Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recent advances in 3D models for multiparametric blood–brain barrier detection in microfluidic systems
Chiara
Boncristiani
 a,
Alessia
Di Gilio
*b,
Federica
De Castro
a,
Alessia
Di Gilio
*b,
Federica
De Castro
 c,
Alessandra
Nardini
ad,
Jolanda
Palmisani
c,
Alessandra
Nardini
ad,
Jolanda
Palmisani
 b,
Rebeca
Martínez Vázquez
b,
Rebeca
Martínez Vázquez
 d,
Gianluigi
de Gennaro
b,
Francesco Paolo
Fanizzi
d,
Gianluigi
de Gennaro
b,
Francesco Paolo
Fanizzi
 c,
Giuseppe
Ciccarella
c,
Giuseppe
Ciccarella
 c and
Viviana
Vergaro
c and
Viviana
Vergaro
 *a
*a
aDepartment of Experimental Medicine, University of Salento c/o College Isufi, Centro Ecoteckne, Via Monteroni 73100, Lecce, Italy. E-mail: viviana.vergaro@unisalento.it
bDepartment of Bioscience, Biotechnology and Environment, University of Bari, Via Edoardo Orabona, 70125 Bari, Italy. E-mail: alessia.digilio@uniba.it
cDepartment of Biological and Environmental Sciences and Technologies (DiSTeBA), University of Salento, Via Monteroni, 73100 Lecce, Italy
dIstituto di Fotonica e Nanotecnologie (IFN)-CNR - Department of Physics - Politecnico di Milano, Piazza Leonardo da Vinci, 32, 20133, Milano, Italy
First published on 5th May 2025
Abstract
Microfluidics has emerged as a valuable technology for modeling the blood–brain barrier (BBB) to study physiological or pathological conditions and plays an important role in neuroscience and pharmaceutical research. Here we discuss the recent advances and the potential application of microfluidic-based systems, because these models, unlike 2D, Transwell and organ-on-chip, accurately mimic the physiological characteristics of the BBB. This review also provides outlooks on the integration of chemical sensors for evaluating BBB models through electrochemical sensors, chemical sensors using MIPs and metabolomic detection, in terms of internal and secreted metabolites. This integration is useful to gain new insights for improving cerebral vascular interventional therapies and discovering new diagnostic tools for clinical practice. Finally, the challenges and future prospects for advancing microfluidics-based BBB systems in neuroscience research are discussed and proposed, particularly regarding new opportunities in multi-disciplinary fundamental research and therapeutic applications for a broader range of disease treatments.
1 Introduction
Microfluidic systems have become a promising tool for modeling the blood–brain barrier (BBB) in vitro. The blood–brain barrier is a highly selective, semi-permeable membrane that separates the blood from the brain's extracellular fluid, maintaining the brain's microenvironment. Understanding its behaviour is crucial for studying neurological diseases and developing drugs, but replicating it in vitro has been challenging due to its complexity. Microfluidics offer the ability to create highly controlled environments at a microscale, mimicking the physiological conditions of the BBB more accurately than traditional cell culture models. Microfluidic platforms can simulate the flow of blood (or blood-mimicking media) and the brain's extracellular fluid allowing a better control of fluid dynamics, shear stress, and nutrient delivery, which are important for replicating the in vivo BBB environment. In this comprehensive review, we first provide a succinct yet informative description of the composition and structure of the blood–brain barrier (BBB), highlighting its critical role in maintaining central nervous system homeostasis. Following this introduction, we delve into an in-depth analysis of the significant advancements that have occurred over time in the field of in vitro modeling of the BBB. We trace the evolution from static systems, such as two-dimensional (2D) monocultures of astrocytes and Transwell systems used for triple co-culture of different cell types, to more sophisticated dynamic and microfluidic conditions that better mimic the physiological environments of the BBB. This section includes a detailed examination of the limitations and advantages associated with existing microfluidic models. We also compare various fabrication methods, discussing their distinct characteristics, including the types of materials used, spatial configurations, overall cost, and size. Such factors are crucial for achieving precise control over device dimensions, surface modifications, and other parameters essential for the realization of three-dimensional (3D) in vitro BBB models. Finally, we explore the broad spectrum of biological applications enabled by microfluidic models, culminating in a discussion on the development of integrated chemical sensors within BBB-on-a-chip systems. These sensors serve as invaluable tools for monitoring, in real-time, various parameters relevant to both physiological and pathological conditions of the BBB, thus paving the way for enhanced understanding and treatment of neurological disorders.2 Advancements in in vitro models of the blood–brain barrier for central nervous system research
Several advancements have been made in the development of microfluidic models to better replicate the physiological conditions of the BBB and improve our understanding of its role in drug delivery, neurovascular health, and disease mechanisms. Recent advancements in in vitro BBB modeling have transitioned from traditional two-dimensional (2D) static systems to more sophisticated three-dimensional (3D) dynamic microfluidic models. These models allow for the co-culture of multiple cell types, including endothelial cells, astrocytes, and pericytes, creating a more physiologically relevant environment that mimics the in vivo conditions of the BBB.1,2 For instance, microfluidic platforms have been designed to replicate the mechanical forces and shear stress experienced by endothelial cells in vivo, which are critical for maintaining BBB integrity and function.3 These innovations have led to the development of “BBB-on-a-chip” systems that not only simulate the structural characteristics of the BBB but also its functional dynamics, including selective permeability and transport mechanisms.4Furthermore, the integration of advanced technologies such as optical and electrophysiological biosensors into these microfluidic systems enhances their utility by enabling real-time monitoring of drug interactions and disease-related markers.1 For example, recent studies have demonstrated the potential of these models in assessing drug transport and barrier integrity under various pathological conditions, thereby providing insights into drug delivery mechanisms and the effects of neurotoxic agents.2,5 Despite these advancements, challenges remain in fully recapitulating the complex interactions within the neurovascular unit. Current models often lack direct cell–cell contact, which is essential for the proper functioning of the BBB.6 However, ongoing research is addressing these limitations by exploring novel approaches, such as incorporating neurons into BBB-on-a-chip models to better simulate the neurovascular interactions that occur in the human brain.6,7 As the field progresses, the continued refinement of these in vitro models holds promise for enhancing drug discovery and improving therapeutic outcomes for CNS disorders. The upcoming subparagraphs will provide a comprehensive overview of the significant advancements that have been made in the field of in vitro microfluidic blood–brain barrier (BBB) chip models. This overview will detail the intricate fabrication processes involved in creating these models, the various methodologies employed during their development, as well as effective troubleshooting techniques that researchers can utilize when faced with challenges. Additionally, we will explore the diverse research applications of these innovative models. To aid in understanding, Fig. 1 summarizes, in a brief yet informative manner, the numerous applications of microfluidics platforms as in vitro models of the blood–brain barrier, specifically highlighting their importance for central nervous system research and the potential they hold for advancing our knowledge in this critical area.
2.1 From bidimensional BBB-models to the first steps of microfluidics
The blood–brain barrier (BBB) is a dynamic and protective membrane that restricts the entry of toxins and pathogens from the bloodstream into the central nervous system (CNS), playing a crucial role in maintaining brain homeostasis by regulating nutrient transport and isolating nervous tissue from potentially harmful substances in circulation, such as hormones and chemicals. Composed of specialized brain microvascular endothelial cells (BMECs), the BBB works alongside supporting cell types like pericytes and astrocytes, forming a neurovascular unit (NVU) (Fig. 2A).8–11 The BMECs are surrounded by a basement membrane rich in proteins and proteoglycans, essential for preserving barrier integrity (Fig. 2B).12 Moreover, the BBB maintains microenvironmental homeostasis and looks after the CNS via multiple different cellular and molecular mechanisms (Fig. 2C). While the BBB is vital for protecting the CNS, it also presents significant challenges for drug delivery, as it limits the entry of therapeutic agents, particularly larger or non-lipid-soluble drugs, due to the presence of efflux transporters like P-glycoprotein, which can hinder treatment efficacy for neurodegenerative diseases and brain cancers.10,13 Disorders such as Alzheimer's disease and Parkinson's disease are linked to BBB dysfunction, and understanding the relationship between barrier integrity and disease progression is essential for developing effective treatments.14 Recent advancements in BBB modeling, particularly through in vitro techniques, are crucial for elucidating the mechanisms of BBB functioning and dysfunction in disease states, offering potential pathways for improved drug delivery strategies. Enhancing our knowledge of BBB and NVU functioning is of uttermost importance for two motives. First, BBB dysfunction is a common feature across almost all CNS disorders.15,16 Impaired barrier function is often accompanied by endothelial inflammation, thus facilitating infiltration of circulating immune cells into the CNS.17,18 The immune cells release inflammatory mediators, such as cytokines, free radicals, and matrix metalloproteinases, which further worsen the barrier function and disease state.19–22 A better understanding of the processes involved in healthy BBB functioning and how these are disturbed in brain diseases will help us find new targets for treatment. Second, while the BBB protects the brain from harmful substances in circulation, it also poses a major challenge when it comes to treating brain diseases. As the BBB only allows small, lipid soluble molecules to pass freely, most drugs require advanced drug delivery strategies to enter the brain.23,24 A better understanding of BBB and NVU functioning will shed light on new techniques and drug delivery strategies to effectively target drugs into the brain to treat CNS disorders. To achieve this goal of improved understanding of NVU functioning in health and disease and advance our knowledge of drug targeting to the brain, we need models that reflect the human NVU in health and disease. While animal models have demonstrated to be helpful in studying the BBB, the employ of animals is expensive, time-consuming, and ethically unwelcome. Furthermore, data gained from animal studies often results in poor translatability to human physiology due to interspecies differences.25 While the cellular component of the NVU is similar between humans and rodents, other relevant features are not. The expression level of many important junctional proteins and transporters diverges between species, which results in differences in drug uptake and efflux. Additionally, drug spreading across the brain may differ due to differences in lipid composition of the brain between species. Importantly, animal models of disease often fail to consider alterations in NVU function related to aging or neurological disease and have reported conflicting results.6,25 While in vitro models of the NVU do not display the level of complexity animal models do, they do allow for the use of human cells, in highly controlled settings, at lower cost, and within shorter time frames. The first effort at in vitro NVU modelling began with the isolation of brain capillaries from rats.26 Since then, many studies of primary rodent, porcine, bovine, and later human brain endothelial cells have been carried out, using both monocultures and co-cultures with supporting cell types.27–31 Later, immortalized cell lines of human brain endothelial cells were established,32,33 followed by protocols for stem-cell derived models34,35 and self-assembling spheroids.36Fig. 3 and Table 1 summarize the evolution over time of the studies conducted for in vitro NVU modelling, which we will analyse in the following paragraphs. Originally, studies were realized using traditional two-dimensional (2D) culture systems. Striving for improved physiological relevance and complexity, the first models using a Transwell system were developed. In this system, brain endothelial cells are cultured on one side of a semi-permeable membrane and astrocytes or pericytes on the other. The Transwell system has been frequently applied to form the BBB structure with appropriate vascular endothelial cells such as human umbilical vein endothelial cells (HUVEC), human cerebral microvascular endothelial cells (hCMEC), and primary human-derived vascular endothelial cells. Although these systems presented a step forward in physiological NVU modelling, the lack of flow and direct cell–cell contact, and the presence of a membrane posed limitations.37–39 In response to those unmet needs, microfluidic platforms made their appearance in the field of NVU modelling.6 Microfluidic platforms need tissue culture chips composed of small channels that permit the development of layered three-dimensional (3D) cell cultures under flux. The first microfluidic NVU models comprised hollow fiber devices to culture bovine aortic endothelial cells and rat glioma cells under shear stress.40,41 These models proved previous papers of advantageous effects of co-culture and for the first time reported that culture under flow improves barrier properties of NVU models. Next the hollow fiber apparatuses, microfluidic polydimethylsiloxane (PDMS) based chips using planar structures were used. Booth and colleagues advanced the first NVU model in such a chip, using murine endothelial cells and astrocytes, creating a much thinner membrane than used earlier in the hollow fiber apparatuses (10 μm versus 150 μm, respectively).42 The thinner membranes allowed for tighter cell–cell contact in co-culture setups, and similar approaches were taken in many subsequent studies using primary cells and cell lines from various species.43–49 The newest microfluidic NVU models again show similarities to the chip reported by Booth et al., but currently special focus is given to all-human models, using primary material, or iPSC-derived cells,1,50 allowing for potential applications in personalized medicine.| Type and period of cultivation | Apparatus | Cells | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| 2D static Transwell system (7–10 days) | Semi-permeable membrane | HUVEC, hCMEC, primary human-derived vascular endothelial cells | High-integrity of BBB using hPSC-derived vascular endothelial cells for BBB formation | No fluidic flow, direct cell–cell contact and shear stress; not for use with astrocytes | 51–53 |
| Microfluidic-integrated BBB model (one week to several weeks) | Horizontal-aligned BBB models | Brain endothelial cells and astrocytes | Easy-to-make BBB model with astrocytes, endothelia, and neurons with a 3D hydrogel structure | A low contact area between neuronal and vascular channels | 49 and 54–56 |
| Vertical-aligned BBB models | Neuronal cells and vascular endothelium | Induction of crosstalk between neuronal cells and vascular endothelium via porous membrane | Relatively hard to make the vertical structure compared with the horizontal model; low contact area between neuronal and vascular channels | 42 and 56–59 | |
| Tubular structure | Vascular EC and neuronal cells | Structural similarity of the blood vessel in the BBB with 3D neuronal structure; induction of biological membrane | Lack of factors to mimic the BBB; difficulty of maintaining for an extended period | 56 and 60–62 | |
| Microfluidics-based dynamic BBB chip model (several weeks) | Laminar microfluidic apparatus | Co-culture of endothelial cells, pericytes and astrocytes | Better transport mechanisms comparable to those observed in vivo | Cannot be applied for high-throughput applications; low throughput; cannot be used to process multiple experiments or screen large drug panels simultaneously | 63 |
| PDMS multiplexed chip with eight parallel channels | hCMEC/D3 mono- or co- cultured with human astrocytes | Run simultaneously different independent reaction units in a single chip | Need to develop standardized tools to operate, monitor, and analyse the cultures inside the device | 64 | |
| Type I collagen hydrogels to produce tubular luminal microchannels through viscous finger patterning | Endothelial cells, astrocytes and pericytes co-cultured in a collagen matrix of a two-lane or three-lane microfluidic platform | Demonstration of successful integration of a human BBB microfluidic model in a high-throughput plate-based format useful for drug screening purposes | It needs to be improved through further experiments | 65 | |
| In vitro mimetic chip divided into two chambers separated by a polyester membrane under shaker force | BMEC in the lower chamber (vascular side) and astrocytes in the upper chamber (neural side) | High- throughput investigations of AAV crossing efficiency in the BBB | It needs to be improved through further experiments | 66 | |
| BBB organoids (several days to weeks) | Organoids | Primary human astrocytes, human brain microvascular pericytes and human cerebral microvascular ECs | Recapitulation of cellular heterogeneity, structure and function of the primary tissue | Fluid control not in account; complex systems, technically challenging, and cannot be reiterated at high throughput | 67–69 |
| 3D printing-based chip (few weeks to several months) | Organoid-on-chip | hPSC-derived pericytes and endothelial cells | Improvement of organoid vascularization process in a 3D network | Complex systems, cannot be reiterated at high throughput | 70 |
2.2 Microfluidic-integrated blood–brain barrier models
Microfluidic-integrated blood–brain barrier (BBB) models have emerged as pivotal tools in the study of the central nervous system (CNS) drug delivery and neurovascular interactions. These models aim to replicate the complex architecture and functionality of the in vivo BBB, which is essential for understanding drug transport mechanisms and the pathophysiology of neurological disorders. Recent advancements in microfluidic technology have led to the development of various configurations, including horizontal-aligned, vertical-aligned, and tubular structures, each designed to enhance the functionality and accuracy of these models.One notable approach is the horizontal-aligned microfluidic model, which features a simple design consisting of two compartments separated by micropillars with a 3 μm gap. This configuration allows for the culture of vascular endothelial cells on the apical side, effectively blocking the permeation of FITC-dextran from the apical to the basolateral side. This model not only facilitates the maintenance of shear flow conditions that mimic in vivo microvessels but also enables interactions between endothelial cells and astrocytes within the middle chamber.71 However, a limitation of this design is the restricted contact area between neuronal and vascular channels, which may affect the overall functionality of the model.
In contrast, vertically aligned microfluidic channels have been utilized to create more complex BBB systems. For instance, Wevers et al. developed a microfluidic model featuring two perpendicular flow channels and transendothelial electrical resistance (TEER) electrodes, utilizing a thin culture membrane of 10 μm. This model allows for real-time monitoring of TEER values, evaluation of drug permeability, and assessment of the cytotoxicity of CNS drug candidates, thereby providing a more dynamic and responsive platform for BBB studies.65 For example, Chung et al. developed a model incorporating two perpendicular flow channels and transendothelial electrical resistance (TEER) electrodes, allowing for real-time monitoring of barrier integrity and drug permeability.72 This model demonstrated enhanced TEER values compared to static systems, enabling simultaneous evaluation of drug cytotoxicity and permeability, which is crucial for assessing CNS drug candidates.1 Further innovations include the integration of pulsed electric fields to improve drug delivery across the BBB.55,73 These approaches underscore the potential of microfluidic models to enhance therapeutic efficacy by manipulating barrier properties.
The creation of tubular structures within microfluidic devices has also gained traction, as these structures more closely mimic the three-dimensional architecture of blood vessels in the CNS. Chung et al. developed a 3D in vitro brain microvasculature system embedded in a collagen matrix, which supports the growth of endothelial cells and facilitates their interaction with surrounding neural tissues.72 Similarly, Silvani et al. employed two-photon lithography to fabricate a 3D microtubular structure that serves as a scaffold for both vascular endothelial cells and glioblastoma cells, allowing for the study of drug transport and interaction within a more physiologically relevant environment.74 The precision offered by two-photon lithography allows for controlled manipulation of pore size and density, which is critical for optimizing cell behaviour and transport dynamics.
Recent studies have highlighted the advantages of using induced pluripotent stem cell (iPSC)-derived endothelial cells in the construction of BBB models. Linville et al., using iPSC-derived human brain microvascular endothelial cells to construct a BBB in templated type I collagen channels, have mimicked the cylindrical geometry, cell–extracellular matrix interactions, and shear flow typical of human brain post-capillary venules.75 This approach not only enhances the physiological relevance of the model but also allows for the investigation of cell–extracellular matrix interactions that are critical for maintaining the BBB integrity.
In addition to endothelial cell monolayers, the incorporation of spheroids into microfluidic platforms has been explored to better simulate the BBB's microenvironment. These spheroids, primarily composed of astrocytes, with brain endothelial cells and pericytes surrounding them, exhibit enhanced expression of tight junction proteins and improved transport regulation compared to traditional 2D models.2 The use of spheroids in microfluidic systems allows for a more realistic representation of the BBB morphology, accounting for blood flow and shear stress, which are crucial for drug testing and optimizing therapeutic designs.76
Troubleshooting in the fabrication and application of microfluidic-integrated BBB models often involves addressing issues related to cell viability, barrier integrity, and reproducibility. Wei et al. described a microfluidic platform with an integrated transparent TEER sensor that allows for continuous monitoring of barrier function, facilitating the identification of conditions that may compromise the BBB.2 Additionally, the selection of appropriate hydrogel matrices for cell culture has been shown to significantly impact the formation and maintenance of a robust BBB on chip, as highlighted by studies focusing on the interactions between endothelial cells and astrocytes within a 3D hydrogel environment.77
The applications of microfluidic-integrated BBB models extend beyond basic research; they are increasingly utilized in drug discovery and development. These models provide a platform for high-throughput screening of drug candidates, allowing researchers to evaluate drug permeability and efficacy in a controlled environment that closely resembles human physiology.78 Moreover, the ability to simulate pathological conditions, such as inflammation or ischemia, within these models enables the investigation of disease mechanisms and the testing of potential therapeutic interventions.
In conclusion, the advancements in microfluidic-integrated blood–brain barrier models represent a significant leap forward in the field of CNS research. By closely mimicking the in vivo environment of the BBB, these models provide valuable insights into drug transport mechanisms, neurovascular interactions, and the pathophysiology of neurological diseases. As fabrication techniques and methodologies continue to evolve, the potential applications of these models in drug development and personalized medicine are bound to expand, paving the way for more effective therapeutic strategies for CNS disorders.
2.3 Microfluidics-based dynamic BBB chip models
Several types of in vitro microfluidic BBB chip models are described in scientific literature, which will be explained below: (a) an early microfluidics-based BBB chip;63 (b) a microfluidics-based high-throughput BBB chip;64 (c) a gravity-driven single-cell channel high-throughput BBB chip;65 (d) a gravity-driven dual-channel high-throughput BBB chip.66 Wang et al. proposed a laminar microfluidic apparatus in which mouse endothelial cells, pericytes, and astrocytes could be co-cultured to create an in vitro 3D BBB model that strongly recapitulated the considerable transport mechanisms observed in vivo.63 However, on account of their low throughput, most single microfluidic systems cannot be used to process multiple experiments or screen large drug panels simultaneously, impeding their adoption for high-throughput applications. To answer this issue, Zakharova et al. tuned a multi-pathway microfluidic chip with eight independent reaction units, in which each individual unit can be worked on simultaneously or separately via a laminar flow effect, without extra pipetting steps. This innovative design allowed eight parallel experiments to run simultaneously in a single chip, while also improving the reproducibility of the results.64 Wevers et al. launched a technique using type I collagen hydrogels to produce tubular luminal microchannels through viscous finger patterning in which endothelial cells, astrocytes, and pericytes could be co-cultured in a collagen matrix of a two-lane or three-lane microfluidic platform that harbors 96 or 40 chips, respectively, in a 384-well plate format. The fluid migration in this plain, cheap and scalable BBB model is directed by gravity, evading the need for an unwieldy continuous perfusion syringe pump.This model can be used to evaluate passage of large biopharmaceuticals, such as therapeutic antibodies, across the BBB.65 Liu et al. created an in vitro biomimetic chip with high-throughput capabilities which was divided into two chambers separated by a polyester membrane. The BMECs have been seeded in the lower chamber to simulate the vascular side and astrocytes in the upper chamber to mimic the neural side. After inoculating cells, the chip is placed in a precision shaker so that fluid shear force simulates in vivo conditions, a fluid flow rate can be controlled by adjusting the tilt angle and oscillation speed, thus bypassing the need for a perfusion device.66Fig. 4 and 5 provides a summary of notable examples of in vitro microfluidic-integrated blood–brain barrier models discussed in the text above. Noteworthy advancement has been made in recreating BBB conditions using primary cells or induced pluripotent stem cells. Validation of this model demonstrated that iPSC-derived brain endothelial cells can be used for mechanistic investigations of antibody traversal of the BBB. In addition, one study examining human iPSC differentiation into BMECs to examine microvascular development under hypoxic conditions, in an in vitro BBB model with greater and more durable barrier function than previous models, found that no microvessels formed following the induction of differentiation under these conditions.1 The miniaturization of microfluidic chip systems thus commits to several advantages over traditional culture systems including efficiency, high throughput scalability, versatility for integrating additional components, and ease of cell manipulation, typically outperforming macroscopic systems in side-by-side comparisons. Although important progress has been made in the field of in vitro BBB modelling recently, the majority of these models are still in their early stages and will be improved through further exploration and experimentation. One problem that cannot be omitted at present is that most of these in vitro BBB chips can only simulate the basic structure of fluid channels in the cerebrovascular network. Thus, hemodynamic simulations are still at odds with the complex, multi-stage in vivo vascular network of the BBB. Hemodynamic conditions in dynamic blood–brain barrier (BBB) chip models are typically simulated using advanced microfluidic technology that closely mimics the physiological environment of the human brain's vascular system. These microfluidic chips are engineered to replicate the intricate architecture and dynamic behaviour of the BBB, enabling researchers to investigate drug transport mechanisms and the impact of various substances on the barrier's integrity. Current methodologies for simulating hemodynamics in these models include several key components: (a) microfluidic design: the architecture of the chip includes microchannels that represent blood vessels, featuring varying geometries, flow rates, and shear stress profiles to accurately simulate blood flow dynamics. This design is crucial for understanding how changes in flow conditions affect BBB function.71,79,80 (b) Endothelial cell culture: human brain endothelial cells are cultured on the chip to form a monolayer that closely resembles the BBB. These cells exhibit tight junctions akin to those found in vivo, which is essential for studying permeability and transport across the barrier.81,82 (c) Perfusion: a peristaltic or syringe pump is employed to perfuse the microchannels with a fluid that mimics blood plasma. This perfusion generates shear stress on the endothelial cells, which is vital for maintaining their physiological functions and barrier properties.79,83 (d) Dynamic conditions: the incorporation of pulsatile flow in the chip design more accurately reflects the conditions of the human circulatory system. This aspect is critical for examining how dynamic flow influences BBB integrity and functionality.1,84 (e) Measurement of hemodynamic parameters: researchers can monitor various parameters such as flow rate, shear stress, and pressure within the microchannels, facilitating real-time assessments of hemodynamic conditions.65,85 (f) Incorporation of other cell types: to better simulate the in vivo environment, additional cell types such as astrocytes, pericytes, and neurons can be integrated into the chip. This multicellular approach enhances the understanding of intercellular interactions and their contributions to BBB function.50,86 Despite the advancements in these dynamic BBB chip models, several limitations exist when compared to in vivo vascular networks: (a) scale and complexity: in vivo vascular networks are highly intricate, featuring diverse diameters, branching patterns, and complex interconnections that are challenging to replicate fully in microfluidic chips.87,88 (b) Cellular microenvironment: the in vivo environment encompasses a variety of biochemical signals, extracellular matrix components, and mechanical forces that are difficult to replicate precisely in vitro. This discrepancy can significantly influence cell behaviour and BBB permeability.80,89 (c) Dynamic biological responses: in vivo conditions involve adaptive biological responses to stimuli such as injury or inflammation, which may not be accurately modelled in static or semi-static chip systems.90,91 (d) Flow dynamics: although microfluidic chips can simulate dynamic flow, the pulsatile nature and pressure gradients of the in vivo circulatory system are complex and may not be fully replicated, potentially affecting drug interactions with the BBB.65,84 (e) Limited time frame: most chip models are utilized for short-term studies, whereas in vivo BBB dynamics can evolve over extended periods due to factors like aging, disease progression, and chronic exposure to substances.79,86 (f) Lack of immune responses: in vivo studies often involve intricate immune responses that are difficult to model in vitro. The interaction of immune cells with the BBB can significantly influence its function and integrity.89,91 In conclusion, while dynamic blood–brain barrier chip models provide valuable insights into BBB function and hemodynamics, researchers must consider these limitations when interpreting results and translating findings to in vivo conditions.
2.4 BBB-organoid models
Research focus has recently started shifting toward more complicated 3D biological systems, enabling greater predictability in preclinical in vitro organoid cultures. BBB organoids are BBB components cultured under low-adhesion conditions which then self-assemble into multicellular structures that recapitulate the cellular heterogeneity, structure, and functions of the primary tissues at the BBB. Capillary networks with surrounding lumen and BM can be formed in spherical models prepared from rat or mouse cortical tissue, although this structure is transient.68 However, these capillary networks have yet to be constructed with human brain endothelial cells. Furthermore, these models typically do not take fluid control into account, making BBB-related transport analyses potentially challenging. Nevertheless, spherical models are well-established as effective methods for screening peptide penetration of the BBB at small scale. In addition, Eilenberger and colleagues developed a microfluidic multi-sized sphere array capable of highly reproducible, high throughput 3D multicellular sphere culture (Fig. 6A). This system enables the repetitive generation of approximately 90 spheres of different sizes on a single chip, with optional gravity driven perfusion.69 Currently, multicellular spheres generated by available 3D culture methods vary in size, shape, and complexity, thus posing a challenge for standardizing data output and analysis. Recent research on blood–brain barrier (BBB) organoids-on-chip has emerged as a pivotal area in biomedical engineering and neuroscience, providing a sophisticated platform for studying the BBB's structure, function, and its implications in drug delivery and disease modeling. These organoids, often derived from human pluripotent stem cells (hPSCs), are integrated into microfluidic systems that mimic the physiological conditions of the human brain, enabling researchers to explore the complex interactions between neural and vascular components. One of the primary advantages of using organoids-on-chip is their ability to closely replicate the in vivo environment of the BBB. For instance, Cho et al. demonstrated that microfluidic devices incorporating brain extracellular matrix components significantly enhance the structural and functional maturation of human brain organoids, leading to improved neuronal differentiation and functionality over extended culture periods.92 This advancement is crucial for developing more accurate models that can simulate human physiological responses, particularly in the context of neurological diseases and drug testing. The applications of BBB organoids-on-chip are diverse and impactful. Wang's research highlights the potential of using cerebral organoids to model breast cancer brain metastasis, showcasing how these systems can be utilized to understand cancer progression and therapeutic responses in the brain.93 Additionally, Sun et al. have successfully generated vascularized brain organoids, which include microglial cells and endothelial cells, to study neurovascular interactions and the immune response within the brain.94 Such models are invaluable for investigating the pathophysiology of various neurological disorders and for screening potential therapeutics. Despite their advantages, there are notable challenges associated with BBB organoids-on-chip. One significant issue is the limited maturity and functionality of organoids compared to native tissues. As highlighted by Martinelli et al., current organoid systems often exhibit a fetal-like gene expression profile, which may not accurately reflect adult brain physiology.95 Furthermore, the complexity of these models can lead to variability in organoid formation and function, complicating data interpretation and reproducibility in experiments.95Moreover, troubleshooting these systems can be intricate. Nzou et al. pointed out that while organoids can effectively model the BBB, factors such as hypoxia and nutrient diffusion can adversely affect their integrity and functionality.96 Researchers must carefully optimize culture conditions, including oxygen levels and nutrient supply, to maintain the viability and performance of these organoids. For instance, the integration of shear stress in organ-on-chip designs has been shown to promote vascularization and enhance the physiological relevance of the models.97
The advantages of BBB organoids-on-chip are compelling. They provide a more accurate representation of human biology than traditional 2D cultures, allowing for better predictions of drug permeability and efficacy. Bergmann et al. emphasized that these organoids can be used to investigate the permeability of central nervous system (CNS) therapeutics, offering insights into how drugs interact with the BBB.98 Additionally, the ability to perform high-throughput screening in these models can accelerate drug discovery processes, as demonstrated by Gazerani's work on migraine therapies.99
However, the high costs associated with maintaining organoid cultures and the technical challenges of measuring outcomes in 3D systems remain significant drawbacks. As noted by Luo et al., the complexity of these models can hinder the assessment of drug delivery mechanisms and the evaluation of therapeutic efficacy.100 Furthermore, the need for specialized equipment and expertise can limit the accessibility of these technologies to many research laboratories.
In conclusion, BBB organoids-on-chip represent a transformative approach to studying the blood–brain barrier (BBB) and its implications in health and disease. By accurately mimicking the complex architecture and cellular composition of the BBB, these organoid models provide a more physiologically relevant platform for investigating fundamental biological processes and the pathophysiology of neurological disorders. This innovative technology not only enhances our understanding of the dynamic interactions between brain endothelial cells, pericytes, astrocytes, and neurons but also opens new avenues for exploring how these interactions can be modulated in various disease contexts. Furthermore, the potential for high-throughput drug screening allows researchers to evaluate the efficacy and safety of therapeutic compounds in a more targeted manner, potentially accelerating the drug development pipeline for conditions such as Alzheimer's disease, multiple sclerosis, and brain tumors. However, the full realization of BBB organoids-on-chip in biomedical research is not without its challenges. Key issues such as limited maturity of the organoid systems, which can affect their functional characteristics and response to pharmacological agents, must be systematically addressed. Variability in organoid generation and performance also poses a significant hurdle, as differences in cellular composition and microenvironment can lead to inconsistent results across experiments. Moreover, the high costs associated with the development and maintenance of organoid cultures, along with the need for specialized equipment and expertise, can limit accessibility for many research laboratories. Overcoming these challenges will require collaborative efforts across disciplines, including advances in biomaterials, microfabrication techniques, and a deeper understanding of the developmental biology of the BBB. As we continue to refine these organoid models and integrate them into broader research frameworks, they have the promise of not only advancing our fundamental knowledge of the brain's protective barriers but also paving the way for novel therapeutic strategies that could significantly improve patient outcomes in a range of neurological disorders.
The vascularization of blood–brain barrier (BBB) organoids-on-chip represents a significant advancement in organoid technology, integrating bioengineering principles to create more physiologically relevant models for studying neurovascular interactions and diseases. The development of vascularized brain organoids is crucial due to the inherent limitations of traditional organoid models, particularly the lack of a vascular network that restricts nutrient and oxygen delivery, leading to cell death and abnormal differentiation.95,101 Recent studies have demonstrated various strategies to achieve vascularization in brain organoids. For instance, the fusion of human umbilical vein endothelial cells (HUVECs) with brain organoids has been shown to generate vascularized structures that mimic the in vivo environment.102,103 Additionally, the expression of the transcription factor ETV2 in pluripotent stem cells (PSCs) has been utilized to induce endothelial cell differentiation within the organoids, facilitating the formation of a vascular network.103,104 These approaches not only enhance the structural integrity of the organoids but also promote the establishment of functional BBB characteristics, which are essential for studying drug delivery and neurovascular dynamics.105 The integration of microfluidic chip technology further enhances the study of vascularized organoids. By employing a 3D spheroid-on-a-chip platform, researchers have been able to create a controlled environment that supports the growth of vascularized neural stem cell spheroids, thereby improving nutrient exchange and cellular interactions.101 This bioengineering approach allows for the precise manipulation of the microenvironment, enabling the study of cellular responses to various stimuli and the investigation of disease mechanisms at a higher resolution.36,106 Moreover, hydrogel-based patterned microcavity arrays have been developed to facilitate the self-assembly of BBB organoids, providing a scalable and reproducible method for generating these complex structures.36,98 The functional assessment of these vascularized organoids has revealed their potential to recapitulate key properties of the BBB, including permeability and transport mechanisms.36,98 For example, studies have shown that the incorporation of astrocytes and pericytes alongside endothelial cells in organoid cultures enhances the expression of tight junction proteins, which are critical for maintaining BBB integrity.107,108 To date, efforts to generate functional vascularized human brain organoids show varying degrees of success.100,109–111 The main reason is that the vasculature generated using these approaches remains non-perfusable as these models do not possess any accessible sites to allow entry into the vasculature. To address the limitations, a recent focus has shifted to the potential of integrating organoid technology and bioengineering.112 Various research groups have utilized on-chip technologies to cultivate brain organoids. For instance, Karzburn et al. grew a brain organoid within a confined compartment of a microfluidic device to explore the mechanisms behind brain wrinkling.113 By limiting the organoid's height within this closed chamber, they were able to conduct in situ whole-organoid fluorescence real-time imaging, a feat difficult to achieve with traditional dish models. Additionally, microfluidics has been employed to enhance the reproducibility and reduce the size variability of brain organoids. Ao et al. developed a comprehensive assembly method for culturing brain organoids entirely within a single microfluidic chip, minimizing disturbances throughout the process.114 This setup not only constrained the organoids to maintain a consistent size of 2 μm but also allowed exposure to atmospheric oxygen, preventing the formation of necrotic cores. The microfluidic device is designed with a bottom-layer perfusable chamber that delivers medium to the upper layer of brain organoids via a polytetrafluoroethylene-coated wire mesh. While this hydrophobic mesh facilitates the formation of embryoid bodies without surface adhesion, it may hinder real-time imaging of the organoids within the device.114 Meanwhile, Seiler et al. created an automated on-chip cell feeding platform that regulates the flow rate and feeding schedule to sustain brain organoid cultures while minimizing the impact of uncontrolled variables during medium changes.115 To further enhance nutrient absorption and facilitate the development of extended neuroepithelial-like zones, Romero-Morales et al. introduced a miniaturized spinner, Spin∞, which supports the long-term culture of brain organoids.116 In another study, Wang et al. explored the impact of prenatal nicotine exposure on a brain organoid through perfusion flow.117 They focused on characterizing the maturity and functionality of the organoid at approximately one month of age, which represents early fetal brain development characterized by immature neurons and a significant absence of oligodendrocytes.118,119 Notably, the effects of perfusion flow on neuronal activities, such as synchronized bursts and spikes, were only observable in organoids older than two months.118 In fact, most established brain organoid protocols allow for maturation periods of up to one year to better replicate later stages of fetal brain development.120 The importance of the culture duration has been thoroughly discussed by Gopurappilly et al.121 Likewise, Ao et al. investigated the infiltration of young and old monocytes into a 45-day-old brain organoid using a 3D-printed microdevice.122 Extending the culture of brain organoids to reflect aging phenotypes is essential for enhancing our understanding of brain aging. In their study, the researchers confined the brain organoid within their platform to promote the development of a pancake-shaped structure, aimed at reducing inner core necrosis. However, it remains unclear whether perfusion flow can effectively address the issue of necrosis in late-stage brain organoids. Most critically, none of these models incorporate vascular structures. Furthermore, the ability of these organoids to respond to immune stimuli, as evidenced by the active engagement of microglial cells, underscores their relevance in modeling neuroinflammatory conditions and other neurological disorders.100,123 In summary, the vascularization of BBB organoids-on-chip represents a promising Frontier in the rapidly evolving field of organoid technology. This innovative approach combines cutting-edge bioengineering techniques with advanced cell culture methods to create models that more accurately reflect the complex architecture and functionality of the human brain. By incorporating vascular networks into these organoids, researchers can simulate the intricate interactions between neuronal cells and the BBB, which is crucial for maintaining homeostasis and protecting the brain from harmful substances. These advancements address the significant limitations associated with traditional organoid systems, such as their inability to mimic the dynamic and interactive environment of the brain effectively, and also open avenues for the development of novel therapeutic strategies. Furthermore, they enhance our understanding of neurovascular dynamics in both health and disease, providing insights into neurodegenerative diseases and brain tumors. As researchers continue to refine these organoid models, we anticipate that they will serve as invaluable tools for drug testing, disease modeling, and personalized medicine, ultimately leading to improved outcomes for patients suffering from a range of neurological disorders. The implications for future research and clinical applications are profound and far-reaching. Table 2 summarizes the fabrication processes crucial for developing robust microfluidics-based organoid-on-chip models that accurately represent the BBB and its interactions with various biological components. The ability to manipulate these systems enables researchers to delve into the intricate dynamics of drug delivery and the multifaceted mechanisms of disease. This manipulation is not merely a technical capability, it represents a significant advancement in our understanding of biomedical processes and therapeutic strategies.
| Fabrication process | Description | Ref. |
|---|---|---|
| Microfluidic device design | Development of microfluidic devices that allow for precise control of fluid flow and environmental conditions. | 1, 50 and 92 |
| Organoid culture | Culturing organoids derived from human pluripotent stem cells (hPSCs) in a 3D environment to mimic in vivo conditions | 99 and 102 |
| Vascularization techniques | Techniques such as embedding endothelial cells within organoids to create vascular networks that enhance nutrient delivery. | 100, 104 and 106 |
| Integration of extracellular matrix | Incorporating ECM components to support cell growth and differentiation, enhancing the physiological relevance of organoids. | 92 and 95 |
| Perfusion systems | Implementing perfusion systems to facilitate nutrient and oxygen delivery, mimicking blood flow in vivo. | 96 and 124 |
| High-throughput screening | Utilizing microfluidic platforms for high-throughput screening of drug permeability and efficacy. | 96 and 98 |
| Real-time imaging | Employing imaging techniques to monitor organoid development and functionality in real-time within microfluidic devices. | 96, 115 and 124 |
| Optimization of culture conditions | Fine-tuning biochemical and biomechanical factors to enhance organoid maturity and functionality. | 97 and 101 |
3 Microfluidic platforms for BBB in vitro modeling: materials and techniques of fabrication
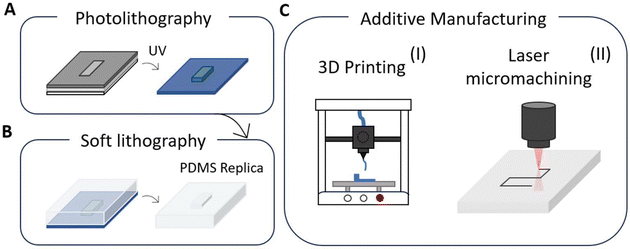
Over the years, a variety of materials have been tested for the manufacturing of microfluidic devices, each with its own set of advantages. Materials are chosen based on parameters such as detection method, device function, reusability, and disposability.125 Silicon, polymers, and glass are the materials most used in microfluidic devices production for the detection and quantification of biomarkers.126,127 Silicon is highly appreciated for its well-defined surface qualities, ease of modification, chemical compatibility, and thermal stability.49,128 However, its high elastic modulus (approximately 130–180 GPa) impedes integration with valves and pumps, restricting its use in biomarker analysis.129 Glass is another ideal material due to its optical clarity, chemical inertness, thermal stability, and ease of reuse after basic cleaning methods. It is optimal for optical detection and enables the simultaneous detection of many biomarkers, including cancer indicators.130,131 However, to complete microchip manufacturing, it is necessary to handle corrosive elements and use heat bonding processes.125 Polymers, particularly polydimethylsiloxane (PDMS), are frequently used due to their flexibility, low-cost manufacturing, and biocompatibility.132 PDMS provides advanced device designs with incorporated micro-valves, gas permeability, and multilayer channels, making it appropriate for a wide range of applications, including cancer biomarker detection and cell culture investigations.128,133 However, the gas permeability makes it sensitive to organic solvents and absorption of hydrophobic compounds, which may affect the precision of test results.127,132,133 Lastly, microfluidic paper-based devices have proven to be efficient for the extraction and detection of numerous biomarkers that facilitate point-of-care applications. In fact, they are particularly used in diagnostic applications, because of their low cost, ease of disposal, simple storage, and portability.134,135 In the framework of microfluidic device fabrication, the most used techniques are lithographic based methods, additive manufacturing approaches and more recently laser based processes (Fig. 7). The choice of the manufacturing technique is closely related to the platform material that will be used and to the different functionalities that the platform should integrate. Actually, in the most advanced devices a hybrid approach is followed, using different techniques for the complete development of a microfluidic platform.Soft lithography is the most widely utilized technique for fabricating microfluidic devices for blood–brain barrier in vitro modeling (Fig. 7B).42,127,131,136,137 It is a versatile technology that can be used with a variety of substrates, including flexible and curved surfaces, achieving resolutions at the micro and nano scale. In contrast to standard lithography which depends on stiff photomasks, soft lithography uses flexible elastomeric materials like PDMS, to create patterns and structures with remarkable fidelity and resolution. Soft lithography is intrinsically a multistep process, that involves firstly the manufacturing of a master mold, by other lithographic methods.63,69,138 The PDMS is then cast onto the master mold to replicate the desired pattern through techniques like microcontact printing, embossing, or injection molding (Fig. 7A and B). Overall, this technique is a high-throughput, cost-effective, and adaptable technology that requires a simple setup for microdevices replication.139 However, it also presents some drawbacks, such as residual stress and shrinkage during the curing process, deformation of the soft material, repeatability concerns, and biocompatibility issues due to residual materials.138,140 Moreover, the need of a master mold for PDMS casting reduces the versatility of the technique when dealing with the optimization of the device scheme. In the development of microfluidic blood–brain barrier models that closely emulate the in vivo environment, until now a great effort has been devoted to the integration of membranes and electrodes inside the same chip, to mimic transportation and measuring resistance during flow. In the early works, commercial membranes and electrodes were directly integrated into the fabricated microfluidic networks. More recently, a more sophisticated approach has been followed, introducing the fabrication of membranes and electrodes in the manufacturing of the lab on a chip devices.
The device developed by Booth et al., is a pioneering microfluidic device for studies of the BBB that integrates different functionalities.42 For instance, they produced a chip by a multi-step process involving the bonding of four patterned PDMS sub-layers, two electrode layers, and a polycarbonate membrane. For the channel feature layers, PDMS was spin-coated, cured, and laser-patterned to form 200 μm thick sheets. Glass slides were embedded in PDMS and cured, followed by the creation of input/output holes. A polycarbonate membrane with 400 nm pores was prepared and cut, and all components were bonded using a PDMS-toluene mixture. Electrical connections were made by bonding copper wires with silver epoxy. Here, the fabrication process allows for precise and integrated construction of the BBB microchip, enabling high functionality and resolution and offering high control over design and performance, crucial for mimicking the blood–brain barrier in vitro. The Takayama group produced a 3D microfluidic device that replicates the selective permeability of the BBB by creating triculture and bi-culture models.63 They accomplished this also through a multilayer PDMS channel encased in a porous membrane. Their fabrication process includes the integration of commercially available wire electrodes, which eliminates the need for specialized microelectrode fabrication. Moreover, the compatibility with microfluidic environments allows for experiments under controlled flow conditions that closely resemble physiological settings. However, the system faces challenges, such as the possibility of leakage due to difficulties in achieving reliable bonding of porous membranes within the layered microfluidic structure,140 as well as material limitations associated with PDMS, such as its tendency to absorb small hydrophobic molecules, which may affect experimental results.141 The approach of Partyka et al. constitutes an alternative for the integration of a membrane in a PDMS device by directly filling a central chamber with a hydrogel. With fine control of pressure along the lateral microfluidic channels they mimic transport across the brain microvasculature, promoted by blood flow.58
With benefits such as easy electronic component integration and design versatility, the devices produced by soft lithography can be further enriched using techniques such as photolithography or micro-nano contact printing. Both techniques are frequently complementary in microfluidic device development, the former one to produce high-resolution master molds and micro/nano contact printing for quick functionalization or integration of features important for biological or chemical applications.
The photolithographic process is also a multistep process (Fig. 7A). It begins with the deposition of a light-sensitive material, known as a photoresist, onto a substrate, followed by exposure to ultraviolet (UV) light through a mask that defines the desired pattern. The exposed areas of the photoresist undergo a chemical transformation, allowing the material to be developed and resulting in the formation of a precise patterned layer, facilitating the creation of micro- and nanoscale features. Photolithography therefore allows for the creation of highly precise microstructures with outstanding resolution up to 5 μm and scalability, making it perfect for generating master molds for soft lithography in microfluidics.131,139 However, it requires complex and expensive equipment, as well as cleanroom facilities, which may limit its applicability to smaller-scale laboratories.142
Micro/nano contact printing, on the other hand, allows for the direct transfer of designs onto substrates utilizing elastomeric stamps, which is simple and cost-effective. This method is very useful for functionalizing surfaces with biomolecules or creating patterns on flexible substrates. Therefore, these techniques are frequently complementary in microfluidic device development producing high-resolution master molds and micro/nano contact printing allowing for quick functionalization or integration of features important for biological or chemical applications. The combination of different manufacturing technologies offers a scalable, rapid, and cost-effective solution for producing high-yield porous PDMS membranes. By combining soft lithography, photolithography and reactive ion etching it is possible to create PDMS membranes with micrometer size pores, facilitating high-throughput drug permeability testing in a controlled BBB model.
For the fabrication of such membranes (Fig. 8A), a silicon wafer is coated with sacrificial photoresist layers that contains a pattern of sub-micrometer column arrays, produced by conventional photolithography. A PDMS solution is then spin-coated over the photoresist structures and cured to form a solid membrane. Reactive ion etching is then employed to create through-holes by selectively removing the PDMS above the photoresist columns. Finally, the sacrificial photoresist layers are dissolved in acetone, yielding the free-standing PDMS membrane.143 For example, in the work by Zahkarova et al.138 a thin PDMS membrane with pores that are 5 μm in diameter is embedded between the two layers of a PDMS-based microchannel chip, arriving even to test transport across eight parallel channels inside the same microfluidic device (Fig. 8B). Recently Ceccarelli et al., presented an advanced microfluidic BBB-on-a-chip device with integrated thin-film electrodes for non-invasive, real-time electrochemical impedance spectroscopy (EIS) monitoring of BBB integrity.144 The fabrication process involved the creation of a microfluidic system and integrated microelectrodes through photolithography and soft-lithography techniques. A 50 μm SU8-50 photoresist layer was spin-coated onto a silicon wafer, soft-baked, UV-exposed, post-baked, and developed to form the master mold. This master was used for replica molding with a PDMS mixture (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 elastomer to crosslinker ratio), which was cast, degassed, cured, and micro-milled to form inlets and outlets. The microelectrodes instead, were fabricated by photolithography on oxygen plasma-treated coverslips, using a 3.5 μm AZ LNR-003 photoresist layer. Following UV exposure, development, and baking, the lift-off process removed excess photoresist, leaving the electrode pattern. Finally, the PDMS microfluidic device was bonded to the electrode substrate using oxygen plasma and baked to complete the BBB-on-a-chip assembly (Fig. 8C). This method enables precise integration of microfluidics and electrodes providing a powerful tool for central nervous system (CNS) drug testing, disease modeling, and personalized medicine applications. For several years now, the trend to include more functionalities in microfluidic devices, while maintaining their compactness, has triggered the use of novel processing techniques, such as additive manufacturing (AM) and laser-based techniques. Additive manufacturing (AM) techniques rely on the sequential addition of materials to fabricate 3D structures, offering significant advantages over traditional subtractive methods (Fig. 7C).139 Many AM techniques are used to fabricate microfluidic devices for biomedical applications, depending on their intended use and materials. These can be classified into stimulus-triggered AM and deposition-based AM, each offering distinct advantages and drawbacks. Stimulus-triggered AM, which relies on external triggers like light or heat to shape materials, provides high processing speeds and resolution but is limited to single-material employment and requires post-fabrication functionalization. In contrast, deposition-based AM enables the direct deposition of materials, potentially including functional materials or particles. This method is typically slower and has lower resolution but supports more complex designs and allows direct 3D printing (Fig. 7C(II)). 3D printing techniques such as fuse deposition molding (FDM), digital light processing (DLP), and in particular, inkjet printing have grown in prominence for producing lab-on-a-chip devices because of their cost-effectiveness and efficiency (Fig. 6B).131,145,146 Inkjet printing enables the deposition of conductive or hydrophobic materials such as alkyl ketone dimers and polystyrene, resulting in microfluidic devices with excellent resolution, repeatability, and speed that do not require masks or extensive post-processing. However, the size of the nozzle, the porosity of the material, and the properties of the ink all have an impact on resolution.136,145 Considering laser-based techniques (Fig. 7C(II)), laser micromachining is one of the most often used for structuring materials such as glass, silicon, and thin foil.125 Laser micromachining, in fact, is based on tightly focused laser beams that provides confined energy and enable material processing at the micron and sub-micron scale allowing the rapid etching of micro-patterns. The main advantage of laser micromachining techniques is the high spatial resolution achieved enhancing resolution and precision meanwhile allowing the incorporation of several microfluidics and sensing components into the same system.139 For example, femtosecond laser irradiation and chemical etching are broadly used to create optically accessible glass lab-on-a-chip with embedded microfluidic channels and microsensors.147–149 As previously stated, glass' inertness to biological molecules and transparency make it a suitable material for complex microfluidics platforms that enable biomarker identification by fluorescence microscopy or fiber-based sensing.125 We envisage that in the future these novel manufacturing techniques will also be implemented in the fabrication of microfluidic BBB in vitro modeling, as they bring the possibility of easily integrating new functionalities in those lab-on-chip platforms.
1 elastomer to crosslinker ratio), which was cast, degassed, cured, and micro-milled to form inlets and outlets. The microelectrodes instead, were fabricated by photolithography on oxygen plasma-treated coverslips, using a 3.5 μm AZ LNR-003 photoresist layer. Following UV exposure, development, and baking, the lift-off process removed excess photoresist, leaving the electrode pattern. Finally, the PDMS microfluidic device was bonded to the electrode substrate using oxygen plasma and baked to complete the BBB-on-a-chip assembly (Fig. 8C). This method enables precise integration of microfluidics and electrodes providing a powerful tool for central nervous system (CNS) drug testing, disease modeling, and personalized medicine applications. For several years now, the trend to include more functionalities in microfluidic devices, while maintaining their compactness, has triggered the use of novel processing techniques, such as additive manufacturing (AM) and laser-based techniques. Additive manufacturing (AM) techniques rely on the sequential addition of materials to fabricate 3D structures, offering significant advantages over traditional subtractive methods (Fig. 7C).139 Many AM techniques are used to fabricate microfluidic devices for biomedical applications, depending on their intended use and materials. These can be classified into stimulus-triggered AM and deposition-based AM, each offering distinct advantages and drawbacks. Stimulus-triggered AM, which relies on external triggers like light or heat to shape materials, provides high processing speeds and resolution but is limited to single-material employment and requires post-fabrication functionalization. In contrast, deposition-based AM enables the direct deposition of materials, potentially including functional materials or particles. This method is typically slower and has lower resolution but supports more complex designs and allows direct 3D printing (Fig. 7C(II)). 3D printing techniques such as fuse deposition molding (FDM), digital light processing (DLP), and in particular, inkjet printing have grown in prominence for producing lab-on-a-chip devices because of their cost-effectiveness and efficiency (Fig. 6B).131,145,146 Inkjet printing enables the deposition of conductive or hydrophobic materials such as alkyl ketone dimers and polystyrene, resulting in microfluidic devices with excellent resolution, repeatability, and speed that do not require masks or extensive post-processing. However, the size of the nozzle, the porosity of the material, and the properties of the ink all have an impact on resolution.136,145 Considering laser-based techniques (Fig. 7C(II)), laser micromachining is one of the most often used for structuring materials such as glass, silicon, and thin foil.125 Laser micromachining, in fact, is based on tightly focused laser beams that provides confined energy and enable material processing at the micron and sub-micron scale allowing the rapid etching of micro-patterns. The main advantage of laser micromachining techniques is the high spatial resolution achieved enhancing resolution and precision meanwhile allowing the incorporation of several microfluidics and sensing components into the same system.139 For example, femtosecond laser irradiation and chemical etching are broadly used to create optically accessible glass lab-on-a-chip with embedded microfluidic channels and microsensors.147–149 As previously stated, glass' inertness to biological molecules and transparency make it a suitable material for complex microfluidics platforms that enable biomarker identification by fluorescence microscopy or fiber-based sensing.125 We envisage that in the future these novel manufacturing techniques will also be implemented in the fabrication of microfluidic BBB in vitro modeling, as they bring the possibility of easily integrating new functionalities in those lab-on-chip platforms.
4 Application of in vitro blood–brain barrier microchips
Some lately evolved in vitro BBB microarray models have a vast range of applications, comprising drug discovery and high-throughput screening, assembly of disease models and continuous measurement in BBB models.150 Highly selective permeability of the BBB also poses a problem for drug delivery to the brain, and may be the primary hindrance, for some drugs, blocking the treatment of neurological diseases. In designing new drugs, increasing lipophilicity affects their water solubility, which can negatively impact their delivery. Thus, considerable research efforts are focused on developing specialized trans-BBB carriers or vehicles for potentially effective drug leads already in existence. In addition, the concentration of drugs successfully delivered to the brain cannot be directly measured in humans, posing a major challenge for determining their efficacy or effects within the CNS. Furthermore, an absence of reliable markers for some CNS diseases restricts the ability to design and assess candidate drugs, which could be advanced through assays using in vitro models of the BBB. Similarly, a poor knowledge of the pathogenic mechanism in many CNS diseases can also severely limit the development of effective pharmacological interventions. As a result, it is fundamental to screen for drugs that can cross the BBB using models that are both trustworthy and sufficiently adaptable to accommodate quantitative investigations of drug toxicity and transport mechanisms. In vitro BBB microarray models have been used to assess drug/compound toxicity, BBB permeation rates, and intracellular molecular transport mechanisms.151 In the last decade, in vitro BBB chips were applied to screen drugs for treating CNS diseases. Hou and co-workers employed a BBB model to screen nanoparticles for use in epilepsy treatments.152 Shao's team designed and constructed a microfluidic 3D BBB model for dynamic culture of human cerebral microvascular endothelial cells (hCMEC/D3) to evaluate the effectiveness of Sunitinib penetration into the brain.62 It is also noteworthy that sensors can be integrated into the in vitro BBB microarray model to monitor culture conditions and fluid dynamics, and also furnishing a viable strategy to high-throughput screening of brain drugs.8 Among others, Wevers et al. combined a human BBB microfluidic model with multi-unit plates to increase drug screening throughput. Throughput is indeed a limitation of BBB models, and the largest compound screen conducted by in vitro BBB models currently reported is only 384 compounds.65 It could occur that throughput can be improved by combining automated liquid handling platforms, improved miniaturization, and the application of deep learning-based image analysis. However, it should be acknowledged that the primary focus of current in vitro model development is improving the accuracy with which they mimic in vivo BBB conditions. Moreover, low throughput is a problem for the large majority of sophisticated organ chip systems. It is possible that advances in throughput for other organs may also be applied to the in vitro BBB, enabling development of urgently needed drugs for diseases of the CNS. Since in vitro BBB chips are typically fabricated by microengineering methods, it is also possible to combine in situ analytical tools to improve temporal resolution and enable continuous measurement with faster readouts, thereby capturing more information more quickly from the model organ or tissue. More recently, electrochemical and optical sensors have also been integrated into organ-on-a-chip systems. In general, sensors generally include three main components: a receptor for event recognition of the analyte or cellular event, a transducer that converts the event into a signal, and a detector coupled with a processing system that provides the data readout. Among sensor applications, label-free transepithelial/trans-endothelial electrical resistance (TEER) is generally used for quantitative assessment of BBB integrity. In addition, BBB chips can be equipped with a light-addressable potentiometric sensor (LAPS) for live detection of changes in pH changes that reflect cellular metabolic activity, thus providing a convenient means of assessing the cell-type specificity of a candidate drug.153 Beyond electrical sensors, implanting optical biosensors in a microfluidic wafer represents a major innovation that enables the development of automated, high-performance in situ monitoring in drug screens for simultaneous detection of multiple analytes.154 Nowadays, different chip-based models of the human BBB have been grown to reproduce neurovascular conditions suited for in vitro study of neurological diseases.155 Amongst these, Parkinson's disease (PD) is a gradual neurodegenerative disorder marked by a shortfall of dopaminergic neurons, which is the cause of defects in motor function. On main reason for the current limits in PD treatment options is poor permeability of the BBB by potential drugs. Cai et al. developed an in vitro model of the BBB using co-culture of rat endothelial cells and glial cells. This model can be used to investigate BBB dysfunction in the pathogenesis and progression of PD, as well as for screening candidate drugs.156 Connected studies have identified inflammatory processes closely linked to various neurodegenerative pathways, thus highlighting the informative value of simulating neuroinflammatory states in microfluidic neurovascular models.91,157–159 To explore the role of cerebral vasculature in Alzheimer's disease, Shin et al. developed a physiologically relevant microfluidic 3D model that recapitulates several key aspects of BBB dysfunction in AD through culture of human neural cells. This tightly controlled platform can be used to investigate BBB function and screen for drugs that can penetrate the BBB to access neural tissues.160 In addition to neurodegenerative diseases, advances in in vitro BBB microarray models have enabled the establishment of pathological tumor models related to the BBB for exhaustive study of treatments targeting brain tumors, such as glioblastoma, glioma,161 and brain metastases by human lung cancer cells (A549), breast cancer cells (MDA-MB-231), melanoma (M624), or liver cancer cells (BEL-7402).162Table 3 summarizes the diverse key applications of in vitro BBB microchips, highlighting their significance in drug development, disease modeling, and continuous monitoring in neuroscience research with description and examples/technologies discussed in the text.
| Application area | Description | Examples/technologies |
|---|---|---|
| Drug discovery | Screening various pharmaceutical compounds for their efficacy in successfully crossing the blood–brain barrier is crucial for developing effective treatments for central nervous system (CNS) diseases | Techniques include microfluidic models,163,164 automated liquid handling165,166 and integration of sensors for monitoring.164,167,168 |
| High-throughput screening | Utilizing advanced microfluidic models allows researchers to significantly increase the number of compounds tested simultaneously, thereby enhancing the efficiency and accuracy of experimental drug discovery processes. | Combination of a human BBB microfluidics model with multi-unit plates to increase drug screening throughput65 |
| Disease modeling | Creating models to study neurodegenerative diseases like Parkinson's and Alzheimer's, and to investigate BBB dysfunction. | Co-culture systems for neurodegenerative diseases and microfluidic 3D models to study BBB dysfunction.155,156,160,169 |
| Continuous measurement | Integrating advanced sensors for comprehensive real-time monitoring of cell behavior, cellular integrity, and the effects of various drug treatments in blood–brain barrier models is vital for research. | Label-free TEER sensors, light-addressable potentiometric sensors (LAPS),153 and optical biosensors for analyte detection.154 |
| Toxicity assessment | Evaluating drug and compound toxicity using sophisticated in vitro blood–brain barrier microarray models provides crucial insights into safety and efficacy for potential therapeutic applications in humans. | In vitro BBB microarray models have been used to assess drug/compound toxicity, BBB permeation rates, and intracellular molecular transport mechanisms151 |
| Permeation rate studies | Assessing BBB permeation rates for various compounds and understanding transport mechanisms. | In vitro BBB microarray models have been used to assess drug/compound toxicity, BBB permeation rates, and intracellular molecular transport mechanisms151 |
| Drug delivery mechanism research | Investigating specialized trans-BBB carriers and mechanisms is crucial for enabling effective and targeted drug delivery directly to the brain, enhancing therapeutic outcomes. | In vitro BBB microarray models have been used to assess drug/compound toxicity, BBB permeation rates, and intracellular molecular transport mechanisms151 |
| Sensor integration | Fine-tuning various biochemical and biomechanical factors is essential to significantly enhance organoid maturity and overall functionality for advanced research applications. | Organ-on-chip systems integrate different sensors153,154 |
| Tumor models | Establishing pathological tumor models to study treatments for brain tumors like glioblastoma and glioma. | A predictive microfluidic model of human glioblastoma to assess trafficking of blood–brain barrier-penetrant nanoparticles161 |
| Neuroinflammatory state simulation | Simulating neuroinflammatory conditions to understand their impact on drug permeability and disease progression. | Inflammatory processes linked to various neurodegenerative pathways can be simulated in microfluidic neurovascular models91,157–159 |
5 Sensors integrated in BBB-on-a-chip systems (BBBoCs)
Although integrated and/or modular biosensors in BBB organ-on-chip have the potential to monitor agents related to physiological and metabolic functions and BBB pathologies, the majority of these sensors detect electrical or electrochemical signals, because these sensors are easy to incorporate in the in vitro models. Furthermore, there are multiple uses of sensors and biosensors for assessing metabolic activity in organ-on-a-chip platforms. Various types of sensors, including amperometric, potentiometric, electrochemical, and optical sensors, operate based on different principles. These sensors are able to track levels of oxygen, pH in the culture solution, and specific metabolites such as glucose or lactate, in addition to identifying biomarker proteins or pathogens.170 Still, the use of sensors in BBB organ-on-chip devices remains limited, with space constraints possibly contributing to this scarcity. Considering this, sensors for BBB organ-on-chip can be designed either integrated or modularly. The integration of sensors into BBB chip models offers various benefits such as convenient sampling, rapid and efficient data collection, immediate evaluation, and versatility in imaging.171 Using biosensors in sampling ensures the BBB chip model's integrity is maintained, provides consistency in experiments, and saves time.In this overview, we discuss existing integrated sensors, focusing the discussion on chemical detection, for evaluating BBB models through electrochemical sensors, chemical sensors using MIPs and metabolomic detection, in terms of internal and secreted molecules (metabolic profiling). To effectively convey the information regarding the sensors integrated into the BBB-on-a-chip system (BBBoCs), we will create comprehensive summarized tables that highlight key aspects of the sensors, their types, functions, and applications. This will help in visualizing the diverse range of sensors and their functionalities within the context of BBB research (Table 4 and Fig. 9).
| Sensor type | Evaluation principles | Parameters | Applications | Functions | Benefits |
|---|---|---|---|---|---|
| Electrochemical | Amperometric, potentiometric, conductometric, impedimetric | TEER, pH, glucose, lactate, O2 levels | Monitoring BBB integrity, metabolic and electrophysiological activity172–174 | Converts analyte interactions into electrical signals | High sensitivity, real-time monitoring and easy integration |
| Evaluating barrier integrity | Quantitative assessment of tight junction dynamics | ||||
| Optical | Absorption, scattering, luminescence | Oxygen, pH, cytokines, insulin, morphological changes | Cellular imaging, monitoring metabolic activities175,176 | Detects changes in light properties | Non-invasive, versatile, easily integrated |
| Non-invasive monitoring, imaging | Monitoring physiological parameters | Real-time data collection, non-invasive sampling | |||
| Chemical (MIPs) | Polymerization with template | Dopamine, β-amyloid oligomers, albumin, sialic acid, astrocyte markers | Biomarker detection,177–179 bioseparations;180 BBB leakage detection, monitoring pathological conditions | Form specific binding sites for target molecules | High selectivity, robustness, cost-effective |
| Metabolomic | Mass spectrometry, NMR spectroscopy | Metabolic profiles (lactate, glucose, etc.), biomarkers | Understanding BBB metabolism, disease markers181 | Analyses metabolic profiles | Enhanced understanding of metabolic pathways |
| Comprehensive profiling of metabolic changes | |||||
| Nanophotonic | Interferometric technology | Viral particles, bacterial infections | Rapid diagnostics in infectious diseases182 | Quantifies biomarker presence | High sensitivity, rapid results |
| Mechanical strain | Piezoelectric or resistive principles | Tissue deformation, mechanical properties | Studying tissue mechanics in BBB models1,84 | Measure mechanical strain in tissues | Real-time measurement of mechanical changes |
| Volatilomics | Gas chromatography, mass spectrometry | Volatile organic compounds (VOCs) | Insights into neurodegenerative disease pathways183 | Investigating neurotoxicity | Real-time monitoring of cellular responses to VOC exposure |
| Reduced costs and resources | |||||
| Early diagnosis and treatment strategies |
5.1 Electrochemical or optochemical sensors
Electrochemical sensors are the primary choice for sensing in microphysiological systems. Electrochemical sensors are typically divided into amperometric, potentiometric, conductometric, and impedimetric types. They offer vital data on barrier integrity, electrophysiological activity, and mechanical strain. On the other hand, electrochemical sensors utilize an electrode surface to convert interactions of an analyte into an electrical signal. Changes in current (amperometry, voltammetry), potential, impedance, or conductivity are detected depending on the measurement mode. In order to improve precision, electrochemical biosensors use biorecognition elements like antibodies, aptamers, and enzymes, which interact specifically with the target analyte. One of the most used electrochemical measurements is the transepithelial/transendothelial electrical resistance (TEER); it is a widely accepted quantitative technique to measure the integrity of tight junction dynamics in cell culture models of endothelial and epithelial monolayers.171 Various in vitro models and microfluidic organs-on-chips (OoCs) are implemented utilizing TEER measurements in some widely studied barrier models. Optical sensors detect changes in properties like absorption, scattering, and luminescence to gather information on the organ-on chip device. Due to their versatility, non-invasiveness, and easy integration, optical sensors show potential in OoC systems.184 The key requirement for incorporating optical sensors like luminescence- or absorbance-based sensors into OoC devices is the device's optical accessibility/transparency.185 Oxygen sensors are commonly used in OoC technology, but optical sensing methods have also been used for pH, secreted molecules (e.g., cytokines, insulin) and morphological changes. For the detection of small molecules, electrical and electrochemical biosensors seem to be the best choice. They are inexpensive, easily integrable into chip devices, and allow rapid sensing. Depending on the particular problem, potentiometric, amperometric, voltammetric or field-effect transistor-based biosensors can be chosen,172–174,186,187 while recent studies have also utilized the advantages of electro-chemiluminescent detection in various bioanalytical applications. In addition, both electric and optical biosensors can make use of biocompatible smart materials, such as graphene oxide derivatives,188 which can be functionalized by molecular grafting techniques.189 Results obtained by optical biosensing and immunoassay approaches, could prove that the spike protein S1 subunit could cross the human brain endothelial cell barrier efficiently. The application of biosensors utilizing the synergistic interaction of electric and optical phenomena can also be advantageous in BBB research, especially when the concentration of penetrating cells or secreted extracellular vesicles are supposed to be quantified.175 For the quick separation of extracellular vesicles according to size, simple filter-based chip modules seem to be the optimal choice.190 A fascinating new concept proposes the use of in vitro companion biomarker diagnostic devices through all the phases of drug development from preclinical tests to clinical studies in Alzheimer's disease precision medicine.191 BBBoCs with patient-derived cells and biosensors could be especially useful in such drug research and development scenarios to promote a more patient-centric approach. Other biosensors were used as an ultrasensitive and specific diagnostic tool by the early and fast detection and quantification of microRNAs for the prediction of diseases (such as cancer) with well-defined microRNA signatures.192 An innovative nanophotonic biosensor, based on bimodal waveguide (BiMW) interferometric technology, was functionalized with novel bioengineered nanobodies targeting the SARS-CoV-2 receptor-binding domain (RBD) in order to quantify viral particles in less than 20 minutes total assay time and with outstanding sensitivity.182 Using the same technology (BiMW), a promising new clinical immunosensor for the user-friendly, cost-effective and real-time microbiological analysis was produced for rapid diagnosis of bacterial infections in cirrhotic patients193 and for rapidly (less than 30 min) identify multidrug-resistance bacteria.194 Biosensors and measurement tools play an essential role in interrogating all biological systems, and are often integrated into microfluidics to enhance their sensitivity, limits of detection, and utility in precious sample processing.195 However, some unique advantages arise from integrating sensing technologies directly into OoCs.196 Typical OoC platforms provide the required mechanical support to study tissues with varying geometries in 3D, highlighting the unique advantage of such systems. In more recent and complex OoCs, this strategy has been used to measure stresses generated by activation of a neuromuscular junction on-chip,197 demonstrating the ability to make real-time and multiplexed measurements in highly realistic engineered model systems. Several recent examples have embedded bulk mechanical characterization of microengineered tissues directly into the design of the OoC device. For example, MacQueen et al. integrated on-chip strain sensors to measure tissue compression in response to deformations applied by pneumatically-actuated compressing micro-platens. This system allowed mechanical characterization of microtissues embedded in a fluidically-controlled device. These systems allow for simultaneous and high-throughput measurement of tissue mechanical properties on-chip, but do require integration of complex electromechanical components for both micro-scale actuation and measurement.198 All in all, modular networks of microfluidic BBBoCs and biosensors are expected to be developed in the near future for both basic- and applied-science utilization. Online control and measurement techniques will provide multiplex time series of data carrying independent information about the barrier properties. For the analysis of such complex data streams, artificial intelligence methods are expected to be especially advantageous.189,1905.2 Chemical sensors using MIPs
Molecularly imprinted polymers (MIPs) are formed through polymerization in the presence of a guest template that, once removed, results in the formation of hollows or indentations that are compatible in both size and function with the template and have the ability to rebind with it non-covalently. MIPs are still being utilized in significant ways in biomarkers,177–179,199 bioseparations,180,200,201 biosensing,200,202–205 biocatalysis and therapeutic delivery applications.206,207 Chemical sensors using molecularly imprinted polymers (MIPs) may be useful for detecting the movement of different substances, such as dopamine, β-amyloid oligomers, albumin, and viruses, through brain endothelial cell layers with the use of BBBoCs. This sensor could potentially also monitor the release of glycocalyx elements, like sialic acid,208 from the luminal surface of brain endothelial cells under pathological conditions, or to identify markers of BBB leakage from the brain, like astrocyte markers glial fibrillary acidic protein and S100B, or neuron-specific enolase, in the bloodstream. Lee et al. have demonstrated that the synthesis of magnetic molecularly imprinted poly(ethylene-co-vinyl alcohols) nanoparticles with creatinine, albumin, and lysozyme and those target molecules rapidly adsorbed (∼1 min). MIPs with magnetic nanoparticles incorporated (magnetic MIPs or MMIPs) showed changes in their magnetization in response to rebinding, indicating that magnetization may be a useful readout for MMIP binding and thus target concentrations. MMIP nanoparticles may be able to be integrated into giant magnetoresistance sensors for biosensing applications. Preliminary studies using real urine samples showed promise for magnetic sensing of albumin concentration, but also demonstrated that interferences from other molecular species in urine must be more fully quantified, or interfering species must be removed.200 Another study on MIP from the same group developed a process for the efficient extraction of resveratrol from polygonum cuspidatum with magnetic orcinol-imprinted poly(ethylene-co-vinyl alcohol) composite particles, whose administration increased the toxicity in human osteogenic sarcoma (HOS) cells.206A recent paper has demonstrated encouraging results for possible future therapeutic applications of MMIPs for the extraction or isolation of undesirable protein aggregates in neurodegenerative disease.209 Moreover, the use of magnetic nanoparticles as a tool to enhance the delivery of therapeutic molecules to the blood–brain barrier is particularly promising. There is special interest in the use of magnetic nanoparticles, as their physical distinguishing features endow them with additional potentially useful properties. Following systemic administration, a magnetic field applied externally can mediate the capacity of magnetic nanoparticles to permeate the blood–brain barrier. Meanwhile, thermal energy liberated by magnetic nanoparticles (iron oxide nanoparticles) under the influence of radiofrequency radiation can modulate blood–brain barrier integrity, increasing its permeability.210 In summary, MIPs certainly have a bright future in biomedical applications due to their potentially high selectivity and versatility, robustness and low costs.
5.3 Understanding the metabolism of BBB: a helpful tool
Metabolomics is a recent area of study within the field of analytical chemistry, which can be extremely useful in the knowledge of molecular mechanisms causing neurodegenerative diseases allowing an early diagnosis thanks to its potential biomarker discovery. As neurodegenerative diseases often do not have a genetic basis and are influenced by various genetic risk factors and environmental triggers, metabolomics have potential for uncovering novel biomarkers and pathways for these disorders that are biologically and clinically significant. Recently, some researchers identified mechanisms responsible for the BBB transport selectivity. These include the shuttling of metabolites via dedicated solute carriers (SLCs) along with the crossing of macromolecules via transcytosis.211,212 The way in which the BBB ensures its own metabolic homeostasis is still unknown. It has been proved that the ECs suffer disease-specific modulations also influencing surrounding cells. However, only few studies have been carried out about the metabolism of brain PCs to date. For human placental pericytes a highly glycolytic metabolism was reported.213 Lee et al demonstrated that the metabolism of ECs and PCs is closely linked.214 Moreover, they observed that the high levels of lactate, produced and secreted by ECs, represents a pivotal metabolic resource for the brain PCs, since it maintains PC homeostasis and BBB integrity. From the literature and research studies conducted so far, it is clear that a better knowledge of how the typical multicellular interactions of the BBB induce individual cell-specific molecular and metabolic changes would provide significant insight.215 Thus, the study of metabolomics could help to better understand the overall behaviour of BBB cells. Metabolomics is a branch of ‘omics’ sciences allowing the high-throughput analysis of the metabolome by using, alone or in a synergistic way, two main detection techniques: mass spectrometry and NMR spectroscopy.216 Both methods are capable of providing extensive information about metabolites without requiring prior selection of analytes, and they can identify metabolite structures while measuring their concentrations. These two methods of analysis bring advantages and limitations, as outlined in Table 5. NMR is non-destructive and may provide, one shot, the overall metabolites content of a complex mixture, without the need for compound separation. It is also noted for its versatility and reliability in determining absolute concentrations (NMR takes advantage of selected nuclei dependent signal responses and does not require a specific standard while MS typically requires calibration curves for quantitation of each metabolite). In contrast, MS generally offers higher sensitivity than NMR. However, MS is normally coupled with a chromatographic technique (GC, HPLC) since it requires previous separation of the compounds present in the sample. Therefore, MS results could be affected by the variability of responses from different compounds in complex mixtures, which can lead to misleading results. Additionally, MS disrupts the structure and interactions of molecular complexes, resulting in the loss of valuable information about metabolite dynamics in tissue samples. While both techniques have their strengths, NMR is particularly useful for intact tissue and clinical research, and in general for studies that require detailed analysis of metabolites in biological samples without compromising their integrity (whereas MS is favoured for its sensitivity in other contexts).217 Each technique (NMR and MS) complements the other, offering a comprehensive approach to molecular analysis.| Methods | Advantages | Disadvantages |
|---|---|---|
| NMR (nuclear magnetic resonance) | Non-destructive analysis | Lower sensitivity |
| Minimal sample preparation | Higher operational costs | |
| Provides detailed structural information | Lower throughput | |
| Higher reproducibility | Requires larger sample volumes | |
| Absolute quantification without standards | ||
| MS (mass spectrometry) | High sensitivity | Extensive sample preparation |
| Can detect low-abundance metabolites | Often destructive | |
| Quantification requires calibration with standards | ||
| Higher throughput | Requires supplementary techniques for structural information | |
| More cost-effective for certain analyses |
The major areas of applications include:
• Biomedical and clinical research aiming at individual profiling for personalized healthcare and the monitoring of the follow up after treatment: metabolic profiling of individuals can help identify their susceptibilities to diseases and predict their responses to medications.
• Molecular epidemiology for level disease risk assessment: metabolic profiling of populations can enable the development of molecular epidemiology, allowing researchers to determine the susceptibilities of specific groups to diseases. This could lead to the identification of metabolite-based biomarkers for diseases, which could impact health screening programs.
• Food-stuff profiling: identifying origin, quality control.
• Environmental science for ecosystem monitoring and bioremediation: studying the impact of pollutants and climate change on biological systems and optimizing microbes for breaking down pollutants by analysing their metabolic pathways respectively.
• Drug discovery: identify biological targets and the mechanisms of drug toxicity and efficiency. The metabolome consists in small molecules (metabolites) which are the intermediate or end products of cellular metabolic reactions, within a biological sample, such as cells, tissues, or biological fluids.216,218 The cell metabolome is represented by a fingerprint of metabolites reflecting the status of cellular physiology, at a specific time point, under defined conditions. In addition, the analysis of the metabolome is very helpful in the study of drug effects (more efficient by using NMR spectroscopy with respect to MS). As described in the previous sections, organ-on-chip and 3D spheroid/organoid models are able to imitate the in vivo BBB cell–cell interactions. Although many metabolomic studies were performed on model 2D cultured cell lines,219,220 including astrocytes and pericytes,221–223 very few have been performed on organ-on-chip and 3D BBB models, opening and suggesting new perspectives for metabolomic studies on this topic. Among these, Brown et al. performed a metabolic approach to examine the effects of inflammation on BBB function ex vivo, by using the OoC model, discussing the metabolic consequences of metabolic responses and repair mechanisms. In detail, the pyrimidine metabolism, a pathway generally involved in systemic inflammation such as neurodevelopmental disorders, was found activated in the vascular chamber.224
Other related metabolomic studies (yet very few) confirmed the perspective great potential of metabolomic techniques in the study and comprehension of the BBB.225,226 This may provide new opportunities for future clinical applications.
Huang et al.226 and Maoz et al.225 used, in their respective studies, organ-on-chip models based on multicellular culture systems to reveal individual cellular responses or connections during environmental changes. Their results provide a very interesting overview of the adaptation and interactions that may occur in vivo at the BBB. Moreover, in a further study Huang et al.226 reported that cells of the BBB exhibit differential metabolomic profiles during physiological or injury conditions and that these changes are also able to modulate the behaviour of the surrounding cells. The metabolomic approach represents a highly significant resource for gaining a comprehensive understanding of the intricate mechanisms underlying blood–brain barrier (BBB) homeostasis. By delving deeper into the metabolic demands and management strategies employed by the BBB, researchers can acquire valuable insights that could ultimately contribute to enhancing barrier health. This enhanced understanding could pave the way for the development of targeted therapeutic strategies aimed at selectively treating a range of diseases that impact the central nervous system (CNS). Such targeted drug treatments could potentially lead to more effective interventions, minimizing side effects and improving patient outcomes in conditions that compromise blood brain barrier integrity and functions.
5.4 From metabolomics to volatilomics
A promising branch of metabolomics is volatilomics that focuses on volatile organic compounds (VOCs), metabolites continuously released in the human body as intermediates or products of cellular metabolic pathways and are easily detectable in biological samples such as blood, urine, faeces, saliva and exhaled breath. VOCs are a large and highly differentiated class of molecules including aliphatic, aromatic and chlorinated hydrocarbons, ketones, aldehydes, alcohols, sulphur compounds, esters, and terpenes, and their distribution in the human body and elimination through biofluids depends on the different physicochemical proprieties of each VOC and, in particular, on blood:air and fat:blood partition coefficients.227,228 Moreover, the VOC composition in biofluids qualitatively and quantitatively changes depending on cellular pathophysiological conditions. In fact, even if the biochemical origin of endogenous VOCs is only poorly studied and the information about the metabolic pathways leading to their production or degradation are missing, it is well-known that physiological and pathophysiological cellular processes result in different patterns of VOCs released from the cells and, consequently, detectable in bloodstream and biofluids.229,230 Therefore, information about cellular VOC profiles as well as VOC patterns in biofluids may be useful to provide a more immediate and dynamic picture of the cell functionality.Recent promising studies highlighted that VOC characterization in biofluids (breath, urine, saliva, faeces, blood, skin emanations and cellular lines) can provide useful insights regarding the mechanisms responsible for the development of neurodegenerative diseases.183 In fact, various neurodegenerative pathways were found closely linked to inflammatory processes responsible for the activation of leukocytes, the release of reactive oxygen species, cytokines and chemokines and, thus, for several metabolic changes in blood and organ tissues.231,232 In these studies, patterns of VOCs rather than a specific biomarker were found associated with Alzheimer's disease, Parkinson's disease, multiple sclerosis, and amyotrophic lateral sclerosis.183,231–243 Anyway, a common VOC pattern characteristic of a specific neurodegenerative disease has been not found. This is probably due to the different methodologies used for sampling and analysis of biofluids and confounding factors affecting VOC composition in biofluids as well as VOCs linked to environmental pollutants, diet, oral cavity and gut microbiome and dysbiosis.233,236,237,244,245 Therefore, the identification of a pattern of VOCs secreted by BBB multicellular spheroids that mimic BBB dysfunction typical of the neurodegenerative diseases using human neural cell cultures, could provide unique insights into ongoing biochemical processes and, then, useful information about VOCs to search for in biofluid samples. In fact, considering that volatilomics data produced by biofluids sampling and analysis contain distinct sources of variance, the analysis of VOCs secreted by cell lines has a benefit over other matrices because the environmental variables as well as confounding factors associated with clinical samples (patients’ diet, age, gender, metabolic state etc.) are more controllable or even negligible, making results more easily interpretable and reproducible.246,247 Additionally, direct detection of VOCs from cells could provide clear-cut association between the obtained findings and the cells per se, rather than with the indirect metabolic pathways in the body.248 Finally, the use of in-vitro cell lines allows us to explore the origins of VOC release by targeting specific processes, conducting controlled molecular biology experiments. For example, in-vitro cell lines experiments could help to identify the hypoxia driven specific volatilomics signatures through the characterization of VOCs released by cell lines under normoxic and hypoxic conditions.249
6 Future perspectives
The outlook for BBB microfluidics is highly promising as advancements in technology continue to open new avenues for research, drug discovery, and personalized medicine. Microfluidic BBB models are increasingly recognized as powerful tools for mimicking the physiological and pathological conditions of the BBB in vitro, providing insights into brain function, disease mechanisms, and therapeutic delivery. The key future outlooks for BBB microfluidics include the realization of a more accurate and complex BBB model applying advanced microfabrication techniques, including 3D bioprinting, and enhanced biomaterials. These developments will enable the recreation of the BBB's multicellular structure, fluid dynamics, and biochemical environment, allowing us to study the BBB's role in health and disease with higher precision. To obtain a more comprehensive model for studying systemic interactions affecting the BBB and its role in neurodegenerative diseases or brain tumors, the integration with other brain models (e.g., neurons, microglia) or peripheral systems (e.g., gut-brain axis) could be useful.250 As microfluidic BBB systems become more scalable and high-throughput, they will play an increasingly central role in drug screening and neurotherapeutic development. In particular, patient-specific BBB models, using induced pluripotent stem cells (iPSCs), will enable personalized drug screening and the development of tailored therapies. This approach is expected to reduce the failure rate of CNS drugs in clinical trials, as it allows for more accurate predictions of how individual patients’ BBBs will respond to treatments. Embedding sensors and biosensors into microfluidic systems will enable continuous, real-time monitoring of BBB properties such as permeability, tight junction integrity, and cellular responses. This real-time data will be invaluable for dynamic studies of how the BBB reacts to drugs, toxins, and disease states. The future of BBB microfluidics will likely involve integrating non-invasive imaging techniques (e.g., fluorescence microscopy, multiphoton microscopy) for observing cellular interactions, BBB disruptions, and drug delivery in real time.251 Automation and miniaturization of microfluidic BBB models will be critical for making them widely accessible and commercially viable. Automated systems could facilitate routine use in drug discovery pipelines, where high-throughput screening of thousands of compounds for BBB permeability and brain drug delivery is necessary. Advancements in BBB microfluidic systems will prompt regulatory bodies like the FDA and EMA to include them in preclinical testing, necessitating standardized design and validation for CNS drug approvals. This evolution will not only enhance the reliability of drug efficacy assessments but also streamline the approval process, ultimately leading to faster delivery of innovative therapies to patients suffering from neurological disorders. As these systems gain acceptance, collaboration between researchers and regulators will be crucial to establish comprehensive guidelines. Microfluidic systems also have a significant economic impact on BBB studies by improving research efficiency, reducing costs, and accelerating drug development. In comparison to traditional BBB models, such as animal studies which are expensive and time-consuming, microfluidic systems offer cost-effective alternatives by using small sample volumes and fewer reagents, minimizing the need for costly in vivo studies, leading to significant savings. Furthermore, microfluidic BBB models enable high-throughput screening, reducing the time and cost of preclinical testing. This scalability allows pharmaceutical companies to integrate these systems into automated workflows, reducing laboratory costs.The standard cost of a single microfluidic chip, considering only the raw materials and the manufacturing cost, can be estimated as a few €/devices. This cost can be considerably reduced in an industrial-scale production, providing significant savings with respect to standard laboratory methods. Moreover, the possibility of integrating multiple biosensors into the same platform will enhance the performance in terms of speed, flexibility, automation and costs.
Conclusions
The institution of microfluidic cell culture platforms has brought about huge evolutions in the field of NVU modelling. Microfluidic NVU models show augmented complexity in comparison to conventional models, permitting co-culture of various cell types, incorporation of cell–matrix interactions, and presence of fluid flow. Furthermore, the latest introduction of microfluidic chips in higher throughput formats now renders NVU on-a-chip models compatible with routine laboratory adoption and assessment of novel drug candidates. As time goes by, NVU on-a-chip models have shown increasing biological complexity. More attention is placed on the use of primary cells and iPSC-derived cells which allow more accurate disease- and patient-specific models. Further betterments in protocols for cell differentiation and continued incorporation will improve future NVU models’ relevance even further. Importantly, the NVU is a highly complex structure, and it is likely that no model will be able to capture all its features. Fit-for-purpose models provide a viable compromise between physiological relevance and ease-of-use and hold the future of NVU modelling: as simple as possible, as complex as needed.The future of BBB microfluidics is bright, with significant advancements on the horizon in terms of model complexity, high-throughput capabilities, disease specificity, and personalized medicine. As these systems continue to evolve, they will become increasingly indispensable tools in neuroscience research, drug development, and the study of brain diseases. Their potential for real-time monitoring, scalability, and integration with other organ-on-chip systems will revolutionize how we understand and treat brain-related conditions. In the future, hybrid systems combining microfluidic BBB models with computational simulations will allow for more predictive and efficient experiments. Computational tools can simulate complex conditions, such as varying blood flow rates or molecular gradients, enabling researchers to optimize their in vitro experiments and understand the BBB's dynamic responses to stimuli. Artificial intelligence and machine learning can assist in the analysis of large datasets generated by microfluidic experiments, accelerating the identification of patterns and predictions of BBB behaviour under different conditions, including drug interactions and disease progressions.252 The future of BBB microfluidics lies in cross-disciplinary collaborations between engineers, neuroscientists, pharmacologists, and clinicians. This will be essential for advancing the design of BBB models and ensuring their relevance to real-world medical challenges.
Author contributions
Boncristiani C. and Vergaro V. designed the review. Boncristiani C. wrote the majority of the manuscript, and prepared figures and tables. De Castro F., Di Gilio A., Palmisani J., Martínez Vázquez R. and Nardini A. contributed to the writing the manuscript. Fanizzi F. P., Ciccarella G. and De Gennaro G. contributed to the revision of the manuscript. Vergaro V. coordinated the study and revised the manuscript. All authors read and approved the final manuscript.Data availability
No primary research results, software or code have been included, and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors are grateful to Project PRIN 2022 - MulticEllular spheRoids model for muLtiparametric blood–brain barrier Injury detectioN in microfluidics (MERLIN) – “Finanziato dall’Unione europea – Next Generation EU, Missione 4 Componente 1 CUP B53D23018380006”.References
- T.-E. Park, N. Mustafaoglu, A. Herland, R. Hasselkus, R. Mannix, E. A. FitzGerald, R. Prantil-Baun, A. Watters, O. Henry, M. Benz, H. Sanchez, H. J. McCrea, L. C. Goumnerova, H. W. Song, S. P. Palecek, E. Shusta and D. E. Ingber, Nat. Commun., 2019, 10, 2621 CrossRef PubMed.
- W. Wei, F. Cardes, A. Hierlemann and M. M. Modena, Adv. Sci., 2023, 10, 2205752 CrossRef CAS PubMed.
- R. M. Linville, M. B. Sklar, G. N. Grifno, R. F. Nerenberg, J. Zhou, R. Ye, J. G. DeStefano, Z. Guo, R. Jha, J. J. Jamieson, N. Zhao and P. C. Searson, Fluids Barriers CNS, 2022, 19, 87 CrossRef CAS PubMed.
- T. D. Chung, R. M. Linville, Z. Guo, R. Ye, R. Jha, G. N. Grifno and P. C. Searson, Fluids Barriers CNS, 2022, 19, 33 CrossRef CAS PubMed.
- N. Bouhrira, B. J. DeOre, K. A. Tran and P. A. Galie, Fluids Barriers CNS, 2022, 19, 94 CrossRef CAS PubMed.
- A. Bhalerao, F. Sivandzade, S. R. Archie, E. A. Chowdhury, B. Noorani and L. Cucullo, Fluids Barriers CNS, 2020, 17, 22 CrossRef PubMed.
- M. Ribecco-Lutkiewicz, C. Sodja, J. Haukenfrers, A. S. Haqqani, D. Ly, P. Zachar, E. Baumann, M. Ball, J. Huang, M. Rukhlova, M. Martina, Q. Liu, D. Stanimirovic, A. Jezierski and M. Bani-Yaghoub, Sci. Rep., 2018, 8, 1873 CrossRef PubMed.
- A. Oddo, B. Peng, Z. Tong, Y. Wei, W. Y. Tong, H. Thissen and N. H. Voelcker, Trends Biotechnol., 2019, 37, 1295–1314 CrossRef CAS PubMed.
- Y. Zhou, Z. Peng, E. S. Seven and R. M. Leblanc, J. Controlled Release, 2018, 270, 290–303 CrossRef CAS PubMed.
- H. Kadry, B. Noorani and L. Cucullo, Fluids Barriers CNS, 2020, 17, 69 CrossRef PubMed.
- J. Xie, Z. Shen, Y. Anraku, K. Kataoka and X. Chen, Biomaterials, 2019, 224, 119491 CrossRef CAS PubMed.
- S. Raut, A. Bhalerao, B. Noorani and L. Cucullo, In Vitro Models of the Blood–Brain Barrier, in The Blood-Brain Barrier: Methods and Protocols, ed. N. Stone, Springer US, New York, NY, 2022, pp. 25–49 Search PubMed.
- A. S. Hanafy, D. Dietrich, G. Fricker and A. Lamprecht, Adv. Drug Delivery Rev., 2021, 176, 113859 CrossRef CAS PubMed.
- L. Yan, R. A. Moriarty and K. M. Stroka, Theranostics, 2021, 11, 10148–10170 CrossRef CAS PubMed.
- C. P. Profaci, R. N. Munji, R. S. Pulido and R. Daneman, J. Exp. Med., 2020, 217(4), e20190062 CrossRef PubMed.
- P. T. Ronaldson and T. P. Davis, J. Cereb. Blood Flow Metab., 2020, 40, S6–S24 CrossRef CAS PubMed.
- L. Marchetti and B. Engelhardt, Vasc. Biol., 2020, 2, H1–H18 CAS.
- F. Zamudio, A. R. Loon, S. Smeltzer, K. Benyamine, N. K. Navalpur Shanmugam, N. J. F. Stewart, D. C. Lee, K. Nash and M.-L. B. Selenica, J. Neuroinflammation, 2020, 17, 283 CrossRef CAS PubMed.
- S. Khaiboullina, T. Uppal, K. Kletenkov, S. C. St Jeor, E. Garanina, A. Rizvanov and S. C. Verma, Front. Pharmacol., 2019, 10, 642 CrossRef CAS PubMed.
- T. P. Buzhdygan, C. R. Rodrigues, H. M. McGary, J. A. Khan, A. M. Andrews, S. M. Rawls and S. H. Ramirez, J. Neuroinflammation, 2021, 18, 63 CrossRef CAS PubMed.
- M. Incerti, L. Crascì, P. Vicini, E. Aki, I. Yalcin, T. Ertan-Bolelli, V. Cardile, A. C. E. Graziano and A. Panico, Molecules, 2018, 23(2), 415 CrossRef PubMed.
- R. L. Jayaraj, S. Azimullah, R. Beiram, F. Y. Jalal and G. A. Rosenberg, J. Neuroinflammation, 2019, 16, 142 CrossRef PubMed.
- W. M. Pardridge, Pharmaceutics, 2022, 14(6), 1283 CrossRef CAS PubMed.
- X. Dong, Theranostics, 2018, 8, 1481–1493 CrossRef CAS PubMed.
- N. M. O’Brown, S. J. Pfau and C. Gu, Genes Dev., 2018, 32, 466–478 CrossRef PubMed.
- F. Joó and I. Karnushina, Cytobios, 1973, 8, 41–48 Search PubMed.
- L. E. DeBault and P. A. Cancilla, Science, 1980, 207, 653–655 CrossRef CAS PubMed.
- J. H. Tao-Cheng, Z. Nagy and M. W. Brightman, J. Neurosci., 1987, 7, 3293–3299 CrossRef CAS PubMed.
- U. Mischeck, J. Meyer and H. J. Galla, Cell Tissue Res., 1989, 256, 221–226 CrossRef CAS PubMed.
- M.-P. Dehouck, S. Méresse, P. Delorme, J.-C. Fruchart and R. Cecchelli, J. Neurochem., 1990, 54, 1798–1801 CrossRef CAS PubMed.
- K. Hayashi, S. Nakao, R. Nakaoke, S. Nakagawa, N. Kitagawa and M. Niwa, Regul. Pept., 2004, 123, 77–83 CrossRef CAS PubMed.
- B. B. Weksler, E. A. Subileau, N. Perrière, P. Charneau, K. Holloway, M. Leveque, H. Tricoire-Leignel, A. Nicotra, S. Bourdoulous, P. Turowski, D. K. Male, F. Roux, J. Greenwood, I. A. Romero and P. O. Couraud, FASEB J., 2005, 19, 1872–1874 CrossRef CAS PubMed.
- Y. Takeshita, B. Obermeier, A. Cotleur, Y. Sano, T. Kanda and R. M. Ransohoff, J. Neurosci. Methods, 2014, 232, 165–172 CrossRef CAS PubMed.
- E. S. Lippmann, S. M. Azarin, J. E. Kay, R. A. Nessler, H. K. Wilson, A. Al-Ahmad, S. P. Palecek and E. V. Shusta, Nat. Biotechnol., 2012, 30, 783–791 CrossRef CAS PubMed.
- R. Cecchelli, S. Aday, E. Sevin, C. Almeida, M. Culot, L. Dehouck, C. Coisne, B. Engelhardt, M.-P. Dehouck and L. Ferreira, PLoS One, 2014, 9, e99733 CrossRef PubMed.
- C. Simonneau, M. Duschmalé, A. Gavrilov, N. Brandenberg, S. Hoehnel, C. Ceroni, E. Lassalle, E. Kassianidou, H. Knoetgen, J. Niewoehner and R. Villaseñor, Fluids Barriers CNS, 2021, 18, 43 CrossRef CAS PubMed.
- G. Fedele, A. Cazzaniga, S. Castiglioni, L. Locatelli, A. Tosoni, M. Nebuloni and J. A. M. Maier, Biochem. Biophys. Res. Commun., 2022, 626, 30–37 CrossRef CAS PubMed.
- G. N. Grifno, A. M. Farrell, R. M. Linville, D. Arevalo, J. H. Kim, L. Gu and P. C. Searson, Sci. Rep., 2019, 9, 13957 CrossRef PubMed.
- J. S. Park, K. Choe, A. Khan, M. H. Jo, H. Y. Park, M. H. Kang, T. J. Park and M. O. Kim, Int. J. Mol. Sci., 2023, 24(6), 5283 CrossRef CAS PubMed.
- L. Cucullo, M. S. McAllister, K. Kight, L. Krizanac-Bengez, M. Marroni, M. R. Mayberg, K. A. Stanness and D. Janigro, Brain Res., 2002, 951, 243–254 CrossRef CAS PubMed.
- W. Neuhaus, R. Lauer, S. Oelzant, U. P. Fringeli, G. F. Ecker and C. R. Noe, J. Biotechnol., 2006, 125, 127–141 CrossRef CAS PubMed.
- R. Booth and H. Kim, Lab Chip, 2012, 12, 1784–1792 RSC.
- L. M. Griep, F. Wolbers, B. de Wagenaar, P. M. ter Braak, B. B. Weksler, I. A. Romero, P. O. Couraud, I. Vermes, A. D. van der Meer and A. van den Berg, Biomed. Microdevices, 2013, 15, 145–150 CrossRef CAS PubMed.
- A. K. H. Achyuta, A. J. Conway, R. B. Crouse, E. C. Bannister, R. N. Lee, C. P. Katnik, A. A. Behensky, J. Cuevas and S. S. Sundaram, Lab Chip, 2013, 13, 542–553 RSC.
- H. Cho, J. H. Seo, K. H. K. Wong, Y. Terasaki, J. Park, K. Bong, K. Arai, E. H. Lo and D. Irimia, Sci. Rep., 2015, 5, 15222 CrossRef CAS PubMed.
- K. L. Sellgren, B. T. Hawkins and S. Grego, Biomicrofluidics, 2015, 9, 61102 CrossRef PubMed.
- A. Herland, A. D. van der Meer, E. A. FitzGerald, T.-E. Park, J. J. F. Sleeboom and D. E. Ingber, PLoS One, 2016, 11, e0150360 CrossRef PubMed.
- G. Adriani, D. Ma, A. Pavesi, R. D. Kamm and E. L. K. Goh, Lab Chip, 2017, 17, 448–459 RSC.
- B. Prabhakarpandian, M.-C. Shen, J. B. Nichols, I. R. Mills, M. Sidoryk-Wegrzynowicz, M. Aschner and K. Pant, Lab Chip, 2013, 13, 1093–1101 RSC.
- G. D. Vatine, R. Barrile, M. J. Workman, S. Sances, B. K. Barriga, M. Rahnama, S. Barthakur, M. Kasendra, C. Lucchesi, J. Kerns, N. Wen, W. R. Spivia, Z. Chen, J. Van Eyk and C. N. Svendsen, Cell Stem Cell, 2019, 24, 995–1005.e6 CrossRef CAS PubMed.
- S. Ohtsuki, C. Ikeda, Y. Uchida, Y. Sakamoto, F. Miller, F. Glacial, X. Decleves, J.-M. Scherrmann, P.-O. Couraud, Y. Kubo, M. Tachikawa and T. Terasaki, Mol. Pharm., 2013, 10, 289–296 CrossRef CAS PubMed.
- B. Weksler, I. A. Romero and P.-O. Couraud, Fluids Barriers CNS, 2013, 10, 16 CrossRef PubMed.
- T. G. D’Aversa, E. A. Eugenin, L. Lopez and J. W. Berman, Neuropathol. Appl. Neurobiol., 2013, 39, 270–283 CrossRef PubMed.
- E. De Jong, D. S. Williams, L. K. E. A. Abdelmohsen, J. C. M. Van Hest and I. S. Zuhorn, J. Controlled Release, 2018, 289, 14–22 CrossRef CAS PubMed.
- M. J. Stebbins, B. D. Gastfriend, S. G. Canfield, M.-S. Lee, D. Richards, M. G. Faubion, W.-J. Li, R. Daneman, S. P. Palecek and E. V. Shusta, Sci. Adv., 2019, 5, eaau7375 CrossRef CAS PubMed.
- J.-H. Choi, M. Santhosh and J.-W. Choi, Micromachines, 2020, 11 Search PubMed.
- S. P. Deosarkar, B. Prabhakarpandian, B. Wang, J. B. Sheffield, B. Krynska and M. F. Kiani, PLoS One, 2015, 10, e0142725 CrossRef PubMed.
- P. P. Partyka, G. A. Godsey, J. R. Galie, M. C. Kosciuk, N. K. Acharya, R. G. Nagele and P. A. Galie, Biomaterials, 2017, 115, 30–39 CrossRef CAS PubMed.
- R. Booth and H. Kim, Ann. Biomed. Eng., 2014, 42, 2379–2391 CrossRef CAS PubMed.
- M. Bonakdar, P. M. Graybill and R. V. Davalos, RSC Adv., 2017, 7, 42811–42818 RSC.
- J. A. Kim, H. N. Kim, S.-K. Im, S. Chung, J. Y. Kang and N. Choi, Biomicrofluidics, 2015, 9, 24115 CrossRef PubMed.
- X. Shao, D. Gao, Y. Chen, F. Jin, G. Hu, Y. Jiang and H. Liu, Anal. Chim. Acta, 2016, 934, 186–193 CrossRef CAS PubMed.
- J. D. Wang, E.-S. Khafagy, K. Khanafer, S. Takayama and M. E. H. ElSayed, Mol. Pharm., 2016, 13, 895–906 CrossRef CAS PubMed.
- M. Zakharova, M. A. Palma do Carmo, M. W. van der Helm, H. Le-The, M. N. S. de Graaf, V. Orlova, A. van den Berg, A. D. van der Meer, K. Broersen and L. I. Segerink, Lab Chip, 2020, 20, 3132–3143 RSC.
- N. R. Wevers, D. G. Kasi, T. Gray, K. J. Wilschut, B. Smith, R. van Vught, F. Shimizu, Y. Sano, T. Kanda, G. Marsh, S. J. Trietsch, P. Vulto, H. L. Lanz and B. Obermeier, Fluids Barriers CNS, 2018, 15, 23 CrossRef PubMed.
- D. Liu, M. Zhu, Y. Lin, M. Li, R. Huang, L. Yang, Y. Song, Y. Diao and C. Yang, Lab Chip, 2022, 22, 4180–4190 RSC.
- C.-F. Cho, J. M. Wolfe, C. M. Fadzen, D. Calligaris, K. Hornburg, E. A. Chiocca, N. Y. R. Agar, B. L. Pentelute and S. E. Lawler, Nat. Commun., 2017, 8, 15623 CrossRef CAS PubMed.
- M. E. Boutin, L. L. Kramer, L. L. Livi, T. Brown, C. Moore and D. Hoffman-Kim, J. Neurosci. Methods, 2018, 299, 55–63 CrossRef PubMed.
- C. Eilenberger, M. Rothbauer, F. Selinger, A. Gerhartl, C. Jordan, M. Harasek, B. Schädl, J. Grillari, J. Weghuber, W. Neuhaus, S. Küpcü and P. Ertl, Adv. Sci., 2021, 8, 2004856 CrossRef CAS PubMed.
- I. Salmon, S. Grebenyuk, A. R. A. Fattah, G. Rustandi, T. Pilkington, C. Verfaillie and A. Ranga, Lab Chip, 2022, 22, 1615–1629 RSC.
- S. I. Ahn, Y. J. Sei, H.-J. Park, J. Kim, Y. Ryu, J. J. Choi, H.-J. Sung, T. J. MacDonald, A. I. Levey and Y. Kim, Nat. Commun., 2020, 11, 175 CrossRef CAS PubMed.
- B. Chung, J. Kim, J. Nam, H. Kim, Y. Jeong, H.-W. Liu, Y. Cho, Y. H. Kim, H. J. Oh and S. Chung, Macromol. Biosci., 2020, 20, e1900425 CrossRef PubMed.
- J. Y. Yang, D.-S. Shin, M. Jeong, S. S. Kim, H. N. Jeong, B. H. Lee, K.-S. Hwang, Y. Son, H.-C. Jeong, C.-H. Choi, K.-R. Lee and M. A. Bae, Pharmaceutics, 2024, 16, 574 CrossRef CAS PubMed.
- G. Silvani, C. Basirun, H. Wu, C. Mehner, K. Poole, P. Bradbury and J. Chou, Adv. Ther., 2021, 4, 2100106 CrossRef.
- R. M. Linville, J. G. DeStefano, M. B. Sklar, Z. Xu, A. M. Farrell, M. I. Bogorad, C. Chu, P. Walczak, L. Cheng, V. Mahairaki, K. A. Whartenby, P. A. Calabresi and P. C. Searson, Biomaterials, 2019, 190–191, 24–37 CrossRef CAS PubMed.
- Z. Vargas-Osorio, A. Da Silva-Candal, Y. Piñeiro, R. Iglesias-Rey, T. Sobrino, F. Campos, J. Castillo and J. Rivas, Nanomaterials, 2019 DOI:10.3390/nano9030449.
- S. Zhang, P. Gong, J. Zhang, X. Mao, Y. Zhao, H. Wang, L. Gan and X. Lin, Front. Neurosci., 2020, 14, 582324 CrossRef PubMed.
- S. Lee, B.-M. Kang, J. H. Kim, J. Min, H. S. Kim, H. Ryu, H. Park, S. Bae, D. Oh, M. Choi and M. Suh, Sci. Rep., 2018, 8, 13064 CrossRef PubMed.
- J. Y. Yang, D.-S. Shin, M. Jeong, S. S. Kim, H. N. Jeong, B. H. Lee, K.-S. Hwang, Y. Son, H.-C. Jeong, C.-H. Choi, K.-R. Lee and M. A. Bae, Pharmaceutics, 2024, 16(5), 574 CrossRef CAS PubMed.
- X. Chen, C. Liu, L. Muok, C. Zeng and Y. Li, Cells, 2021, 10(11), 3183 CrossRef CAS PubMed.
- D. D. Sahtoe, A. Coscia, N. Mustafaoglu, L. M. Miller, D. Olal, I. Vulovic, T.-Y. Yu, I. Goreshnik, Y.-R. Lin, L. Clark, F. Busch, L. Stewart, V. H. Wysocki, D. E. Ingber, J. Abraham and D. Baker, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(17), e2021569118 CrossRef CAS PubMed.
- R. Ito, K. Umehara, S. Suzuki, K. Kitamura, K.-I. Nunoya, Y. Yamaura, H. Imawaka, S. Izumi, N. Wakayama, T. Komori, N. Anzai, H. Akita and T. Furihata, Mol. Pharm., 2019, 16, 4461–4471 CrossRef CAS PubMed.
- T. P. Buzhdygan, B. J. DeOre, A. Baldwin-Leclair, T. A. Bullock, H. M. McGary, J. A. Khan, R. Razmpour, J. F. Hale, P. A. Galie, R. Potula, A. M. Andrews and S. H. Ramirez, Neurobiol. Dis., 2020, 146, 105131 CrossRef CAS PubMed.
- I. Papademetriou, E. Vedula, J. Charest and T. Porter, PLoS One, 2018, 13, e0205158 CrossRef PubMed.
- B. Noorani, A. Bhalerao, S. Raut, E. Nozohouri, U. Bickel and L. Cucullo, Pharmaceutics, 2021, 13(9), 1474 CrossRef CAS PubMed.
- N. A. Boghdeh, K. H. Risner, M. D. Barrera, C. M. Britt, D. K. Schaffer, F. Alem, J. A. Brown, J. P. Wikswo and A. Narayanan, Viruses, 2022, 14(12), 2799 CrossRef CAS PubMed.
- A. Williams-Medina, M. Deblock and D. Janigro, Front. Med. Technol., 2020, 2, 623950 CrossRef PubMed.
- A. Sood, A. Kumar, A. Dev, V. K. Gupta and S. S. Han, Pharmaceutics, 2022, 14(5), 993 CrossRef CAS PubMed.
- M. A. Erickson and W. A. Banks, Pharmacol. Rev., 2018, 70, 278–314 CrossRef CAS PubMed.
- Y. Song, X. Cai, D. Du, P. Dutta and Y. Lin, ACS Appl. Bio Mater., 2019, 2, 1050–1055 CrossRef CAS PubMed.
- I. Galea, Cell. Mol. Immunol., 2021, 18, 2489–2501 CrossRef CAS PubMed.
- A.-N. Cho, Y. Jin, Y. An, J. Kim, Y. S. Choi, J. S. Lee, J. Kim, W.-Y. Choi, D.-J. Koo, W. Yu, G.-E. Chang, D.-Y. Kim, S.-H. Jo, J. Kim, S.-Y. Kim, Y.-G. Kim, J. Y. Kim, N. Choi, E. Cheong, Y.-J. Kim, H. S. Je, H.-C. Kang and S.-W. Cho, Nat. Commun., 2021, 12, 4730 CrossRef CAS PubMed.
- C. Wang, A. Nagayach, H. Patel, L. Dao, H. Zhu, A. R. Wasylishen, Y. Fan, A. Kendler and Z. Guo, Breast Cancer Res., 2024, 26, 108 CrossRef CAS PubMed.
- X.-Y. Sun, X.-C. Ju, H.-F. Zhao, Z.-W. You, R.-R. Han and Z.-G. Luo, Bio-protocol, 2023, 13, e4870 CrossRef CAS PubMed.
- I. Martinelli, S. K. Tayebati, D. Tomassoni, G. Nittari, P. Roy and F. Amenta, Cells, 2022, 11(7), 1120 CrossRef CAS PubMed.
- G. Nzou, R. T. Wicks, E. E. Wicks, S. A. Seale, C. H. Sane, A. Chen, S. V. Murphy, J. D. Jackson and A. J. Atala, Sci. Rep., 2018, 8, 7413 CrossRef PubMed.
- H. N. Lee, Y. Y. Choi, J. W. Kim, Y. S. Lee, J. W. Choi, T. Kang, Y. K. Kim and B. G. Chung, Nano Converg., 2021, 8, 35 CrossRef CAS PubMed.
- S. Bergmann, S. E. Lawler, Y. Qu, C. M. Fadzen, J. M. Wolfe, M. S. Regan, B. L. Pentelute, N. Y. R. Agar and C.-F. Cho, Nat. Protoc., 2018, 13, 2827–2843 CrossRef CAS PubMed.
- P. Gazerani, Med. Res. Arch., 2024, 12(2), 3113 Search PubMed.
- X.-Y. Sun, X.-C. Ju, Y. Li, P.-M. Zeng, J. Wu, L.-B. Shen, Y.-J. Chen and Z.-G. Luo, bioRxiv, 2022, preprint DOI:10.7554/eLife.76707.
- N. Shin, Y. Kim, J. Ko, S. W. Choi, S. Hyung, S.-E. Lee, S. Park, J. Song, N. L. Jeon and K.-S. Kang, Biotechnol. Bioeng., 2022, 119, 566–574 CrossRef CAS PubMed.
- M. G. Kook, S.-E. Lee, N. Shin, D. Kong, D.-H. Kim, M.-S. Kim, H. K. Kang, S. W. Choi and K.-S. Kang, Int. J. Stem Cells, 2022, 15, 85–94 CrossRef PubMed.
- B. Cakir and I.-H. Park, eLife, 2022, 11, e80373 CrossRef PubMed.
- M. T. Pham, K. M. Pollock, M. D. Rose, W. A. Cary, H. R. Stewart, P. Zhou, J. A. Nolta and B. Waldau, NeuroReport, 2018, 29, 588–593 CrossRef PubMed.
- T. K. Matsui, Y. Tsuru, K. Hasegawa and K.-I. Kuwako, Stem Cells, 2021, 39, 1017–1024 CrossRef PubMed.
- Y. Ahn, J.-H. An, H.-J. Yang, D. G. Lee, J. Kim, H. Koh, Y.-H. Park, B.-S. Song, B.-W. Sim, H. J. Lee, J.-H. Lee and S.-U. Kim, Cells, 2021, 10(8), 2036 CrossRef CAS PubMed.
- M. M. Wang, X. Zhang, S. J. Lee, S. Maripudi, R. F. Keep, A. M. Johnson, S. M. Stamatovic and A. V. Andjelkovic, Sci. Rep., 2018, 8, 10042 CrossRef PubMed.
- H. W. Song, K. L. Foreman, B. D. Gastfriend, J. S. Kuo, S. P. Palecek and E. V. Shusta, Sci. Rep., 2020, 10, 12358 CrossRef CAS PubMed.
- A. A. Mansour, J. T. Gonçalves, C. W. Bloyd, H. Li, S. Fernandes, D. Quang, S. Johnston, S. L. Parylak, X. Jin and F. H. Gage, Nat. Biotechnol., 2018, 36, 432–441 CrossRef CAS PubMed.
- Y. Shi, L. Sun, M. Wang, J. Liu, S. Zhong, R. Li, P. Li, L. Guo, A. Fang, R. Chen, W.-P. Ge, Q. Wu and X. Wang, PLoS Biol., 2020, 18, e3000705 CrossRef CAS PubMed.
- B. Cakir, Y. Xiang, Y. Tanaka, M. H. Kural, M. Parent, Y.-J. Kang, K. Chapeton, B. Patterson, Y. Yuan, C.-S. He, M. S. B. Raredon, J. Dengelegi, K.-Y. Kim, P. Sun, M. Zhong, S. Lee, P. Patra, F. Hyder, L. E. Niklason, S.-H. Lee, Y.-S. Yoon and I.-H. Park, Nat. Methods, 2019, 16, 1169–1175 CrossRef CAS PubMed.
- E. Garreta, R. D. Kamm, S. M. Chuva de Sousa Lopes, M. A. Lancaster, R. Weiss, X. Trepat, I. Hyun and N. Montserrat, Nat. Mater., 2021, 20, 145–155 CrossRef CAS PubMed.
- E. Karzbrun, A. Kshirsagar, S. R. Cohen, J. H. Hanna and O. Reiner, Nat. Phys., 2018, 14, 515–522 Search PubMed.
- Z. Ao, H. Cai, D. J. Havert, Z. Wu, Z. Gong, J. M. Beggs, K. Mackie and F. Guo, Anal. Chem., 2020, 92, 4630–4638 CrossRef CAS PubMed.
- S. T. Seiler, G. L. Mantalas, J. Selberg, S. Cordero, S. Torres-Montoya, P. V. Baudin, V. T. Ly, F. Amend, L. Tran, R. N. Hoffman, M. Rolandi, R. E. Green, D. Haussler, S. R. Salama and M. Teodorescu, Sci. Rep., 2022, 12, 20173 CrossRef CAS PubMed.
- A. I. Romero-Morales, B. J. O’Grady, K. M. Balotin, L. M. Bellan, E. S. Lippmann and V. Gama, HardwareX, 2019, 6, e00084 CrossRef PubMed.
- Y. Wang, L. Wang, Y. Zhu and J. Qin, Lab Chip, 2018, 18, 851–860 RSC.
- L. O. Porciúncula, L. Goto-Silva, P. F. Ledur and S. K. Rehen, Front. Neurosci., 2021, 15, 674563 CrossRef PubMed.
- P.-S. Cheah, J. O. Mason and K. H. Ling, Neurosci. Res. Notes, 2019, 2, 1–6 CrossRef.
- S. L. Giandomenico, M. Sutcliffe and M. A. Lancaster, Nat. Protoc., 2021, 16, 579–602 CrossRef CAS PubMed.
- R. Gopurappilly and R. Pal, Chapter 16 - Bioengineering of brain organoids: Advancements and challenges, ed. C. P. Sharma, T. Chandy, V. Thomas and F. G. B. T.-T. E.Thankam, Academic Press, 2022, pp. 399–414 Search PubMed.
- Z. Ao, S. Song, C. Tian, H. Cai, X. Li, Y. Miao, Z. Wu, J. Krzesniak, B. Ning, M. Gu, L. P. Lee and F. Guo, Adv. Sci., 2022, 9, e2200475 CrossRef PubMed.
- Z. Bao, K. Fang, Z. Miao, C. Li, C. Yang, Q. Yu, C. Zhang, Z. Miao, Y. Liu and J. Ji, Oxid. Med. Cell. Longev., 2021, 2021, 6338722 CrossRef PubMed.
- X. Wang, B. Bijonowski and N. Kurniawan, Organoids, 2023, 2, 239–255 CrossRef.
- S. Aralekallu, R. Boddula and V. Singh, Mater. Des., 2023, 225, 111517 CrossRef CAS.
- J. B. Nielsen, R. L. Hanson, H. M. Almughamsi, C. Pang, T. R. Fish and A. T. Woolley, Anal. Chem., 2020, 92, 150–168 CrossRef CAS PubMed.
- K. Chandnani, N. Rajput, T. Jadav, M. Pillai, P. Dhakne, R. K. Tekade and P. Sengupta, Microchem. J., 2023, 195, 109532 CrossRef CAS.
- A. G. Niculescu, C. Chircov, A. C. Bîrcă and A. M. Grumezescu, Int. J. Mol. Sci., 2021, 22, 1–26 Search PubMed.
- K. Ren, J. Zhou and H. Wu, Acc. Chem. Res., 2013, 46, 2396–2406 CrossRef CAS PubMed.
- Y. Xie, X. Zhi, H. Su, K. Wang, Z. Yan, N. He, J. Zhang, D. Chen and D. Cui, Nanoscale Res. Lett., 2015, 10, 1–9 CrossRef CAS PubMed.
- D. S. Dkhar, R. Kumari, S. J. Malode, N. P. Shetti and P. Chandra, J. Pharm. Biomed. Anal., 2023, 223, 115120 CrossRef CAS PubMed.
- E. Sollier, C. Murray, P. Maoddi and D. Di Carlo, Lab Chip, 2011, 11, 3752–3765 RSC.
- P. N. Nge, C. I. Rogers and A. T. Woolley, Chem. Rev., 2013, 113(4), 2550 CrossRef CAS PubMed.
- A. W. Martinez, S. T. Phillips, G. M. Whitesides and E. Carrilho, Anal. Chem., 2010, 82, 3–10 CrossRef CAS PubMed.
- X. Tong, L. Ga, R. Zhao, J. Ai, S. Das, R. Gagandeep, D. Bhatia, B. Lin, J. Li, X. Qi, S. Ji, W. Yang and L. C. Wang, Sens. Actuators, B, 2022, 303, 8793–8820 Search PubMed.
- A. Tony, I. Badea, C. Yang, Y. Liu, G. Wells, K. Wang, R. Yin, H. Zhang and W. Zhang, Polymers, 2023, 15, 1–18 Search PubMed.
- H. Wu, S. Shi, Y. Liu, Q. Zhang, R. H. W. Lam, C. T. Lim and J. Hu, Biofabrication, 2023, 15(4) DOI:10.1088/1758-5090/acdaf9.
- M. Zakharova, M. A. Palma Do Carmo, M. W. Van Der Helm, H. Le-The, M. N. S. De Graaf, V. Orlova, A. Van Den Berg, A. D. Van Der Meer, K. Broersen and L. I. Segerink, Lab Chip, 2020, 20, 3132–3143 RSC.
- F. Rey, B. Barzaghini, A. Nardini, M. Bordoni, G. V. Zuccotti, C. Cereda, M. T. Raimondi and S. Carelli, Cells, 2020, 9(7), 1636 CrossRef CAS PubMed.
- B. Chueh, D. Huh, C. R. Kyrtsos, T. Houssin, N. Futai and S. Takayama, Anal. Chem., 2007, 79, 3504–3508 CrossRef CAS PubMed.
- N. J. Douville, Y.-C. Tung, R. Li, J. D. Wang, M. E. H. El-Sayed and S. Takayama, Anal. Chem., 2010, 82, 2505–2511 CrossRef CAS PubMed.
- S. A. Campbell, Fabrication Engineering at the Micro- and Nanoscale, Oxford University Press, 2013 Search PubMed.
- H. Le-The, M. Tibbe, J. Loessberg-Zahl, M. Palma do Carmo, M. van der Helm, J. Bomer, A. van den Berg, A. Leferink, L. Segerink and J. Eijkel, Nanoscale, 2018, 10, 7711–7718 RSC.
- M. C. Ceccarelli, M. C. Lefevre, A. Marino, F. Pignatelli, K. Krukiewicz, M. Battaglini and G. Ciofani, Lab Chip, 2024, 24(22), 5085–5100 RSC.
- C. M. B. Ho, S. H. Ng, K. H. H. Li and Y. J. Yoon, Lab Chip, 2015, 15, 3627–3637 RSC.
- I. Salmon, S. Grebenyuk, A. R. Abdel Fattah, G. Rustandi, T. Pilkington, C. Verfaillie and A. Ranga, Lab Chip, 2022, 22, 1615–1629 RSC.
- P. Paiè, F. Bragheri, R. M. Vazquez and R. Osellame, Lab Chip, 2014, 14, 1826–1833 RSC.
- F. Sala, C. Ficorella, R. Martínez Vázquez, H. M. Eichholz, J. A. Käs and R. Osellame, Front. Bioeng. Biotechnol., 2021, 9, 1–10 Search PubMed.
- F. Sima, K. Sugioka, R. M. Vázquez, R. Osellame, L. Kelemen and P. Ormos, Nanophotonics, 2018, 7, 613–634 CAS.
- M. Li, M. Zhu, R. Huang, K. Wang, Z. Zeng, L. Xiao, Y. Lin and D. Liu, Organs-on-a-Chip, 2023, 5, 100027 CrossRef CAS.
- E. L. J. Moya, E. Vandenhaute, E. Rizzi, M.-C. Boucau, J. Hachani, N. Maubon, F. Gosselet and M.-P. Dehouck, Pharmaceutics, 2021, 13(6), 892 CrossRef CAS PubMed.
- Q. Hou, L. Zhu, L. Wang, X. Liu, F. Xiao, Y. Xie, W. Zheng and X. Jiang, Nanoscale, 2022, 14, 3971 RSC.
- T. Liang, C. Gu, Y. Gan, Q. Wu, C. He, J. Tu, Y. Pan, Y. Qiu, L. Kong, H. Wan and P. Wang, Sens. Actuators, B, 2019, 301, 127004 CrossRef CAS.
- Z. Liao, Y. Zhang, Y. Li, Y. Miao, S. Gao, F. Lin, Y. Deng and L. Geng, Biosens. Bioelectron., 2019, 126, 697–706 CrossRef CAS PubMed.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed.
- P. Cai, Y. Zheng, Y. Sun, C. Zhang, Q. Zhang and Q. Liu, ACS Chem. Neurosci., 2021, 12, 3829–3837 CrossRef CAS PubMed.
- J.-H. Choi, M. Santhosh and J.-W. Choi, Micromachines, 2019, 11(1), 21 CrossRef PubMed.
- P. Mora, P.-L. Hollier, S. Guimbal, A. Abelanet, A. Diop, L. Cornuault, T. Couffinhal, S. Horng, A.-P. Gadeau, M.-A. Renault and C. Chapouly, PLoS Biol., 2020, 18, e3000946 CrossRef CAS PubMed.
- I. Pediaditakis, K. R. Kodella, D. V. Manatakis, C. Y. Le, C. D. Hinojosa, W. Tien-Street, E. S. Manolakos, K. Vekrellis, G. A. Hamilton, L. Ewart, L. L. Rubin and K. Karalis, Nat. Commun., 2021, 12, 5907 CrossRef CAS PubMed.
- Y. Shin, S. H. Choi, E. Kim, E. Bylykbashi, J. A. Kim, S. Chung, D. Y. Kim, R. D. Kamm and R. E. Tanzi, Adv. Sci., 2019, 6, 1900962 CrossRef CAS PubMed.
- J. P. Straehla, C. Hajal, H. C. Safford, G. S. Offeddu, N. Boehnke, T. G. Dacoba, J. Wyckoff, R. D. Kamm and P. T. Hammond, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2118697119 CrossRef CAS PubMed.
- H. Xu, Z. Li, Y. Yu, S. Sizdahkhani, W. S. Ho, F. Yin, L. Wang, G. Zhu, M. Zhang, L. Jiang, Z. Zhuang and J. Qin, Sci. Rep., 2016, 6, 36670 CrossRef CAS PubMed.
- S. E. Park, A. Georgescu and D. Huh, Science, 2019, 364, 960–965 CrossRef CAS PubMed.
- G. Costamagna, G. Pietro Comi and S. Corti, Int. J. Mol. Sci., 2021, 22(5), 2659 CrossRef CAS PubMed.
- J. Dinesh, R. K. Pathinarupothi and K. P. Soman DOI:10.21203/rs.3.rs-2503574/v1.
- C. E. Brocklehurst, E. Altmann, J. André, C. Bon, T. Caya, H. Davis, O. Decoret, D. Dunstan, P. Ertl, C. Ginsburg-Moraff, J. Grob, D. J. Gosling, G. Lapointe, D. Majumdar, A. N. Marziale, H. Mues, M. Palmieri, S. Racine, R. I. Robinson, C. Springer, K. Tan, W. Ulmer, L. West and R. Wyler, J. Med. Chem., 2024, 67, 5111–5112 CrossRef CAS PubMed.
- S. Van Dorpe, L. Lippens, R. Boiy, C. Pinheiro, G. Vergauwen, P. Rappu, I. Miinalainen, P. Tummers, H. Denys, O. De Wever and A. Hendrix, J. Nanobiotechnol., 2023, 21, 157 CrossRef CAS PubMed.
- R. Madaj, B. Geoffrey, A. Sanker and P. P. Valluri, J. Biomol. Struct. Dyn., 2022, 40, 7511–7516 CrossRef CAS PubMed.
- E. C. Ko, S. Spitz, F. M. Pramotton, O. M. Barr, C. Xu, G. Pavlou, S. Zhang, A. Tsai, A. Maaser-Hecker, M. Jorfi, S. H. Choi, R. E. Tanzi and R. D. Kamm, Front. Bioeng. Biotechnol., 2023, 11, 1251195 CrossRef PubMed.
- M. A. Kaisar, R. K. Sajja, S. Prasad, V. V. Abhyankar, T. Liles and L. Cucullo, Expert Opin. Drug Discovery, 2017, 12, 89–103 CrossRef PubMed.
- B. Srinivasan, A. R. Kolli, M. B. Esch, H. E. Abaci, M. L. Shuler and J. J. Hickman, J. Lab. Autom., 2015, 20, 107–126 CrossRef CAS PubMed.
- J. Ding and W. Qin, TrAC, Trends Anal. Chem., 2020, 124, 115803 CrossRef CAS.
- J. Liu, Y. Xu, S. Liu, S. Yu, Z. Yu and S. S. Low, Biosensors, 2022, 12(7), 494 CrossRef CAS PubMed.
- C.-A. Vu and W.-Y. Chen, Sensors, 2019, 19, 4214 CrossRef CAS PubMed.
- D. Petrovszki, F. R. Walter, J. P. Vigh, A. Kocsis, S. Valkai, M. A. Deli and A. Dér, Biomedicines, 2022, 10(1), 188 CrossRef CAS PubMed.
- Y. Liang and J.-Y. Yoon, Sens. Actuators Rep., 2021, 3, 100031 CrossRef.
- M. You, S. Yang, Y. An, F. Zhang and P. He, J. Electroanal. Chem., 2020, 862, 114017 CrossRef CAS.
- K. Dashtian, S. Hajati and M. Ghaedi, Sens. Actuators, B, 2021, 326, 128824 CrossRef CAS.
- N. Li, C. Nan, X. Mei, Y. Sun, H. Feng and Y. Li, Microchim. Acta, 2020, 187, 496 CrossRef CAS PubMed.
- M.-H. Lee, C.-C. Lin, J. L. Thomas, C.-K. Chan and H.-Y. Lin, Nanotechnology, 2021, 32, 18LT02 CrossRef CAS PubMed.
- A. Donatti, A. M. Canto, A. B. Godoi, D. C. da Rosa and I. Lopes-Cendes, Metabolites, 2020, 10(10), 389 CrossRef CAS PubMed.
- G. Ruiz-Vega, M. Soler, M. C. Estevez, P. Ramirez-Priego, M. D. Pazos, M. A. Noriega, Y. Margolles, C. Francés-Gómez, R. Geller, G. Matusali, F. Colavita, A. di Caro, J. M. Casasnovas, L. A. Fernández and L. M. Lechuga, Sens. Diagn., 2022, 1, 983–993 RSC.
- S. Patsiris, A. Karpouza, T. Exarchos and P. Vlamos, Exhaled Breath Analysis in Neurodegenerative Diseases, in Handbook of Computational Neurodegeneration, ed. P. Vlamos, I. S. Kotsireas and I. Tarnanas, Springer International Publishing, Cham, 2020, pp. 1–12 Search PubMed.
- M.-H. Lee, J. L. Thomas, Z.-L. Su, W.-K. Yeh, A. S. Monzel, S. Bolognin, J. C. Schwamborn, C.-H. Yang and H.-Y. Lin, Biosens. Bioelectron., 2021, 175, 112852 CrossRef CAS PubMed.
- Y.-T. Chen, Y.-C. Lee, Y.-H. Lai, J.-C. Lim, N.-T. Huang, C.-T. Lin and J.-J. Huang, Biosensors, 2020, 10(12), 209 CrossRef CAS PubMed.
- P. Bollella and L. Gorton, Curr. Opin. Electrochem., 2018, 10, 157–173 CrossRef CAS.
- D. Dhanjai, A. Sinha, X. Lu, L. Wu, D. Tan, Y. Li, J. Chen and R. Jain, TrAC, Trends Anal. Chem., 2018, 98, 174–189 CrossRef.
- S. G. Taneva, S. Krumova, F. Bogár, A. Kincses, S. Stoichev, S. Todinova, A. Danailova, J. Horváth, Z. Násztor, L. Kelemen and A. Dér, Int. J. Biol. Macromol., 2021, 175, 19–29 CrossRef CAS PubMed.
- R. Vashistha, A. K. Dangi, A. Kumar, D. Chhabra and P. Shukla, 3 Biotech, 2018, 8, 358 CrossRef PubMed.
- F. Inci, Langmuir, 2022, 38, 1897–1909 CrossRef CAS PubMed.
- H. Hampel, E. J. Goetzl, D. Kapogiannis, S. Lista and A. Vergallo, Front. Pharmacol., 2019, 10, 310 CrossRef CAS PubMed.
- C. S. Huertas, D. Fariña and L. M. Lechuga, ACS Sens., 2016, 1, 748–756 CrossRef CAS.
- J. Maldonado, A. B. González-Guerrero, C. Domínguez and L. M. Lechuga, Biosens. Bioelectron., 2016, 85, 310–316 CrossRef CAS PubMed.
- J. Maldonado, A. B. González-Guerrero, A. Fernández-Gavela, J. J. González-López and L. M. Lechuga, Ultrasensitive Label-Free Detection of Unamplified Multidrug-Resistance Bacteria Genes with a Bimodal Waveguide Interferometric Biosensor, Diagnostics, 2020 DOI:10.3390/diagnostics10100845.
- E. W. K. Young and C. Moraes, Integr. Biol., 2015, 7, 962–966 CrossRef PubMed.
- I. A. Morales, C.-M. Boghdady, B. E. Campbell and C. Moraes, Front. Bioeng. Biotechnol., 2022, 10, 1060895 CrossRef PubMed.
- O. F. Vila, M. Chavez, S. P. Ma, K. Yeager, L. V. Zholudeva, J. M. Colón-Mercado, Y. Qu, T. R. Nash, C. Lai, C. M. Feliciano, M. Carter, R. D. Kamm, L. M. Judge, B. R. Conklin, M. E. Ward, T. C. McDevitt and G. Vunjak-Novakovic, Biomaterials, 2021, 276, 121033 CrossRef CAS PubMed.
- L. MacQueen, O. Chebotarev, C. A. Simmons and Y. Sun, Lab Chip, 2012, 12, 4178–4184 RSC.
- G. Siciliano, M. S. Chiriacò, F. Ferrara, A. Turco, L. Velardi, M. A. Signore, M. Esposito, G. Gigli and E. Primiceri, Analyst, 2023, 148, 4447–4455 RSC.
- M.-H. Lee, J. L. Thomas, M.-H. Ho, C. Yuan and H.-Y. Lin, ACS Appl. Mater. Interfaces, 2010, 2, 1729–1736 CrossRef CAS PubMed.
- M.-H. Lee, J. L. Thomas, C.-L. Liao, S. Jurcevic, T. Crnogorac-Jurcevic and H.-Y. Lin, Sep. Purif. Technol., 2018, 192, 213–219 CrossRef CAS.
- H.-Y. Lin, M.-S. Ho and M.-H. Lee, Biosens. Bioelectron., 2009, 25, 579–586 CrossRef CAS PubMed.
- M.-H. Lee, J. L. Thomas, H.-Y. Tseng, W.-C. Lin, B.-D. Liu and H.-Y. Lin, ACS Appl. Mater. Interfaces, 2011, 3, 3064–3071 CrossRef CAS PubMed.
- M.-H. Lee, K.-H. Liu, J. L. Thomas, C.-Y. Chen, C.-Y. Chen, C.-H. Yang and H.-Y. Lin, Biosens. Bioelectron., 2022, 200, 113930 CrossRef CAS PubMed.
- K. Bartold, Z. Iskierko, P. Borowicz, K. Noworyta, C.-Y. Lin, J. Kalecki, P. S. Sharma, H.-Y. Lin and W. Kutner, Biosens. Bioelectron., 2022, 208, 114203 CrossRef CAS PubMed.
- M.-H. Lee, J. L. Thomas, H.-Y. Wang, C.-C. Chang, C.-C. Lin and H.-Y. Lin, J. Mater. Chem., 2012, 22, 24644–24651 RSC.
- M.-H. Lee, J. L. Thomas, J.-Z. Chen, J.-S. Jan and H.-Y. Lin, Chem. Commun., 2016, 52, 2137–2140 RSC.
- A. D. Batista, W. R. Silva and B. Mizaikoff, Med. Devices Sens., 2021, 4, e10166 CrossRef CAS.
- M.-H. Lee, J.-S. Jan, J. L. Thomas, Y.-P. Shih, J.-A. Li, C.-Y. Lin, T. Ooya, L. Barna, M. Mészáros, A. Harazin, G. Porkoláb, S. Veszelka, M. A. Deli and H.-Y. Lin, Cells, 2022, 11(16), 2584 CrossRef CAS PubMed.
- M. A. Busquets, A. Espargaró, R. Sabaté and J. Estelrich, Nanomaterials, 2015, 5, 2231–2248 CrossRef CAS PubMed.
- Z. Zhao, A. R. Nelson, C. Betsholtz and B. V. Zlokovic, Cell, 2015, 163, 1064–1078 CrossRef CAS PubMed.
- L. Kaplan, B. W. Chow and C. Gu, Nat. Rev. Neurosci., 2020, 21, 416–432 CrossRef CAS PubMed.
- A. R. Cantelmo, L.-C. Conradi, A. Brajic, J. Goveia, J. Kalucka, A. Pircher, P. Chaturvedi, J. Hol, B. Thienpont, L.-A. Teuwen, S. Schoors, B. Boeckx, J. Vriens, A. Kuchnio, K. Veys, B. Cruys, L. Finotto, L. Treps, T. E. Stav-Noraas, F. Bifari, P. Stapor, I. Decimo, K. Kampen, K. De Bock, G. Haraldsen, L. Schoonjans, T. Rabelink, G. Eelen, B. Ghesquière, J. Rehman, D. Lambrechts, A. B. Malik, M. Dewerchin and P. Carmeliet, Cancer Cell, 2016, 30, 968–985 CrossRef CAS PubMed.
- H.-G. Lee, M. A. Wheeler and F. J. Quintana, Nat. Rev. Drug Discovery, 2022, 21, 339–358 CrossRef CAS PubMed.
- S.-F. Huang and O. O. Ogunshola, Neural Regener. Res., 2021, 16, 1786–1787 CrossRef CAS PubMed.
- F. De Castro, M. Benedetti, L. Del Coco and F. P. Fanizzi, Molecules, 2019, 24(12), 2240 CrossRef CAS PubMed.
- J. K. Nicholson and J. C. Lindon, Nature, 2008, 455, 1054–1056 CrossRef CAS PubMed.
- F. De Castro, V. Vergaro, M. Benedetti, F. Baldassarre, L. Del Coco, M. M. Dell’Anna, P. Mastrorilli, F. P. Fanizzi and G. Ciccarella, ACS Appl. Bio Mater., 2020, 3, 6836–6851 CrossRef CAS PubMed.
- E. Stefàno, A. Muscella, M. Benedetti, F. De Castro, F. P. Fanizzi and S. Marsigliante, Biochem. Pharmacol., 2022, 202, 115124 CrossRef PubMed.
- F. De Castro, E. Stefàno, E. De Luca, A. Muscella, S. Marsigliante, M. Benedetti and F. P. Fanizzi, Bioinorg. Chem. Appl., 2022, 2022, 8932137 CrossRef PubMed.
- D. B. Castellanos, C. A. Martín-Jiménez, A. Pinzón, G. E. Barreto, G. F. Padilla-González, A. Aristizábal, M. Zuluaga and J. González Santos, Biomolecules, 2022, 12(7), 986 CrossRef CAS PubMed.
- E. Nwadozi, M. Rudnicki and T. L. Haas, Front. Cell Dev. Biol., 2020, 8, 77 CrossRef PubMed.
- A. Herland, B. M. Maoz, E. A. FitzGerald, T. Grevesse, C. Vidoudez, S. P. Sheehy, N. Budnik, S. Dauth, R. Mannix, B. Budnik, K. K. Parker and D. E. Ingber, Adv. Biosyst., 2020, 4, 1900230 CrossRef CAS PubMed.
- J. A. Brown, S. G. Codreanu, M. Shi, S. D. Sherrod, D. A. Markov, M. D. Neely, C. M. Britt, O. S. Hoilett, R. S. Reiserer, P. C. Samson, L. J. McCawley, D. J. Webb, A. B. Bowman, J. A. McLean and J. P. Wikswo, J. Neuroinflammation, 2016, 13, 306 CrossRef PubMed.
- B. M. Maoz, A. Herland, E. A. FitzGerald, T. Grevesse, C. Vidoudez, A. R. Pacheco, S. P. Sheehy, T.-E. Park, S. Dauth, R. Mannix, N. Budnik, K. Shores, A. Cho, J. C. Nawroth, D. Segrè, B. Budnik, D. E. Ingber and K. K. Parker, Nat. Biotechnol., 2018, 36, 865–874 CrossRef CAS PubMed.
- S.-F. Huang, S. Fischer, A. Koshkin, E. Laczko, D. Fischer and O. O. Ogunshola, Sci. Rep., 2020, 10, 7760 CrossRef CAS PubMed.
- K. Unterkofler, J. King, P. Mochalski, M. Jandacka, H. Koc, S. Teschl, A. Amann and G. Teschl, J. Breath Res., 2015, 9, 36002 CrossRef PubMed.
- H. Koc, J. King, G. Teschl, K. Unterkofler, S. Teschl, P. Mochalski, H. Hinterhuber and A. Amann, J. Breath Res., 2011, 5, 37102 CrossRef CAS PubMed.
- W. Filipiak, P. Mochalski, A. Filipiak, C. Ager, R. Cumeras, C. E. Davis, A. Agapiou, K. Unterkofler and J. Troppmair, Curr. Med. Chem., 2016, 23, 2112–2131 CrossRef CAS PubMed.
- A. Catino, G. de Gennaro, A. Di Gilio, L. Facchini, D. Galetta, J. Palmisani, F. Porcelli and N. Varesano, Cancers, 2019, 11(6), 831 CrossRef CAS PubMed.
- T. Hüppe, D. Lorenz, F. Maurer, F. W. Albrecht, K. Schnauber, B. Wolf, D. I. Sessler, T. Volk, T. Fink and S. Kreuer, J. Breath Res., 2016, 10, 16016 CrossRef PubMed.
- M. Malek and M. Nematbakhsh, J. Renal Inj. Prev., 2015, 4, 20–27 CAS.
- J.-P. Bach, M. Gold, D. Mengel, A. Hattesohl, D. Lubbe, S. Schmid, B. Tackenberg, J. Rieke, S. Maddula, J. I. Baumbach, C. Nell, T. Boeselt, J. Michelis, J. Alferink, M. Heneka, W. Oertel, F. Jessen, S. Janciauskiene, C. Vogelmeier, R. Dodel and A. R. Koczulla, PLoS One, 2015, 10, e0132227 CrossRef PubMed.
- A. Tiele, A. Wicaksono, E. Daulton, E. Ifeachor, V. Eyre, S. Clarke, L. Timings, S. Pearson, J. A. Covington and X. Li, J. Breath Res., 2020, 14, 26003 CrossRef CAS PubMed.
- S. Emam, M. Nasrollahpour, B. Colarusso, X. Cai, S. Grant, P. Kulkarni, A. Ekenseair, C. Gharagouzloo, C. F. Ferris and N.-X. Sun, Alzheimer's Dement., 2020, 12, e12088 Search PubMed.
- H.-C. Lau, J.-B. Yu, H.-W. Lee, J.-S. Huh and J.-O. Lim, Sensors, 2017, 17(8), 1783 CrossRef PubMed.
- U. Tisch, I. Schlesinger, R. Ionescu, M. Nassar, N. Axelrod, D. Robertman, Y. Tessler, F. Azar, A. Marmur, J. Aharon-Peretz and H. Haick, Nanomedicine, 2013, 8(8), 43–56 CrossRef CAS PubMed.
- C. Wang, M. Li, H. Jiang, H. Tong, Y. Feng, Y. Wang, X. Pi, L. Guo, M. Nie, H. Feng and E. Li, Sci. Rep., 2016, 6, 26120 CrossRef CAS PubMed.
- W. Li, R. Pan, Z. Qi and K. J. Liu, Brain Circ., 2018, 4, 145–152 CrossRef PubMed.
- E. Sinclair, C. Walton-Doyle, D. Sarkar, K. A. Hollywood, J. Milne, S. H. Lim, T. Kunath, A. M. Rijs, R. M. A. de Bie, M. Silverdale, D. K. Trivedi and P. Barran, ACS Cent. Sci., 2021, 7, 300–306 CrossRef CAS PubMed.
- D. K. Trivedi, E. Sinclair, Y. Xu, D. Sarkar, C. Walton-Doyle, C. Liscio, P. Banks, J. Milne, M. Silverdale, T. Kunath, R. Goodacre and P. Barran, ACS Cent. Sci., 2019, 5, 599–606 CrossRef CAS PubMed.
- T. Tsuda, T. Nonome, S. Goto, J. Takeda, M. Tsunoda, M. Hirayama and K. Ohno, Chromatography, 2019, 40, 149–155 CrossRef CAS.
- H. Tian, S. Li, H. Wen, X. Zhang and J. Li, J. Chromatogr. A, 2020, 1614, 460717 CrossRef CAS PubMed.
- A. Mazzatenta, M. Pokorski, F. Sartucci, L. Domenici and C. Di Giulio, Respir. Physiol. Neurobiol., 2015, 209, 81–84 CrossRef CAS PubMed.
- M. Lueno, H. Dobrowolny, D. Gescher, L. Gbaoui, G. Meyer-Lotz, C. Hoeschen and T. Frodl, Front. Psychiatry, 2022, 13, 819607 CrossRef PubMed.
- M. Leemans, P. Bauër, V. Cuzuel, E. Audureau and I. Fromantin, Biomarker Insights, 2022, 17, 11772719221100709 CrossRef PubMed.
- M. Skawinski, F. J. van Schooten and A. Smolinska, J. Breath Res., 2025, 19(1), 15001 CrossRef PubMed.
- N. Peled, O. Barash, U. Tisch, R. Ionescu, Y. Y. Broza, M. Ilouze, J. Mattei, P. A. J. Bunn, F. R. Hirsch and H. Haick, Nanomedicine, 2013, 9, 758–766 CrossRef CAS PubMed.
- R. Taware, K. Taunk, T. V. S. Kumar, J. A. M. Pereira, J. S. Câmara, H. A. Nagarajaram, G. C. Kundu and S. Rapole, Metabolomics, 2020, 16, 21 CrossRef CAS PubMed.
- W. Bi, S. Cai, T. Lei and L. Wang, Ageing Res. Rev., 2023, 87, 101921 CrossRef CAS PubMed.
- E. Jagtiani, M. Yeolekar, S. Naik and V. Patravale, J. Controlled Release, 2022, 343, 13–30 CrossRef CAS PubMed.
- S. Deng, C. Li, J. Cao, Z. Cui, J. Du, Z. Fu, H. Yang and P. Chen, Theranostics, 2023, 13, 4526–4558 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |