clinical samples were obtained from D. Y. Patil Hospital, Kolhapur, MS, India. DNA quantification and polymerase chain reaction PCR-associated agarose gel electrophoresis were performed to evaluate the DNA sensing efficiency. DNA–glycine@Fe3O4 interactions were optimized using the following parameters: concentration of DNA, pH of the electrolyte, and incubation time. A detailed description is provided in the ESI.†2.3. Fabrication of the biosensor
2.4. Computational analysis
Interactions between K. pneumoniae DNA and glycine molecules present on glycine@Fe3O4 were studied at a computational level as a basis of primary analysis. Docking analysis was carried out with PyRX 0.8 software. The PDB structure of glycine (PDB ID-DB00145) was obtained from the protein data bank (PDB). The K. pneumoniae DNA model was built using Discovery studio 2021 software. Docking was performed with PyRx 0.8. Visualization of docking was done with Pymol.3. Results and discussion
3.1. Characterization of glycine@Fe3O4 nanoparticles
The glycine@Fe3O4 nanoparticles were characterized by X-ray diffractometer (XRD), X-ray photoelectron spectroscopy (XPS), Raman spectroscopy, scanning electron microscopy (SEM), and transmission electron microscopy (TEM).The nature of the nanoparticles was confirmed by XRD of pristine Fe3O4 and glycine@Fe3O4 with the JCPDS card value 75-0449 shown in Fig. 1a. Possibly, hydrogen bonding occurred between 2 glycine and 1 Fe3O4 molecules by eliminating a water molecule.
 | ||
| Fig. 1 Characterization of the pristine Fe3O4 and glycine@Fe3O4 nanoparticles; (a) XRD, (b) Raman, (c) SEM of pristine Fe3O4, (d) pristine glycine and (e) the glycine@ Fe3O4 composite. | ||
Raman spectroscopy analysis depicted alterations in composition, internal structures, and bonding as shown in Fig. 1b. Although there was no distinguishable change in the morphology of the pristine or composite materials, significant alterations at compositional and functional levels in the composite compared with pristine materials were observed. The given exciting wavelength was 532 nm with a power of 25 mW. The time of laser exposure was 5 minutes. It remained the same for all spectra. Pristine Fe3O4 had a characteristic peak near 670 nm as shown in Fig. 1b, which confirmed the magnetite phase of the synthesized material (Fe3O4).38 The absence of other peaks depicted that Fe3O4 was a poor Raman scatterer. Pristine glycine indicated a Raman scattering effect in a wide range with the presence of NH, OH, and CH2 stretching as shown in Fig. 1b. Peaks above 2000 cm−1 (3007, 2972, 2806, and 2595 cm−1) depicted NH and OH-stretchings. The peak at 1576 cm−1 represented distorted carbon. The peaks at 1412, 1320, 1143, 1033, and 899 cm−1 represented CH2 stretchings, whereas the peaks at 700, 604, 501, 369, 200, 120, and 82 cm−1 indicated interactions between water molecules (solvent) and outer surface atoms of glycine molecules.39 As shown in Fig. 1b, the peak at 2146 cm−1 depicted OH-stretching. The peak at 1318 cm−1 showed CH2 stretching. Peaks at 672 and 613 cm−1 depicted the presence of Fe3O4. Some traces of Fe2O3 were also present in the composite sample based on peaks located at 404, 305, and 232 cm−1 in the spectrum. The Glycine@Fe3O4 composite showed a clear red shift, signifying that light scattering shifted to lower wavelengths as shown in Fig. 1b. Hence, it could be interpreted that the formation of glycine@Fe3O4 could change the light-scattering properties of the pristine material. Raman analysis showed the non-elastic nature of the collisions between the light particles and glycine@Fe3O4. As a property of a non-elastic collision, colliding light particles will either gain or emit electromagnetic radiation from the collided matter, here glycine@Fe3O4. Here, a red shift was observed for glycine@Fe3O4. When wavelength shift occurs from higher to lower, it is called a Raman redshift. The Raman spectrum provides the basis of bond vibrations. The spectra revealed the main phase of the nanomaterial, i.e., magnetite Fe3O4. Changes in Raman spectra of glycine@Fe3O4 acted as a function of doping or surface modification to the pristine Fe3O4. These obtained peaks were material-specific. The advantage of characterizing particles using Raman spectroscopy is that the composition of amino acids such as glycine would not alter40 because the analysis is carried out at room temperature. In Fig. 1(c–e), pristine Fe3O4 shows a hybrid morphology for the majority of particles. Also, aggregation is seen as the particles are in solution. Pristine glycine showed a smooth nearly triangular morphology, which might have acted as a base for DNA attachments. The Glycine@Fe3O4 composite shows similar morphology to pristine Fe3O4. It can be interpreted that no morphological alteration has occurred due to the addition of glycine in Fe3O4. The SEM image of pristine Fe3O4 showed a nearly cuboidal (or hybrid) morphology for many particles. However, some particles had distorted shapes. It also revealed agglomeration of particles in a solution form. Glycine is considered the smallest and simplest amino acid with a simple carbon chain. SEM analysis showed fused triangular particles with smooth surfaces. The image of glycine@Fe3O4 depicted a similar morphology to pristine Fe3O4. Glycine modification did not affect the morphology of the particles. It could be interpreted that no morphological alteration occurred due to the addition of glycine in Fe3O4. The average size of glycine@Fe3O4 was calculated to be 16 ± 20.54 nm based on the TEM micrographs (Fig. S2, ESI†).
XPS analysis was performed to confirm the functionalization or modification of the surface of Fe3O4, as shown in Fig. 2a and b for pristine Fe3O4 and glycine@Fe3O4, respectively. Fig. 2a(a′) displays the Fe 2p region, showing the presence of Fe, with Fe 2p1/2 and Fe 2p3/2 peaks. In pristine Fe3O4, only Fe and O elements are present. In contrast, Fig. 2b(b′) highlights the glycine@Fe3O4 sample, which also displays Fe 2p peaks, along with the presence of nitrogen, indicating the successful incorporation of glycine. Additionally, Fig. 2a(a′′) and b(b′′) shows the O 1s peak, confirming the presence of oxygen in both the modified and unmodified samples.
 | ||
| Fig. 2 Characterization of pristine Fe3O4 and glycine@Fe3O4 nanoparticles; (a), (a′) and (a′′) XPS of pristine Fe3O4; and (b), (b′) and (b′′) glycine@Fe3O4. | ||
Therefore, from all the above characterizations, it was concluded that the synthesized nanoparticles are glycine-modified iron oxide nanoparticles glycine@Fe3O4.
3.2. Electrochemical biosensor
The three-electrode system-based electrochemical biosensor was employed to detect K. pneumoniae DNA based on the abovementioned optimized parameters. Previously, in pilot experiments, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 glycine@Fe3O4 nanoparticle
1 glycine@Fe3O4 nanoparticle![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA ratio provided the best optimum results for the conjugation (Fig. S3–S5, ESI†). However, to improve the ease of application and achieve a wider detection range, the 100 μg mL−1 glycine@Fe3O4 nanoparticles were drop cast onto the working electrode. To determine if the biosensor detects smaller DNA quantities, its efficiency was analyzed in nanomolar ranges for early-stage disease detection.
DNA ratio provided the best optimum results for the conjugation (Fig. S3–S5, ESI†). However, to improve the ease of application and achieve a wider detection range, the 100 μg mL−1 glycine@Fe3O4 nanoparticles were drop cast onto the working electrode. To determine if the biosensor detects smaller DNA quantities, its efficiency was analyzed in nanomolar ranges for early-stage disease detection.
3.3. Electrochemical analysis of the developed biosensor with CV, EIS, and SWV
The following are the techniques used to study the efficiency of the proposed biosensor. | ||
| Fig. 3 Cyclic voltammetry curves of glycine@Fe3O4–GCE electrochemically studied at various scan rates of standard DNA in PBS (1 M) electrolyte; (a) 10–100 mV s−1, and (b) 110–200 mV s−1. | ||
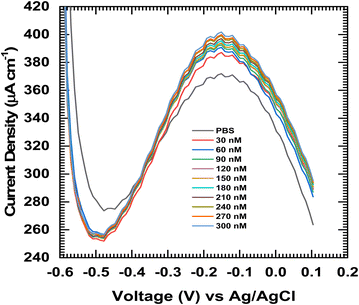
Fig. 4 shows the cyclic voltammetry (CV) curves of the glycine@Fe3O4-modified electrodes in the presence of DNA at concentrations ranging from 0 to 300 nM, recorded at scan rates of (a) 20 mV s−1, (b) 40 mV s−1, and (c) 60 mV s−1. As the DNA concentration increases, a gradual change in the redox peak currents is observed, indicating effective interaction between the DNA molecules and the modified electrode surface. Fig. 4(d) presents the electrochemical impedance spectroscopy (EIS) data for the glycine@Fe3O4-modified electrode at 60 mV s−1 following exposure to varying DNA concentrations. The Nyquist plots reveal a systematic change in the charge transfer resistance (Rct), confirming the successful immobilization and surface binding of DNA onto the electrode.
For further investigation, we used a Nyquist plot frequently employed for impedimetric detection. Fig. 5c and d show Nyquist plots for DNA concentrations from 0 nM (bare) to 300 nM. The corresponding equivalent circuit is depicted in Fig. 5d. The equivalent circuit consisted of solution resistance (Rs) in series with a parallel combination of constant phase element (Q2), which represents charge transfer processes at the electrode-solution interface, charge transfer resistance (Rct), and constant phase element (Q3), with interpretations for additional interactions within the system, such as diffusion effects.
Values of circuit elements for bare DNA to 300 nM DNA are shown in Table 2. Values of solution resistance (Rs) decreased from 8480 Ω to 47.9 Ω with an increase in DNA concentration, suggesting an increase of electrolyte conductivity. The curvature of the semicircle, which was correlated with the charge transfer resistance (Rct), was decreased from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 374 to 48 as DNA content increased, indicating that the electrode (glycine@Fe3O4) was interacting with DNA more when the DNA content was higher.41 The value of α parameter for CPE indicates the dominance of a capacitive or resistive effect within an electrochemical cell. The value of α is 0 for pure resistive behavior. It is 1 for optimum capacitive behavior. All values for Q2 and Q3 were above 0.75, indicating that the capacitive effect was dominant at lower frequencies.42
374 to 48 as DNA content increased, indicating that the electrode (glycine@Fe3O4) was interacting with DNA more when the DNA content was higher.41 The value of α parameter for CPE indicates the dominance of a capacitive or resistive effect within an electrochemical cell. The value of α is 0 for pure resistive behavior. It is 1 for optimum capacitive behavior. All values for Q2 and Q3 were above 0.75, indicating that the capacitive effect was dominant at lower frequencies.42
| Sample | R s (Ω) | Q 2 (F s−1) | α 2 | R ct (Ω) | Q 3 (F s−1) | α 3 |
|---|---|---|---|---|---|---|
| Bare | 8480 | 3.499 × 10−6 | 0.9498 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 372 372 |
5.036 × 10−6 | 0.8623 |
| 20 nM DNA | 263 | 0.1489 × 10−6 | 0.9546 | 343.8 | 6.08 × 10−6 | 0.8345 |
| 30 nM DNA | 263 | 0.1508 × 10−6 | 0.9544 | 322.7 | 6.698 × 10−6 | 0.8144 |
| 40 nM DNA | 133.7 | 0.2944 × 10−6 | 0.9272 | 191.2 | 6.715 × 10−6 | 0.8153 |
| 80 nM DNA | 95.04 | 0.4164 × 10−6 | 0.9270 | 121.2 | 7.397 × 10−6 | 0.8024 |
| 150 nM DNA | 68.23 | 0.3201 × 10−6 | 1 | 68.7 | 8.285 × 10−6 | 0.7873 |
| 200 nM DNA | 50.59 | 0.3734 × 10−6 | 1 | 40.7 | 10.19 × 10−6 | 0.7560 |
| 300 nM DNA | 47.9 | 0.589 × 10−6 | 0.9598 | 48.0 | 7.627 × 10−6 | 0.8004 |
| LOD = (3.3 × SD)/slope | (1) |
| LOQ = (10 × SD)/slope | (2) |
As presented in Fig. 8b, the LOD and LOQ values were calculated. For DNA, for the range 30–90 nM, LOD = 3.27 nM, R2 = 0.9972, LOQ = 3.90 nM. For the higher range of 120–270 nM, LOD = 3.94 nM, R2 = 0.9992, and LOQ = 11.95 nM. These values emphasize the precision and accuracy of the sensor at both low and moderate levels of concentration, which reflects quick and sensitive surface hybridization. The values of LOQ dynamic range of 0–300 nM achieved by our biosensor align well with sensitivity targets commonly required in molecular diagnostics.43–46 However, clinical standards often vary depending on the specific application and assay type. These features make the biosensor suitable for point-of-care applications and resource-limited settings. To further validate its clinical relevance, comparative testing with patient-derived samples and established diagnostic methods would be a valuable future step. Sensitivity was calculated from the slope of the calibration curves obtained at 0.2 V using the formula:
| Slope = (3.3 × SD)/LOD | (3) |
For DNA (30–90 nM) range, = 0.1009 μA nM−1
For DNA (120–270 nM) range, = 0.0838 μA nM−1
With the working electrode surface area 0.07 cm2 (GCE):
(1) DNA 30–90 nM (low range): 1.441 μA nM−1 cm−2
(2) DNA 120–270 nM (high range): 1.197 μA nM−1 cm−2
These results indicate that the sensor has greater sensitivity in DNA detection, particularly at low concentrations, which is beneficial for the detection of bacteria at early stages or trace-level nucleic acid monitoring. The glycine@Fe3O4 interface is providing both biocompatibility and enhanced electron transfer properties.
3.4. Analysis of the biosensor using clinical samples
Based on the Bode plot obtained in Fig. 5a, we obtained a calibration curve as shown in Fig. S6 (ESI†). Here, we considered the peak position frequency of the 0 nM Bode plot as fi. Peak position frequencies for 10, 20, 40, 80, 150, 200, and 300 nM were considered as f1–f7, respectively. The difference between fi and f1–7 was considered as Δf for each concentration. A linear fitting of the calibration plot indicated a linear increment in peak position frequency with an increase in DNA concentration (0–300 nM). For isolated unknown clinical sample concentrations, we simply measured electrochemical impedance spectroscopy (EIS) and plotted a Bode plot for it. The peak position frequency difference (Δf) was calculated by comparing with the bare peak position frequency, which was found to be, Δf = 993.13. Finally, we compared this Δf with the calibration curve and found that an isolated unknown concentration of Klebsiella pneumoniae DNA from the clinical culture was around 20 nM in the presence of other biomolecules in the clinical culture. The above-mentioned DNA sensing was validated with a currently used DNA detection method i.e., UV-visible spectroscopy and DNA quantification, which showed an increase in DNA concentration with an increase in bacterial growth (Fig. S6c, ESI†). The sensed DNA was separated from glycine@Fe3O4 by ceasing the applied current, confirming that the sensed material was DNA. Conventional PCR-based agarose gel electrophoresis is shown in Fig. S6d (ESI†). Separated DNA was further PCR-amplified to detect the specific gene sequence of DNA. Qualitative PCR estimation of the sensed DNA [K. pneumoniae] from clinical culture was performed by 0.8% agarose gel electrophoresis. The 1st well represented a reference 100–1000 base pairs (bp) DNA ladder. The 2nd well was the SHV gene with 294 bp. The 3rd well represented the TEM gene with 404 bp. The 4th and 6th wells represented the OXA-1 gene with 464 bp. The 5th well represented the CTX-M gene with 754 bp. The 7th well represented a negative control with nuclease-free water. The obtained gene sequences in the present study are presented in Table 3.47 Detailed conditions given for K. pneumoniae DNA detection are summarized in Table 4. Other PCR details are attached to the ESI.†| S. no. | DNA (gene) | Primer | Amplicon (bp) |
|---|---|---|---|
| 1 | SHV | SHV-F: CGCCTGTGTATTATCTCCCT | 294 |
| SHV-R: CGAGTAGTCCACCAGATCCT | |||
| 2 | TEM | TEM-F: TTTCGTGTCGCCCTTATTCC | 404 |
| TEM-R: ATCGTTGTCAGAAGTAAGTTGG | |||
| 3 | CTX-M | CTX-M-F: CGCTGTTGTTAGGAAGTGTG | 754 |
| CTX-M-R: GGCTGGGTGAAGTAAGTGAC | |||
| 4 | OXA-1 | OXA-1-F: ACACAATACATATCAACTTCGC | 464 |
| OXA-1-R: AGTGTGTTTAGAATGGTGATC |
| Steps | TEM | SHV | CTX-M | OXA-48 |
|---|---|---|---|---|
| Initial denaturation | 94 °C for 5 min | 94 °C for 5 min | 94 °C for 5 min | 94 °C for 3 min |
| Denaturation | 94 °C for 30 s | 94 °C for 30 s | 94 °C for 30 s | 61.7 °C for 5 min |
| Annealing | 65.5 °C for 30 s | 60 °C for 30 s | 60 °C for 30 s | 61.7 °C for 30 s |
| Extension | 72 °C for 50 s | 72 °C for 50 s | 72 °C for 50 s | 72 °C for 1 min |
| Final extension | 72 °C for 5 min | 72 °C for 5 min | 72 °C for 5 min | 72 °C for 7 min |
| Cycles | 35 | 35 | 35 | 35 |
| Hold | 4 °C | 4 °C | 4 °C | 4 °C |
3.5. Mechanism of the proposed biosensor
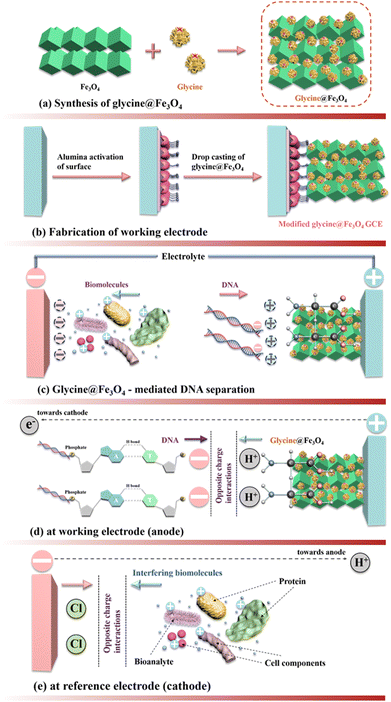
Development of a glycine@Fe3O4 nanoparticle-based electrochemical biosensor for specific detection of K. pneumoniae DNA was carried out in six stages as given in Fig. S1 (ESI†): {1} mechanism of proposed biosensor, {2} synthesis of glycine@Fe3O4 nanoparticles, {3} characterization of glycine@Fe3O4 nanoparticles, {4} optimization of glycine@Fe3O4–DNA conjugation, {5} development of electrochemical biosensor, and {6} electrochemical analysis with CV, EIS, DPV, and SWV. All the stages of the study have been explained in detail below.K. pneumoniae DNA was detected by a biosensing platform which is fabricated using glycine@Fe3O4 nanoparticles as shown in Scheme 1 in a stepwise manner. Initially, glycine@Fe3O4 nanoparticles were synthesized through the co-precipitation method and simultaneously functionalized with glycine biomolecules (Scheme 1a). Then they are immobilized on a glassy carbon electrode (GCE) using a drop casting method and it is considered as a working electrode (Scheme 1b). Two other electrodes, viz. counter (platinum wire) and reference (Ag/AgCl), were utilized to set up a three-electrode system as described in the methodology section.
The resulting biosensing platform when exposed to K. pneumoniae DNA, and the glycine@Fe3O4–GCE could capture the DNA present in the electrolyte, here 1 M PBS. Detection and quantification of DNA targets were achieved using suitable analytical techniques such as electrochemical measurements.
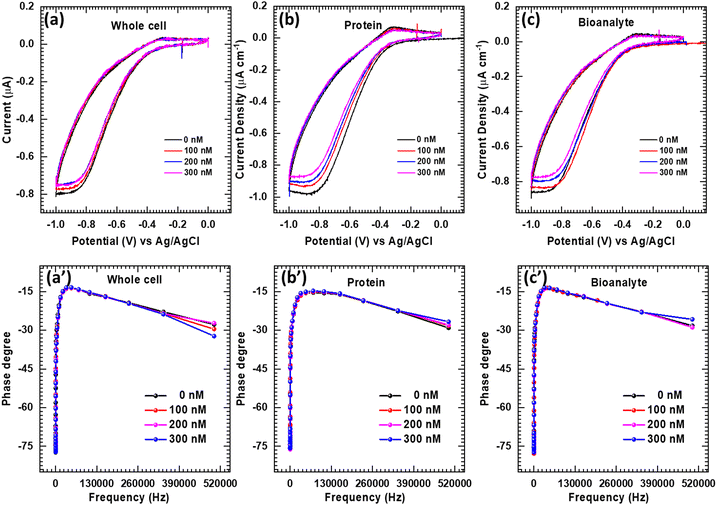
In the three electrode system, glycine@Fe3O4–GCE acts as an anode which possesses positive charge (+) and hence negatively charged DNA (−) attracts towards it (Scheme 1c). Glycine present on the nanoparticles achieved the actual binding to the DNA. It is an amino acid with a simple carbon chain. It conducts a positive charge on the surface and hence attracts negatively charged DNA present in the electrolyte (Scheme 1d). As a result, specific electrostatic ionic interactions take place on the electrode surface that could enable electrochemical detection of the K. pneumoniae DNA.37 The above-mentioned reaction forms on the electrode surface without the addition of any enzyme or aptamers, resulting in greater selectivity towards K. pneumoniae DNA compared to traditional DNA sensing methods, while effectively repelling other non-specific biomolecules such as proteins and cells from the electrode surface. In contrast, the reference electrode possesses negative charge (−) and therefore negatively charged DNA (−) gets repelled by this electrode and other positively charged interfering biomolecules present in the electrolyte may get attracted towards this electrode (Scheme 1e). Therefore, the present work is described as a non-enzymatic label-free DNA detection methodology with the help of glycine@Fe3O4 nanoparticles.
3.6. DNA sensing by computational analysis
Computational analysis was performed to support interactions between K. pneumoniae DNA and glycine molecules, as shown in Fig. 9a and b. Glycine, a simple chain amino acid, exhibits a property of cationic surface charge in an acidic environment. Chemically, it can be interpreted that cationic glycine and anionic DNA make a conjugate. However, glycine–DNA interactions were seen to develop a bond or conjugate when studied computationally. Computer-aided interactions of different biomolecules are then predicted. A computational study (docking analysis) of the DNA–glycine conjugate was carried out using different software programs such as PyRx, Discovery studio 2021, Pymol, and so on. Docking analysis can be used for computational biology where possible bond formations are predicted when two biomolecules come in proximity. Here, DNA of Klebsiella pneumoniae and the glycine biomolecule were studied as a function of their conjugate formation. A DNA model was built by providing Klebsiella pneumoniae DNA sequence (5′TTTCGTGTCGCCCTTATTCC3′) using Discovery studio 2021. The protein data bank (PDB) model of glycine was taken from the PDB database. With PyRx 0.8, glycine and DNA macromolecule models were prepared for docking by minimizing their energy. These molecules were then docked. The docked image was directly obtained after the process was done as in Fig. 9a, where the glycine molecule was seen to be docked inside the DNA with a binding energy of around −6 kcal mol−1. Negative binding energy provides natural binding interactions between molecules. The obtained model was visualized with Pymol software as shown in Fig. 9b. The figure clearly depicts the formation of the glycine–DNA conjugate.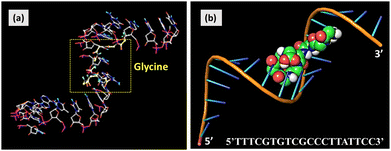 | ||
| Fig. 9 Computational analysis of interactions between glycine and K. pneumoniae DNA. (a) DNA–glycine docking with PyRx 0.8, and (b) DNA–glycine conjugate visualization with Pymol. | ||
4. Conclusions
This study presents a glycine-modified iron oxide (glycine@Fe3O4) electrochemical biosensor for highly sensitive and selective detection of Klebsiella pneumoniae DNA, offering a rapid, cost-effective, and reliable alternative to conventional methods. The biosensor functions through label-free and non-enzymatic interactions, leveraging electrostatic attraction for efficient DNA binding.Electrochemical characterization via CV, EIS, and SWV demonstrated DNA for the range 30–90 nM, LOD = 3.27 nM, R2 = 0.9972, LOQ = 3.90 nM. For the higher range of 120–270 nM, LOD = 3.94 nM, R2 = 0.9992, and LOQ = 11.95 nM, with a dynamic range of 0 to 300 nM, confirming high sensitivity. Sensitivity analysis revealed values of 1.441 μA nM−1 cm−2 for DNA, ensuring robust signal detection. Computational docking validated strong glycine–DNA binding interactions with a binding energy of approximately −6 kcal mol−1, reinforcing the feasibility of the sensing mechanism. Selectivity tests confirmed negligible interference from whole bacterial cells, proteins such as albumin, and bioanalytes including folic acid, ensuring accurate pathogen detection in complex biological samples. Clinical validation through PCR-based gel electrophoresis successfully identified key antimicrobial resistance genes SHV, TEM, CTX-M, and OXA-1, verifying the biosensor's real-world applicability for hospital infection surveillance.
This biosensor integrates high sensitivity, operational simplicity, and selectivity, making it an ideal candidate for point-of-care diagnostics and real-time infection monitoring. Future research should focus on expanding detection capabilities to other bacterial pathogens, validating performance with diverse clinical samples, and optimizing real-world implementation, further solidifying its role in combating antimicrobial resistance and hospital-acquired infections.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declared no conflict of interest in the present work.Acknowledgements
This work was supported by D. Y. Patil Education Society (Deemed to be University), Kolhapur through providing the required infrastructure, experimental facility, and financial support through an intramural project (No. DYPES/DU/R&D/2023/1164). This research work was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. RS-2019-NR040081). The authors are thankful to the D. Y. Patil Hospital, Kolhapur, MS, India for gifting MDR Klebsiella pneumoniae cultures and the University of Madras, Chennai, India for helping with PCR validation. Dr T. D. Dongale and Prof. R. K. Kamat would like to thank RUSA Maharashtra for providing financial assistance under the ‘RUSA-Industry Sponsored Centre for VLSI System Design’.References
- H. Namikawa, K. I. Oinuma, K. Yamada, Y. Kaneko, H. Kakeya and T. Shuto, Differences in severity of bacteremia caused by hypermucoviscous and non-hypermucoviscous Klebsiella pneumoniae, Int. J. Antimicrob. Agents, 2023, 61(5), 106767 CrossRef CAS PubMed.
- C. L. Gorrie, M. Mirčeta, R. R. Wick, L. M. Judd, M. M. Lam, R. Gomi, I. J. Abbott, N. R. Thomson, R. A. Strugnell, N. F. Pratt and J. S. Garlick, Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen, Nat. Commun., 2022, 13(1), 3017 CrossRef CAS PubMed.
- K. K. Tsang, M. M. Lam, R. R. Wick, K. L. Wyres, M. Bachman, S. Baker, K. Barry, S. Brisse, S. Campino, A. Chiaverini and D. M. Cirillo, Diversity, functional classification and genotyping of SHV β-lactamases in Klebsiella pneumoniae, Microb. Genomics, 2024, 10(10), 001294 CAS.
- P. Manohar, S. Leptihn, B. S. Lopes and R. Nachimuthu, Dissemination of carbapenem resistance and plasmids encoding carbapenemases in Gram-negative bacteria isolated in India, JAC-Antimicrob. Resist., 2021, 3(1), dlab015 CrossRef PubMed.
- J. Henderson, H. Ciesielczuk, S. M. Nelson and M. Wilks, Community prevalence of carbapenemase-producing organisms in East London, J. Hosp. Infect., 2019, 103(2), 142–146 CrossRef CAS PubMed.
- A. Abuawad, Y. Ashhab, A. Offenhäusser and H. J. Krause, DNA Sensor for the Detection of Brucella spp. Based on Magnetic Nanoparticle Markers, Int. J. Mol. Sci., 2023, 24(24), 17272 CrossRef CAS PubMed.
- C. Tang, Z. He, H. Liu, Y. Xu, H. Huang, G. Yang, Z. Xiao, S. Li, H. Liu, Y. Deng and Z. Chen, Application of magnetic nanoparticles in nucleic acid detection, J. Nanobiotechnol., 2020, 18, 1–9 CrossRef PubMed.
- G. Oza, K. Krishnajyothi, V. I. Merupo, K. A. Bracamontes, P. C. Olmos, E. Garrido, S. Velumani, M. Sridharan, A. Sharma, L. G. Arriaga and J. T. Ramirez, Gold-Iron oxide yolk-shell nanoparticles (YSNPs) as magnetic probe for fluorescence-based detection of 3 base mismatch DNA, Colloids Surf., B, 2019, 176, 431–438 CrossRef CAS PubMed.
- C. Wan, A. Qu, M. Li, R. Tang, L. Fu, X. Liu, P. Wang and C. Wu, Electrochemical sensor for directional recognition and measurement of antibiotic resistance genes in water, Anal. Chem., 2021, 94(2), 732–739 CrossRef PubMed.
- C. Wan, A. Qu, L. Deng, X. Liu and C. Wu, Preparation of electrochemical sensor based on glassy carbon electrode and its specificity and sensitivity for directional detection of antibiotic resistance genes spreading in the water environment, Environ. Sci. Pollut. Res., 2023, 30(3), 7904–7913 CrossRef CAS PubMed.
- K. Marchlewicz, I. Ostrowska, S. Oszwałdowski, A. Zasada, R. Ziółkowski and E. Malinowska, Molecular diagnostic of toxigenic Corynebacterium diphtheriae strain by DNA sensor potentially suitable for electrochemical point-of-care diagnostic, Talanta, 2021, 227, 122161 CrossRef CAS PubMed.
- M. I. Hassan, K. R. Alkarshah, A. J. Alzarani, O. E. Obeied, A. H. Khamis and A. Diab, Detection of extended spectrum beta-lactamases-producing isolates and effect of AmpC overlapping, J. Infect. Dev. Ctries., 2013, 7(8), 618 CrossRef PubMed.
- I. Carvalho, J. A. Carvalho, S. M. Alvarez, M. Sadi, R. Capita, C. A. Calleja, F. Rabbi, M. L. Dapkevicius, G. Igrejas, C. Torres and P. Poeta, Characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniaeisolated from clinical samples in a northern Portuguese hospital: predominance of CTX-M-15 and high genetic diversity, Microorganisms, 2021, 9(9), 1914 CrossRef CAS PubMed.
- C. S. S, V. Kini, M. Singh, C. Mukhopadhyay, P. Nag and K. Sadani, Disposable electrochemical biosensors for the detection of bacteria in the light of antimicrobial resistance, Biotechnol. Bioeng., 2024, 121(9), 2549–2584 CrossRef PubMed.
- J. Wang, X. Cui, L. Liang, J. Li, B. Pang and J. Li, Advances in DNA-based electrochemical biosensors for the detection of foodborne pathogenic bacteria, Talanta, 2024, 275, 126072 CAS.
- M. Pan, Y. Zhao, J. Qiao and X. Meng, Electrochemical biosensors for pathogenic microorganisms detection based on recognition elements, Folia Microbiol., 2024, 69(2), 283–304 CrossRef CAS PubMed.
- A. G. da Silva Junior, I. A. Frias, R. G. Lima-Neto, O. L. Franco, M. D. Oliveira and C. A. Andrade, Electrochemical detection of Gram-negative bacteria through mastoparan-capped magnetic nanoparticle, Enzyme Microb. Technol., 2022, 160, 110088 CAS.
- A. Khoshroo, M. Mavaei, M. Rostami, B. Valinezhad-Saghezi and A. Fattahi, Recent advances in electrochemical strategies for bacteria detection, BioImpacts, 2022, 12(6), 567–588 CAS.
- P. V. V. Romanholo, C. A. Razzino, P. A. Raymundo-Pereira, T. M. Prado, S. A. S. Machado and L. F. Sgobbi, Biomimetic electrochemical sensors: New horizons and challenges in biosensing applications, Biosens. Bioelectron., 2021, 185(1), 113242 CAS.
- I. H. Cho, D. H. Kim and S. Park, Electrochemical biosensors: Perspective on functional nanomaterials for on-site analysis, Biomater. Res., 2020, 24(1), 1 Search PubMed.
- A. Villalonga, R. Villalonga and D. Vilela, Hybrid magnetic nanoparticles for electrochemical biosensors, Magn. Nanopart.-Hybrid Mater., 2021, 1(1), 679 Search PubMed.
- A. Sengupta, V. Sharma and B. Roy, Development of enzyme-based electrochemical biosensors: Advancements and limitations, Biosensors, 2025, 15(3), 127 CrossRef PubMed.
- R. Ahuja and N. Patel, Glycoprotein sensing through potentiometric methods: Challenges and innovations, Sensors, 2025, 25(7), 2064 CrossRef PubMed.
- H. Chen, Y. Liu and J. Zhou, Advances in DNA aptamer-based biosensors for electrochemical applications, Lab Chip, 2024, 15(2), 67 Search PubMed.
- X. Zhang, L. Wang and J. Wei, Electrochemical analysis of protein biomarkers: A linear sweep voltammetry approach, Biosensors, 2023, 15(3), 140 Search PubMed.
- S. Kumar and R. Gupta, Detection of RNA biomarkers using electrochemical techniques: Recent advancements, Sensors, 2023, 25(5), 1212 Search PubMed.
- C. Lee and D. Han, Electrochemical glucose detection through amperometric biosensors, Sensors, 2022, 22(4), 345 Search PubMed.
- T. Ahmed and M. Khan, Environmental applications of electrochemical sensors for heavy metal detection, Sensors, 2025, 25(6), 2124 Search PubMed.
- N. Nagdeve and P. Kulkarni, miRNA biosensing using electrochemical techniques: Graphene-modified electrodes, J. Biol. Eng., 2025, 14(7), 98 Search PubMed.
- R. Singh and D. Chauhan, Pathogen detection using antibody-functionalized biosensors: EIS-based analysis, SpringerLink, 2024, 12(3), 207 Search PubMed.
- D. Grieshaber and R. MacKenzie, Nanowire-based biosensors for enzyme detection: Opportunities and limitations, Biosensors, 2023, 13(2), 187 CrossRef PubMed.
- Q. Wang and Z. Zhang, DNA aptamer-functionalized electrochemical sensors: Advances in DPV and EIS techniques, Sensors, 2022, 15(5), 92 Search PubMed.
- M. Patel and S. Kaur, Electrochemical biosensors for environmental toxin detection: Amperometric methodologies, Sensors, 2024, 20(6), 287 Search PubMed.
- R. Lima and M. Pedro, Immunosensors for COVID-19 detection: Analytical performance and real-world applications, Biosensors, 2025, 15(4), 300 Search PubMed.
- S. Varnakavi and H. Kim, Biomolecule detection using nanostructured materials: A review of SWV methods, Sensors, 2021, 22(3), 1400 Search PubMed.
- A. Papreja and K. Saxena, Enzyme-based biosensors for rapid electrochemical analysis: Cyclic voltammetry focus, AIP Publ., 2023, vol. 45, 5, p. 78 Search PubMed.
- R. P. Gambhir, A. K. Parthasarathy, S. Sharma, S. Kale, V. V. Magdum and A. P. Tiwari, pH-responsive glycine functionalized magnetic iron oxide nanoparticles for SARS-CoV-2 RNA extraction from clinical sample, J. Mater. Sci., 2022, 57, 13620–13631 CrossRef CAS PubMed.
- P. C. Panta and C. P. Bergmann, Raman spectroscopy of iron oxide of nanoparticles (Fe3O4), J. Mater. Sci. Eng., 2015, 5(3), 1 Search PubMed.
- I. V. Krauklis, A. V. Tulub, A. V. Golovin and V. P. Chelibanov, Raman Spectra of Glycine and Their Modeling in Terms of the Discrete–Continuum Model of Their Water Solvation Shell, Opt. Spectrosc., 2020, 128(10), 1598 CrossRef CAS.
- C. Sudakar, P. Kharel, G. Lawes, R. Suryanarayanan, R. naik and V. M. Naik, Raman spectroscopic studies of oxygen defects in Co-doped ZnO films exhibiting room-temperature ferromagnetism, J. Phys.: Condens. Matter, 2006, 19(2), 026212 CrossRef.
- A. M. Abouzied, A. I. Al-falouji, S. I. Khalifa, H. T. Omran and H. A. Azab, Potential affinity binding of sarcophine to DNA, Egypt. J. Nat. Toxins., 2007, 4(1), 51 Search PubMed.
- A. Villalonga, R. Villalonga and D. Vilela, Hybrid magnetic nanoparticles for electrochemical biosensors, Magn. Nanopart.-Hybrid Mater., 2021, 1(1), 679 Search PubMed.
- Y. Yuan, F. Guillon, S. Griveau, F. Bedioui, M. Lazerges and C. Slim, Evolution of nucleic acids biosensors detection limit III, Anal. Bioanal. Chem., 2022, 414, 943–968 CrossRef PubMed.
- M. Santhanam, I. Algov and L. Alfonta, DNA/RNA Electrochemical Biosensing Devices: A Future Replacement of PCR Methods for a Fast Epidemic Containment, Sensors, 2020, 20(16), 4648 CrossRef CAS PubMed.
- G. Coletta and V. Amendola, Numerical Modelling of the Optical Properties of Plasmonic and Latex Nanoparticles to Improve the Detection Limit of Immuno-Turbidimetric Assays, Nanomaterials, 2021, 11(5), 1147 CrossRef CAS PubMed.
- H. Wang, H. Wang, Y. Huang, H. Zhang, Y. Fu, Z. Yang, Y. Chen, X. Qiu, D. Yu and L. Zhang, Multi-parameter surface plasmon resonance instrument for multiple nucleic acid quantitative detection, Biomed. Microdevices, 2023, 25, 1–24 CrossRef PubMed.
- P. Vadhva, J. Hu, M. J. Johnson, R. Stocker, M. Braglia, D. J. L. Brett and A. J. E. Rettie, Electrochemical impedance spectroscopy for all-solid-state batteries: Theory, methods and future outlook, ChemElectroChem, 2021, 8(11), 1930 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5tb00377f |
| This journal is © The Royal Society of Chemistry 2025 |