Open Access Article
Open Access ArticleAstragalus polysaccharide-containing 3D-printed scaffolds promote cartilage regeneration by correcting metabolic disorders and promoting cellular matrix remodeling†
Weibin
Du‡
 *ab,
Zhenwei
Wang‡
ab,
Wenxiang
Zeng‡
ab,
Huahui
Hu
ab,
Yanghua
Tang
ab,
Guoping
Cao
ab,
Gang
Qu
ab and
Rongliang
Chen
*ab
*ab,
Zhenwei
Wang‡
ab,
Wenxiang
Zeng‡
ab,
Huahui
Hu
ab,
Yanghua
Tang
ab,
Guoping
Cao
ab,
Gang
Qu
ab and
Rongliang
Chen
*ab
aResearch Institute of Orthopedics, the Jiangnan Hospital affiliated to Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China. E-mail: chrl123456@163.com; dwbbdm@163.com
bHangzhou Xiaoshan Hospital of Traditional Chinese Medicine, Hangzhou, Zhejiang, China
First published on 11th June 2025
Abstract
Background: Astragalus polysaccharide-containing 3D-printed scaffolds show great potential for cartilage defect repair. The aim of this study is to investigate their repairing role, combine them with metabolomics technology to deeply analyze the related metabolite changes, and provide a new strategy for the treatment of cartilage defects. Methods: Biocompatible astragalus polysaccharide-containing 3D-printed scaffolds were prepared. Thirty New Zealand rabbits were divided into normal, model and scaffold groups, with 10 rabbits in each group. The repair of cartilage defects by the scaffolds was evaluated by gross observation, micro-CT, HE and ABH staining after 12 weeks of intervention. The expression of VEGFA, Col2a1 and Biglycan was detected by immunofluorescence. Newly formed cartilage tissues were collected for metabolomics analysis to comprehensively evaluate the mechanism of action of astragalus polysaccharide-containing 3D-printed scaffolds in cartilage repair. Results: The recovery of cartilage defects in the scaffold group was found to be significantly better than that in the model group and comparable to that of the normal group by gross observation, micro-CT, HE and ABH staining. Immunofluorescence results showed that the expression of VEGFA, Col2a1 and Biglycan in the scaffold group was higher than that in the model group (all P < 0.05), comparable to that in the normal group. Metabolomics revealed that 29 metabolites were reversed in the scaffold group, with a reversal rate of 58%. The reversal mainly included groups of phospholipids, sphingolipids, purines, amino acids and energy metabolism-related changes. Fifteen metabolic pathways may be involved, and phospholipid and sphingolipid metabolism, fatty acid metabolism and purine metabolism are the major differential metabolic pathway change groups. Conclusion: Astragalus polysaccharide-containing 3D-printed scaffolds may accelerate cartilage collagen matrix remodeling, correct cartilage tissue metabolic disorders by promoting the expression of vascular-related factors, and ultimately promote cartilage repair.
1 Introduction
Articular cartilage plays a vital role in maintaining joint function and mobility in the human body. As a special connective tissue, cartilage not only provides the smoothness of joint surfaces and reduces friction, but is also able to withstand and disperse the forces exerted on the joints.1 However, with aging and various types of trauma, cartilage defects are becoming increasingly common and have become an important factor in people's quality of life. Available studies have shown that epidemiological data on cartilage defects are on the rise, especially in athletes and elderly populations.2,3 Although cartilage has some self-repairing ability, its regenerative capacity is limited, leading to difficulties in complete recovery after injury.4 Current treatments mainly include conservative therapy, surgical repair and biomaterial implantation, but these methods often face limitations such as insufficient effectiveness, long recovery time and postoperative complications.5,6 Therefore, there is an urgent need to explore new treatment strategies to improve the clinical management of cartilage defects.3D printed scaffolds offer significant advantages in bone defect repair, as they can be personalized according to the shape and size of the defect and ensure that the scaffold fits perfectly with the defect site. Secondly, the pore structure of 3D printed scaffolds can be adjusted according to the needs, which helps to promote cell attachment and proliferation, and exhibits better biocompatibility and antibacterial and anti-inflammatory effects.7–9 In addition, the scaffold material can be chosen flexibly, and commonly used biocompatible materials such as polylactic acid and hyaluronic acid are favorable for chondrocyte survival and functional recovery. This method reduces surgical trauma, shortens the recovery cycle, and enhances cartilage repair.10–12 Astragalus polysaccharide, as a natural polysaccharide, has gradually gained attention due to its property of promoting cell proliferation and differentiation.13,14 Meanwhile, biomaterials such as collagen, sodium alginate and filipin protein have been widely used in the development of tissue engineering scaffolds due to their excellent biocompatibility and biodegradability.15 These materials not only provide the necessary support and growth environment for cells, but also promote the differentiation of cells to specific phenotypes by regulating the microenvironment.
In our previous study, a biodegradable 3D-printed skin scaffold loaded with astragalus polysaccharide was applied to treat skin defects. This scaffold demonstrated the ability to promote regeneration of blood vessels, collagen fibers, and hair follicles, correct protein metabolic disorders, and achieve more complete structural and functional regeneration of skin.15,16 In another study, a 3D-printed acinar-mimetic silk fibroin–collagen–astragalus polysaccharide scaffold was developed for parotid gland repair. This scaffold effectively supported tissue reconstruction and functional recovery of damaged salivary glands by mimicking the natural acinar microstructure and enhancing cell adhesion and proliferation.17 To date, there are limited reports on 3D-printed scaffolds incorporating astragalus polysaccharide.
Pre-experimentation by our group and literature review revealed that repair of cartilage defects in rabbit knee joints reaches a critical stage at three months. The contrast in cartilage repair status at this time is significant and most studies have used it as a baseline for assessment for between-group comparisons.18–20 Therefore, in this study, three months was taken as the intervention time point, and astragalus polysaccharide-containing 3D-printed scaffolds were prepared and transplanted to the defective sites of cartilage in rabbits to explore their repairing effects. In this study, we innovatively combined astragalus polysaccharide-containing 3D-printed scaffolds with metabolomics technology. The novel mechanism of astragalus polysaccharide-containing 3D-printed scaffolds regulating dual cartilage structure–function repair was elucidated for the first time by in vivo and ex vivo experiments, providing a new way of thinking for clinical treatment.
2 Methods
2.1 Experimental animals
Thirty male New Zealand rabbits weighing 2–3 kg were grouped and acclimatized at the Animal Experiment Center of Zhejiang University of Traditional Chinese Medicine for 1 week for experiments. The purchase and feeding of New Zealand rabbits, as well as other animal procedures, followed the animal research guidelines of the National Institutes of Health and the Animal Research Council. And it was approved by the Experimental Animal Ethics Committee of Zhejiang University of Traditional Chinese Medicine (No. IACUC-20231030-13).2.2 Preparation of astragalus polysaccharide-containing 3D-printed scaffolds
The printed materials were brought to a sol–gel state by dissolving 0.8 g of sodium alginate and 0.8 g of filipin protein in 8 liters of sterile PBS and stirring until completely dissolved. The material was transferred to an airtight container and sterilized at 70 °C for 30 minutes at a time, and the process was repeated every 24 hours for a total of three times. The material should then be stored at 4 °C for later use. This treatment produces the 3D printing matrix material. Subsequently, the sol was continuously printed using a 3D bioprinter controlled by computer-aided design (CAD) and driven by pneumatic pressure and extruded into a scaffold with a diameter of 6 mm and a layer height of 2.4 mm. A printing needle with a diameter of 0.41 mm was used, the extrusion air pressure was 120 kPa, and the heating temperature was 37 °C. The scaffolds were printed by a computer-aided design (CAD) controlled and driven by pneumatic pressure. Each scaffold consisted of 6 layers with 6 strips per layer and was printed with a 90° turn. Immediately after the printing process, 5% perchloric acid was added drop by drop to cross the scaffolds to solidify and harden them. The scaffolds were then cleaned with PBS and transferred to Petri dishes. A mixture containing 0.2% type I collagen and 200 μg mL−1 astragalus polysaccharide was added dropwise to the scaffolds, followed by placing the scaffolds in an incubator at 37 °C for 2 hours to promote collagen gelation to prepare astragalus polysaccharide drug scaffolds. All steps were performed under sterile conditions.2.3 Model preparation
With 0.4 mL min−1 isoflurane inhalation anesthesia, an incision was made at the inner edge of the patella, the subcutaneous tissues were carefully separated and the medial joint capsule was cut off, and the patella was dislocated to the lateral side, and the center of the intertrochanteric talar surface of the femur of the knee joint of rabbits was drilled using an electric drill, resulting in the model of cartilage defect, and the cartilage defects were sutured layer by layer. Based on previous literature studies and preliminary research findings from our group, a defect with a 5 mm diameter and a 3 mm depth was selected.21–232.4 Grouping and treatment
Thirty New Zealand rabbits were randomly divided into three groups of 10 rabbits each according to the random number table method as follows: normal group, modeling group and scaffolding group.1 In the normal group, the modeling area was only clipped and no cartilage defect model was prepared.2 In the model group, the defective model of knee cartilage was prepared and sutured layer by layer to keep the wound dry and prevent infection.3 In the scaffold group, astragalus polysaccharide-containing 3D-printed scaffolds were implanted on the day of modeling. (The scaffold preparation scheme follows the pre-proven preparation techniques of our group).15 Briefly, sodium alginate and filipin protein were mixed proportionally, then 3D printing parameters were set to prepare a cylindrical scaffold using a 3D printer, and finally 0.2% type I collagen and 200 μM astragalus polysaccharide solution were added dropwise to the scaffold), and sutured layer by layer to keep the wound dry and prevent wound infection. Penicillin 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U was given daily for 3 days after surgery to fight infection in the model and stent groups.
000 U was given daily for 3 days after surgery to fight infection in the model and stent groups.
2.5 Detection of cell viability by the CCK-8 method
Chondrocytes were evenly spread on 96-well plates with 1 × 104 cells per well and cultured for 24 h. Different concentrations (50 μM, 100 μM, 200 μM, and 400 μM) of astragalus polysaccharide were added, and 10 μL of CCK-8 solution was added to each well after 24 h. The cells were then incubated at 37 °C with 5 μM of astragalus polysaccharide. Then the plates were placed in an incubator at 37 °C, 5% CO2 for 2 h. The absorbance was measured using an enzyme marker at 450 nm.2.6 Determination of the water-absorption swelling rate of the stent
Three freeze-dried scaffolds were taken, 1 mL of saline was added into each scaffold, and they were placed in a shaker at 37 °C at 135 rpm min−1. The scaffolds were removed every 30 min, the surface water was dried, the mass of the scaffolds was weighed after water absorption, and the scaffolds were monitored continuously for 3 h. The expansion rate of the scaffolds was calculated according to the following formula: expansion rate of the scaffolds (%) = (mass of the scaffolds after absorption of water − dry scaffolds)/dry scaffold mass × 100%.2.7 TUNEL assay
The scaffold was placed in a 24-well plate, and 1 × 104 cells were inoculated on the scaffold. The scaffold was taken out after 48 h and observed and photographed under a microscope. Then, the scaffolds were fixed in 4% paraformaldehyde for 20 min and permeabilized with 0.5% Triton X-100 for 10 min. After that, the TUNEL apoptosis assay was performed according to the instructions, and the nuclei of the cells were stained with DAPI. Finally, stereoscopic images were taken under a laser confocal microscope.2.8 Scanning electron microscopy
Astragalus polysaccharide-containing 3D-printed scaffolds were fixed in 2.5% glutaraldehyde solution at 4 °C overnight. After the steps of fixation in 1% starvation acid solution, dehydration of the samples in ethanol solution with graded concentrations (including six concentrations of 30%, 50%, 70%, 80%, 90% and 100%) was performed, followed by the treatment of the samples with a mixture of ethanol and isoamyl acetate. After drying at the critical point and platinum spraying, the internal structure of the scaffolds was observed under a scanning electron microscope.2.9 Micro-CT
The rabbit femur was collected from each group after 12 weeks, and the soft tissues of the distal end were removed, fixed with paraformaldehyde for 48 h and then stored in 75% ethanol solution. Micro-CT was used to scan the distal end of the femur to create a three-dimensional image and to observe the changes of bone microstructure in the modeling area.2.10 Histological analysis
The newborn cartilage tissue in the modeling area was collected from each group after 12 weeks, fixed with 4% PFA solution for more than 24 h, dehydrated, paraffin embedded and cut into 4 μm sections. H&E and ABH staining was performed, and the sections were dehydrated and sealed after completing the steps, observed under a microscope and photographed for comparison.2.11 Immunofluorescence
After 12 weeks in each group, newborn cartilage tissues were collected from the modeling area and subjected to frozen sectioning. 4% PFA solution was used for fixation, endogenous peroxidase was removed, 5% BSA was used for sealing, and the cells were incubated with the primary antibody against VEGFA, Col2a1, and Biglycan at 4 °C overnight, washed three times with PBS and then incubated with the secondary antibody at room temperature, and the nuclei of the cells were restained with DAPI, and the final rinsing and sealing of the slices were done with PBS, and then the cells were observed under a fluorescence microscope and images were taken.2.12 UHPLC-QTOF/MS detection of cartilage tissue pretreatment
After 12 weeks, cartilage tissue from the modeling area was collected from each group and cut into pieces, about 60 mg was weighed and placed in a 2 mL centrifuge tube, then 480 μL of methanol and magnetic beads were added, and placed in a grinder for rapid grinding, with the following parameters: total number of grinding = 10, frequency = 60 Hz, interruption time = 10 s, and total running time = 10 min. After grinding, centrifugation was conducted at high speed for 15 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (4 °C), then 200 μL of the supernatant was pipetted and blow dried under nitrogen. The samples were re-dissolved and quality control (QC) samples were prepared. The samples were analyzed by UHPLC-QTOF/MS liquid chromatography.
000 rpm (4 °C), then 200 μL of the supernatant was pipetted and blow dried under nitrogen. The samples were re-dissolved and quality control (QC) samples were prepared. The samples were analyzed by UHPLC-QTOF/MS liquid chromatography.
2.13 UHPLC-QTOF/MS analysis
Chromatographic separation was performed on an ExionLC system (AB Sciex, Foster City, CA, USA). A Waters Acquity HSS T3 column (2.1 mm × 150 mm, 1.8 μm) was applied at a temperature of 35 °C. Mobile phase A was water with 0.1% formic acid (v/v) and B was acetonitrile. The gradient was optimized as follows: 0–5 min from 3 to 8% B, 5–11 min from 8 to 30% B, 11–20 min from 30 to 80% B, 20–21 min from 80 to 95% B, 21–27 min at 95% B, then back to the initial ratio of 3% B and maintained with additional 6 min for re-equilibration. The injection volume for all samples was 2 μL.2.14 Mass spectrometry analysis
To enable high-resolution detection, the samples were analyzed using a SCIEX Exion LC United X500B QTOF mass spectrometer (AB Sciex, Foster City, CA, USA). Mass spectrometry detection was performed in both negative and positive ion modes with a mass range of m/z 50–1500. To monitor the reproducibility and stability of the collection system, QC samples were prepared by pooling small shares of each sample. QC samples were analyzed every 5 samples throughout the analysis. For metabolite annotation, databases such as HMDB (https://hmdb.ca/) and LipidMaps (https://www.lipidmaps.org/) were referenced.2.15 Data processing
The raw data were subjected to noise reduction and filtering, peak identification and alignment, and data matrix conversion using MS-DIAL software (ver.5.1.230912), which mainly includes information such as the mass-to-charge ratio (m/z), retention time (Rt) and peak area (intensity). All data were normalized by total peak area to generate an Excel sheet for subsequent metabolomic analysis. To minimize signal interference from chance errors, variables with RSD ≥ 40% in QC were first eliminated in Excel. The Excel files were imported into SIMCA 14.1 (Umetrics, Umeå, Sweden) software for multivariate mathematical statistical analysis. Principal component analysis (PCA) is an unsupervised mode of analysis that allows observation of the overall distribution of the samples. Partial least squares discriminant analysis (PLS-DA) was used to analyze all groups and to observe trends in the overall metabolic profiles of all samples, and the permutation test was used to verify that the model was not overfitted.2.16 Analysis of differential metabolites and metabolic pathways
OPLS-DA (orthogonal partial least squares discriminant analysis) was used to identify differential variables between normal and model groups. Screening principle: VIP ≥ 1, |pcorr| ≥ 0.4. Screened variables were tested for significant differences using the Mann–Whitney test, and a variable with a P value ≤ 0.05 was considered significantly different. Fold change (FC) was calculated, and a volcano plot was drawn with FC (model/control) as the horizontal coordinate and the P value as the vertical coordinate, the further a variable is from the origin, the greater its contribution to the difference, with red indicating a significant up-regulation after modeling, green indicating a significant decrease after modeling, and gray indicating no difference. The differential metabolite lists of Normal vs. Model were imported into Metabo Analyst 5.0 (https://www.MetaboAnalyst.ca/) for metabolite pathway analysis.2.17 Statistical analysis
All the experimental results were expressed as mean ± SD (standard deviation). All statistical analyses were performed using SPSS 21.0 software. The significance of differences between groups was determined by a 2-tailed unpaired Student's t-test or one-way ANOVA with Dunnett's post hoc test when samples were not distributed normally. A value of P < 0.05 was considered to be statistically significant.3 Results
3.1 Results of the effect of astragalus polysaccharide on the proliferation ability of chondrocytes
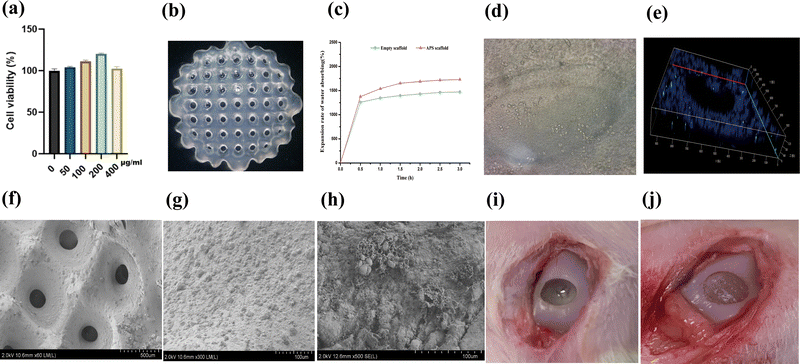
When the concentration of astragalus polysaccharide was between 50 and 400 μg mL−1, the proliferation of chondrocytes was obvious, and the efficiency of cell proliferation was positively correlated with the concentration of astragalus polysaccharide, especially when the proliferation reached the peak at 200 μg mL−1. The above results indicate that astragaloside can effectively promote chondrocyte proliferation, and 200 μg mL−1 of astragaloside is a relatively optimal concentration for the preparation of astragaloside-containing 3D-printed biodegradable skin scaffolds (Fig. 1a).3.2 Preparation of astragalus polysaccharide-containing 3D-printed scaffolds and swelling rate results
The whole scaffold was transparent, well molded, with consistent scaffold pore size, uniform spacing, consistent alignment, uniform thickness, and a good water-absorbing swelling rate (Fig. 1b and c). This indicates that we successfully prepared 3D-printed biodegradable skin scaffolds containing astragalus polysaccharide.3.3 TUNEL assay and SEM results
The chondrocytes grew all over the scaffold, and TUNEL assay revealed that apoptosis was present in only a small number of cells (Fig. 1d and e). The scaffolds were in the shape of a white transparent grid, the scaffold apertures were of uniform size, uniform spacing, and uniform thickness, and the surface of the scaffolds was in the shape of paving stone honeycomb, which was suitable for cell adhesion (Fig. 1f and g). After implantation of chondrocytes, it was found that the cells could adhere to and grow stably on the scaffolds (Fig. 1h). The above results indicate that we have successfully prepared astragalus polysaccharide-containing 3D-printed scaffolds, which are suitable for cell growth and have no obvious toxicity to cells.3.4 Model preparation and micro-CT results
We successfully established a rabbit distal femur cartilage defect model and implanted the scaffold into the defect site (Fig. 1i and j). Three months after implantation, we removed the rabbit distal femur for observation, and parallel micro-CT detection revealed that, compared with the model group, the cartilage defect site achieved better repair with the scaffold and was comparable to the normal group (Fig. 2a and b).3.5 Histology and immunofluorescence detection results
Effective repair of cartilage after implantation of scaffolds was observed under both HE and ABH staining (Fig. 2c and d). The fluorescence expression of VEGFA, Col2a1 and Biglycan was significantly better in the scaffold group than in the model group, and it was comparable to that in the normal group (P < 0.05), and the results are shown in Fig. 2e–h. The above results suggest that the astragalus polysaccharide-containing 3D-printed scaffolds can promote structural repair of cartilage, which may be associated with accelerated vascularization and collagen neogenesis.3.6 Repeatability and stability of the UHPLC-QTOF/MS method
Fig. 3a and b show the QC stacked plots in positive and negative ion modes. The plots overlap almost completely, indicating that the obtained data are stable and reliable. Fig. 3c and g show the PCA plots of all the samples in negative and positive ion modes, respectively, and the QC samples are more tightly clustered, indicating good stability and reproducibility of this experiment. In addition, from the results in Fig. 3c–j, it can be seen that the distinction between the normal group, model group and treatment group is more obvious, and the scatter point of the treatment group is located in the middle, suggesting that the overall migration of cartilage metabolism is obvious after modeling in the model group, and it can be adjusted back to a certain extent after treatment.3.7 Results of differential metabolite analysis
Fig. 4a–h show the OPLS-DA of normal vs. model, indicating that there is a difference between the two groups and the model is valid. Fig. 4d and h show the volcano plots of the normal vs. model, with red indicating variables up-regulated after modeling, blue indicating variables down-regulated after modeling, and gray indicating variables with no difference. A total of 50 different metabolites were screened and identified in normal vs. model, of which 24 were up-regulated and 26 were down-regulated. They were mainly related to phospholipid metabolism (e.g., choline, lysophosphatidylethanolamine (LPE), lysophosphatidylcholine (LPC), sphingosine), fatty acid and energy metabolism (e.g., fatty acids, amide, stearoylcarnitine, L-carnitine, lactic acid), bile acid metabolism (e.g., cholic acid, deoxycholic acid), amino acid metabolism (e.g., L-tryptophan, hippuric acid, pyroglutamic acid, choline, L-carnitine), and purine metabolism (e.g., xanthine, allantoin), and the results are shown in Table 1. The fragments of MS/MS are shown in Table S1 (ESI†).| No. | Metabolites | Formula | RT (min) | VIP | p(corr) | P value | FC | P value | FC | Sig. regulated | Class |
|---|---|---|---|---|---|---|---|---|---|---|---|
| N–M | M/N | M–S | S/M | ||||||||
| MG: monoglycerides, LPC: lysophosphatidylcholine, LPE: lysophosphatidylethanolamine, PE: phosphatidylethanolamine. | |||||||||||
| 1 | Pyroglutamic acid | C5H7NO3 | 2.24 | 1.40 | −0.68 | 0.0090 | 0.68 | 0.6015 | 1.00 | N | Amino acids |
| 2 | L-Tryptophan | C11H12N2O2 | 7.86 | 1.16 | 0.69 | 0.0472 | 1.71 | 0.6015 | 0.96 | — | Amino acids |
| 3 | Hippuric acid | C9H9NO3 | 8.96 | 3.15 | 0.84 | 0.0090 | 7.22 | 0.0090 | 0.22 | Y | Amino acids |
| 4 | Choline | C5H14NO+ | 1.20 | 1.64 | 0.89 | 0.0090 | 1.43 | 0.1745 | 0.90 | — | Amino acids |
| 5 | Allantoin | C4H6N4O3 | 1.32 | 1.06 | 0.88 | 0.0090 | 1.72 | 0.0283 | 0.77 | Y | Purines |
| 6 | Xanthine | C5H4N4O2 | 2.31 | 2.16 | 0.73 | 0.0283 | 2.45 | 0.3472 | 0.82 | Y | Purines |
| 7 | Cholic acid | C24H40O5 | 11.10 | 1.33 | 0.69 | 0.0090 | 3.84 | 0.0090 | 0.26 | Y | Bile acids |
| 8 | 3b-Hydroxy-5-cholenoic acid | C24H38O3 | 12.35 | 1.71 | 0.60 | 0.0163 | 3.19 | 0.1172 | 0.46 | — | Bile acids |
| 9 | Deoxycholic acid | C24H40O4 | 12.35 | 4.50 | 0.60 | 0.0090 | 3.24 | 0.0758 | 0.45 | — | Bile acids |
| 10 | L-Carnitine | C7H15NO3 | 1.25 | 1.55 | 0.76 | 0.0090 | 2.05 | 0.1745 | 0.82 | — | Carnitines |
| 11 | Palmitoylcarnitine | C23H45NO4 | 11.86 | 1.86 | 0.81 | 0.0163 | 2.42 | 0.1745 | 0.76 | — | Carnitines |
| 12 | Stearoylcarnitine | C25H49NO4 | 12.97 | 1.90 | 0.87 | 0.0090 | 2.36 | 0.0472 | 0.67 | Y | Carnitines |
| 13 | Tetradecenoylcarnitine | C21H39NO4 | 14.73 | 2.19 | 0.87 | 0.0090 | 2.37 | 0.0472 | 0.69 | Y | Carnitines |
| 14 | Sebacic acid | C10H18O4 | 10.42 | 1.14 | 0.70 | 0.0090 | 2.54 | 0.0283 | 0.47 | Y | Fatty acids |
| 15 | Dodecadienoic acid | C12H20O2 | 10.73 | 1.62 | 0.86 | 0.0090 | 1.99 | 0.2506 | 0.87 | — | Fatty acids |
| 16 | Prostaglandin F2a | C20H34O5 | 10.80 | 1.36 | −0.64 | 0.0472 | 0.39 | 0.1745 | 1.43 | — | Fatty acids |
| 17 | 13-OxoODE | C18H30O3 | 13.31 | 2.82 | −0.55 | 0.0472 | 0.11 | 0.4647 | 0.80 | N | Fatty acids |
| 18 | 5,6-Epoxy-8,11,14-eicosatrienoic acid | C20H32O3 | 13.41 | 1.43 | −0.61 | 0.0283 | 0.44 | 0.1745 | 1.47 | — | Fatty acids |
| 19 | Linoleamide | C18H33NO | 15.43 | 1.44 | 0.76 | 0.0283 | 2.03 | 0.0758 | 0.65 | — | Fatty acids |
| 20 | Dihomo-gamma-linolenic acid | C20H34O2 | 18.12 | 2.57 | −0.88 | 0.0090 | 0.41 | 0.0163 | 2.20 | Y | Fatty acids |
| 21 | LysoPC(14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C22H46NO7P | 11.53 | 3.48 | −0.92 | 0.0090 | 0.41 | 0.0163 | 2.29 | Y | LPC |
| 22 | LysoPC(16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C24H48NO7P | 11.81 | 2.89 | −0.86 | 0.0163 | 0.60 | 0.0163 | 1.74 | Y | LPC |
| 23 | LysoPC(0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20 0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
C28H50NO7P | 12.02 | 2.78 | 0.87 | 0.0090 | 1.98 | 0.3472 | 0.89 | — | LPC |
| 24 | LysoPC(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4/0 4/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C28H50NO7P | 12.02 | 2.64 | 0.91 | 0.0090 | 2.06 | 0.1745 | 0.82 | — | LPC |
| 25 | LysoPC(15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C23H48NO7P | 12.06 | 5.01 | −0.84 | 0.0090 | 0.41 | 0.0163 | 2.03 | Y | LPC |
| 26 | LysoPC(14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C22H44NO7P | 12.31 | 1.50 | −0.87 | 0.0090 | 0.36 | 0.0283 | 2.26 | Y | LPC |
| 27 | LysoPC(17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C25H50NO7P | 12.40 | 2.81 | −0.91 | 0.0090 | 0.33 | 0.0090 | 2.41 | Y | LPC |
| 28 | LysoPC(0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16 0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C24H50NO7P | 12.47 | 3.96 | −0.65 | 0.0472 | 0.80 | 0.0283 | 1.35 | Y | LPC |
| 29 | LysoPC(16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C24H50NO7P | 12.73 | 12.59 | −0.87 | 0.0090 | 0.75 | 0.0163 | 1.29 | Y | LPC |
| 30 | LysoPC(22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4/0 4/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C30H54NO7P | 13.03 | 1.17 | 0.86 | 0.0163 | 1.82 | 0.7540 | 1.01 | N | LPC |
| 31 | LysoPC(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C26H52NO7P | 13.13 | 6.04 | −0.90 | 0.0090 | 0.64 | 0.0163 | 1.46 | Y | LPC |
| 32 | LysoPC(17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C25H52NO7P | 13.55 | 4.18 | −0.87 | 0.0090 | 0.50 | 0.0090 | 1.64 | Y | LPC |
| 33 | LysoPC(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/0 2/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C28H54NO7P | 13.73 | 1.32 | −0.94 | 0.0090 | 0.36 | 0.0090 | 1.76 | Y | LPC |
| 34 | LysoPC(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C28H56NO7P | 14.92 | 1.11 | −0.84 | 0.0090 | 0.61 | 0.0283 | 1.52 | Y | LPC |
| 35 | LysoPE(14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C19H40NO7P | 11.47 | 1.01 | −0.80 | 0.0283 | 0.49 | 0.0283 | 1.94 | Y | LPE |
| 36 | LysoPE(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4/0 4/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C25H44NO7P | 11.95 | 6.24 | 0.72 | 0.0283 | 1.91 | 0.2506 | 0.87 | — | LPE |
| 37 | LysoPE(15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C20H42NO7P | 11.98 | 1.90 | −0.92 | 0.0090 | 0.45 | 0.0163 | 1.69 | Y | LPE |
| 38 | LysoPE(22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5/0 5/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C27H46NO7P | 12.18 | 2.70 | 0.83 | 0.0090 | 2.88 | 0.2506 | 0.76 | — | LPE |
| 39 | LysoPE(22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4/0 4/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C27H48NO7P | 12.92 | 3.35 | 0.79 | 0.0283 | 2.60 | 0.2506 | 0.87 | — | LPE |
| 40 | LysoPE(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C23H46NO7P | 13.01 | 7.26 | −0.84 | 0.0163 | 0.64 | 0.0472 | 1.45 | Y | LPE |
| 41 | LysoPE(P-18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C23H46NO6P | 13.47 | 9.67 | −0.84 | 0.0163 | 0.61 | 0.0090 | 1.73 | Y | LPE |
| 42 | LysoPE(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/0 2/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C25H48NO7P | 13.59 | 1.11 | −0.83 | 0.0090 | 0.43 | 0.0472 | 1.66 | Y | LPE |
| 43 | LysoPE(19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C24H48NO7P | 13.76 | 1.63 | −0.91 | 0.0090 | 0.34 | 0.0090 | 1.70 | Y | LPE |
| 44 | LysoPE(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/0 1/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C25H50NO7P | 14.73 | 1.42 | −0.67 | 0.0163 | 0.69 | 0.0758 | 1.36 | — | LPE |
| 45 | LysoPE(P-18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
C23H48NO6P | 14.98 | 6.25 | −0.69 | 0.0283 | 0.67 | 0.0472 | 1.31 | Y | LPE |
| 46 | Monoelaidin | C21H40O4 | 17.10 | 2.19 | −0.93 | 0.0090 | 0.29 | 0.0283 | 1.71 | Y | MG |
| 47 | PE(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20 0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
C43H78NO8P | 10.27 | 1.04 | −0.69 | 0.0090 | 0.41 | 0.0283 | 1.41 | Y | PE |
| 48 | Heme | C34H32FeN4O4 | 11.13 | 7.43 | 0.65 | 0.0163 | 12.68 | 0.1745 | 0.30 | — | Porphyrins |
| 49 | Lactic acid | C3H6O3 | 1.96 | 2.73 | 0.68 | 0.0472 | 1.69 | 0.6015 | 0.88 | — | Organic acids |
| 50 | Sphingosine | C18H37NO2 | 10.97 | 1.07 | 0.85 | 0.0090 | 1.42 | 0.7540 | 0.95 | Y | Sphingolipids |
The heatmap of the mass spectral response of the differential metabolites in the normal, model and scaffold groups is shown in Fig. 5a, where red color indicates high response and green color indicates low response. The obvious difference between the normal and model groups and the partial restoration of scaffolds can be seen. PLS-DA was used to analyze each group and observe the trend of the overall metabolic profile of all samples. The results are shown in Fig. 5b and c. The three groups were clearly distinguished, the treatment group was located between the normal and model groups, and this scatter distribution indicated that overall the metabolome of the treatment group was between the normal and the modeling damage, i.e., the cartilage metabolome tended to recover after treatment, and this result was further verified by the restoration of the differential metabolites. Of the 50 differential metabolites, 29 metabolites exhibited reversal, and 58% of the metabolites were reversed. The metabolites that were reversed included: PE (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/20
0/20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) and lysophospholipids associated with phospholipid metabolism, sphingosine associated with sphingolipid metabolism, xanthine and allantoin associated with purine metabolism, lipoarylcarnitine and a small amount of fatty acids associated with energy metabolism (fatty acid metabolism), equine uric acid associated with amino acid metabolism, and hippuric acid and cholic acid associated with bile acid metabolism.
4) and lysophospholipids associated with phospholipid metabolism, sphingosine associated with sphingolipid metabolism, xanthine and allantoin associated with purine metabolism, lipoarylcarnitine and a small amount of fatty acids associated with energy metabolism (fatty acid metabolism), equine uric acid associated with amino acid metabolism, and hippuric acid and cholic acid associated with bile acid metabolism.
3.8 Results of metabolic pathway analysis
The metabolomic pathway analysis of rabbit cartilage tissues was visualized using both bubble and streamer plots. Cartilage injury may involve 15 metabolic pathways, of which the metabolites that were reversed after treatment mainly involved phospholipid and sphingolipid metabolism, fatty acid metabolism and purine metabolism, which are prominently highlighted in orange in Fig. 6a and b, respectively. Combined with KEGG, HMDB, and general lipid structure and function classification, the pathways were grouped and organized, and the results are shown in Table 2. The retuned metabolites were grouped and summarized, and combined with the pathway and mass spectrometry response results were plotted in a combinatorial map, and the results are shown in Fig. 6c.| Pathways | P | −log(P) | Impact | Metabolites | |
|---|---|---|---|---|---|
| Glycerophospholipid metabolism (lipid metabolism) | Glycerophospholipid metabolism | 0.0055 | 2.2580 | 0.13 | Choline, PE, LPE, LPC |
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 0.1473 | 0.8318 | 0.01 | PE | |
| Energy metabolism (lipid metabolism) | Pyruvate metabolism | 0.2173 | 0.6629 | 0.00 | Lactic acid |
| Glycolysis/gluconeogenesis | 0.2421 | 0.6159 | 0.00 | Lactic acid | |
| Fatty acid degradation | 0.3414 | 0.4667 | 0.00 | Palmitoylcarnitine, stearoylcarnitine, tetradecenoylcarnitine | |
| Arachidonic acid metabolism | 0.0700 | 1.1552 | 0.03 | Prostaglandin F2a, 5,6-epoxy-8,11,14-eicosatrienoic acid | |
| Porphyrin metabolism | 0.2819 | 0.5499 | 0.16 | Heme | |
| Biosynthesis of unsaturated fatty acids | 0.3197 | 0.4953 | 0.00 | Dodecadienoic acid, 13-oxoODE, 5,6-epoxy-8,11,14-eicosatrienoic acid, dihomo-gamma-linolenic acid | |
| Amino acid metabolism | Glutathione metabolism | 0.2583 | 0.5879 | 0.01 | Pyroglutamic acid |
| Lysine degradation | 0.2741 | 0.5621 | 0.00 | L-Carnitine | |
| Glycine, serine and threonine metabolism | 0.2972 | 0.5269 | 0.00 | Choline | |
| Tryptophan metabolism | 0.3486 | 0.4577 | 0.16 | L-Tryptophan | |
| Bile acid metabolism (lipid metabolism) | Primary bile acid biosynthesis | 0.3897 | 0.4093 | 0.00 | Cholic acid, 3b-hydroxy-5-cholenoic acid, |
| Sphingolipid metabolism (lipid metabolism) | Sphingolipid metabolism | 0.2896 | 0.5382 | 0.06 | Sphingosine |
| Purine metabolism | Purine metabolism | 0.1656 | 0.7808 | 0.03 | Allantoin, xanthine |
4 Discussion
Cartilage and bone are organs that form the scaffolding of the body, and they have cartilaginous and bony tissues as their main structural components, respectively. Articular cartilage is mainly composed of chondrocytes and extracellular matrix.24 Chondrocytes secrete cartilage matrix components and are the active center of articular cartilage. The extracellular matrix (ECM) consists mainly of water, collagen fibers and proteoglycan polymers.25,26 We prepared collagen–sodium alginate–silk fibroin 3D-printed scaffolds to promote cartilage regeneration. Prior to this, our research group has studied these matrix materials for a longer period of time, and preliminary studies have shown that these materials exhibit good biocompatibility in vivo with no significant immune rejection or inflammatory response.15 In the present study, we found that astragalus polysaccharide-containing 3D-printed scaffolds promote the expression of vascular factors and accelerates cartilage matrix remodeling. The scaffolds also corrected cartilage tissue metabolic disorders, especially lipid metabolism, amino acid metabolism and purine metabolism, which are metabolic pathways closely related to chondrocyte differentiation. The implantation of the scaffold promoted a virtuous cycle in which chondrocytes synthesized and replenished the matrix, which in turn held chondrocytes in place and provided them with nutrients (Fig. 7).As one of the most important growth factors regulating vascular development and postnatal angiogenesis,27,28 VEGF plays a key role in cartilage repair. During endochondral osteogenesis in bone repair, VEGF promotes the recruitment of osteochondral progenitor cells, induces cartilage formation, and stimulates the resorption of cartilage and its replacement by bone.29 Type II collagen (Col2a1) consists of three identical α-1(II) chains and is the main specific component of the extracellular matrix of cartilage secreted by chondrocytes, which is a major marker of mature chondrocyte differentiation and is present in articular cartilage.30–32 Biglycan is a member of the SLRP family and is abundant in the ECM of a variety of tissues including bone, cartilage and tendon.33,34 Proteoglycans are important components of the extracellular matrix of articular cartilage, providing biomechanical properties essential for its proper functioning.35 Lack of Biglycan affects the differentiation of embryonic osteoblasts, subsequently leading to structural changes in bone, including reduced integrity and strength. And this process may be achieved by regulating the cartilage stage prior to bone formation.36 In our results, we found that the expression of VEGFA, Col2a1 and Biglycan in the scaffold group was significantly higher than that in the model group. This suggests that astragalus polysaccharide-containing 3D printed scaffolds may promote cartilage tissue repair by promoting the expression of vascular-related factors and accelerating cartilage collagen matrix remodeling.
Lipid metabolism is an important part of energy metabolism, and fatty acids enter the TCA cycle to provide energy for the body through β-oxidation, which takes place in the mitochondria, and fatty acids complete the shuttle with the help of the carnitine system, which generates stearoylcarnitine. In this experiment, accumulation of lipoylcarnitine was observed in the model group, and it was hypothesized that cartilage injury led to mitochondrial damage in chondrocytes. Mitochondrial dysfunction is often accompanied by ROS accumulation and inflammatory infiltration,37 while insufficient energy supply will further lead to degeneration of the cartilage matrix, inducing apoptosis and autophagy, among others.38,39 Phospholipids and sphingolipids have a variety of biological functions in cartilage such as maintaining cell membrane homeostasis and transmitting information, and an imbalance in the metabolism of phospholipids and sphingolipids would signal a disruption of the basic cellular morphology and function. Any change in phospholipid levels may affect the inflammatory state of joints;40 in addition, it has also been shown that phospholipids are closely linked to chondrocyte differentiation, LPC and LPE can regulate chondrocyte growth and differentiation by directly or indirectly activating transforming growth factor β-1 stored in the extracellular matrix;41 sphingolipids also regulate the migration of osteoblast precursor cells and bone metabolic homeostasis.42,43 Cholesterol metabolism is also an important metabolic pathway for the body to regulate a variety of biological processes, and it has been shown that when cartilage is destroyed, chondrocyte uptake is enhanced, leading to increased cholesterol levels, which results in an up-regulation of cholesterol hydroxylase expression and an increase in oxysterol metabolites such as bile acids, which is consistent with the results of the elevated bile acid levels observed in the present experiment. Cholesterol is also one of the inducers of inflammation.44,45 The results of metabolomic analysis showed that the modulation of the above lipids by the present intervention was significant.
Chondrocytes store lipids in the form of lipid droplets (LDs) and maintain cartilage lipid metabolic balance by consuming or regenerating LDs. Among the regulatory factors, fibroblast growth factor 8 (FGF8) is one of the critical mediators involved in chondrocyte proliferation, differentiation, and migration, and has garnered increasing attention in both physiological and pathological studies of cartilage.46,47 Huang et al. demonstrated that FGF8 enhances Plin1 expression through the FGFR1/p38 signaling pathway, thereby promoting accumulation of intracellular LDs in chondrocytes.48 Additionally, NSD1 has been identified as a key regulator of chondrogenic differentiation and cartilage metabolic homeostasis. In vitro experiments have revealed that NSD1 knockout leads to disruptions in cell proliferation, differentiation, and metabolism, while NSD1-knockout mice developed arthritis.49 These regulatory factors may play pivotal roles in cartilage repair and metabolism, particularly in lipid-related metabolic processes. However, their underlying mechanisms remain to be further elucidated.
Amino acids are closely related to metabolism, such as serine, which can be converted to choline in vivo, thus participating in the synthesis of phospholipids; carnitine can be synthesized with fatty acids to lipoylcarnitine to participate in β-oxidation.50–52 In addition, amino acids are the basic units of proteins, and type-II collagen is the main component of the extracellular matrix of articular cartilage, mainly composed of L-phenylalanine, proline and hydroxyproline.53 Cartilage injury causes loss of the extracellular matrix and destruction of chondrocytes, which disrupts the balance of amino acid metabolism and protein synthesis. However, the modulation of amino acid metabolism by the astragalus polysaccharide-containing 3D-printed scaffolds in this experiment was less pronounced.
An imbalance in purine metabolism was also observed after cartilage injury, which was manifested by elevated xanthine and allantoin. Xanthine is a product of hypoxanthine oxidation, which is often one of the indicators of gout,54 and other studies have reported a significant increase in allantoin concentration in RA patients.55 Therefore, combining the literature and experimental results, we hypothesized that the cartilage damage further caused inflammation in the joints, and after implantation of the scaffold, the relevant metabolites were regressed, suggesting that the present intervention may have ameliorated the inflammation in the joints and provided a homeostatic environment for the differentiation and growth of chondrocytes.
In summary, astragalus polysaccharide-containing 3D-printed scaffolds may accelerate cartilage collagen matrix remodeling and correct cartilage tissue metabolic disorders by promoting the expression of vascular-related factors, which ultimately promote cartilage repair. However, there are some limitations in the study: the mechanism of action of astragalus polysaccharide-containing 3D-printed scaffolds in cartilage metabolomics and promotion of vascular-related factor expression need to be verified by further experimental studies, and there has not yet been a clinical trial to validate their efficacy. In the future, we will further validate its mechanism of action experimentally; optimize the production process to ensure stable and controllable stent pore size and drug release rate; conduct a systematic evaluation of the stent's biocompatibility with human cells and tissues, with a particular focus on long-term safety for clinical applications; provide scientific experimental evidence for the clinical treatment of cartilage defects; and actively promote the translation of research findings into clinical practice. These efforts will provide new ideas for the treatment of cartilage defects.
Ethics approval and consent to participate
Purchase and feeding, and other animal procedures followed the animal research guidelines of the National Institute of Health and the Animal Research Committee. All animal experiments were approved by the Experimental Animal Ethics Committee of Zhejiang Chinese Medical University (No. IACUC-20231030-13).Author contributions
Conceptualization: Weibin Du, Zhenwei Wang. Data curation: Weibin Du, Zhenwei Wang. Funding acquisition: Weibin Du. Methodology: Weibin Du, Wenxiang Zeng, Huahui Hu, Yanghua Tang, Gang Qu. Writing – original draft: Weibin Du, Zhenwei Wang. Formal analysis: Wenxiang Zeng, Zhenwei Wang, Huahui Hu, Yanghua Tang. Validation: Wenxiang Zeng, Huahui Hu, Yanghua Tang. Supervision: Guoping Cao, Gang Qu, Rongliang Chen. Writing – review & editing: Guoping Cao, Gang Qu, Rongliang Chen. Project administration: Rongliang Chen.Data availability
All data generated and/or analyzed during this study are included in this article.Conflicts of interest
All the authors declare that they have no conflicts of interest.Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 81904053), the Zhejiang Provincial Natural Science Foundation of China (Grant No. LTGY24H290006), the Zhejiang Province Medical and Health Science and Technology Project (No. 2025KY191), the Special Research Project of the Affiliated Hospital of Zhejiang Chinese Medical University (No. 2021FSYYZY43), the Hangzhou Science and Technology Planning Project (No. 20220919Y084), the Zhejiang Province Traditional Chinese Medicine Science and Technology Project (No. 2023ZR046), and the Hangzhou Bio-medicine and Health Industry Development Support Science and Technology Project (No. 2023WJC243 and 2023WJC249). The authors thank Professor Yadong Yang from the School of Laboratory Medicine and Bioengineering, Hangzhou Medical College (Hangzhou, China) for her guidance in the field of 3D printing.References
- Y. Fujii, L. Liu and L. Yagasaki, et al., Cartilage Homeostasis and Osteoarthritis, Int. J. Mol. Sci., 2022, 23(11), 6316 CrossRef CAS PubMed.
- T. Ruediger, V. Horbert and A. Reuther, et al., Thickness of the Stifle Joint Articular Cartilage in Different Large Animal Models of Cartilage Repair and Regeneration, Cartilage, 2021, 13(2), 438s–452s CrossRef CAS PubMed.
- M. Moradi, F. Parvizpour and Z. Arabpour, et al., Articular Cartilage Injury; Current Status and Future Direction, Curr. Stem Cell Res. Ther., 2024, 19(5), 653–661 CrossRef CAS PubMed.
- H. S. Kan, P. K. Chan and K. Y. Chiu, et al., Non-surgical treatment of knee osteoarthritis, Hong Kong Med. J., 2019, 25(2), 127–133 CAS.
- A. Eccleston, Cartilage regeneration for osteoarthritis, Nat. Rev. Drug Discovery, 2023, 22(2), 96 CrossRef CAS PubMed.
- X. N. Xiang, S. Y. Zhu and H. C. He, et al., Mesenchymal stromal cell-based therapy for cartilage regeneration in knee osteoarthritis, Stem Cell Res. Ther., 2022, 13(1), 14 CrossRef.
- P. Feng, H. Tian and F. Yang, et al., Reduced graphene oxide-mediated electron-hole separation using titanium dioxide increases the photocatalytic antibacterial activity of bone Scaffolds, Bio-Des. Manuf., 2025, 8(1), 100–115 CrossRef.
- P. Feng, R. Zhao and W. Tang, et al., Structural and Functional Adaptive Artificial Bone: Materials, Fabrications, and Properties, Adv. Funct. Mater., 2023, 33(23), 2214726 CrossRef CAS.
- C. Shuai, X. Shi and F. Yang, et al., Oxygen vacancy boosting Fenton reaction in bone Scaffold towards fighting bacterial infection, Int. J. Extreme Manuf., 2024, 6(1), 015101 CrossRef CAS.
- D. G. O’Shea, T. Hodgkinson and C. M. Curtin, et al., An injectable and 3D printable pro-chondrogenic hyaluronic acid and collagen type II composite hydrogel for the repair of articular cartilage defects, Biofabrication, 2023, 16(1) DOI:10.1088/1758-5090/ad047a.
- Y. Huan, D. Zhou and X. Wu, et al., 3D bioprinted autologous bone particle Scaffolds for cranioplasty promote bone regeneration with both implanted and native BMSCs, Biofabrication, 2023, 15(2) DOI:10.1088/1758-5090/acbe21.
- M. Rahimnejad, R. Rezvaninejad and R. Rezvaninejad, et al., Biomaterials in bone and mineralized tissue engineering using 3D printing and bioprinting technologies, Biomed. Phys. Eng. Express, 2021, 7(6) DOI:10.1088/2057-1976/ac21ab.
- S. Tu, A. Shao and L. Ren, et al., Angiogenesis effect of Astragalus polysaccharide combined with endothelial progenitor cells therapy in diabetic male rat following experimental hind limb ischemia, Chin. Med. J., 2014, 127(11), 2121–2128 CrossRef CAS.
- Y. Peng, F. Ma and L. Hu, et al., Strontium based Astragalus polysaccharides promote osteoblasts differentiation and mineralization, Int. J. Biol. Macromol., 2022, 205, 761–771 CrossRef CAS PubMed.
- W. Du, J. Hu and X. Huang, et al., Feasibility of repairing skin defects by VEGF(165) gene-modified iPS-HFSCs seeded on a 3D printed Scaffold containing astragalus polysaccharide, J. Cell. Mol. Med., 2023, 27(15), 2136–2149 CrossRef CAS PubMed.
- W. Du, Z. Wang and M. Han, et al., Astragalus polysaccharide-containing 3D-printed Scaffold for traumatized skin repair and proteomic study, J. Cell. Mol. Med., 2024, 28(16), e70023 CrossRef CAS.
- H. Liu, L. Qiu and H. Li, et al., A 3D-printed acinar-mimetic silk fibroin-collagen-astragalus polysaccharide Scaffold for tissue reconstruction and functional repair of damaged parotid glands, Int. J. Biol. Macromol., 2024, 277(Pt 3), 134427 CrossRef CAS PubMed.
- Z. Zhao, Y. Wang and B. Yin, et al., Defect-adaptive Stem-cell-microcarrier Construct Promotes Tissue Repair in Rabbits with Knee Cartilage Defects, Stem Cell Rev. Rep., 2023, 19(1), 201–212 CrossRef CAS PubMed.
- C. Ye, J. Chen and Y. Qu, et al., Naringin in the repair of knee cartilage injury via the TGF-β/ALK5/Smad2/3 signal transduction pathway combined with an acellular dermal matrix, J. Orthop. Translat., 2022, 32, 1–11 CrossRef PubMed.
- H. Liu, H. Liu and Q. Yang, et al., LncRNA SNHG1 enhances cartilage regeneration by modulating chondrogenic differentiation and angiogenesis potentials of JBMMSCs via mitochondrial function regulation, Stem Cell Res. Ther., 2024, 15(1), 177 CrossRef CAS PubMed.
- Y. Hu, C. Lyu and L. Teng, et al., Glycopolypeptide hydrogels with adjustable enzyme-triggered degradation: A novel proteoglycans analogue to repair articular-cartilage defects, Mater. Today Bio, 2023, 20, 100659 CrossRef CAS.
- X. Liu, S. Jiang and T. Jiang, et al., Bioenergetic-active exosomes for cartilage regeneration and homeostasis maintenance, Sci. Adv., 2024, 10(42), eadp7872 CrossRef CAS.
- W. Du, W. Zeng and Z. Wang, et al., Collagen-sodium alginate-silk fibroin 3D-printed Scaffold loaded with polydatin promotes cartilage regeneration by improving lipid metabolism and cell apoptosis, IJB, 2024, 11(1), 453–469 Search PubMed.
- R. Deng, R. Zhao and Z. Zhang, et al., Chondrocyte membrane-coated nanoparticles promote drug retention and halt cartilage damage in rat and canine osteoarthritis, Sci. Transl. Med., 2024, 16(735), eadh9751 CrossRef CAS.
- G. Maiti, S. Ashworth and T. Choi, et al., Molecular cues for immune cells from small leucine-rich repeat proteoglycans in their extracellular matrix-associated and free forms, Matrix Biol., 2023, 123, 48–58 CrossRef CAS.
- S. G. Rees, C. M. Dent and B. Caterson, Metabolism of proteoglycans in tendon, Scand. J. Med. Sci. Sports, 2009, 19(4), 470–478 CrossRef CAS PubMed.
- L. Coultas, K. Chawengsaksophak and J. Rossant, Endothelial cells and VEGF in vascular development, Nature, 2005, 438(7070), 937–945 CrossRef CAS PubMed.
- J. Lawler, Counter regulation of tumor angiogenesis by vascular endothelial growth factor and thrombospondin-1, Semin. Cancer Biol., 2022, 86(Pt 2), 126–135 CrossRef CAS PubMed.
- K. Hu and B. R. Olsen, Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair, J. Clin. Invest., 2016, 126(2), 509–526 CrossRef PubMed.
- H. Deng, X. Huang and L. Yuan, Molecular genetics of the COL2A1-related disorders, Mutat. Res., Rev. Mutat. Res., 2016, 768, 1–13 CrossRef CAS PubMed.
- Q. Liu, X. Z. Zhu and R. B. Feng, et al., Crude triterpenoid saponins from Anemone flaccida (Di Wu) exert anti-arthritic effects on type II collagen-induced arthritis in rats, Chin. Med., 2015, 10, 20 CrossRef PubMed.
- S. N. Bessonov, A. V. Korablev and P. A. Korablev, [Chondroblast proliferative activity study in forming big alar cartilage], Stomatologiia, 2009, 88(3), 29–33 CAS.
- P. Bianco, L. W. Fisher and M. F. Young, et al., Expression and localization of the two small proteoglycans biglycan and decorin in developing human skeletal and non-skeletal tissues, J. Histochem. Cytochem., 1990, 38(11), 1549–1563 CrossRef CAS.
- M. V. Nastase, M. F. Young and L. Schaefer, Biglycan: a multivalent proteoglycan providing structure and signals, J. Histochem. Cytochem., 2012, 60(12), 963–975 CrossRef PubMed.
- L. Alcaide-Ruggiero, R. Cugat and J. M. Domínguez, Proteoglycans in Articular Cartilage and Their Contribution to Chondral Injury and Repair Mechanisms, Int. J. Mol. Sci., 2023, 24(13), 10824 CrossRef CAS PubMed.
- R. Shainer, V. Kram and T. M. Kilts, et al., Biglycan regulates bone development and regeneration, Front. Physiol., 2023, 14, 1119368 CrossRef PubMed.
- M. Fernández-Moreno, I. Rego-Pérez and F. J. Blanco, Is osteoarthritis a mitochondrial disease? What is the evidence, Curr. Opin. Rheumatol., 2022, 34(1), 46–53 CrossRef.
- P. Chen, X. Liu and C. Gu, et al., A plant-derived natural photosynthetic system for improving cell anabolism, Nature, 2022, 612(7940), 546–554 CrossRef CAS.
- K. Sun, Y. Wu and Y. Zeng, et al., The role of the sirtuin family in cartilage and osteoarthritis: molecular mechanisms and therapeutic targets, Arthritis Res. Ther., 2022, 24(1), 286 CrossRef CAS PubMed.
- M. K. Kosinska, G. Liebisch and G. Lochnit, et al., A lipidomic study of phospholipid classes and species in human synovial fluid, Arthritis Rheum., 2013, 65(9), 2323–2333 CrossRef CAS PubMed.
- I. Gay, Z. Schwartz and V. L. Sylvia, et al., Lysophospholipid regulates release and activation of latent TGF-beta1 from chondrocyte extracellular matrix, Biochim. Biophys. Acta, 2004, 1684(1–3), 18–28 CrossRef CAS.
- D. A. Baker, J. Eudaly and C. D. Smith, et al., Impact of sphingosine kinase 2 deficiency on the development of TNF-alpha-induced inflammatory arthritis, Rheumatol. Int., 2013, 33(10), 2677–2681 CrossRef CAS PubMed.
- J. M. Grewe, P. R. Knapstein and A. Donat, et al., The role of sphingosine-1-phosphate in bone remodeling and osteoporosis, Bone Res., 2022, 10(1), 34 CrossRef CAS.
- W. S. Choi, G. Lee and W. H. Song, et al., The CH25H-CYP7B1-RORα axis of cholesterol metabolism regulates osteoarthritis, Nature, 2019, 566(7743), 254–258 CrossRef CAS PubMed.
- Y. Yang, J. Wei and J. Li, et al., Lipid metabolism in cartilage and its diseases: a concise review of the research progress, Acta Biochim. Biophys. Sin., 2021, 53(5), 517–527 CrossRef CAS PubMed.
- M. B. Ellman, D. Yan and K. Ahmadinia, et al., Fibroblast growth factor control of cartilage homeostasis, J. Cell. Biochem., 2013, 114(4), 735–742 CrossRef CAS PubMed.
- H. Chen, Y. Cui and D. Zhang, et al., The role of fibroblast growth factor 8 in cartilage development and disease, J. Cell. Mol. Med., 2022, 26(4), 990–999 CrossRef CAS PubMed.
- M. Huang, H. Chen and J. Wei, et al., FGF8 promotes lipid droplet accumulation via the FGFR1/p-p38 axis in chondrocytes, Acta Biochim. Biophys. Sin., 2025 DOI:10.3724/abbs.2025075.
- R. Shao, J. Suo and Z. Zhang, et al., H3K36 methyltransferase NSD1 protects against osteoarthritis through regulating chondrocyte differentiation and cartilage homeostasis, Cell Death Differ., 2024, 31(1), 106–118 CrossRef CAS.
- Y. Gao, J. T. Li and X. Li, et al., Tetrahydroxy stilbene glycoside attenuates acetaminophen-induced hepatotoxicity by UHPLC-Q-TOF/MS-based metabolomics and multivariate data analysis, J. Cell. Physiol., 2021, 236(5), 3832–3862 CrossRef CAS PubMed.
- L. Yuan, F. Jiang and X. Cao, et al., Metabolomics reveals the toxicological effects of polar compounds from frying palm oil, Food Funct., 2020, 11(2), 1611–1623 RSC.
- Y. Zhao, X. Dong and C. R. Guo, et al., Dietary isoleucine affects muscle fatty acid and amino acid profiles through regulating lipid metabolism and autophagy in hybrid catfish (Pelteobagrus vachelli ♀ × Leiocassis longirostris ♂), Anim. Nutr., 2022, 11, 369–380 CrossRef CAS PubMed.
- E. J. Miller, A review of biochemical studies on the genetically distinct collagens of the skeletal system, Clin. Orthop. Relat. Res., 1973, 92, 260–280 CrossRef PubMed.
- Y. Liu, P. Yu and X. Sun, et al., Metabolite target analysis of human urine combined with pattern recognition techniques for the study of symptomatic gout, Mol. BioSyst., 2012, 8(11), 2956–2963 RSC.
- A. Zinellu and A. A. Mangoni, A Systematic Review and Meta-Analysis of the Association between Uric Acid and Allantoin and Rheumatoid Arthritis, Antioxidants, 2023, 12(8), 1569 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: See DOI: https://doi.org/10.1039/d5tb00362h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |