Open Access Article
Open Access ArticleAdipose derived mesenchymal stem cell-seeded regenerated silk fibroin scaffolds reverse liver fibrosis in mice†
Weilong
Li‡
a,
Xiaonan
Shi‡
a,
Daxu
Zhang‡
b,
Jingjing
Hu
a,
Shuo
Zhao
f,
Shujun
Ye
a,
Jingyi
Wang
a,
Xiaojiao
Liu
c,
Qian
Zhang
d,
Zhanbo
Wang
*e,
Yaopeng
Zhang
 *c and
Li
Yan
*c and
Li
Yan
 *a
*a
aThe Second Medical Center and National Clinical Research Center of Geriatric Diseases, Chinese PLA General Hospital, Beijing 100853, PR China. E-mail: yanlifmu@126.com
bDepartment of General Surgery, The First Medical Center, Chinese PLA General Hospital, Beijing 100853, PR China
cState Key Laboratory of Advanced Fiber Materials, College of Materials Science and Engineering, Shanghai Engineering Research Center of Nano-Biomaterials and Regenerative Medicine, Donghua University, Shanghai, 201620, PR China. E-mail: zyp@dhu.edu.cn
dSchool of nursing, Lanzhou University, Gansu 730000, PR China
eDepartment of Pathology, Chinese PLA General Hospital, Beijing 100853, P. R. China. E-mail: wzb4000@126.com
fDepartment of Critical Care Medicine,Aerospace Central Hospital,Beijing,, PR China
First published on 26th February 2025
Abstract
Liver fibrosis (LF) is an important process in the progression of chronic liver disease to cirrhosis. We have previously demonstrated that a regenerated silk fibroin scaffold loaded with adipose-derived stem cells (RSF + ADSCs) can repair acute liver injury. In this study, we established a chronic LF animal model using carbon tetrachloride (CCl4) and a high-fat diet. We then investigated the liver repair capacity after transplanting RSF + ADSC scaffolds and RSF scaffolds onto the liver surface of mice. Compared with the control group, the concentrations of ALT and AST in the serum were significantly reduced in the RSF and RSF + ADSC groups. HE staining and Masson trichrome staining revealed a decrease in the SAF score in both the RSF and RSF + ADSC groups. Meanwhile, the biomarkers of blood vessels and bile ducts, such as CD34, ERG, muc1, and CK19, were significantly elevated in the RSF + ADSC group. Finally, transcriptome analysis showed that the PPAR signaling pathway, which inhibits liver fibrosis, was significantly upregulated in both the RSF and RSF + ADSC groups. Our study suggests that, compared with RSF scaffolds alone, RSF + ADSCs have a significant repair effect on chronic LF in mice.
1. Introduction
Currently, the global prevalence of fatty liver disease exceeds 30%.1,2 In China, the number of people with severe fatty liver disease has reached approximately 36 million, and these individuals are at risk of liver fibrosis (LF).3 Without effective intervention, patients with liver fibrosis may face the risk of progression to cirrhosis or even liver cancer. At present, once liver fibrosis progresses to cirrhosis, there are no effective drugs available. Therefore, reversing the progression of liver fibrosis is crucial.In recent years, stem cell regeneration research has provided new insights for treating liver fibrosis.4–6 Recent studies have shown that stem cells can promote liver regeneration through transdifferentiation or paracrine mechanisms.7 In some clinical trials, stem cells have been used to treat hepatitis B-related cirrhosis. However, issues such as the difficulty in controlling stem cell quality, poor stem cell engraftment capacity, and tumorigenicity make the therapeutic effects unpredictable.4,8,9 Fortunately, if stem cells are combined with biomaterials that can mimic the liver microenvironment, their potential to promote liver fibrosis repair can be greatly enhanced.10 Our preliminary studies suggest that the combination of regenerated silk fibroin (RSF) and adipose-derived stem cells (ADSCs) can effectively repair liver damage.11,12 Additionally, we have observed the presence of vascular structures in RSF,12 though their characteristics need further validation.
In this study, we explored the role of RSF + ADSCs in reversing liver fibrosis and their effect on the formation of blood vessels and bile ducts. First, we established a liver fibrosis animal model with carbon tetrachloride and a high-fat diet. Subsequently, we transplanted RSF and RSF + ADSC scaffolds onto the liver surface of the animal model. On the 7th, 14th, 30th, and 60th day after transplantation, we observed the recovery of liver function. With the help of HE staining, we discovered good compatibility between the scaffold and the liver. Furthermore, the result of Masson's trichrome staining showed that RSF + ADSCs could reverse liver fibrosis. Immunohistochemical staining helped us to confirm the formation of blood vessels and bile ducts. Finally, according to transcriptome analysis, the molecules related to the inhibition of liver fibrosis (PPAR signaling pathway), including Acad1, Cpt1a, Dbi, Ppar, and Slc27a5 genes, were upregulated in both RSF and RSF + ADSC scaffold groups. We performed quantitative validation using PCR technology. The results suggest that inducing the formation of blood vessels and bile ducts through RSF + ADSCs to promote liver repair and reverse liver fibrosis may become an alternative therapeutic method for liver fibrosis.
2. Materials and methods
2.1. Preparation of electrospun RSF matrices
Based on the literature,13 the RSF matrix was prepared from all aqueous solutions. In short, Bombyx mori silkworm cocoons were degummed and subsequently dissolved in a 9.0 M aqueous solution of LiBr. The solution was diluted, centrifuged, and filtered, and then dialyzed in deionized water to remove salts. Finally, a 33% RSF aqueous solution was obtained by forced air cooling. Using conventional electrospinning technology, RSF mats were prepared on an aluminum collection plate with an electric potential of 20 kV, a flow rate of 1.2 mL h−1, and a span of 10 centimeters between the sample and the spinneret. Then, the obtained mat with a thickness of 130 μm was soaked in a 90 vol% ethanol aqueous solution for 30 minutes to convert the RSF to an insoluble state in water.2.2. Cell culture assay
After collecting adipose tissue from the inguinal region of mice, it is transferred to Dulbecco's modified Eagle's medium (DMEM) containing antibiotics (100 mg mL−1 penicillin and 100 mg mL−1 streptomycin), and 2 mM Glutamax under sterile conditions. The adipose layer is cleaned with phosphate-buffered saline (PBS). Then, these small pieces are digested with 15 mL of 0.2% collagenase type 1 at 37 °C for 2 hours. DMEM containing 10% fetal bovine serum (FBS) is used to terminate the collagenase activity, and the cells are centrifuged at 400 × g for 10 minutes to separate the floating cells from the vascular matrix. The pellet is resuspended in the complete medium containing 10% FBS, 5% penicillin/streptomycin, and 1% Glutamax, and passed through a 100 μm nylon mesh filter to remove undigested tissue. The filtered cells are carefully transferred to a 50 mL tube containing 1.077 g mL−1 Percoll, and subjected to density gradient centrifugation at 400 × g for 30 minutes. Enriched cells are collected from the interface, and washed twice with serum-free medium. Finally, the pellet is resuspended in DMEM containing 10% FBS, 100 mg mL−1 penicillin/streptomycin, and 2 mM Glutamax, and cultured in an incubator at 25 °C with 5% CO2/95% air and 90% relative humidity. The medium is changed every 24 hours for the first 3 days to remove non-adherent hematopoietic cells, and then changed every 3 days. After the adherent mesenchymal stem cells reach confluence, they are digested with 0.25% trypsin-EDTA, and transferred to new 25 cm2 culture flasks for further culture. All experiments are performed using mesenchymal stem cells after three to six passages. Ultimately, these cells are seeded onto RSF scaffolds.2.3. Cell induction
The sterilized bioscaffolds were placed in 24-well culture plates (Corning, USA). Then, 1 × 106 ADSCs were seeded into each 3D-PSFS. The ADSCs were cultured at 37 °C with 5% CO2. Once the ADSCs adhered to 3D-PSFS, the basic medium (DMEM with 10% FBS) was replaced with hepatocyte induction medium which has been used in our previous research.14 Then, 1.5 mL of hepatocyte induction medium was added to each well of the 24-well culture plate and replaced every 48 hours.2.4. Animal model construction
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Chinese People's Liberation Army General Hospital. Mice aged three days were utilized to prepare mesenchymal stem cells, and 6 to 8 weeks were used for animal experiments. To establish a chronic LF animal model, 6- to 8-week-old mice were intraperitoneally injected with an olive oil solution containing 40% CCl4 at a dose of 2 mL kg−1, three times a week. They were also fed a high-fat diet containing 60% fat for 6 to 8 weeks.2.5. Transplantation and sample collection
All surgeries were performed by the same surgeon. Mice were anesthetized through an intraperitoneal injection of pentobarbital (1%, 50 mg per kg). The left lateral lobe of the liver was exposed and the scaffold was sutured in place. Standard layered closure of the wound was performed. Mouse survival was recorded and monitored for 60 days post-transplantation. Liver tissue and blood samples were obtained on the 7th, 14th, 30th, and 60th days after transplantation for subsequent experimental analysis. n = 3 mice per group.2.6. Evaluation of liver function and C-reactive protein (CRP)
On days 7, 14, 30, and 60 post-transplantation, serum levels of alanine aminotransferase (ALT), aspartate transferase (AST), alkaline phosphatase (ALP), albumin (ALB), total bilirubin (TBIL), triglycerides (TG), total cholesterol (TC), and CRP were measured using an automated analyzer (Mindray, BS-240 Vet).2.7. Histological staining and scoring
The collected liver tissue was fixed in 4% paraformaldehyde for 24 hours to obtain paraffin sections. Following embedding, sections were cut into 6 μm slices using a Leica SM2000R microtome. HE staining, as well as Masson's trichrome staining, was carried out following the manufacturer's instructions (Solarbio, n = 3).Histological scoring was conducted using the SAF scoring system: hepatocyte steatosis was scored from 0 to 3, ballooning degeneration from 0 to 2, inflammation from 0 to 2, and fibrosis from 0 to 4. The fibrosis scoring follows the same criteria as the European SAF and American NAS scoring systems. When fibrosis is graded as F1, it is further categorized into F1a, F1b, and F1c. The total SAF score is obtained by summing the individual scores.
2.8. Immunohistochemical staining
The frozen liver tissue sections were rehydrated and fixed with 4% PFA. After peroxidase treatment, the sections were blocked with PBS containing 5% goat serum and 2% BSA. Then the sections were incubated with primary antibodies, including anti-CD34 (Abcam), anti-ERG (Abcam), anti-MUC1 (Abcam), and keratin 17/19 (cell signaling) overnight at 4 °C. Secondary antibodies were applied for 1 hour at room temperature. The sections were then incubated with 3,3′-diaminobenzidine and retained with hematoxylin. Finally, the sections were coverslipped with xylene and neutral resin. Images were taken with a microscope (Nikon, Japan). n = 3 samples in each group.2.9. Reverse transcription polymerase chain reaction
After 7 days of transplanting the material, the liver of the mouse was lysed using the Trizol reagent (from Invitrogen Life Technologies). The RNA was then reverse-transcribed into cDNA under the following conditions: 50 °C for 15 minutes and 85 °C for 5 seconds. After the synthesis of cDNA, it was stored at 4 °C before being used for real-time quantitative PCR. The PCR reaction conditions were set as follows: pre-denaturation at 95 °C for 30 seconds, annealing at 95 °C for 10 seconds, and extension at 60 °C for 30 seconds, with a total of 40 cycles. Three replicate wells were set, with actin as the internal reference, and the relative expression level was calculated using the 2−ΔΔCt method. To verify the expression levels of CPt1a, Dbi, Ppar, Slc27a5, Tnfrsf1a, and Tnfrsf1b genes in liver tissue. The sequences for the primers used are listed in Table S1 (ESI†).2.10. mRNA transcriptome sequencing analysis
Liver tissue samples were collected after 7 days, and immediately frozen and stored in a −80 °C refrigerator. TRIzol reagent or a commercial RNA extraction kit was used to extract total RNA, and the concentration, purity, and integrity of the RNA were detected using a NanoDrop spectrophotometer and Bioanalyzer. Then, mRNA was enriched using poly(A) selection or ribosomal RNA depletion, followed by reverse transcription to synthesize cDNA and library preparation, which includes fragmentation of the cDNA, end repair, addition of an A-tail, and ligation of adapters. Finally, PCR amplification and purification were performed. High-throughput sequencing was performed on the Illumina NovaSeq 6000 or HiSeq 4000 platform, selecting the appropriate read length and sequencing depth. The quality of the raw data was evaluated using FastQC, the data were filtered using Trimmomatic or Cutadapt, and the filtered reads were aligned to the reference genome using HISAT2 or STAR. Gene expression levels were calculated using HTSeq or featureCounts, and differential expression gene analysis was performed using DESeq2 or edgeR. Finally, functional annotation and pathway enrichment analysis of the differentially expressed genes were performed through databases such as GO and KEGG.2.11. Statistical analysis
Statistical analysis of two groups of parameter data was performed using the Student's t-test. Normality tests were conducted, and all analyses were carried out using GraphPad Prism 9 software. Data are reported as mean ± standard deviation, with significance defined as a p-value < 0.05.3. Results
3.1. Construction of the liver fibrosis animal model
We constructed a mouse model of LF using intraperitoneal injection of CCl4 combined with a high-fat diet. After constructing the LF mouse model through intraperitoneal injection of CCl4 combined with a high-fat diet, we transplanted either a regenerated silk fibroin scaffold loaded with adipose-derived stem cells (RSF + ADSCs) or a pure regenerated silk fibroin scaffold onto the liver surface of the fibrotic mice. Upon macroscopic inspection, the liver surface of mice in the control group appeared bright red and smooth. In contrast, the liver surface of mice, after 8 weeks of intraperitoneal injection of CCl4 combined with a high-fat diet, appeared dull (Fig. 1C). We performed Masson and HE staining (Fig. 1A and B) on the liver tissues of the mice to determine the progression of fibrosis: After eight weeks, the normal lobular structure of the liver had disappeared. We observed excessive lipid droplet accumulation, ballooning degeneration, hepatocyte swelling and necrosis, and extensive inflammatory cell infiltration. Under Masson staining, the density and coverage of collagen fibers (stained blue or dark blue) were significantly higher than in the normal group. This resulted in dense fiber bundles or clusters, forming a complex “chicken wire” network structure. The SAF score reached 10 (Fig. 1D), indicating significant fibrosis characteristics in the liver by the eighth week. All of the above pathological diagnoses have been evaluated by pathology specialists to ensure accuracy and reliability. The survival rates of the RSF + ADSC group compared to the control groups (LF or Control) in mice showed no significant differences at various time points (P > 0.05). At the endpoint of the experiment (day 60), the survival rate of the RSF + ADSC group was 97% (±2%), the RSF group was 98% (±2)%, and the LF group was 95% (±4%). The differences were not statistically significant (P = 0.339) (Fig. 1E). This suggests that neither RSF + ADSCs nor RSF had a significant impact on the survival rate of mice after transplantation.3.2. Biocompatibility and degradation of RSF + ADSCs and RSF on liver surface
HE staining showed that the degree of injury in mice transplanted with RSF and RSF + ADSCs was much weaker than that in non-transplanted injured mice (Fig. 2A). Seven days after transplantation, there was still liver injury, accompanied by liver cell edema, fatty liver degeneration, inflammatory cell infiltration, and cytoplasmic degeneration. On the 14th day, compared with the RSF group, the RSF + ADSC group had less large vacuolar fatty degeneration. At 60 days, all experimental groups were basically recovered in terms of histology. However, significant damage areas could always be observed in the LF group. These observations suggest that RSF and RSF + ADSCs have similar protective effects on liver injury induced by CCl4 combined with high-fat diet, but the RSF + ADSC group has a faster repair effect than the RSF group (Fig. 2B).3.3. Dynamics changes of the histopathology of liver fibrosis
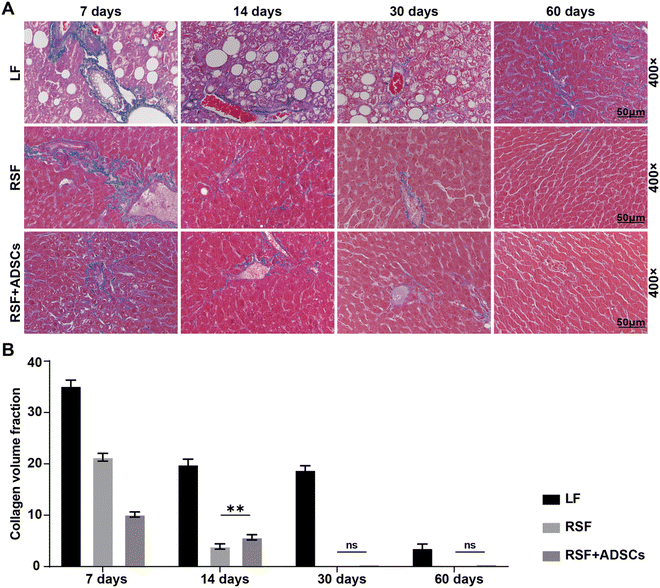
To observe the changes in the degree of fibrosis in the liver tissue of mice after implantation, we performed Masson staining on the liver of mice at 7 days, 14 days, 30 days, and 60 days (Fig. 3A). At 7 days, the results showed the presence of collagen fibers in liver tissue, indicating a high degree of fibrosis. At 14 days, the collagen area in all experimental groups was significantly reduced. At 30 days, the collagen area in the experimental group was further reduced, indicating the potential of RSF implantation to improve fibrosis. At 60 days, the results of Masson staining showed a significant reduction in the area of collagen fibers compared with the initial state, indicating that the repair effect of RSF implantation on LF gradually increased (Fig. 3B).3.4. Dynamics of liver function and inflammatory marker CRP
To assess the recovery of liver function and the levels of inflammatory markers, we conducted blood biochemical analysis. The results showed that 7 days after stent transplantation, the biomarker levels representing liver cell injury (AST, ALT), liver metabolic function (ALP, Tbil, TC, TG), and liver inflammation (CRP) were high. The expression levels in the RSF + ADSC group were lower at different time intervals than those in the LF group and RSF group. In addition, both the RSF group and the RSF + ADSC group showed the lowest expression of liver injury and inflammatory biomarkers on the 60th day (Fig. 4). Furthermore, the RSF group showed a certain degree of therapeutic ability, but it was much weaker than the RSF + ADSC group.3.5. Formation of new vascular and bile duct structures in the RSF scaffold
To observe the neotissue formed on the liver surface by RSF + ADSCs, we performed HE and immunohistochemical staining on the neotissue 7 days after transplantation. HE staining showed that tube-like structures had formed on the scaffolds in the RSF + ADSC group (Fig. 5A). Immunohistochemical staining of these tube-like structures revealed significant expression of angiogenesis markers (ERG, CD34) and biliary markers (MUC1, CK19) (Fig. 5B). These results suggest that RSF + ADSCs play an important role in liver regeneration and angiogenesis.3.6. RNA transcriptome analysis
To investigate the potential mechanism of RSF in repairing LF, we performed transcriptomic analysis of liver tissue on the 7th day after transplantation. Cluster analysis showed that the gene expression modules were similar in the RSF group and the RSF + ADSC group, but significantly different from the control group (Fig. 6A). The Venn diagram results showed that the RSF + ADSC group expressed 7070 deferentially expressed genes, while the RSF group expressed 7345 deferentially expressed genes (Fig. 6B). Through KEGG pathway enrichment analysis, compared with the normal group, the RSF group and the RSF + ADSC group significantly up-regulated cell proliferation, fat degradation, redox, protein synthesis, drug metabolism and other pathways. We found that both the RSF and RSF + ADSC groups significantly up-regulated the PPAR pathway, which helps maintain the balance of fat metabolism in the body by regulating fatty acid uptake, synthesis, and oxidation. In addition, both the RSF and RSF + ADSC group significantly down-regulated immune response, inflammation, and apoptosis pathways, mainly including TNF, NF-kappa B signaling pathways (Fig. 6C and D).3.7. RSF + ADSC scaffolds upregulate the PPAR signaling pathway and the downregulate TNF signaling pathway
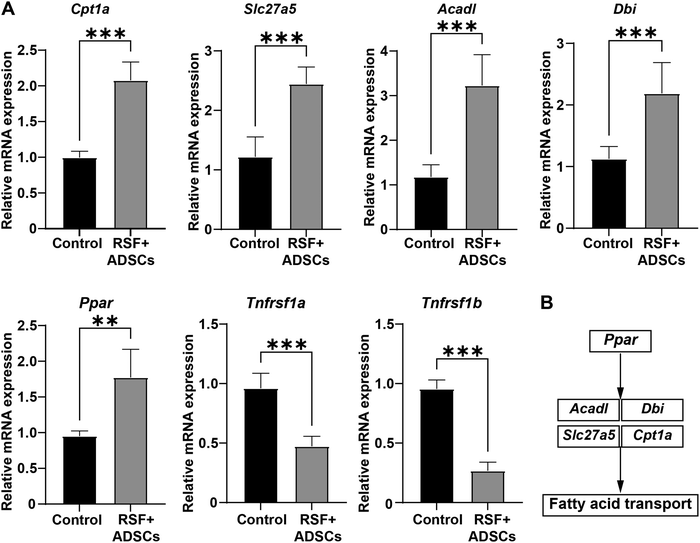
To further verify the specific mechanism of RSF + ADSCs in repairing LF, we used the RT-qPCR method to evaluate the gene expression of the PPAR signaling pathway and TNF signaling pathway in the liver tissue-scaffold connection of treated mice. The results showed that compared with the control group, the expression of CPt1a, Slc27a5, Acadl, Dbi, and Ppar genes in the RSF + ADSC group was significantly up-regulated (Fig. 7A). These genes are involved in the process of fatty acid uptake, transport and oxidation. On the other hand, the expression levels of Tnfrsf1a and Tnfrsf1b genes in the RSF + ADSC group were significantly decreased, which is related to inflammatory response and fibrosis. RSF + ADSC scaffolds regulate the PPAR signaling pathway: a simplified diagram is shown in Fig. 7B. The results suggest that the PPAR signaling pathway and the TNF signaling pathway may be involved in the fibrosis repair of LF mice after RSF + ADSC transplantation.4. Discussion
In recent years, research on stem cells and biomaterials has provided new insights into the treatment of liver fibrosis. Some clinical trial reports show that stem cells have been used to treat LF, but the application of stem cells still has limitations.15 Fortunately, our previous research has indicated that biomaterials such as regenerated silk fibroin,11 apple extract,16 and nucleic acid tetrahedra17 contribute to the repair of acute liver injury. However, these materials have not yet been applied to the repair of chronic liver injury.In this study, we combined CCl4 and a high-fat diet to establish a mouse model of chronic liver injury. Additionally, we explored the mechanisms of liver fibrosis repair using RSF and RSF + ADSCs and found that the RSF + ADSC group had a stronger ability to repair chronic liver injury than the RSF group. This group also formed a large number of new blood vessels and bile ducts in the transplanted materials. Finally, we used transcriptome analysis and PCR technology to validate that the expression of anti-fibrosis-related molecules in the PPAR pathway was higher in the RSF + ADSC group compared with the RSF group.
In this study, by combining CCl4 with high-fat diet, we established a mouse model of liver fibrosis model that closely resembles the human living environment.18,19 Unlike conventional non-alcoholic fatty liver disease models, which are established using diets rich in fats, fructose (or sucrose), and cholesterol,20,21 our model showed late-stage histological features of liver fibrosis by the 8th week, with a SAF score of 10. This model reflects both lifestyle-induced liver fibrosis and drug-induced liver fibrosis,22–24 aligning with the mechanisms of liver fibrosis caused by the high-paced lifestyle and drug abuse that are prevalent today.
The liver possesses a strong regenerative ability, which varies depending on the extent of injury and its underlying cause.7 In previous studies, various biomaterials, such as hydrogels;25 polydimethylsiloxane;26,27 and various natural biomaterials,28 including, hyaluronic acid,29 and animal extracellular matrix,30,31 have been used, but none have been able to form liver-like tissues with complex structures that include functional vascular and bile duct networks. The reconstruction of complex vascular and bile duct systems remains a common challenge in tissue engineering. Compared with previous studies,11 our innovation lies in the confirmation of vascular and bile duct formation in the RSF + ADSC group with the help of immunohistochemical staining.
RSF is a natural biomaterial with good biocompatibility and biodegradability, widely used in clinical applications.32,33 The degradation process of the RSF materials is a complex biological degradation process, and its properties directly affect its application in the biomedical field.34 As a natural protein-based biomaterial, the degradation of RSF primarily occurs through enzymatic action, particularly by proteinase XIV, which exhibits high efficiency in degrading RSF both in vivo and in vitro.35,36 The degradation of RSF typically begins in its hydrophilic unordered regions, such as the C-terminus, N-terminus, connecting segments, and light chains, and then gradually infiltrates the crystalline regions, such as the β-structured areas.37 Finally, RSF is degraded into small peptide fragments and amino acids, which are non-toxic and can be metabolized or cleared by the host without accumulating in the tissue.38–40In vivo degradation is closely related to the host immune system and is primarily mediated by macrophages and foreign body giant cells (FBGCs).41,42 It is worth noting that the degradation process of RSF materials typically does not cause significant inflammation or immune reactions.43 Its natural components (e.g., silk protein) exhibit good biocompatibility, and its degradation products (e.g., peptides and amino acids) are natural metabolites that will not trigger immune reactions.44 These characteristics make RSF materials highly safe and reliable for clinical applications.
As a material with a three-dimensional spatial structure, RSF provides a microenvironment for cell adhesion, proliferation, and differentiation, and also contributes to the formation of new blood vessels, making it a popular material in the field of liver regeneration. In this study, transcriptomic technology was used to identify that RSF scaffolds may promote protein synthesis, regulate fatty acid metabolism balance, and reverse liver fibrosis by upregulating the valine, leucine, and isoleucine degradation signaling pathway, the P450 signaling pathway (metabolism of xenobiotics by cytochrome P450), and the PPAR signaling pathway.
A potential limitation of this study is the lack of a treatment group that uses ADSCs alone or other materials combined with ADSCs for comparison.45 However, ADSCs alone may not be sufficient for effective liver fibrosis repair due to their inability to specifically target the fibrotic area.46 Additionally, the choice of RSF as a scaffold is based on its unique properties,47 making it difficult to find an equivalent control group with similar characteristics. The aim of our research is to investigate whether RSF, as a carrier for stem cells, can provide an appropriate microenvironment for ADSC proliferation and differentiation, and whether it has the potential to promote ADSC-mediated liver injury repair. The RSF material itself only plays an auxiliary role in liver injury repair, similar to other synthetic scaffolds such as polycaprolactone48,49 and PEG-based scaffolds,50 but these differ significantly from RSF scaffolds and are not suitable as controls. Therefore, we emphasize the combination of RSF and ADSCs, investigating their combined potential for promoting liver injury repair. This study focuses on exploring the mechanism of local liver transplantation of regenerative biomaterials combined with stem cells for liver injury repair. Future studies could explore the addition of other materials or different scaffold types to further optimize the repair process.
To further verify the mechanism of RSF on ADSCs, this study performed transcriptomic analysis between the RSF + ADSC group and the RSF group. The results showed that the RSF + ADSCs scaffold significantly upregulated the expression of genes associated with the PPAR signaling pathway,51 such as Cpt1a, Slc27a5, Acadl, Dbi, and Fabp1 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC ≥ 2, P < 0.05). PPAR52 is a transcription factor involved in inflammation and lipid metabolism, and its activation can inhibit liver fibrosis. In addition, the RSF + ADSC scaffold can inhibit the TNF signaling pathway. TNF53 can bind to its receptors TNFR1 (Tnfrsf1a) and TNFR2 (Tnfrsf1b),54 activating downstream pathways such as NF-κB,55 thereby triggering inflammatory responses and cell apoptosis.56
2FC ≥ 2, P < 0.05). PPAR52 is a transcription factor involved in inflammation and lipid metabolism, and its activation can inhibit liver fibrosis. In addition, the RSF + ADSC scaffold can inhibit the TNF signaling pathway. TNF53 can bind to its receptors TNFR1 (Tnfrsf1a) and TNFR2 (Tnfrsf1b),54 activating downstream pathways such as NF-κB,55 thereby triggering inflammatory responses and cell apoptosis.56
5. Conclusions
In summary, RSF + ADSCs have great potential in liver regeneration and LF treatment. In the future, we will continue to explore the effects of RSF + ADSCs on the upstream and downstream molecules of the PPAR and TNF signaling pathways in reversing liver fibrosis.Abbreviations
| RSF | Regenerated silk fibroin |
| ADSCs | Adipose-derived mesenchymal stem cells |
| LF | Liver fibrosis |
| CCl4 | Carbon tetrachloride |
| DMEM | Dulbecco's modified Eagle medium |
| PBS | Phosphate-buffered saline |
| FBS | Fetal bovine serum |
| ALT | Alanine aminotransferase |
| AST | Aspartate transferase |
| ALP | Alkaline phosphatase |
| ALB | Albumin |
| TBIL | Total bilirubin |
| TG | Triglycerides |
| TC | Total cholesterol |
| CRP | C-reactive protein |
| NASH | Non-alcoholic steatohepatitis |
| CPT1A | Carnitine palmitoyltransferase 1A |
| SLC27A5 | Solute carrier 27A5 |
| ACADL | Acetyl-CoA dehydrogenase long chain |
| DBI | DNA-binding inhibitor |
| PPAR | Peroxisome proliferator-activated receptor |
| Tnfrsf1a | Tumor necrosis factor superfamily member 1a |
| Tnfrsf1b | Tumor necrosis factor superfamily member 1b |
| MAPK | Mitogen-activated protein kinase |
| NF-κB | Nuclear factor kappa B |
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. Weilong Li: writing – review & editing, writing – original draft, software, methodology, formal analysis, data curation, and conceptualization. Xiaonan Shi: writing – review & editing, writing – original draft, validation, software, methodology, formal analysis, data curation, and conceptualization. Daxu Zhang: methodology, formal analysis, and data curation. Jingjing Hu: methodology. Shuo Zhao: methodology. Shujun Ye: methodology. Jingyi Wang: methodology. Xiaojiao Liu: methodology. Qian Zhang: methodology. Yaopeng Zhang: supervision. Zhanbo Wang: methodology, formal analysis, and supervision. Li Yan: supervision, resources, funding acquisition, and conceptualization.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This study was supported by the National Key Research and Development Program of China (2019yfa0110600) and the National Natural Science Foundation of China (31971263).References
- J. C. Cohen, J. D. Horton and H. H. Hobbs, Human fatty liver disease: old questions and new insights, Science, 2011, 332(6037), 1519–1523 CrossRef CAS PubMed.
- P. Angulo, Nonalcoholic fatty liver disease, N. Engl. J. Med., 2002, 346(16), 1221–1231 CrossRef CAS PubMed.
- S. Liu, H. Wan, L. Yang, J. Shen and X. Qi, High prevalence of steatotic liver disease and fibrosis in the general population: A large prospective study in China, J. Hepatol., 2025, 82(1), e23–e25 CrossRef PubMed.
- X. Yang, Q. Li, W. Liu, C. Zong, L. Wei, Y. Shi and Z. Han, Mesenchymal stromal cells in hepatic fibrosis/cirrhosis: from pathogenesis to treatment, Cell. Mol. Immunol., 2023, 20(6), 583–599 CrossRef CAS PubMed.
- G. Lou, Z. Chen, M. Zheng and Y. Liu, Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases, Exp. Mol. Med., 2017, 49(6), e346 CrossRef PubMed.
- P. Liu, Y. Mao, Y. Xie, J. Wei and J. Yao, Stem cells for treatment of liver fibrosis/cirrhosis: clinical progress and therapeutic potential, Stem Cell Res. Ther., 2022, 13(1), 356 CrossRef PubMed.
- G. K. Michalopoulos and B. Bhushan, Liver regeneration: biological and pathological mechanisms and implications, Nat. Rev. Gastroenterol. Hepatol., 2021, 18(1), 40–55 CrossRef PubMed.
- M. Shi, Y.-Y. Li, R.-N. Xu, F.-P. Meng, S.-J. Yu, J.-L. Fu, J.-H. Hu, J.-X. Li, L.-F. Wang, L. Jin and F.-S. Wang, Mesenchymal stem cell therapy in decompensated liver cirrhosis: a long-term follow-up analysis of the randomized controlled clinical trial, Hepatol. Int., 2021, 15(6), 1431–1441 CrossRef PubMed.
- M. Shi, Z. Zhang, R. Xu, H. Lin, J. Fu, Z. Zou, A. Zhang, J. Shi, L. Chen, S. Lv, W. He, H. Geng, L. Jin, Z. Liu and F.-S. Wang, Human mesenchymal stem cell transfusion is safe and improves liver function in acute-on-chronic liver failure patients, Stem Cells Transl. Med., 2012, 1(10), 725–731 CrossRef CAS PubMed.
- J. Guo, G.-s Lin, C.-y Bao, Z.-m Hu and M.-y Hu, Anti-inflammation role for mesenchymal stem cells transplantation in myocardial infarction, Inflammation, 2007, 30(3–4), 97–104 CrossRef CAS PubMed.
- L. Xu, S. Wang, X. Sui, Y. Wang, Y. Su, L. Huang, Y. Zhang, Z. Chen, Q. Chen, H. Du, Y. Zhang and L. Yan, Mesenchymal Stem Cell-Seeded Regenerated Silk Fibroin Complex Matrices for Liver Regeneration in an Animal Model of Acute Liver Failure, ACS Appl. Mater. Interfaces, 2017, 9(17), 14716–14723 CrossRef CAS PubMed.
- Q. Chang, L. Yan, C.-Z. Wang, W.-H. Zhang, Y.-Z. Hu and B.-Y. Wu, In vivo transplantation of bone marrow mesenchymal stem cells accelerates repair of injured gastric mucosa in rats, China Med. J., 2012, 125(6), 1169–1174 Search PubMed.
- S. Fan, Y. Zhang, H. Shao and X. Hu, Electrospun regenerated silk fibroin mats with enhanced mechanical properties, Int. J. Biol. Macromol., 2013, 56, 83–88 CrossRef CAS PubMed.
- M. Ali and S. L. Payne, Biomaterial-based cell delivery strategies to promote liver regeneration, Biomater. Res., 2021, 25(1), 5 CrossRef PubMed.
- S. Yamanaka, Pluripotent Stem Cell-Based Cell Therapy-Promise and Challenges, Cell Stem Cell, 2020, 27(4), 523–531 CrossRef CAS PubMed.
- J. Hu, S. He, D. Zhang, Z. Wang, S. Zhao, X. Shi, W. Li, Q. Guo, W. Guan and L. Yan, Constructing liver-like tissue in situ based on plant-derived cellulose scaffolds alleviating acute liver injury, Mater. Des., 2024, 240, 112856 CrossRef CAS.
- D. Zhang, L. Fu, Y. Yang, Q. Guo, J. Hu, P. Li, S. Zhao, X. Shi, W. Li, Y. Lin, W. Lu and L. Yan, Tetrahedral framework nucleic acids improve the effectiveness of adipose-derived mesenchymal stem cells in the repair of acute liver failure, Mater. Today Nano, 2024, 25, 100454 CrossRef CAS.
- L. Recena Aydos, L. Aparecida do Amaral, R. Serafim de Souza, A. C. Jacobowski, E. Freitas Dos Santos and M. L. Rodrigues Macedo, Nonalcoholic Fatty Liver Disease Induced by High-Fat Diet in C57bl/6 Models, Nutrients, 2019, 11(12), 3067 CrossRef PubMed.
- N. Hariri and L. Thibault, High-fat diet-induced obesity in animal models, Nutr. Res. Rev., 2010, 23(2), 270–299 CrossRef CAS PubMed.
- G. Farrell, J. M. Schattenberg, I. Leclercq, M. M. Yeh, R. Goldin, N. Teoh and D. Schuppan, Mouse Models of Nonalcoholic Steatohepatitis: Toward Optimization of Their Relevance to Human Nonalcoholic Steatohepatitis, Hepatology, 2019, 69(5), 2241–2257 CrossRef PubMed.
- M. A. Van Herck, L. Vonghia and S. M. Francque, Animal Models of Nonalcoholic Fatty Liver Disease-A Starter’s Guide, Nutrients, 2017, 9(10), 1072 CrossRef PubMed.
- R. Pérez Tamayo, Is cirrhosis of the liver experimentally produced by CCl4 and adequate model of human cirrhosis?, Hepatology, 1983, 3(1), 112–120 CrossRef PubMed.
- J. G. Abraldes, A. Rodríguez-Vilarrupla, M. Graupera, C. Zafra, H. García-Calderó, J. C. García-Pagán and J. Bosch, Simvastatin treatment improves liver sinusoidal endothelial dysfunction in CCl4 cirrhotic rats, J. Hepatol., 2007, 46(6), 1040–1046 CrossRef CAS PubMed.
- Y.-S. Lee and E. Seki, In Vivo and In Vitro Models to Study Liver Fibrosis: Mechanisms and Limitations, Cell. Mol. Gastroenterol. Hepatol., 2023, 16(3), 355–367 CrossRef CAS PubMed.
- Z. Li, Y. Zhao, X. Ouyang, Y. Yang, Y. Chen, Q. Luo, Y. Zhang, D. Zhu, X. Yu and L. Li, Biomimetic hybrid hydrogel for hemostasis, adhesion prevention and promoting regeneration after partial liver resection, Bioact. Mater., 2022, 11, 41–51 CAS.
- P. Ma, L. Jiang, X. Luo, J. Chen, Q. Wang, Y. Chen, E. Ye, X. J. Loh, C. Wu, Y.-L. Wu and Z. Li, Hybrid Polydimethylsiloxane (PDMS) Incorporated Thermogelling System for Effective Liver Cancer Treatment, Pharmaceutics, 2022, 14(12), 2623 CrossRef CAS PubMed.
- J. Peng, A. P. Tomsia, L. Jiang, B. Z. Tang and Q. Cheng, Stiff and tough PDMS-MMT layered nanocomposites visualized by AIE luminogens, Nat. Commun., 2021, 12(1), 4539 CrossRef CAS PubMed.
- X. Yuan, J. Wu, Z. Sun, J. Cen, Y. Shu, C. Wang, H. Li, D. Lin, K. Zhang, B. Wu, A. Dhawan, L. Zhang and L. Hui, Preclinical efficacy and safety of encapsulated proliferating human hepatocyte organoids in treating liver failure, Cell Stem Cell, 2024, 31(4), 484–498.e5 CrossRef CAS PubMed.
- F. Yu, Z. Liu, J. Feng, Y. Man, H. Zhang, J. Shi, X. Pang, Y. Yu and Y. Bi, Hyaluronic acid modified extracellular vesicles targeting hepatic stellate cells to attenuate hepatic fibrosis, Eur. J. Pharm. Sci., 2024, 198, 106783 CrossRef CAS PubMed.
- W. Fan, T. Liu, W. Chen, S. Hammad, T. Longerich, I. Hausser, Y. Fu, N. Li, Y. He, C. Liu, Y. Zhang, Q. Lian, X. Zhao, C. Yan, L. Li, C. Yi, Z. Ling, L. Ma, X. Zhao, H. Xu, P. Wang, M. Cong, H. You, Z. Liu, Y. Wang, J. Chen, D. Li, L. Hui, S. Dooley, J. Hou, J. Jia and B. Sun, ECM1 Prevents Activation of Transforming Growth Factor β, Hepatic Stellate Cells, and Fibrogenesis in Mice, Gastroenterology, 2019, 157(5), 1352–1367.e13 CrossRef CAS PubMed.
- K. H. Hussein, K.-M. Park, L. Yu, H.-H. Kwak and H.-M. Woo, Decellularized hepatic extracellular matrix hydrogel attenuates hepatic stellate cell activation and liver fibrosis, Mater. Sci. Eng., C, 2020, 116, 111160 CrossRef CAS PubMed.
- X. Yao, S. Zou, S. Fan, Q. Niu and Y. Zhang, Bioinspired silk fibroin materials: From silk building blocks extraction and reconstruction to advanced biomedical applications, Mater. Today Bio, 2022, 16, 100381 CrossRef CAS PubMed.
- J. K. Sahoo, O. Hasturk, T. Falcucci and D. L. Kaplan, Silk chemistry and biomedical material designs, Nat. Rev. Chem., 2023, 7(5), 302–318 CrossRef CAS PubMed.
- C. Guo, C. Li and D. L. Kaplan, Enzymatic Degradation of Bombyx mori Silk Materials: A Review, Biomacromolecules, 2020, 21(5), 1678–1686 CrossRef CAS PubMed.
- R. L. Horan, K. Antle, A. L. Collette, Y. Wang, J. Huang, J. E. Moreau, V. Volloch, D. L. Kaplan and G. H. Altman, In vitro degradation of silk fibroin, Biomaterials, 2005, 26(17), 3385–3393 CrossRef CAS PubMed.
- W. Xiao, J. Zhang, X. Qu, K. Chen, H. Gao, J. He, T. Ma, B. Li and X. Liao, Fabrication of protease XIV-loaded microspheres for cell spreading in silk fibroin hydrogels, J. Mater. Sci.: Mater. Med., 2020, 31(12), 128 CrossRef CAS PubMed.
- H.-Y. Wang, Y.-Q. Zhang and Z.-G. Wei, Dissolution and processing of silk fibroin for materials science, Crit. Rev. Biotechnol., 2021, 41(3), 406–424 CrossRef CAS PubMed.
- T. Zhu, G. Cai, W. Zhao, X. Yao and Y. Zhang, Effects of Silk Fibroin Hydrogel Degradation on the Proliferation and Chondrogenesis of Encapsulated Stem Cells, Biomacromolecules, 2025, 26(2), 1305–1319 CrossRef CAS PubMed.
- K. Liu, Z. Shi, S. Zhang, Z. Zhou, L. Sun, T. Xu, Y. Zhang, G. Zhang, X. Li, L. Chen, Y. Mao and T. H. Tao, A Silk Cranial Fixation System for Neurosurgery, Adv. Healthcare Mater., 2018, 7(6), 1701359 CrossRef PubMed.
- X. Yang, Y. Li, X. Liu, R. Zhang and Q. Feng, In Vitro Uptake of Hydroxyapatite Nanoparticles and Their Effect on Osteogenic Differentiation of Human Mesenchymal Stem Cells, Stem Cells Int., 2018, 1, 1 Search PubMed.
- Q. Lu, X. Hu, X. Wang, J. A. Kluge, S. Lu, P. Cebe and D. L. Kaplan, Water-insoluble silk films with silk I structure, Acta Biomater., 2010, 6(4), 1380–1387 CrossRef CAS PubMed.
- X. Hu, K. Shmelev, L. Sun, E.-S. Gil, S.-H. Park, P. Cebe and D. L. Kaplan, Regulation of silk material structure by temperature-controlled water vapor annealing, Biomacromolecules, 2011, 12(5), 1686–1696 CrossRef CAS PubMed.
- J. Zhu, Y. Du, L. J. Backman, J. Chen, H. Ouyang and W. Zhang, Cellular Interactions and Biological Effects of Silk Fibroin: Implications for Tissue Engineering and Regenerative Medicine, Small, 2025, 21(4), e2409739 CrossRef PubMed.
- Z. Tian, H. Chen and P. Zhao, Compliant immune response of silk-based biomaterials broadens application in wound treatment, Front. Pharmacol., 2025, 16, 1548837 CrossRef PubMed.
- C. Hu, L. Zhao and L. Li, Current understanding of adipose-derived mesenchymal stem cell-based therapies in liver diseases, Stem Cell Res. Ther., 2019, 10(1), 199 CrossRef PubMed.
- Y. He, X. Guo, T. Lan, J. Xia, J. Wang, B. Li, C. Peng, Y. Chen, X. Hu and Z. Meng, Human umbilical cord-derived mesenchymal stem cells improve the function of liver in rats with acute-on-chronic liver failure via downregulating Notch and Stat1/Stat3 signaling, Stem Cell Res. Ther., 2021, 12(1), 396 CrossRef CAS PubMed.
- F. Sun, D. Xiao, H. Su, Z. Chen, B. Wang, X. Feng, Z. Mao and X. Sui, Highly stretchable porous regenerated silk fibroin film for enhanced wound healing, J. Mater. Chem. B, 2023, 11(7), 1486–1494 RSC.
- V. Rahimkhoei, M. Padervand, M. Hedayat, F. Seidi, E. A. Dawi and A. Akbari, Biomedical applications of electrospun polycaprolactone-based carbohydrate polymers: A review, Int. J. Biol. Macromol., 2023, 253(1), 126642 CrossRef CAS PubMed.
- M. Labet and W. Thielemans, Synthesis of polycaprolactone: a review, Chem. Soc. Rev., 2009, 38(12), 3484–3504 RSC.
- J. Zhu, Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering, Biomaterials, 2010, 31(17), 4639–4656 CrossRef CAS PubMed.
- M. Pawlak, P. Lefebvre and B. Staels, Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease, J. Hepatol., 2015, 62(3), 720–733 CrossRef CAS PubMed.
- S. Francque, A. Verrijken, S. Caron, J. Prawitt, R. Paumelle, B. Derudas, P. Lefebvre, M.-R. Taskinen, W. Van Hul, I. Mertens, G. Hubens, E. Van Marck, P. Michielsen, L. Van Gaal and B. Staels, PPARα gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis, J. Hepatol., 2015, 63(1), 164–173 CrossRef CAS PubMed.
- H. Tilg and A. M. Diehl, Cytokines in alcoholic and nonalcoholic steatohepatitis, N. Engl. J. Med., 2000, 343(20), 1467–1476 CrossRef CAS PubMed.
- A. Wullaert, G. van Loo, K. Heyninck and R. Beyaert, Hepatic tumor necrosis factor signaling and nuclear factor-kappaB: effects on liver homeostasis and beyond, Endocr. Rev., 2007, 28(4), 365–386 CrossRef CAS PubMed.
- W.-X. Ding and X.-M. Yin, Dissection of the multiple mechanisms of TNF-alpha-induced apoptosis in liver injury, J. Cell. Mol. Med., 2004, 8(4), 445–454 Search PubMed.
- J. C. A. Broen and J. M. van Laar, Mycophenolate mofetil, azathioprine and tacrolimus: mechanisms in rheumatology, Nat. Rev. Rheumatol., 2020, 16(3), 167–178 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The sequences of the primers used (PDF). See DOI: https://doi.org/10.1039/d5tb00275c |
| ‡ Weilong Li, Xiaonan Shi and Daxu Zhang contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |