Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Diverse nanostructures and antimicrobial activity of lipopeptides bearing lysine-rich tripeptide sequences†
Ian W.
Hamley
 *a,
Valeria
Castelletto
*a,
Valeria
Castelletto
 a,
Callum
Rowding
a,
Callum
Wilkinson
a,
Lucas R.
de Mello
a,
Bruno
Mendes
b,
Glyn
Barrett
b and
Jani
Seitsonen
c
a,
Callum
Rowding
a,
Callum
Wilkinson
a,
Lucas R.
de Mello
a,
Bruno
Mendes
b,
Glyn
Barrett
b and
Jani
Seitsonen
c
aSchool of Chemistry, Food Biosciences and Pharmacy, University of Reading, Whiteknights, Reading, RG6 6AD, UK. E-mail: I.W.Hamley@reading.ac.uk
bSchool of Biological Sciences, University of Reading, Whiteknights, Reading, RG6 6AS, UK
cNanomicroscopy Center, Aalto University, Puumiehenkuja 2, FIN-02150, Espoo, Finland
First published on 25th June 2025
Abstract
The self-assembly and conformation in aqueous solution and bioactivity of three lipopeptides bearing lysine-rich tripeptide sequences are compared for C16-KFK, C16-KWK, and C16-KYK, where C16 denotes an N-terminal hexadecyl (palmitoyl) chain. The central aromatic residue has a significant effect on the self-assembled nanostructures, since C16-KFK forms nanotubes, whereas the other two lipopeptides form nanotapes. The nanotubes and nanotapes are built from lipopeptide bilayers, as confirmed by small-angle X-ray scattering. Circular dichroism (CD) spectroscopy and thioflavin T dye fluorescence show the presence of β-sheet structures, and the latter technique was used to determine critical aggregation concentrations (CACs). Fibre X-ray diffraction for C16-KFK shows a well-defined helical diffraction pattern arising from the helically wrapped bilayers in the nanotube walls. The lipopeptides act as surfactants, as confirmed by surface tension measurements (also used to determine CAC values). All three lipopeptides show minimal cytotoxicity to human fibroblasts but also, unexpectedly, low activity against Gram-negative and Gram-positive bacteria, in contrast to previously studied analogues (with switch of two residues) C16-WKK and C16-YKK that show significant antimicrobial action with low minimum inhibitory concentration (MIC) values [A. Adak et al., ACS Appl. Bio Mater., 2024, 7, 5553–5565]. Also in contrast to these molecules (which show a transition from micelles to fibrils upon increasing the pH), C16-KFK, C16-KWK, and C16-KYK form extended β-sheet structures over the whole pH range examined (pH 2–8). These observations point to the remarkable sensitivity to the tripeptide pattern of lipopeptide self-assembly and antibacterial activity. Whereas the C16-XKK (X = W or Y) lipopeptides form cylindrical fibrils, the C16-KXK analogues form bilayer nanotapes. The former show significant toxicity to bacteria in contrast to the latter, which we propose is due to the effect of the lipopeptide assembly curvature on induced bacterial membrane deformation.
Introduction
Lipopeptide (LPP) molecules comprise a lipid hydrophobic chain attached to a peptide sequence leading to diverse properties resulting from a bio-derived or bio-inspired peptide sequence.1 They have demonstrated selective anticancer activity,2 collagen stimulation by fibroblasts,3 selective antimicrobial activity,4–12 and many other properties key to biomedicine and tissue engineering.10,13–17LPPs self-assemble in solution above a critical aggregation concentration. Different lipid chain types, peptide sequences or solution conditions, can result in distinct self-assembly motifs,1,13–15,18–22 which include nanofibrils, nanosheets and nanotubes. Our group and others have previously reported the self-assembly of several LPPs into nanofibrils, with a β-sheet secondary structure,12,14,23–26 or micelles with generally unordered peptide conformation.27–35 Although less frequent, LPPs can also form nanotubes based on wrapping of β-sheet tapes.26,36–42
The present study builds on our prior research on LPPs bearing short bioactive lysine-rich peptide sequences. We studied the self-assembly and biological activity of a LPP bearing a lysine-rich collagen-stimulating pentapeptide KTTKS, used in MatrixylTM formulations,3,28,43 as well as the self-assembly of LPPs including shorter sequences such as C16-KT and C16-GHK.44 We also investigated co-assembly with the LPP C16-ETTES bearing oppositely charged glutamic acid residues replacing lysines.45 The self-assembly of the related lipopeptides (peptide amphiphiles) C14-KTTKS and C18-KTTKS with distinct lipid chain lengths was also examined,46 and most recently we have investigated the self-assembly and bioactivity and responsiveness to environmental triggers (UV radiation or chemical reductants) of lipoyl-KTTKS bearing a lipoic acid moiety at the N-terminus.47 The K-rich LPP (containing a core fragment of the amyloid β peptide, KLVFF) C16-KKFFVLK, self-assembles into nanotubes and helical ribbons which undergo a reversible unwinding transition upon increasing the temperature from 20 to 55 °C.36 In a demonstration of enzyme-responsive nanostructural changes, this LPP can be cleaved by α-chymotrypsin at two sites leading to products C16-KKF and C16-KKFF, which do not self-assemble into nanotubes but instead form spherical micelles.48 Later, C16-KKFF was used to prepare alginate/graphene oxide capsules with selective antimicrobial activity towards the Gram-positive bacterium Listeria.49 More recently, we modified KTTKS by attaching one or two cycloalkane chains (cycloheptadecyl or cyclododecyl) to the peptide sequence.50 The cycloalkane-modified LPPs self-assemble to form globular structures or twisted nanotapes, depending on the number of cycloalkane chains, and the molecules show excellent compatibility with dermal fibroblasts with additional pro-vascularization capability.
We have recently studied the self-assembly, colloidal properties and antimicrobial activity of LPPs comprising a palmitoyl (hexadecyl, C16) lipid chain attached to a lysine (K)-rich tripeptide.11,12,35 Specifically, we compared the aggregation and properties of C16-YKK, C16-Ykk, C16-WKK and C16-Wkk. In these molecules, the cationic lysine (K) or D-lysine (k) residues promote self-assembly and are responsible for antimicrobial activity; whereas π-stacking interactions between tyrosine (Y) and tryptophan (W) residues promote aggregation and enable other interactions such as π–π or cation–π interactions. At acidic pH, all these LPPs self-assemble into micelles.11,35 At basic pH, K-containing LPPs self-assemble into nanofibers, while k-containing lipopeptides self-assemble into nanotapes or nanotubes.12 All four LPPs have antimicrobial activity against both Gram-negative and Gram-positive bacteria, with low values of the minimum inhibitory concentration (MIC) and favorable values of the selectivity index (haemolysis compared to the MIC), especially for the tryosine-containing lipopeptides.11,12 The antimicrobial activities against Gram-positive and negative bacteria and fungi of lysine-rich LPPs including C16-K, C16-KKK and other tripeptide sequences and selected sequences containing k have been compared, and particularly potent activity (for all strains examined) was noted for C16-KKK, although it also exhibits the highest haemolysis.5 These authors also present a preliminary examination of nanostructures of selected lysine-rich LPPs (the tripeptide compounds) through transmission electron microscopy (TEM). The self-assembly of C16-KK and C16-KKK has been investigated by theoretical and experimental methods,51 and these molecules can be used to prepare antimicrobial hydrogels.52 The lipidated amino acid C16-K (minimal lysine-based sequence, i.e. a single amino acid) shows a salt-dependent transition from self-assembled planar bilayers into scroll-like cochleates.53 The antimicrobial and haemolysis activity of a series of lipidated conjugates of tripeptides KKK and KGK have been reported, with a range of lipid chains including C16 and other hydrocarbon and fluorocarbon chains.54 A notable reduction in antimicrobial activity was observed in the presence of serum albumin, due to non-specific binding. The antimicrobial activities, lipid binding and micelle formation of C16-KK, C16-KGK and C16-KKKK (all with an amidated C-terminus) have been compared by another group.7 Later, other analogues with varied lipid chain lengths C12–C18 and lysine-rich di- to tetra-peptide sequences were studied, and C16-KKKK-NH2 was found to have an optimal balance of hydrophobic and polar fragments leading to a good balance between antimicrobial and hemolytic activity.55 Molecules bearing “double headgroups” of lysine residues (Y-shaped amphiphilic molecules with one or two lysines on each branch and a C16-alkyl chain) have been introduced by the same group, and they also show good antimicrobial activity.56 In another recent example, the self-assembly and antimicrobial and angiogenic activities of LPPs containing K-rich sequences from the amyloid β peptide have been demonstrated, along with the formation of injectable hydrogels.9
Here, we investigate the self assembly, cytocompatibility and antimicrobial activity of three LPPs, namely C16-KFK, C16-KWK and C16-KYK (Scheme 1). This work builds on our previous study of related K-rich LPPs discussed in the paragraphs above. The secondary structure of the LPPs was studied using circular dichroism (CD) and fibre X-ray diffraction. Values of the critical aggregation concentration (CAC) were determined from surface tension experiments and fluorescence assays. The self-assembly motif was characterised using small-angle X-ray scattering (SAXS), cryogenic-transmission electron microscopy (cryo-TEM) and X-ray diffraction (XRD). Cell viability was studied through mitochondrial activity (MTT) assays and antimicrobial activity was assessed by determination of the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values.
Methods
Materials and sample preparation
Lipopeptides were purchased from BioservUK (Rotherham, UK) and supplied as TFA salts. The molar masses measured by ESI-MS are as follows: C16-KFK 659.50 g mol−1 (660.60 g mol−1 expected), C16-KWK 699.55 g mol−1 (698.51 g mol−1 expected) and C16-KYK 676.50 g mol−1 (675.49 g mol−1 expected). The purity by HPLC (0.1% TFA in an acetonitrile/water gradient) is 97.2% for C16-KFK, 96.5% for C16-KWK and 95.5% for C16-KYK. Solutions were prepared by dissolution in ultrapure water, leading to a pH of 2.3 for 1 wt% solutions of C16-KWK and C16-KYK and a pH of 2.5 for C16-KFK. Samples at pH 8 were prepared by the addition of suitable amounts of 2M NaOH solution.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin
sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ/λ (2θ: scattering angle). The detector was a Sapphire CCD.
θ/λ (2θ: scattering angle). The detector was a Sapphire CCD.
 | (1) |
 | (2) |
Results and discussion
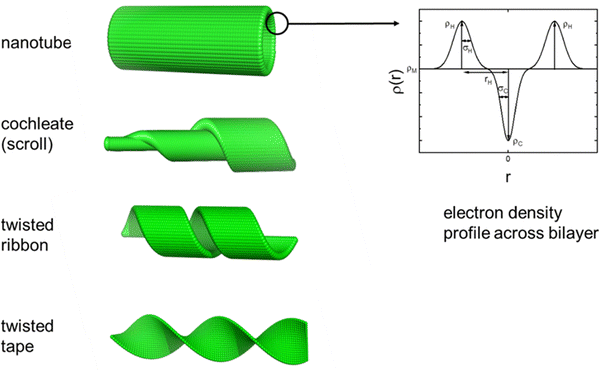
The self-assembled nanostructures of the LPPs were examined using cryo-TEM. Representative images for solutions at native pH (pH ≈ 2) and pH 8 are shown in Fig. 1 and additional images are shown in Fig. S1 (ESI†). The images show that C16-KFK predominantly forms nanotube structures, although nested nanotube/cochleate structures as well as a small population of helical ribbon and twisted tape structures are observed in other regions of the grids. In contrast, C16-KWK forms a dense array of twisted tape structures, whereas C16-KYK shows a much less dense population of short (finite length) twisted tapes, resembling in some regions farfalle (bow-tie pasta)-like shapes. These morphologies are schematically illustrated in Fig. 2, along with the electron density across the bilayer used in the fitting of small-angle X-ray scattering (SAXS) data. | ||
| Fig. 1 Cryo-TEM images for 1 wt% samples at native pH (pH 2): (a) C16-KFK, (b) C16-KWK, (c) C16-KYK, and at pH 8: (d) C16-KFK, (e) C16-KWK, (f) C16-KYK. | ||
SAXS was used to provide an in situ complement to cryo-TEM, to probe the self-assembled nanostructure shapes and dimensions. The data for native pH (pH ≈ 2) are shown in Fig. 3a, along with form factor fits based on the lipopeptide bilayer structures in nanotapes (for C16-KWK and C16-KYK) or nanotubes/cochleates (for C16-KFK); these morphologies being those imaged by cryo-TEM. The data for all samples show an intensity scaling at a low wavenumber q, I ∼ q−2, consistent with the formation of planar bilayer structures.62,63 In the previous work,26,64,65 we have fitted SAXS data for LPP nanotube-forming systems to a form factor comprising that for a cylindrical shell and a Gaussian bilayer to allow for the variation in the LPP electron density across the bilayer. The latter is a combination of three Gaussian functions, representing the electron-poor lipid core of the bilayer and the electron-rich headgroup regions (Fig. 2). The sum of the hollow cylinder and Gaussian bilayer form factors represents an approximate convolution of the two terms and also allows for the presence of unclosed or partially closed nanotubes in many peptide and lipopeptide-based systems, for which twisted tape bilayers, scrolls (‘cochleates’53) and ribbon structures are observed.36,53,66,67 This model can be seen to fit the data satisfactorily, including the amplitude and position of the oscillations over much of the q range (except the peak at the lowest q, which may be influenced by structure factor), which arise due to the interference from the nanotube wall diameter and the form factor maximum at high q, which is due to the wall thickness. Consistent with the cryo-TEM results, which show twisted tape structures, the SAXS data for C16-KYK and C16-KWK can be fitted just using a Gaussian bilayer form factor to represent the structure within the nanotapes. The SAXS data at pH 8, shown in Fig. 3b, are very similar to those at native pH (and in fact nearly superpose and have the same low q scaling behaviour of a bilayer structure), consistent with the cryo-TEM images (Fig. 1), which show that the morphologies are stable across this pH range. Additional SAXS data were obtained at other pH values up to pH 12 and the data (Fig. S2, ESI†) show that the nanotape structure is retained across the range from the native pH to the highest pH measured (pH 10 or pH 12). Although there are, in some cases, changes in the sharpness of form factor maxima, and in some cases nanotube form factor maxima at low q; the intensity scaling at low q is I ∼ q−2.
To further probe the ordering within the remarkable nanotube structures formed by C16-KFK, fibre XRD was performed. The pattern shown in Fig. 4 shows clear features resulting from the helical wrapping of the LPP in the nanotube walls, in particular the characteristic X-pattern of off-axis reflections from a helical structure (at ±38° with respect to the equator).36,38,68,69 The equatorial reflections at 16.84 and 7.96 Å are assigned as the second and fourth orders of reflection from a lamellar structure with layer spacing 33 Å (the first order reflection can just be seen on the right-hand side of the 2D image), close to the layer spacing estimated from the fitting of the solution SAXS data (Table S1, ESI†). The presence of well-defined equatorial spacings along with a strong meridional reflection at d = 4.57 Å provides clear evidence for β-sheet structures,36,45,70–72 while the other smaller d-spacings can also be assigned to spacings within β-sheet structures.45,70,71
 | ||
| Fig. 4 Fiber XRD pattern from an aligned stalk prepared from a 1 wt% solution of C16-KFK (pH 4) along with one-dimensional intensity profile showing peak positions. | ||
The critical aggregation concentration (CAC) was next estimated using fluorescence probe measurements. Since cryo-TEM and SAXS indicate the formation of extended fibrillar types of nanostructures, we used Thioflavin T (ThT), a dye sensitive to the formation of ‘amyloid’ β-sheet structures.59,73,74 The dependence of the peak fluorescence I (relative to the fluorescence of a reference ThT solution without lipopeptide, I0) is plotted in Fig. 5 for the three LPPs (original fluorescence spectra are shown in Fig. S3, ESI†). The CAC for all three lipopeptides is the same within uncertainty, (0.01 ± 0.01) wt%. The data for C16-KWK show lower intensities and more noise than the other two samples, which is ascribed to the possible mixing of fluorescence from the tryptophan residue and ThT. This may also affect the gradient of the fluorescence intensity versus concentration at low concentration values. Similar values for CAC for the LPPs are obtained at pH 8 (Fig. S4, ESI†).
The conformation and chirality of the tripeptides in the LPP assemblies (above the CAC) were investigated using circular dichroism (CD) spectroscopy. The spectra of 0.25 wt% samples at native pH (pH ≈ 2) are shown in Fig. 6, with additional spectra of samples at pH 8 in Fig. S5 (ESI†). The spectra at both pH values show similar features. The spectra of C16-KFK and C16-KYK show peaks in the range 210–240 nm, which are due to the n–π* and π–π* transitions in the aromatic Phe and Tyr groups;75–78 however, such features are absent for C16-KWK that shows a spectrum with a single broad negative maximum near 220 nm, which can be assigned in part to a red-shifted spectrum arising from β-sheet structures,79 although there may also be a contribution from excitonic coupling. This sample forms a notably more dense network of fibrillar structures according to cryo-TEM and SAXS. The spectra of C16-KWK and C16-KYK show features similar to those reported for LPPs with the same residues in different sequences C16-WKK and C16-YKK reported previously.12 The spectrum of C16-KFK shows features previously analysed in detail for Fmoc-FF.80 This includes some contribution from the excitonic coupling of Lα transitions, but it is heavily positively biased. The spectrum of C16-KWK seems to have an excitonic couplet character with a slight negative bias, whereas this contribution for C16-KYK is only positive. In summary, the CD spectra contain features of the excitonic coupling of the aromatic residues. For C16-KFK and C16-KYK, these features arising from the chiral environment of the Phe and Trp residues in the nanotube and twisted tape structures formed by these LPPs dominate, whereas C16-KWK shows some features resembling those in classical β-sheet CD spectra.
To examine whether these cationic molecules act as biosurfactants, and to determine the CAC from an independent method, surface tension measurements were performed. The data shown in Fig. 7 show that all three LPPs behave as surfactants, reducing the surface tension from the value for pure water (72 mN m−1), to less than 50 mN m−1, with C16-KWK showing the greatest activity, with a surface tension around 40 mN m−1 at the highest concentration studied. The CAC values indicated are in the range 0.01–0.02 wt% and are consistent with the values from the ThT fluorescence assays. The areas per molecule were also obtained from the Langmuir adsorption isotherm equation (eqn (1) and (2)) and are in the range 32.8–39.7 Å2, which is reasonable for a lipopeptide packed in a monolayer at the air–water interface.35
 | ||
| Fig. 7 Surface tension data (native pH). Langmuir adsorption isotherms for (a) C16-KFK, (b) C16-KYK and (c) C16-KWK. Indicated are values of the CAC from the break points and the areas per molecule A from the slope of the lines below the CAC using the Langmuir adsorption isotherm equation (eqn (1) and (2)). | ||
The cytocompatibility of all three LPPs was determined using an MTT assay, a colorimetric assay based in the capacity of the eukaryotic cells to reduce tetrazolium salts into formazan crystals using mainly mitochondrial enzymes that can be used to evaluate the overall cell viability based on the mitochondrial metabolic activity.81,82 Samples with concentrations from 0.0001 wt% to 0.05 wt% were evaluated and good cytocompatibility was found. As shown in Fig. 8, for most concentrations of any of the three LPPs, a non-significant decrease in cell viability is found, according to the Welch ANOVA. The only exceptions were cells incubated with C16-KFK and C16-KYK at 0.0125 wt% (Fig. 8a and b) and C16-KWK at 0.0625 wt% (Fig. 8c).
The LPPs generally show good cytocompatibility to human cells (fibroblasts), which is one requirement to be useful as an antibiotic. We then investigated antimicrobial activity against Gram-positive and -negative species via determination of MIC and MBC values for the three LPPs. Contrary to the expectation for molecules bearing tripeptide sequences containing two cationic lysine residues, none of the molecules show substantial antibacterial activity against either Gram-positive S. aureus and S. enterica or Gram-negative E. coli (Table 1). This can be contrasted with our previous results12 for C16-YKK that shows an MIC of 31.25–62.5 μg ml−1 against the Gram-positive and Gram-negative strains studied (Table 1) or C16-WKK that has a slightly higher MIC of 62.5 μg ml−1 except for S. enterica (Table 1). C16-KFK, C16-KWK and C16-KYK were also tested against P. aeruginosa (PA01) and MRSA (methicillin-resistant S. aureus) and showed little activity, with MBC and MIC values > 1000 μg ml−1 for all three LPPs.
| Lipopeptide | MIC (μg mL−1) | MBC (μg mL−1) | ||||
|---|---|---|---|---|---|---|
| E. coli (K12-ATCC 25922) | S. enterica (NCTC 5188) | S. aureus (ATCC 12600) | E. coli (K12-ATCC 25922) | S. enterica (NCTC 5188) | S. aureus (ATCC 12600) | |
| MIC = minimum inhibitory concentration is the lowest concentration that inhibits visible bacterial growth after incubation. MBC = minimal bactericidal concentration is the lowest concentration required to kill colony-forming units of bacterial population. ND = value not determined. | ||||||
| C16-KFK | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 |
| C16-KYK | 500 | >1000 | 500 | 1000 | >1000 | 1000 |
| C16-YKK12 | 31.25-ND | 62.5 | 62.5 | 62.5-ND | 125 | 62.5 |
| C16-KWK | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 |
| C16-WKK12 | 62.5-ND | 250 | 62.5 | 250 | 250 | 250 |
Discussion and conclusions
In summary, C16-KFK, C16-KWK, and C16-KYK form extended bilayer structures across a pH range from native acidic pH conditions to pH 8 and above. This can be contrasted with C16-WKK and C16-YKK (and analogues with D-lysine residues) that form micelles at native pH 4.6 but cylindrical fibrils at pH 8.12 This points to the significant effects of the sequence in tuning the intermolecular interactions. To further examine whether the sequence influences the electrostatics, pKa values were calculated using web software H++, which is based on continuum electrostatics or Poisson–Boltzmann calculations.83,84 Insignificant differences in the predicted pKa values for the lysine ε-amino group (and tyrosine –OH and termini) are apparent on comparing C16-KYK with C16-YKK and C16-KWK with C16-WKK (Table S2, ESI†). Therefore electrostatic effects alone cannot explain the difference in self-assembly which is pH-sensitive for C16-XKK but not C16-KXK (X = W or Y and F in the latter case). The C16-KXK LPPs form planar bilayer structures, whereas the C16-XKK analogues form cylindrical fibrils at pH 8.12 This is due to the differences in the packing and hydrogen bonding of the tripeptides for the two types of sequences.The surface tension values for concentration values above the CAC, below 50 mM m−1 or 40 mM m−1 for C16-KWK, are similar to those previously reported by us for C16-WKK and C16-YKK (and analogous lipopeptides with D-amino acid lysines).35 However, it is important to note that here the lipopeptides self-assemble as nanotapes, whereas the previous studies on C16-WKK and C16-YKK were performed under conditions (native pH 4.6) where the lipopeptides form micelles. In fact, the determination of the CAC of fibrillar self-assembled peptide systems from surface tension measurements has been rarely reported (in one example, it has been demonstrated that amyloid β peptide fragments show surfactant properties85). The distinct one-dimensional fibrillization process may be responsible for the observation that the surface tension shows an inflection point (at the same concentration as the CAC from ThT dye fluorescence) rather than a point at which the surface tension levels off as in the case of a critical micelle concentration (CMC),60,61 which corresponds to a closed association process into zero-dimensional spherical aggregates. The one-dimensional fibrillization is a nucleation and growth process with potential secondary nucleation and other events as described, for example, by the Oosawa model for homogeneous nucleation and one-dimensional growth.86,87
All three LPPs show good cytocompatibility to L929 fibroblasts; however, they showed unexpectedly low antimicrobial activity to either Gram-positive or Gram-negative species, as assessed via MIC and MBC determination. Clearly the sequence swap of two residues significantly influences the antimicrobial activity, as shown by the much lower activity for C16-KXK compared to C16-XKK (X = Y or W) reported in our previous paper12 evident from the data in Table 1. With such cationic lipopeptides, this activity is believed to be mediated by interactions with oppositely charged membranes in the bacterial cell walls (Gram-positive bacteria lack an outer lipid membrane but have a peptidoglycan coating with a negative charge) and the sequences with two C-terminal lysines show much greater activity than those with the KXK pattern, where X is an aromatic residue. This is ascribed to the distinct modes of assembly of C16-XKK and C16-KXK at pH 8 (and across neutral pH relevant to the antimicrobial activity studies, see the SAXS data in Fig. S2, ESI†). Lipopeptides C16-WKK and C16-YKK form cylindrical fibrils, as evident from the SAXS data intensity scaling at a low q, I ∼ q−1,12 whereas C16-KWK and C16-KYK form bilayer nanotapes (SAXS intensity scaling I ∼ q−2). We propose that the difference in the curvature of the nanostructures influences the antimicrobial activity. Specifically, we suggest that the cylindrical fibrils (wormlike micelles or ‘filomicelles’88,89) of C16-XKK are able to induce a greater curvature in the bacterial membrane than the planar bilayer of C16-KXK. The ability of cationic peptides to induce a curvature in lipid membranes is demonstrated,90 and indeed proposed as one mechanism for the interaction of cationic cell-penetrating peptides with lipid membranes.91,92 This is an interesting topic for future research on these systems, using model lipid membranes as in our previous work on surfactants—like arginine-based peptides.4,93–95 The high-density presentation of peptide units at the surface of fibrils has been shown to promote delivery of active compounds and binding to targets in vivo, and thus there can be profound effects of the shape of self-assembled nanostructures on bioactivity, as discussed with examples, in a previous review.96 Our results provide clear evidence that even subtle changes in sequences can profoundly influence both self-assembly (in particular, pH-dependence) and antimicrobial activity of designer lipopeptides with short cationic peptide motifs.
Conflicts of interest
The authors declare no competing interests.Data availability
The data supporting this article have been included as part of the ESI.† Other data are available upon request from the authors.Acknowledgements
This work was supported by the EPSRC Fellowship grant (reference EP/V053396/1) to IWH. We thank Diamond for the award of SAXS beamtime on B21 (ref. SM35585-1) and Nikul Khunti for assistance. We acknowledge the Warwick Advanced Bioimaging Research Technology Platform, for the use of the JEOL 2100Plus, supported by the MRC award reference MC_PC_17136, and Dr Saskia Bakker supported by EPSRC EP/V007688/1 for the images. We acknowledge the use of facilities in the Chemical Analysis Facility (CAF) at the University of Reading and Nick Spencer for assistance with XRD.References
- I. W. Hamley, Chem. Commun., 2015, 51, 8574–8583 RSC.
- V. Castelletto, C. J. C. Edwards-Gayle, F. Greco, I. W. Hamley, J. Seitsonen and J. Ruokolainen, ACS Appl. Mater. Interfaces, 2019, 11, 33573–33580 CrossRef CAS PubMed.
- R. R. Jones, V. Castelletto, C. J. Connon and I. W. Hamley, Mol. Pharmaceutics, 2013, 10, 1063–1069 CrossRef CAS PubMed.
- V. Castelletto, R. H. Barnes, K. A. Karatzas, C. J. C. Edwards-Gayle, F. Greco, I. W. Hamley, R. Rambo, J. Seitsonen and J. Ruokolainen, Biomacromolecules, 2018, 19, 2782–7294 CrossRef CAS PubMed.
- A. Makovitzki, J. Baram and Y. Shai, Biochemistry, 2008, 47, 10630–10636 CrossRef CAS PubMed.
- G. Laverty, M. McLaughlin, C. Shaw, S. P. Gorman and B. F. Gilmore, Chem. Biol. Drug Des., 2010, 75, 563–569 CrossRef CAS PubMed.
- E. Sikorska, M. Dawgul, K. Greber, E. Ilowska, A. Pogorzelska and W. Kamysz, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 2625–2634 CrossRef CAS PubMed.
- E. Kamysz, E. Sikorska, M. Jaskiewicz, M. Bauer, D. Neubauer, S. Bartoszewska, W. Baranska-Rybak and W. Kamysz, Int. J. Mol. Sci., 2020, 21, 887 CrossRef CAS PubMed.
- N. Mukherjee, S. Ghosh, J. Sarkar, R. Roy, D. Nandi and S. Ghosh, ACS Appl. Mater. Interfaces, 2023, 15, 33457–33479 CrossRef CAS PubMed.
- C. Vicente-Garcia and I. Colomer, Nat. Rev. Chem., 2023, 7, 710–731 CrossRef CAS PubMed.
- A. Adak, I. W. Hamley, V. Castelletto, A. S. L. de Sousa, K.-A. Karatsas and C. Wilkinson, Biomacromolecules, 2024, 25, 1205–1213 CrossRef CAS PubMed.
- A. Adak, V. Castelletto, B. Mendes, G. Barrett, J. Seitsonen and I. W. Hamley, ACS Appl. Bio. Mater., 2024, 7, 5553–5565 CrossRef CAS PubMed.
- H. G. Cui, M. J. Webber and S. I. Stupp, Biopolymers, 2010, 94, 1–18 CrossRef CAS PubMed.
- J. B. Matson, R. H. Zha and S. I. Stupp, Curr. Opin. Solid State Mater. Sci., 2011, 15, 225–235 CrossRef CAS PubMed.
- J. B. Matson and S. I. Stupp, Chem. Commun., 2012, 48, 26–33 RSC.
- M. J. Webber, E. J. Berns and S. I. Stupp, Isr. J. Chem., 2013, 53, 530–554 CrossRef CAS PubMed.
- E. Arslan, I. C. Garip, G. Gulseren, A. B. Tekinay and M. O. Guler, Adv. Healthcare Mater., 2014, 3, 1357–1376 CrossRef CAS PubMed.
- D. W. P. M. Löwik and J. C. M. van Hest, Chem. Soc. Rev., 2004, 33, 234–245 RSC.
- F. Versluis, H. R. Marsden and A. Kros, Chem. Soc. Rev., 2010, 39, 3434–3444 RSC.
- I. W. Hamley, Soft Matter, 2011, 7, 4122–4138 RSC.
- A. Trent, R. Marullo, B. Lin, M. Black and M. Tirrell, Soft Matter, 2011, 7, 9572–9582 RSC.
- M. P. Hendricks, K. Sato, L. C. Palmer and S. I. Stupp, Acc. Chem. Res., 2017, 50, 2440–2448 CrossRef CAS PubMed.
- J. D. Hartgerink, E. Beniash and S. I. Stupp, Science, 2001, 294, 1684–1688 CrossRef CAS PubMed.
- E. T. Pashuck, H. Cui and S. I. Stupp, J. Am. Chem. Soc., 2010, 132, 6041–6046 CrossRef CAS PubMed.
- A. D. Ozkan, A. B. Tekinay, M. O. Guler and E. D. Tekin, RSC Adv., 2016, 6, 104201–104214 RSC.
- E. Rosa, L. de Mello, V. Castelletto, M. L. Dallas, A. Accardo, J. Seitsonen and I. W. Hamley, Biomacromolecules, 2023, 24, 213–224 CrossRef CAS PubMed.
- X. D. Xu, Y. Jin, Y. Liu, X. Z. Zhang and R. X. Zhuo, Colloids Surf., B, 2010, 81, 329–335 CrossRef CAS PubMed.
- J. F. Miravet, B. Escuder, M. D. Segarra-Maset, M. Tena-Solsona, I. W. Hamley, A. Dehsorkhi and V. Castelletto, Soft Matter, 2013, 9, 3558–3564 RSC.
- I. W. Hamley, S. Kirkham, A. Dehsorkhi, V. Castelletto, M. Reza and J. Ruokolainen, Chem. Commun., 2014, 50, 15948–15951 RSC.
- A. Dehsorkhi, V. Castelletto, I. W. Hamley, J. Adamcik and R. Mezzenga, Soft Matter, 2013, 9, 6033–6036 RSC.
- H. C. Fry, L. A. Solomon, B. T. Diroll, Y. Z. Liu, D. J. Gosztola and H. M. Cohn, J. Am. Chem. Soc., 2020, 142, 233–241 CrossRef CAS PubMed.
- I. S. Oliveira, M. Lo, M. J. Araújo and E. F. Marques, Soft Matter, 2019, 15, 3700–3711 RSC.
- J. A. Hutchinson, I. W. Hamley, J. Torras, C. Aleman, J. Seitsonen and J. Ruokolainen, J. Phys. Chem. B, 2019, 123, 614–621 CrossRef CAS PubMed.
- J. N. B. D. Pelin, C. J. C. Edwards-Gayle, A. M. Aguilar, A. Kaur, I. W. Hamley and W. A. Alves, Soft Matter, 2020, 16, 4615–4624 RSC.
- I. W. Hamley, A. Adak and V. Castelletto, Nat. Commun., 2024, 15, 6785 CrossRef CAS PubMed.
- I. W. Hamley, A. Dehsorkhi, V. Castelletto, S. Furzeland, D. Atkins, J. Seitsonen and J. Ruokolainen, Soft Matter, 2013, 9, 9290–9293 RSC.
- I. W. Hamley, A. Dehsorkhi and V. Castelletto, Chem. Commun., 2013, 49, 1850–1852 Search PubMed.
- K. L. Morris, S. Zibaee, L. Chen, M. Goedert, P. Sikorski and L. C. Serpell, Angew. Chem., Int. Ed., 2013, 52, 2279–2283 CrossRef CAS PubMed.
- J. Madine, V. Castelletto, I. W. Hamley and D. A. Middleton, Angew. Chem., Int. Ed., 2013, 52, 10537–10540 CrossRef PubMed.
- M. H. Koc, G. C. Ciftci, S. Baday, V. Castelletto, I. W. Hamley and M. O. Guler, Langmuir, 2017, 33, 7947–7956 CrossRef PubMed.
- A. Reja, S. P. Afrose and D. Das, Angew. Chem., Int. Ed., 2020, 59, 4329–4334 CrossRef CAS PubMed.
- I. W. Hamley, Angew. Chem., Int. Ed., 2014, 53, 6866–6881 CrossRef CAS PubMed.
- V. Castelletto, I. W. Hamley, J. Perez, L. Abezgauz and D. Danino, Chem. Commun., 2010, 46, 9185–9187 Search PubMed.
- V. Castelletto, I. W. Hamley, C. Whitehouse, P. Matts, R. Osborne and E. S. Baker, Langmuir, 2013, 29, 9149–9155 CrossRef CAS PubMed.
- I. W. Hamley, A. Dehsorkhi and V. Castelletto, Langmuir, 2013, 29, 5050–5059 Search PubMed.
- P. Palladino, V. Castelletto, A. Dehsorkhi, D. Stetsenko and I. W. Hamley, Langmuir, 2012, 28, 12209–12215 CrossRef CAS PubMed.
- L. R. de Mello, V. Castelletto, L. Cavalcanti, J. Seitsonen and I. W. Hamley, J. Pept. Sci., 2025, 31, e70002 CrossRef CAS PubMed.
- A. Dehsorkhi, I. W. Hamley, J. Seitsonen and J. Ruokolainen, Langmuir, 2013, 29, 6665–6672 CrossRef CAS PubMed.
- V. Castelletto, A. Kaur, I. W. Hamley, R. H. Barnes, K. A. Karatzas, D. Hermida-Merino, S. Swioklo, C. J. Connon, J. Stasiak, M. Reza and J. Ruokolainen, RSC Adv., 2017, 7, 8366–8375 RSC.
- A. Adak, V. Castelletto, I. W. Hamley, J. Seitsonen, A. Jana, S. Ghosh, N. Mukherjee and S. Ghosh, ACS Appl. Mater. Interfaces, 2024, 16, 58417–58426 CrossRef CAS PubMed.
- G. Zaldivar, S. Vernulapalli, V. Udumula, M. Conda-Sheridan and M. Tagliazucchi, J. Phys. Chem. C, 2019, 123, 17606–17615 CrossRef CAS.
- G. Zaldivar, J. C. Feng, L. Lizarraga, Y. F. Yu, L. de Campos, K. M. P. de Oliveira, K. H. Piepenbrink, M. Conda-Sheridan and M. Tagliazucchi, Adv. Mater. Interfaces, 2023, 10, 2300046 CrossRef CAS.
- C. R. Gao, S. Kewalramani, D. M. Valencia, H. H. Li, J. M. McCourt, M. O. de la Cruz and M. J. Bedzyk, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 22030–22036 CrossRef CAS PubMed.
- B. Findlay, G. G. Zhanel and F. Schweizer, Int. J. Antimicrob. Agents, 2012, 40, 36–42 CrossRef CAS PubMed.
- O. Stachurski, D. Neubauer, A. Walewska, E. Ilowska, M. Bauer, S. Bartoszewska, K. Sikora, A. Hac, D. Wyrzykowski, A. Prahl, W. Kamysz and E. Sikorska, Antibiotics, 2022, 11, 1491 CrossRef CAS PubMed.
- I. Maluch, O. Stachurski, P. Kosikowska-Adamus, M. Makowska, M. Bauer, D. Wyrzykowski, A. Hac, W. Kamysz, M. Deptula, M. Pikula and E. Sikorska, Int. J. Mol. Sci., 2020, 21, 8944 CrossRef CAS PubMed.
- N. P. Cowieson, C. J. C. Edwards-Gayle, K. Inoue, N. S. Khunti, J. Doutch, E. Williams, S. Daniels, G. Preece, N. A. Krumpa, J. P. Sutter, M. D. Tully, N. J. Terrill and R. P. Rambo, J. Synchrotron Radiat., 2020, 27, 1438–1446 CrossRef CAS PubMed.
- I. W. Hamley, Angew. Chem., Int. Ed., 2007, 46, 8128–8147 CrossRef CAS PubMed.
- H. Levine III, Protein Sci., 1993, 2, 404–410 CrossRef PubMed.
- D. F. Evans and H. Wennerström, The colloidal domain. Where physics, chemistry, biology and technology meet, Wiley, New York, 1999 Search PubMed.
- I. W. Hamley, Introduction to Soft Matter, Revised Edition, Wiley, Chichester, 2007 Search PubMed.
- I. W. Hamley, Small-Angle Scattering: Theory, Instrumentation, Data and Applications, Wiley, Chichester, 2021 Search PubMed.
- A. Adak, V. Castelletto, L. de Mello, B. Mendes, G. Barrett, J. Seitsonen and I. W. Hamley, ACS Appl. Bio. Mater., 2025, 8, 803–813 CrossRef CAS PubMed.
- V. Castelletto, R. J. Gouveia, C. J. Connon and I. W. Hamley, Faraday Discuss., 2013, 166, 381–397 RSC.
- V. Castelletto and I. W. Hamley, in Peptide Self-Assembly: Methods and Protocols, ed. B. L. Nilsson and T. M. Doran, Humana Press Inc, Totowa, 2018, vol. 1777, pp. 3–21 Search PubMed.
- J. Adamcik, V. Castelletto, I. W. Hamley and R. Mezzenga, Angew. Chem., Int. Ed., 2011, 50, 5495–5498 CrossRef CAS PubMed.
- L. Ziserman, H. Y. Lee, S. R. Raghavan, A. Mor and D. Danino, J. Am. Chem. Soc., 2011, 133, 2511–2517 CrossRef CAS PubMed.
- K. Lu, J. Jacob, P. Thiyagarajan, V. P. Conticello and D. G. Lynn, J. Am. Chem. Soc., 2003, 125, 6391–6393 CrossRef CAS PubMed.
- A. K. Mehta, K. Lu, W. S. Childers, S. Liang, J. Dong, J. P. Snyder, S. V. Pingali, P. Thiyagarajan and D. G. Lynn, J. Am. Chem. Soc., 2008, 130, 9829–9835 CrossRef CAS PubMed.
- M. Sunde, L. C. Serpell, M. Bartlam, P. E. Fraser, M. B. Pepys and C. C. F. Blake, J. Mol. Biol., 1997, 273, 729–739 CrossRef CAS PubMed.
- L. C. Serpell, Biochim. Biophys. Acta, Biomembr., 2000, 1502, 16–30 CrossRef CAS PubMed.
- I. W. Hamley, Angew. Chem., Int. Ed., 2007, 46, 8128–8147 CrossRef CAS PubMed.
- H. Levine, in Methods in Enzymology, ed. R. Wetzel, Academic Press, San Diego, 1999, vol. 309, pp. 274–284 Search PubMed.
- A. K. Buell, C. M. Dobson, T. P. J. Knowles and M. E. Welland, Biophys. J., 2010, 99, 3492–3497 CrossRef CAS PubMed.
- A. Rodger and B. Nordén, Circular dichroism and linear dichroism, Oxford University Press, Oxford, 1997 Search PubMed.
- M. Gupta, A. Bagaria, A. Mishra, P. Mathur, A. Basu, S. Ramakumar and V. S. Chauhan, Adv. Mater., 2007, 19, 858–861 CrossRef CAS.
- M. J. Krysmann, V. Castelletto and I. W. Hamley, Soft Matter, 2007, 3, 1401–1406 RSC.
- N. Amdursky and M. M. Stevens, Chem. Phys. Chem., 2015, 16, 2768–2774 CrossRef CAS PubMed.
- I. W. Hamley, D. R. Nutt, G. D. Brown, J. F. Miravet, B. Escuder and F. Rodríguez-Llansola, J. Phys. Chem. B, 2010, 114, 940–951 CrossRef CAS PubMed.
- E. Hughes, N. S. O’Neill and R. Schweitzer-Stenner, J. Phys. Chem. B, 2024, 129, 260–272 CrossRef PubMed.
- T. Bernas and J. Dobrucki, Cytometry, 2002, 47, 236–242 CrossRef CAS PubMed.
- M. Ghasemi, T. Turnbull, S. Sebastian and I. Kempson, Int. J. Mol. Sci., 2021, 22, 12827 CrossRef CAS PubMed.
- J. C. Gordon, J. B. Myers, T. Folta, V. Shoja, L. S. Heath and A. Onufriev, Nucleic Acids Res., 2005, 33, W368–W371 CrossRef CAS PubMed.
- R. Anandakrishnan, B. Aguilar and A. V. Onufriev, Nucleic Acids Res., 2012, 40, W537–W541 CrossRef CAS PubMed.
- B. Soreghan, J. Kosmoski and C. G. Glabe, J. Biol. Chem., 1994, 269, 28551–28554 CrossRef CAS PubMed.
- F. Oosawa and M. Kasai, J. Mol. Biol., 1962, 4, 10–21 CrossRef CAS PubMed.
- F. Oosawa and S. Asakura, Thermodynamics of the polymerization of protein, Academic Press, New York, 1975 Search PubMed.
- Y. Geng, P. Dalhaimer, S. S. Cai, R. Tsai, M. Tewari, T. Minko and D. E. Discher, Nat. Nanotechnol., 2007, 2, 249–255 CrossRef CAS PubMed.
- S. S. Cai, K. Vijayan, D. Cheng, E. M. Lima and D. E. Discher, Pharm. Res., 2007, 24, 2099–2109 CrossRef CAS PubMed.
- A. Yaghmur, P. Laggner, S. G. Zhang and M. Rappolt, PLoS One, 2007, 2, e479 CrossRef PubMed.
- N. Schmidt, A. Mishra, G. H. Lai and G. C. L. Wong, FEBS Lett., 2009, 584, 1806–1813 CrossRef PubMed.
- A. Mishra, G. H. Lai, N. W. Schmidt, V. Z. Sun, A. R. Rodriguez, R. Tong, L. Tang, J. J. Cheng, T. J. Deming, D. T. Kamei and G. C. L. Wong, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16883–16888 Search PubMed.
- V. Castelletto, R. H. Barnes, K.-A. Karatsas, C. J. C. Edwards-Gayle, F. Greco, I. W. Hamley, J. Seitsonen and J. Ruokolainen, Langmuir, 2019, 35, 1302–1311 CrossRef CAS PubMed.
- C. J. C. Edwards-Gayle, V. Castelletto, I. W. Hamley, G. Barrett, F. Greco, D. Hermida-Merino, R. Rambo, J. Seitsonen and J. Ruokolainen, ACS Appl. Bio. Mater., 2019, 2, 2208–2218 CrossRef CAS PubMed.
- C. J. C. Edwards-Gayle, G. Barrett, S. Roy, V. Castelletto, J. Seitsonen, J. Ruokolainen and I. W. Hamley, ACS Appl. Bio Mater., 2020, 3, 1165–1175 CrossRef CAS PubMed.
- I. W. Hamley and V. Castelletto, Bioconjugate Chem., 2017, 28, 731–739 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional cryo-TEM images and SAXS data, ThT fluorescence data, CD spectra, Tables of SAXS fit parameters and calculated pKa values. See DOI: https://doi.org/10.1039/d5sm00432b |
| This journal is © The Royal Society of Chemistry 2025 |