Open Access Article
Open Access ArticleCan large language models predict the hydrophobicity of metal–organic frameworks?†
Xiaoyu
Wu
 and
Jianwen
Jiang
and
Jianwen
Jiang
 *
*
Department of Chemical and Biomolecular Engineering, National University of Singapore, Singapore, 117576, Singapore. E-mail: chejj@nus.edu.sg
First published on 4th April 2025
Abstract
Recent advances in large language models (LLMs) offer a transformative paradigm for data-driven materials discovery. Herein, we exploit the potential of LLMs in predicting the hydrophobicity of metal–organic frameworks (MOFs). By fine-tuning the state-of-the-art Gemini-1.5 model exclusively on the chemical language of MOFs, we demonstrate its capacity to deliver weighted accuracies that surpass those of traditional machine learning approaches based on sophisticated descriptors. To further interpret the chemical “understanding” embedded within the Gemini model, we conduct systematic moiety masking experiments, where our fine-tuned Gemini model consistently retains robust predictive performance even with partial information loss. Finally, we show the practical applicability of the Gemini model via a blind test on solvent- and ion-containing MOFs. The results illustrate that Gemini, combined with lightweight fine-tuning on chemically annotated texts, can serve as a powerful tool for rapidly screening MOFs in pursuit of hydrophobic candidates. Taking a step forward, our work underscores the potential of LLMs in offering robust and data-efficient approaches to accelerate the discovery of functional materials.
1. Introduction
Metal–organic Frameworks (MOFs) are a versatile class of nanoporous materials typically synthesized under relatively mild hydrothermal or solvothermal conditions from metal ions and organic ligands.1 Their modular architectures enable precise tunability of pore structures and functional properties, rendering them attractive for a wide range of applications, including gas storage, separation and catalysis.2 A subset of MOFs exhibit low affinity for water and this class of hydrophobic MOFs has garnered increasing attention for their potential utility under humid conditions.3 To date, over 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MOFs have been experimentally produced; however, many of them were synthesized without reporting their hydrophobicity.4 To streamline the identification of hydrophobic MOFs, Henry's constant of water (KH) was adopted as a metric in a computational workflow5 to duly benchmark against representative hydrophobic structures (e.g., ZIF-8
000 MOFs have been experimentally produced; however, many of them were synthesized without reporting their hydrophobicity.4 To streamline the identification of hydrophobic MOFs, Henry's constant of water (KH) was adopted as a metric in a computational workflow5 to duly benchmark against representative hydrophobic structures (e.g., ZIF-8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6). This approach has been integrated with high-throughput computational screening to shortlist promising candidates for various applications (e.g., CO2 capture and removal of toxic gases).7,8 Nevertheless, in principle, the combinatorial design space of MOFs is infinite due to the myriad coupling chemistries of building units as well as their underlying topologies. As such, KH calculations to identify hydrophobic MOFs on a trial-and-error basis are exceedingly laborious.9
6). This approach has been integrated with high-throughput computational screening to shortlist promising candidates for various applications (e.g., CO2 capture and removal of toxic gases).7,8 Nevertheless, in principle, the combinatorial design space of MOFs is infinite due to the myriad coupling chemistries of building units as well as their underlying topologies. As such, KH calculations to identify hydrophobic MOFs on a trial-and-error basis are exceedingly laborious.9
Machine learning (ML) has emerged as a powerful alternative in this regard, as it has already proven to be indispensable in the design of functional materials.10 One of the most compelling advances in ML is the advent of large language models (LLMs) such as ChatGPT11 and Gemini,12 which have been trained on massive text corpora spanning diverse disciplines.13 These foundational LLMs excel at generating language textual responses from simple prompts, which in many instances, are indistinguishable from human articulations. Such generative capabilities have unleashed exciting opportunities for digital chemistry including chemical synthesis,14–16 dataset mining,17–19 and pattern recognition.20
One of the fascinating aspects of LLMs lies in their predictive capacity in both forward and inverse chemical discovery, relying solely on chemical language instead of engineered molecular descriptors.21 Though typically pretrained for general purposes, LLMs can significantly enhance their predictive accuracy for chemistry-specific tasks through fine-tuning with domain knowledge even based on light-weight LLMs (i.e., LLMs pretrained with fewer parameters, e.g., 8B, 70B).22 Molecular representations including simplified molecular input line entry systems (SMILES)23 and self-referencing embedded strings (SELFIES)24 have facilitated chemical language modeling.25,26 These chemical notations can be further augmented with metal information, thereby capturing both the inorganic and organic constituents of MOFs.27,28 As exemplified in Scheme 1, a typical MOF named Cu-BTC29 can be rendered in augmented SMILES and SELFIES notations, each meticulously tokenized into smaller units carrying unique IDs for LLM ingestion. Notably, variably pretrained LLMs may adopt different tokenization approaches, and the default tokenizer of Gemini is employed here.
 | ||
| Scheme 1 Tokenized SMILES and SELFIES strings encoding Cu-BTC for Gemini. The tokenization is visualized via LLM-text-tokenization: https://github.com/glaforge/llm-text-tokenization. | ||
In this work, we embark on the fine-tuning of a cutting-edge LLM, Gemini-1.5,30 leveraging the latest CoRE-2024 database with thousands of “computation-ready” experimental MOFs.28 Particularly, we focus on MOFs from the all-solvents-removed (ASR) subset of single metal and single linker type to fine-tune the base model. For both binary and quaternary classifications of hydrophobicity, the fine-tuned Gemini achieves comparable overall accuracy and notably excels in weighted accuracy—a critical advantage given the class imbalance inherent in the large dataset. Furthermore, we assess the robustness and transferability of the model through moiety masking experiments and a rigorous blind test on distinct MOFs. These findings demonstrate the coherence between LLMs and digital chemistry, potentially shedding light on harnessing the power of Gemini as a useful agent for open questions in the broader physical sciences.
2. Methodology
2.1. Dataset
We started with the ASR subset from the CoRE-MOF-2024 database, which contains 8857 meticulously curated experimental MOFs with computed water affinity (i.e., KH) via the Widom insertion method.28 Considering synthetic accessibility, we adapted a similar decomposition-based protocol by Pouya et al.,31 which narrowed down the dataset to 2642 MOFs of single metal and single linker type. As depicted in Fig. S1a and 1a†, 2642 MOFs can be categorized using binary classification with two labels: strong hydrophobic (Strong) and weak hydrophobic (Weak), as well as quaternary classification with four labels: super strong hydrophobic (SS), strong hydrophobic (S), weak hydrophobic (W), and super weak hydrophobic (SW). The resulting dataset spans diverse pore geometric properties (Fig. S2 and S3†). For both binary and quaternary classifications, the dataset was split into an 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio, with the former as a training set for model development and the latter as a hold-out test set for model evaluation. Despite non-significant bias across any other labels in the training set compared to the test set, we noticed a notable imbalance in the distribution of labels, with SS being the least populated (Fig. 1b). This is not unexpected due to the challenging water-repelling nature of MOFs,32 which may pose difficulty in the model prediction. Generally, class imbalance remains a core challenge in applying ML to diverse research topics in physical sciences, including MOF discovery33 and photocatalyst design,34 as such tasks are not always exhaustively addressed with solely hand-crafted descriptors.35
20 ratio, with the former as a training set for model development and the latter as a hold-out test set for model evaluation. Despite non-significant bias across any other labels in the training set compared to the test set, we noticed a notable imbalance in the distribution of labels, with SS being the least populated (Fig. 1b). This is not unexpected due to the challenging water-repelling nature of MOFs,32 which may pose difficulty in the model prediction. Generally, class imbalance remains a core challenge in applying ML to diverse research topics in physical sciences, including MOF discovery33 and photocatalyst design,34 as such tasks are not always exhaustively addressed with solely hand-crafted descriptors.35
 | ||
| Fig. 1 (a) Kernel density estimations of quaternary classification (SS, S, W and SW) versus density. The vertical axis denotes probability. (b) Data distribution in training and test sets. | ||
2.2. Fine-tuning gemini
Fine-tuning refers to the process of adapting a base model, which has already been pre-trained on a vast corpora of generalized data, to perform better on a more specific task. During this process, parameters in the model are adjusted to minimize errors for a new task. This allows the model to tailor its domain knowledge and enhances its task-specific performance. Gemini-1.5 is a state-of-the-art foundational LLM developed by Google.30 It is characterized by its significantly enhanced processing speed and long-text effectiveness, making it a valuable LLM for fine-tuning where iterative adjustments and accurate predictions are beneficial. Gemini has demonstrated fine-tuning capability in the medical domain.36In this work, we fine-tuned the “gemini-1.5-flash-001-tuning” base model in Google AI Studio. The training dataset comprised labeled MOFs, alongside two molecular representations: SMILES and SELFIES, both augmented with inorganic motifs (hereafter referred to simply as SMILES and SELFIES). During fine-tuning, the training data were structured as prompt and response pairs: (“Representation”, “Label”). In such a format, SMILES or SELFIES served as prompts and were completed with corresponding hydrophobicity labels, which were categorized as [0, 1] and [0, 1, 2, 3] for binary and quaternary classifications, respectively. For computational feasibility and model stability, the model was tuned with 3 epochs and a batch size of 16 with a learning rate of 2 × 10−4 to reach a minimum loss. Due to the general knowledge stored in Gemini, a predicted response from an out-of-sample prompted MOF may not always be expected to predict an ideal label. For these MOFs, augmented prompts were utilized, as detailed in Section S2 in the ESI.†
The fine-tuned Gemini for prediction of MOF hydrophobicity was benchmarked against descriptor-based supervised ML models. Global pore descriptors (e.g., pore size) computed via Zeo++37 and the revised autocorrelations (RACs)38,39 were adopted for featurizing MOFs (detailed in Tables S1 and S2†). All the baseline models were trained using Support Vector Machine (SVM), with hyperparameters: {‘C’: [0.1, 1, 10], ‘kernel’: [‘linear’, ‘rbf’]} tuned through 10-fold cross-validations on the same training set as in the fine-tuned Gemini, with the identical hold-out test set and blind test set to ensure fair comparison.
3. Results and discussion
3.1. Fine-tuning results
The fine-tuned Gemini was evaluated on the hold-out test set using the overall accuracy and weighted F1-score, which accounts for data imbalance as discussed in Section 2.1. As summarized in Table 1, for binary classification, the fine-tuned Gemini based on SMILES achieves exemplary performance, with an overall accuracy of 0.78 and a weighted F1-score of 0.74, respectively. However, when shifted to quaternary classification, its predictive capacity drops slightly, achieving 0.73 overall accuracy and 0.70 weighted F1-score, respectively. A similar trend is observed for the fine-tuned Gemini with SELFIES, albeit with less predictive performance. Though SELFIES has demonstrated a certain capacity in representing reticular chemistry,40 its generic tokenization appears to dilute additional chemical information compared to SMILES.41 Moreover, while Gemini is a closed-source LLM, its stored cut-off knowledge likely encompasses public datasets with diverse chemical information, including SMILES notations.42 In contrast, datasets incorporating SELFIES remain scanty, which leads to less compatibility of Gemini with property prediction based on SELFIES. We anticipate that as chemically-rich datasets including SELFIES notations become more prevalent, LLMs like Gemini may demonstrate improved performance in future SELFIES-based classification tasks. Here, the fine-tuned Gemini with SMILES stands out as the optimal approximator for both binary and quaternary classifications.| Model | Binary classification | Quaternary classification | ||
|---|---|---|---|---|
| Accuracya | F1-scoreb | Accuracy | F1-score | |
| a For binary classification, the accuracy metric reflects the model's ability to correctly classify instances into two labels (Strong and Weak). For quaternary classification, the accuracy metric aggregates the overall correct four labels (SS, S, W and SW). b The weighted F1-score combines precision and recall into a single metric, accounting for class imbalance by weighing each class's F1-score by the proportion of instances in that class. c The descriptor-based models were trained using the SVM classifier in scikit-learn, with hyperparameters: [‘C’: [0.1, 1, 10], ‘kernel’: [‘linear’, ‘rbf’]] tuned through 10-fold cross-validation. | ||||
| Gemini (SMILES) | 0.78 | 0.74 | 0.73 | 0.70 |
| Gemini (SELFIES) | 0.73 | 0.73 | 0.71 | 0.67 |
| Porec | 0.76 | 0.66 | 0.72 | 0.62 |
| Pore + RACs | 0.77 | 0.71 | 0.75 | 0.64 |
Descriptor-based ML models were used to benchmark the performance of the fine-tuned Gemini. For effective featurization, we adopted pore descriptors and RACs. Leveraging non-weighted molecular graphs to derive the products or differences of atomic heuristics, RACs have been used to effectively map the chemical space of MOFs, encompassing linker and metal chemistry.43,44 Combined with pore descriptors, RACs have been shown to be effective and robust in predicting various properties of MOFs.45–49 As illustrated in Table 1, despite being trained upon a simple text-based representation like SMILES, the fine-tuned Gemini has predictive capability comparable with that of sophisticated descriptor-based models, with a slight underestimation of overall accuracy for quaternary classification. This is expected, to a certain extent, as RACs embed both similarities and dissimilarities, thus better encapsulating chemical topology and connectivity as well.47 Significantly, our fine-tuned Gemini exhibits commendable weighted accuracy, maintaining a weighted F1-score of 0.70 even as the classification complexity increases from binary to quaternary. As depicted in Fig. 2a, the fine-tuned Gemini correctly distinguishes most of the “Strong” labels (376 in the bottom-left cell), which is slightly less predicted than Pore + RACs (Fig. S5a†). Conversely, Pore + RACs fail to predict their counterparts labelled as “Weak”, with only 15 correct predictions and 111 samples overpredicted as “Strong”. Such mislabeling in “Weak” hydrophobic MOFs is more pronounced for quaternary classification (Fig. S5b†). Despite offering a finer-grained picture of misclassification across the four labels, the fine-tuned Gemini outperforms in predictions of “SS”, “W” and “SW” (Fig. 2b), leading to a higher weighted F1-score. To further evaluate the consistency of fine-tuning results, we examined the effect of a random state in the training/test split of the dataset, all of which yielded stable model performance (±0.01), as summarized in Table S3.† These results position the fine-tuned Gemini as an efficient tool for sorting a large number of MOFs for subsequent, time-consuming computations (e.g., via first-principles molecular dynamics simulation50) to more precisely determine hydrophobicity. While descriptor-based ML models demonstrate better predictive capability through advanced feature engineering, they typically necessitate extensive preprocessing or the derivation of specialized property-specific descriptors that rely heavily on precise atomic positions. In contrast, our LLM-based method exclusively leverages text-based chemical representations (i.e., SMILES/SELFIES) that encode the building units in MOFs. These engineering-free string-based representations require no structural optimization, thus facilitating rapid screening across a diverse and potentially unexplored topology space without exhaustive structural validation. We should note that our method is not intended to replace direct KH calculations (e.g., via the Widom insertion method); rather, it serves as a rapid, surrogate screening tool capable of efficiently narrowing down a large candidate pool. Nevertheless, we note that such a LLM based model may not be ideal for near-quantitative predictions, where regression models might deliver higher accuracy.51,52
 | ||
| Fig. 2 Confusion matrices on the test set by the fine-tuned Gemini. (a) Binary classification and (b) quaternary classification. | ||
Acquiring high-fidelity data through an experimental or computational approach can be time-consuming and costly. Ideally, a model should maintain data efficiency, even trained on a limited budget of data.53,54 In this context, we fine-tuned Gemini using various training set ratios, ranging from 0.2 to 0.8 out of 2112 total training data points to assess its data efficiency. For fair comparison, the ML model with Pore + RACs was also trained on the same classification tasks using the same training data as in Gemini. The learning curves on the test set by the fine-tuned Gemini and the ML model are presented in Fig. 3a. Intriguingly, both models demonstrate similar accuracy across a wide range of training set ratios, with predictive performance significantly compromised when trained on less than 845 data points (i.e., 0.4 training set ratio). Such an effect is markedly amplified for quaternary classification, where the model performance drops below random guessing (<0.5 accuracy). The accuracy of both models saturates at a training set ratio of 0.6, with marginal improvement afterwards. The optimal accuracy scores of 0.78 and 0.73 are achieved by the fine-tuned Gemini for binary and quaternary classifications, respectively. Such an early performance ceiling likely stems from the complexity of the hydrophobic nature in reticular chemistry, a subtle property governed by a complex interplay that is difficult to fully encapsulate. Discretizing hydrophobicity into binary or quaternary classification may also introduce discontinuous boundaries for KH that challenge the capability of model prediction.55
Prompted by the proven data efficiency, we examined the dissimilarity of the feature space encoded by Pore + RACs and SMILES, respectively, through t-distributed stochastic neighbor embedding (t-SNE).56 In a t-SNE map, points are spatially arranged such that the closer the two points, the more similar the two structures are, as described by the encoding fingerprints. The SMILES representation, as text-based input, was tokenized through the BERT model57 to emulate the enclosed dimensions captured by the fine-tuned Gemini. As evidenced in Fig. 3b and c, increasing the training set ratio leads to a progressively denser and more saturated feature space. The dense pattern stabilizes after a training set ratio of 0.6, highlighting an optimal balance between training set size and predictive performance. We acknowledge the challenge presented by the SS label in the quaternary classification even with a full training set (1.0), which notably yields several misclassifications in the fine-tuned Gemini model predictions. To interpret this difficulty, we examined the chemical space in both trained and tested MOFs labeled as SS and S. As clearly illustrated in Fig. S6,† the chemical space coverage for SS in the training set is significantly diluted compared to that for S. This limited chemical diversity contributes to the difficulty of the fine-tuned Gemini in accurately distinguishing closely overlapping chemical syntax, leading to multiple SS-labeled MOFs being misclassified as S in the test set. We anticipate that enriching the chemical diversity, particularly for the underrepresented SS label, could substantially enhance the predictive accuracy of the fine-tuned Gemini for these challenging instances. As such, the fine-tuned Gemini appears to be an effective approach for data-driven discovery of MOFs, as it requires minimal efforts in preparing input (simple as a string-based representation as SMILES), compared to laborious feature engineering efforts for descriptor-based ML models. Despite this simplicity, the fine-tuned Gemini is on par with descriptor-based ML in terms of overall accuracy but achieves higher weighted accuracy, underscoring its potential to balance predictive power with practical implementations.
3.2. Moiety masking experiments
We have demonstrated the data-efficiency and prediction capability of the fine-tuned Gemini. Nevertheless, LLMs like Gemini, often perceived as “black boxes”, do not facilitate interpretability and tunable hyperparameters like ML models trained on carefully engineered descriptors. To interpret the captured inner sense as well as the robustness against information loss of the fine-tuned Gemini, we conducted a series of moiety masking experiments, systematically masking or ‘ablating’ sections representing specific chemical moieties within the SMILES prompts. As exemplified in such a moiety masking experiment for Cu-BTC (Fig. 4), we replaced one of the Cu identities with a <missing> annotation. By deliberately attacking chemical substructures within the SMILES representation, we assessed whether the fine-tuned Gemini could distinguish solid and meaningful chemical patterns, rather than merely “memorized” training data. | ||
| Fig. 4 Schematic illustration of a moiety masking experiment, where Cu is replaced with a <missing> annotation. Color scheme: Cu, brown; O, red; C, grey; H, white. | ||
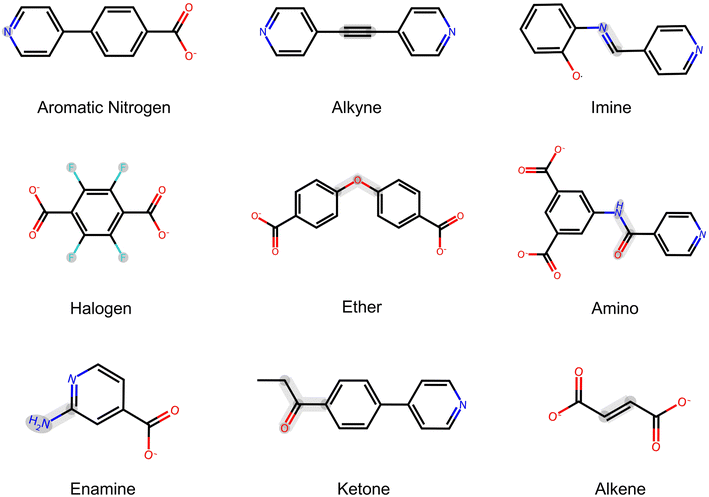
Unlike a metal identity, a linker chemical moiety may exhibit varied forms in a SMILES representation; we thus adopted SMILES arbitrary target specification (SMARTS)58 to locate and identify the substructures of intended linker chemical moieties in SMILES strings. As illustrated in Table 2 and Fig. 5, we considered a list of 9 linkers, which were identified by SMARTS substructure matching using the RDKit package.59 Each of these linker chemical moieties was subsequently subjected to a moiety masking experiment to assess model robustness. As previously discussed, the fine-tuned Gemini maintains both data efficiency and accuracy at a training set ratio of 0.6. To ensure a diverse coverage of linker chemistry, the base model was fine-tuned anew using 1268 data points from the full training data set (i.e., at a training set ratio of 0.6), with the remaining 845 data points combined with 529 data points from the original test set for out-of-sample predictions. The moiety masking experiments were conducted on the data points correctly predicted in this new out-of-sample test, resulting in 1019 and 962 SMILES representations to be tested for binary and quaternary classifications, respectively.
| Moiety | SMARTS | Binary | Quaternary | ||
|---|---|---|---|---|---|
| No. of tests | Accuracy | No. of tests | Accuracy | ||
| Metal | — | 1019 | 0.96 | 962 | 0.92 |
| Aromatic N | [n; a] | 317 | 0.97 | 299 | 0.94 |
| Alkyne | [C#C] | 10 | 1.00 | 10 | 1.00 |
| Imine | [$([CX3]([#6])[#6]),$([CX3H][#6])] = [$([NX2][#6]),$([NX2H])] | 18 | 1.00 | 14 | 1.00 |
| Halogen | [F,Cl,Br,I] | 69 | 0.99 | 63 | 0.95 |
| Ether | [$([CX4]),$([cX3])]O[$([CX4]),$([cX3])] | 75 | 0.97 | 70 | 0.92 |
| Amino | [OX1] = CN | 30 | 1.00 | 22 | 0.87 |
| Enamine | [NX3][$(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C),$(cc)] C),$(cc)] |
76 | 0.93 | 71 | 0.89 |
| Ketone | [#6][CX3](=O)[#6] | 16 | 1.00 | 15 | 1.00 |
| Alkene | [C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C] C] |
59 | 0.92 | 49 | 0.88 |
| Total | — | 1689 | 0.96 | 1576 | 0.92 |
As captured in Table 2, across 10 probed chemical moieties, the fine-tuned Gemini retains the majority of its predictive capacity, achieving a total accuracy of 0.96 for binary classification. For quaternary classification, a slightly lower accuracy of 0.92 is observed, which aligns with the challenging nature of skewed data distributions as discussed in Section 2.1. Among the chemical moieties, masking amino and alkene groups results in the greatest prediction disagreement for quaternary classification, indicating a relatively high importance attributed by the fine-tuned Gemini. However, these moieties have a relatively small sample size (22 and 49 for amino and alkene groups, respectively), suggesting that the low agreement could partially stem from the limited data diversity of the test set. One plausible interpretation is that functionalities such as amino groups exert a notable influence on MOF hydrophobicity. It is essential to acknowledge that the outcome of moiety masking experiments might vary depending on the dataset utilized for fine-tuning, thus making our conclusions dataset-specific. Nevertheless, these findings imply that the fine-tuned Gemini is capable of withstanding minor information disruptions in text prompts, reflecting its robustness.
3.3. Blind test
We further assess the potential of the fine-tuned Gemini in a real-world application by predicting hydrophobicity in an unseen (or blind test) set of MOFs, randomly selected from the free-solvent-removed (FSR) and ion-containing (ION) subsets in the CoRE-MOF-2024 database, comprising 150 MOFs in each subset. To facilitate full external validation, we retained only unique MOFs in the blind test set by discarding those with identical molecular identifiers (as represented by SMILES) compared to the trained MOFs. As illustrated in Fig. 6, the fine-tuned Gemini achieves predictive accuracy scores of 0.66 and 0.57 for binary and quaternary classifications, respectively, in the blind test set. It is worthwhile to note that the residual solvents and ions included in these MOFs pose significant interference in simulating host–guest interactions.60,61 Zhao et al. also highlighted that the removal of strongly bound solvent molecules in the ASR subset (employed as the training set in fine-tuning) could yield MOFs with significantly stronger water affinity, causing substantial variations in predicted hydrophobicity in FSR and ION subsets.28 This effect is reflected by the predictions in individual subsets (Fig. S7–S8†), with less accuracy in the ION subset compared to the FSR subset. Nevertheless, this more rigorous test reinforces that fine-tuning Gemini provides a reliable and effective approach for predicting MOF properties, thus facilitating large-scale, high-throughput computational screening. For instance, the fine-tuned Gemini can be effortlessly deployed with solely SMILES input to swiftly deprioritize potentially less hydrophobic MOFs from time-consuming computations. | ||
| Fig. 6 Confusion matrices by the fine-tuned Gemini on 300 randomly selected MOFs from (a) FSR subset and (b) ION subset. | ||
4. Conclusions
This study illustrates the promise of LLMs in effectively predicting the hydrophobicity of MOFs. By fine-tuning Gemini solely on SMILES and label pairs, we bypass the laborious process of feature engineering and yet achieve accuracy on par with or exceeding descriptor-based ML benchmarks. Moiety masking experiments and a stringent blind test demonstrate the robustness and transferability of the fine-tuned Gemini, which can serve as a powerful screening tool for rapidly identifying hydrophobic MOFs. We anticipate that continued refinement of LLM architectures, expanded training sets, and closer integration with domain-specific knowledge will further advance data-driven discovery of MOFs and other emerging materials.Data availability
All datasets and python codes including moiety masking experiments are available at the GitHub repository: https://github.com/xiaoyu961031/Fine-tuned-Gemini.Conflicts of interest
There is no conflict of interest to declare.Acknowledgements
We gratefully acknowledge the A*STAR LCER-FI projects (LCERFI01-0015 U2102d2004 and LCERFI01-0033 U2102d2006) and the National Research Foundation Singapore (NRF-CRP26-2021RS-0002) for financial support, the National University of Singapore and the National Supercomputing Centre (NSCC) Singapore for computational resources.References
- M. Eddaoudi, J. Kim, N. Rosi, D. Vodak, J. Wachter, M. O'Keeffe and O. M. Yaghi, Science, 2002, 295, 469–472 CrossRef CAS PubMed.
- H. Furukawa, K. E. Cordova, M. O'Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef PubMed.
- K. Jayaramulu, F. Geyer, A. Schneemann, Š. Kment, M. Otyepka, R. Zboril, D. Vollmer and R. A. Fischer, Adv. Mater., 2019, 31, 1900820 CrossRef PubMed.
- L.-H. Xie, M.-M. Xu, X.-M. Liu, M.-J. Zhao and J.-R. Li, Adv. Sci., 2020, 7, 1901758 CrossRef CAS PubMed.
- P. Z. Moghadam, D. Fairen-Jimenez and R. Q. Snurr, J. Mater. Chem. A, 2015, 4, 529–536 RSC.
- P. Küsgens, M. Rose, I. Senkovska, H. Fröde, A. Henschel, S. Siegle and S. Kaskel, Microporous Mesoporous Mater., 2009, 120, 325–330 CrossRef.
- Z. Qiao, Q. Xu and J. Jiang, J. Mater. Chem. A, 2018, 6, 18898–18905 RSC.
- Z. Qiao, Q. Xu, A. K. Cheetham and J. Jiang, J. Phys. Chem. C, 2017, 121, 22208–22215 CrossRef CAS.
- H. Lyu, Z. Ji, S. Wuttke and O. M. Yaghi, Chem, 2020, 6, 2219–2241 CAS.
- H. Tang, L. Duan and J. Jiang, Langmuir, 2023, 39, 15849–15863 CrossRef CAS PubMed.
- T. B. Brown, B. Mann, N. Ryder, M. Subbiah, J. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry, A. Askell, S. Agarwal, A. Herbert-Voss, G. Krueger, T. Henighan, R. Child, A. Ramesh, D. M. Ziegler, J. Wu, C. Winter, C. Hesse, M. Chen, E. Sigler, M. Litwin, S. Gray, B. Chess, J. Clark, C. Berner, S. McCandlish, A. Radford, I. Sutskever and D. Amodei, arXiv, 2020, preprint, arXiv:2005.14165, DOI:10.48550/arXiv.2005.14165.
- Gemini Team Google, arXiv, 2023, preprint, arXiv:2312.11805, DOI:10.48550/arXiv.2312.11805.
- R. Bommasani, D. A. Hudson, E. Adeli, R. Altman, S. Arora, S. von Arx, M. S. Bernstein, J. Bohg, A. Bosselut, E. Brunskill, E. Brynjolfsson, S. Buch, D. Card, R. Castellon, N. Chatterji, A. Chen, K. Creel, J. Q. Davis, D. Demszky, C. Donahue, M. Doumbouya, E. Durmus, S. Ermon, J. Etchemendy, K. Ethayarajh, L. Fei-Fei, C. Finn, T. Gale, L. Gillespie, K. Goel, N. Goodman, S. Grossman, N. Guha, T. Hashimoto, P. Henderson, J. Hewitt, D. E. Ho, J. Hong, K. Hsu, J. Huang, T. Icard, S. Jain, D. Jurafsky, P. Kalluri, S. Karamcheti, G. Keeling, F. Khani, O. Khattab, P. W. Koh, M. Krass, R. Krishna, R. Kuditipudi, A. Kumar, F. Ladhak, M. Lee, T. Lee, J. Leskovec, I. Levent, X. L. Li, X. Li, T. Ma, A. Malik, C. D. Manning, S. Mirchandani, E. Mitchell, Z. Munyikwa, S. Nair, A. Narayan, D. Narayanan, B. Newman, A. Nie, J. C. Niebles, H. Nilforoshan, J. Nyarko, G. Ogut, L. Orr, I. Papadimitriou, J. S. Park, C. Piech, E. Portelance, C. Potts, A. Raghunathan, R. Reich, H. Ren, F. Rong, Y. Roohani, C. Ruiz, J. Ryan, C. Ré, D. Sadigh, S. Sagawa, K. Santhanam, A. Shih, K. Srinivasan, A. Tamkin, R. Taori, A. W. Thomas, F. Tramèr, R. E. Wang, W. Wang, B. Wu, J. Wu, Y. Wu, S. M. Xie, M. Yasunaga, J. You, M. Zaharia, M. Zhang, T. Zhang, X. Zhang, Y. Zhang, L. Zheng, K. Zhou and P. Liang, arXiv, 2021, preprint, arXiv:2108.07258, DOI:10.48550/arXiv.2108.07258.
- Z. Zheng, Z. Rong, N. Rampal, C. Borgs, J. T. Chayes and O. M. Yaghi, Angew. Chem., Int. Ed., 2023, 62, e202311983 CrossRef CAS PubMed.
- Z. Zheng, A. H. Alawadhi, S. Chheda, S. E. Neumann, N. Rampal, S. Liu, H. L. Nguyen, Y.-h. Lin, Z. Rong, J. I. Siepmann, L. Gagliardi, A. Anandkumar, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 28284–28295 CrossRef CAS PubMed.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef CAS PubMed.
- W. Zhang, Q. Wang, X. Kong, J. Xiong, S. Ni, D. Cao, B. Niu, M. Chen, Y. Li, R. Zhang, Y. Wang, L. Zhang, X. Li, Z. Xiong, Q. Shi, Z. Huang, Z. Fu and M. Zheng, Chem. Sci., 2024, 15, 10600–10611 RSC.
- M. S. Wilhelmi, M. R. García, S. Shabih, M. V. Gil, S. Miret, C. T. Koch, J. A. Márquez and K. M. Jablonka, Chem. Soc. Rev., 2025, 54, 1125–1150 RSC.
- M. Suvarna, A. C. Vaucher, S. Mitchell, T. Laino and J. Pérez-Ramírez, Nat. Commun., 2023, 14, 7964 CrossRef CAS PubMed.
- Z. Zheng, Z. He, O. Khattab, N. Rampal, M. A. Zaharia, C. Borgs, J. T. Chayes and O. M. Yaghi, Digital Discovery, 2024, 3, 491–501 RSC.
- K. M. Jablonka, P. Schwaller, A. Ortega-Guerrero and B. Smit, Nat. Mach. Intell., 2024, 6, 161–169 CrossRef.
- J. V. Herck, M. V. Gil, K. M. Jablonka, A. Abrudan, A. S. Anker, M. Asgari, B. Blaiszik, A. Buffo, L. Choudhury, C. Corminboeuf, H. Daglar, A. M. Elahi, I. T. Foster, S. Garcia, M. Garvin, G. Godin, L. L. Good, J. Gu, N. X. Hu, X. Jin, T. Junkers, S. Keskin, T. P. J. Knowles, R. Laplaza, M. Lessona, S. Majumdar, H. Mashhadimoslem, R. D. McIntosh, S. M. Moosavi, B. Mouriño, F. Nerli, C. Pevida, N. Poudineh, M. R. Kochi, K. L. Saar, F. H. Saboor, M. Sagharichiha, K. J. Schmidt, J. Shi, E. Simone, D. Svatunek, M. Taddei, I. Tetko, D. Tolnai, S. Vahdatifar, J. Whitmer, D. C. F. Wieland, R. Willumeit-Römer, A. Züttel and B. Smit, Chem. Sci., 2025, 16, 670–684 RSC.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36 CrossRef CAS.
- M. Krenn, F. Häse, A. Nigam, P. Friederich and A. Aspuru-Guzik, Mach. Learn.: Sci. Technol., 2020, 1, 045024 Search PubMed.
- M. Krenn, Q. Ai, S. Barthel, N. Carson, A. Frei, N. C. Frey, P. Friederich, T. Gaudin, A. A. Gayle, K. M. Jablonka, R. F. Lameiro, D. Lemm, A. Lo, S. M. Moosavi, J. M. Nápoles-Duarte, A. Nigam, R. Pollice, K. Rajan, U. Schatzschneider, P. Schwaller, M. Skreta, B. Smit, F. Strieth-Kalthoff, C. Sun, G. Tom, G. F. v. Rudorff, A. Wang, A. D. White, A. Young, R. Yu and A. Aspuru-Guzik, Patterns, 2022, 3, 100588 CrossRef CAS PubMed.
- M. Leon, Y. Perezhohin, F. Peres, A. Popovič and M. Castelli, Sci. Rep., 2024, 14, 25016 CrossRef CAS PubMed.
- B. J. Bucior, A. S. Rosen, M. Haranczyk, Z. Yao, M. E. Ziebel, O. K. Farha, J. T. Hupp, J. I. Siepmann, A. Aspuru-Guzik and R. Q. Snurr, Cryst. Growth Des., 2019, 19, 6682–6697 CrossRef CAS.
- G. Zhao, L. Brabson, S. Chheda, J. Huang, H. Kim, K. Liu, K. Mochida, T. Pham, P. Prerna, G. Terrones, S. Yoon, L. Zoubritzky, F.-X. Coudert, M. Haranczyk, H. Kulik, M. Moosavi, D. Sholl, I. Siepmann, R. Snurr and Y. Chung, ChemRxiv, 2024, preprint arXiv:2024-nvmnr Search PubMed.
- S. S. Y. Chui, S. M. F. Lo, J. P. H. Charmant, A. G. Orpen and I. D. Williams, Science, 1999, 283, 1148–1150 CrossRef CAS PubMed.
- Gemini Team Google, arXiv, 2024, preprint arXiv:2403.05530, DOI:10.48550/arXiv.2403.05530.
- R. Pétuya, S. Durdy, D. Antypov, M. W. Gaultois, N. G. Berry, G. R. Darling, A. P. Katsoulidis, M. S. Dyer and M. J. Rosseinsky, Angew. Chem., Int. Ed., 2022, 61, e202114573 CrossRef PubMed.
- S. Pal, S. Kulandaivel, Y.-C. Yeh and C.-H. Lin, Coord. Chem. Rev., 2024, 518, 216108 CrossRef CAS.
- I. B. Orhan, H. Daglar, S. Keskin, T. C. Le and R. Babarao, ACS Appl. Mater. Interfaces, 2021, 14, 736–749 CrossRef PubMed.
- X. Li, P. M. Maffettone, Y. Che, T. Liu, L. Chen and A. I. Cooper, Chem. Sci., 2021, 12, 10742–10754 RSC.
- K. M. Jablonka, D. Ongari, S. M. Moosavi and B. Smit, Chem. Rev., 2020, 120, 8066–8129 CrossRef CAS PubMed.
- K. Saab, T. Tu, W.-H. Weng, R. Tanno, D. Stutz, E. Wulczyn, F. Zhang, T. Strother, C. Park, E. Vedadi, J. Z. Chaves, S.-Y. Hu, M. Schaekermann, A. Kamath, Y. Cheng, D. G. T. Barrett, C. Cheung, B. Mustafa, A. Palepu, D. McDuff, L. Hou, T. Golany, L. Liu, J.-b. Alayrac, N. Houlsby, N. Tomasev, J. Freyberg, C. Lau, J. Kemp, J. Lai, S. Azizi, K. Kanada, S. Man, K. Kulkarni, R. Sun, S. Shakeri, L. He, B. Caine, A. Webson, N. Latysheva, M. Johnson, P. Mansfield, J. Lu, E. Rivlin, J. Anderson, B. Green, R. Wong, J. Krause, J. Shlens, E. Dominowska, S. M. A. Eslami, K. Chou, C. Cui, O. Vinyals, K. Kavukcuoglu, J. Manyika, J. Dean, D. Hassabis, Y. Matias, D. Webster, J. Barral, G. Corrado, C. Semturs, S. S. Mahdavi, J. Gottweis, A. Karthikesalingam and V. Natarajan, arXiv, 2024, preprint, arXiv:2404.18416, DOI:10.48550/arXiv.2404.18416.
- T. F. Willems, C. H. Rycroft, M. Kazi, J. C. Meza and M. Haranczyk, Microporous Mesoporous Mater., 2012, 149, 134–141 CrossRef CAS.
- A. Nandy, C. Duan, J. P. Janet, S. Gugler and H. J. Kulik, Ind. Eng. Chem. Res., 2018, 57, 13973–13986 CrossRef CAS.
- A. Nandy, C. Duan, M. G. Taylor, F. Liu, A. H. Steeves and H. J. Kulik, Chem. Rev., 2021, 121, 9927–10000 CrossRef CAS PubMed.
- Z. Yao, B. Sánchez-Lengeling, N. S. Bobbitt, B. J. Bucior, S. G. H. Kumar, S. P. Collins, T. Burns, T. K. Woo, O. K. Farha, R. Q. Snurr and A. Aspuru-Guzik, Nat. Mach. Intell., 2021, 3, 76–86 CrossRef.
- Z. Xie, X. Evangelopoulos, Ö. H. Omar, A. Troisi, A. I. Cooper and L. Chen, Chem. Sci., 2024, 15, 500–510 RSC.
- S. Zhong and X. Guan, Environ. Sci. Technol. Lett., 2023, 10, 872–877 CrossRef CAS.
- S. M. Moosavi, A. Nandy, K. M. Jablonka, D. Ongari, J. P. Janet, P. G. Boyd, Y. Lee, B. Smit and H. J. Kulik, Nat. Commun., 2020, 11, 1–10 RSC.
- S. Majumdar, S. M. Moosavi, K. M. Jablonka, D. Ongari and B. Smit, ACS Appl. Mater. Interfaces, 2021, 13, 61004–61014 CrossRef CAS PubMed.
- A. Nandy, G. Terrones, N. Arunachalam, C. Duan, D. W. Kastner, H. J. Kulik, A. Nandy, G. Terrones, N. Arunachalam, C. Duan, D. W. Kastner and H. J. Kulik, Sci. Data, 2022, 9, 74 CrossRef CAS PubMed.
- G. G. Terrones, S.-P. Huang, M. P. Rivera, S. Yue, A. Hernandez and H. J. Kulik, J. Am. Chem. Soc., 2024, 146, 20333–20348 CrossRef CAS PubMed.
- Y. Yue, S. A. Mohamed and J. Jiang, J. Chem. Inf. Model., 2024, 64, 4966–4979 CrossRef CAS PubMed.
- K. M. Jablonka, S. M. Moosavi, M. Asgari, C. Ireland, L. Patiny and B. Smit, Chem. Sci., 2021, 12, 3587–3598 RSC.
- X. Wu, R. Zheng and J. Jiang, J. Chem. Theory Comput., 2025, 21, 900–911 CrossRef CAS PubMed.
- A. S. Palakkal, S. A. Mohamed and J. Jiang, Chem Bio Eng., 2024, 1, 970–978 CrossRef CAS PubMed.
- Y. Qiu, L. Chen, X. Zhang, D. Ping, Y. Tian and Z. Zhou, AIChE J., 2024, 70, e18575 CrossRef CAS.
- Z. Zhang, F. Pan, S. A. Mohamed, C. Ji, K. Zhang, J. Jiang and Z. Jiang, Small, 2024, 20, 2405087 CrossRef CAS PubMed.
- A. Jose, E. Devijver, N. Jakse and R. Poloni, J. Am. Chem. Soc., 2024, 146, 6134–6144 CrossRef CAS PubMed.
- Z. Cao, R. Magar, Y. Wang and A. B. Farimani, J. Am. Chem. Soc., 2023, 145, 2958–2967 CrossRef CAS PubMed.
- T.-W. Liu, Q. Nguyen, A. Bousso Dieng, D. A. Gómez-Gualdrón, T.-W. Liu, Q. Nguyen, A. Bousso Dieng and D. A. Gómez-Gualdrón, Chem. Sci., 2024, 15, 18903–18919 RSC.
- L. Van Der Maaten and G. Hinton, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
- J. Devlin, M.-W. Chang, K. Lee and K. Toutanova, arXiv, 2018, preprint, arXiv:1810.04805, DOI:10.48550/arXiv.1810.04805.
- Daylight Theory: SMARTS - A Language for Describing Molecular Patterns, https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html, (accessed 13 Jan, 2025) Search PubMed.
- RDKit: Open-Source Cheminformatics. https://www.rdkit.org Search PubMed.
- I. Cooley and E. Besley, Chem. Mater., 2023, 36, 219–231 CrossRef.
- T. D. Pham, F. Joodaki, F. Formalik and R. Q. Snurr, J. Phys. Chem. C, 2024, 128, 17165–17174 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ta01139f |
| This journal is © The Royal Society of Chemistry 2025 |