 Open Access Article
Open Access ArticlePathogenic bacteria characterization through portable optical scatter device and machine learning
Ramana
Pidaparti
 *a,
Sanjay
Oruganti
*a,
Sanjay
Oruganti
 bc,
Naveen
Kurra
d,
Patrick
Maffe
e,
Everett
Grizzle
*e,
Arnab
Mondal
bc,
Naveen
Kurra
d,
Patrick
Maffe
e,
Everett
Grizzle
*e,
Arnab
Mondal
 e,
Rebecca
Johnson
e,
Rebecca
Johnson
 f,
Hitesh
Handa
f,
Hitesh
Handa
 e and
Rao
Tatavarti
g
e and
Rao
Tatavarti
g
aSchool of ECAM, University of Georgia, Athens, 30602, Georgia, USA. E-mail: rmparti@uga.edu
bSchool of ECE, University of Georgia, Athens, 30602, Georgia, USA
cCognitive Science Department, Rensselaer Polytechnic Institute, Troy, 12180, New York, USA
dSchool of Computing, University of Georgia, Athens, 30602, Georgia, USA
eSchool of CMBE, University of Georgia, Athens, 30602, Georgia, USA
fDepartment of Bioengineering, University of Maryland, College Park, 20742, Maryland, USA
gGVP-SIRC, GVP College of Engineering, Vishakhapatnam, 530048 Andhra Pradesh, India
First published on 30th September 2025
Abstract
Rapid and accurate detection and characterization of pathogenic bacteria is critical for clinical diagnosis. Most selective clinical procedures are limited by their diagnostic speed, accuracy, and sensitivity challenges. In order to overcome these, we introduce a novel photonics-based, point-of-care device designed for rapid and accurate characterization of bacteria. The device is designed to capture optical scatter signatures unique to various pathogenic bacteria, which are analyzed using advanced clustering and machine learning techniques for characterization. Our preliminary results from controlled experiments show that our device successfully distinguishes bacteria genus with reasonable accuracy.
1 Introduction
Pathogenic bacteria continue to impose a substantial global burden across clinical,1,2 food safety,3 and environmental domains.4 Traditional methods for identifying and characterizing bacteria encompass a variety of approaches, including molecular methods,5,6 phenotypic assays,7–9 microscopy,10 and proteomics-based techniques,11 each with its unique advantages and limitations. Culture-based techniques are considered the gold standard in bacterial quantification.7 However, these methods are often time-consuming and may pose challenges with culturing certain bacteria, making them less suitable for rapid diagnostics and quantification. Also, polymerase chain reaction (PCR)12 is a widely used technique that excels in detecting and quantifying specific DNA sequences of pathogenic bacteria, yet it may not always accurately reflect bacterial counts. Microscopy methods,13 such as phase-contrast and fluorescence microscopy, are effective for visualizing and counting bacteria. However, these techniques are generally not conducive to high-throughput quantification and are labor-intensive. Flow cytometry14 approach allows for high-throughput quantification by analyzing bacteria based on their optical properties and fluorescence signals, but involves complex processes and infrastructure.14,15 While microbiology-based diagnostic procedures and devices have traditionally played a vital role in identifying pathogens,16,17 enhancing their speed and accuracy reliably remains challenging. Major challenges include the complexity of obtaining biological samples,18 varying sample volumes, and the need for expensive and specialized equipment combined with the need for the knowledge of the diverse technological approaches currently in use.17,19Over the past few years, optical methods have emerged as compelling alternatives or complements to traditional approaches because they can interrogate pathogen–light interactions with high sensitivity, label-free operation, and minimal sample preparation. Techniques span colorimetric and fluorescence readouts, surface plasmon resonance (SPR), Raman and surface-enhanced Raman spectroscopy (SERS), interferometry, and elastic/scattering-based modalities, many of which can be miniaturized and multiplexed via nanophotonic and microfluidic integration.3,6,20 These platforms have demonstrated the capacity to detect and differentiate clinically relevant bacteria (e.g., Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa) and foodborne pathogens with fast turnaround and potential portability.3,20,21 The integration of optical sensing with machine learning (ML) has emerged as an approach for innovative applications. Optical measurements generate high-dimensional signatures including vibrational spectra, scattering patterns, or image textures that are challenging to interpret by hand or with simple heuristics. Modern ML (including convolutional neural networks and object-detection models) automates feature discovery and classification, improving accuracy and robustness while enabling near-real-time analysis.5,22 For example, alignment-free optical scattering with commodity LEDs coupled to YOLOv8 achieved high performance for colony-level identification, illustrating how simplified optics plus deep learning can deliver practical, low-cost systems that integrate into existing workflows.5,23 Likewise, microfluidic systems that embed sample preparation and optical detection are maturing toward POC deployment, offering compact form factors, reduced reagent consumption, and faster time-to-answer for infectious-disease diagnostics.24,25
Recent reviews on AI-enabled Raman/SERS and photonic POC devices and applications report high classification accuracies and progress on resistance phenotyping when paired with deep learning, while also highlighting dataset curation and interpretability challenges.2,5 Also, photonic POC devices that co-integrate optics, nanostructured substrates, and microfluidics are enabling portable, low-cost platforms for rapid, on-site pathogen detection and, increasingly, for accelerated susceptibility testing.6,24,26 Authors27 developed a compact, non-invasive photonic system that utilizes backscattering for real-time, remote detection of airborne microbes, demonstrating its ability to characterize various pathogenic bacterial and their mixtures. Collectively, these trends point to miniaturized, AI-driven optical diagnostics that bridge laboratory precision and field practicality. Despite these advances, gaps remain in standardized datasets, clinically interpretable models, calibration/transfer across instruments and sites, and end-to-end validation within real clinical workflows.2,5 This work addresses those gaps by presenting an optical scatter based characterization framework coupled with machine-learning classifiers designed for rapid, and high-throughput bacterial characterization.
2 Portable bacteria characterization device
The portable bacteria characterization device is a photonic system based on forward scatter principles to characterize bacteria using scatter data and machine learning models. This photonic system is composed of a laser source, a duo-lateral quadrant photodetector (PD), a data acquisition system (DAQ), an onboard computing module, and an enclosure that houses these components, as shown in Fig. 1. The laser source is precisely aligned to project its beam onto the center of the PD's active sensing area, enabling the detection of bacteria present in the beam path. The PD's outputs are connected to an amplifier circuit that calculates the beam's deflection on the (x, y) plane based on the differential currents from the four quadrants and the total beam power, which is proportional to the sum of the currents. This amplified data signal is then digitized by a high-speed DAQ connected to a compute module. The device is designed with a compact form factor to ensure its portability while also efficiently meeting the requirements for computing power, storage, and data transmission. A service-based firmware was developed for the device that enables wireless connectivity to an interface with software running on a personal computer. This device operates on the principles of optical scattering and employs a continuous, coherent, and collimated laser beam aimed at the surface of the photodetector (PD). The intensity and position of the laser beam, as received by the PD, alter due to scattering and refraction events caused by the presence and composition of bacteria in the intervening space. The working principles of this device are elaborated in our patent28 and it was observed that the scattering signatures are distinct and vary depending on the composition of the aerosols containing pathogens. The present study involves the collection and characterization of scatter data from various bacteria, analyzing them based on their distinct optical scatter features. A detailed representation of the device, in its most recent version, is depicted in Fig. 1, and a sectional view along with scatter configuration is presented in Fig. 3. The device supports the deposition of particles onto glass slides, as illustrated in 1, enabling the accumulation of pathogenic bacteria for subsequent static analysis. | ||
| Fig. 1 (a) Overview of the bacteria characterization device. (b) An exploded view showing various components of the device. | ||
3 Materials and methods
We conducted controlled experiments by depositing specific bacteria at known concentrations onto glass slides to examine their unique scatter characteristics. This section elaborates on the bacteria preparation and deposition process, followed by an in-depth discussion of data collection and analysis methodologies.3.1 Materials
Staphylococcus aureus (ATCC® 6538™), Escherichia coli (ATCC® 25922™), Klebsiella pneumoniae (ATCC® 700603™), Staphylococcus epidermidis (ATCC® 35984™), Staphylococcus mutans (ATCC® 25175™), and Pseudomonas aeruginosa (ATCC® 27853™) were purchased from American Type Culture Collection (ATCC, Manassas, VA USA). Table 1 presents the size and shape characteristics of various bacteria considered in this study. Phosphate buffered saline (PBS) at 7.4 pH and Luria Bertani (LB) broth were purchased from Sigma-Aldrich (St. Louis, MO USA).| Bacteria | Type | Size | Shape |
|---|---|---|---|
| Staphylococcus aureus (SA) – ATCC 6538 | Gram positive | 0.5–1.5 μm diameter | Round |
| Escherichia coli (EC) – ATCC 25922 | Gram negative | 0.5 μm wide 1.5 μm long | Rod |
| Pseudomonas aeruginosa (PA) – ATCC 27853 | Gram negative | 0.5–1.0 μm wide 1.0–5.0 μm long | Rod |
| Staphylococcus epidermidis (SE) – ATCC 35984 | Gram positive | 0.5–1.5 μm diameter | Round |
| Klebsiella pneumoniae (KP) – ATCC 700603 | Gram negative | 0.3–1.0 μm wide 0.6–6.0 μm long | Rod |
| Streptococcus mutans (SM) – ATCC 25175 | Gram positive | 0.5–0.75 μm diameter | Round |
| EC + SA | Combination | ||
| PA + SE | Combination | ||
| SM + KP | Combination |
3.2 Methods
The methods for preparing bacterial solutions were derived from the well-established ISO 10993 standards29 and performed under aseptic conditions in a laminar airflow cabinet. All media and buffer solutions were sterilized in an autoclave at 121 °C for 30 minutes. Preparation of the bacterial solution began by inoculating a single bacteria colony into either Luria Bertani (LB) or brain heart infusion (BHI) broth for all bacteria used. These solutions were placed in an incubated shaker at 37 °C and 150 rpm until the log-phase of growth for each bacterium was reached. The bacteria were then centrifuged at 3500 rpm for 7.5 minutes using an Allegra X-30R Centrifuge (Beckman Coulter, Indianapolis, IN USA). The remaining supernatant fluid was removed and then the bacterial pellet was resuspended with 0.01 M sterile phosphate-buffered saline (PBS) for rinsing at 3500 rpm for 7.5 minutes. Supernatant fluid was removed once more, and the bacterial pellet resuspended with fresh 0.01 M PBS. At this point, the bacteria solutions were measured for absorbance at 600 nm using a Cary 60 UV-vis spectrophotometer (Agilent, Santa Clara, CA USA) and diluted to 108 CFU mL−1. The optical density at 600 nm (OD600nm) was measured using a Genesys 10S ultraviolet-visible spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), with the assumption that an OD of 0.1 corresponds to approximately 108 cells per mL. A summary of the bacteria solutions prepared is presented in Table 1.After the bacterial solution was prepared it was pipetted directly onto a glass slide inside of a BSL-2 approved biosafety cabinet to maintain sterility. The droplets were then uniformly spread and contained using a cover slip before testing. The entire sample preparation is carried out inside of the biosafety cabinet. Only once the sample is finalized is it removed from the cabinet for testing. Usually the testing took 60 s for each trail. An overview of bacteria solution preparation and testing is presented in Fig. 2.
4 Testing and data analysis
Testing began 1–2 hours after placing the glass slides onto the device. The slide with the bacteria coating was inserted in a slot closer to the laser source in the P2 position, and a blank slide was inserted on the detector end in the P1 position, as shown in Fig. 3. We conducted several trials involving combinations of concentrations (0.05, 0.1, and 0.2) and volumes (5 μL, 10 μL, and 20 μL) to capture scatter signatures of light passing through various bacteria. In each trial, optical scatter data was collected for 60 seconds. The data processing and classification workflow began with controlled experiments aimed at gathering optical scatter data, particularly focusing on beam power (Power) and position measurements as outlined in Fig. 4. The raw data underwent pre-processing steps, including demeaning and outlier removal through sigma clipping. Following this, key metrics such as beam deflection efficiency (BDE) and scatter intensity variability (SIV) was computed. These metrics were then refined using principal component analysis (PCA). The processed data was subsequently fed into machine learning models to accurately classify and quantify various bacterial genus.During data collection and pre-processing steps to ensure optimal alignment of the laser and detector, we analyzed the Xpos and Ypos difference signals. Ideally, these signals should be close to zero, indicating proper alignment. However, even with good alignment, deviations from zero can occur due to various environmental factors, such as dust on the detector surface, air currents, vibrations, or slight misalignments during assembly. To mitigate these effects and eliminate baseline errors, we conducted multiple trials using blank slides in positions P1 and P2, without any samples. The beam power and position values from these trials were averaged to establish a baseline representing the inherent system noise and drift. This baseline data was then deducted from the experimental data collected with actual samples as shown in eqn (1) and (2), effectively isolating the bacterial signal while minimizing the impact of environmental factors and system noise. This process, known as de-meaning, enhances the data's sensitivity to the unique scatter profiles of bacteria by removing the mean values obtained from the control trials across all experimental runs.
| Xpos = Xraw − Mean(Xcontrol) | (1) |
| Ypos = Yraw − Mean(Ycontrol) | (2) |
From the computed Xpos and Ypos, the magnitude of the beam's displacement according to eqn (3) and (4).
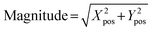 | (3) |
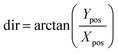 | (4) |
The following step in the procedure entails the outlier removal, which is a critical pre-processing step in our current data analysis, aimed at identifying and excluding anomalous data points from a dataset. In our study, outliers are defined as those points lying beyond three standard deviations from the mean. This threshold is based on the empirical rule,30 which suggests that approximately 99.7% of data in a normal distribution should fall within three standard deviations from the mean, rendering points outside this range exceedingly uncommon. Upon examination, we found approximately 0–0.5 seconds' worth of outlier data per trial, equating to 0–500 data points. Consequently, any samples exceeding this predefined boundary were deemed outliers and subsequently omitted from the dataset. For the ease of reading, we continue the usage of labels Xpos, Ypos and Power that from this point represent data with the outliers removed.
4.1 Characterization metrics
In this section, we explore two characterization metrics: beam deflection efficiency (BDE) and scattering intensity variability (SIV). These features have been specifically designed to enhance the differentiation and classification of bacteria based on their optical scatter patterns.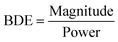 | (5) |
4.2 Scattering intensity variability
The scattering intensity variability (SIV), as shown in eqn (6), is the coefficient of variation that measures the variation in light scattering responses from bacterial samples by assessing the degree of fluctuation in scattering intensity across different detection angles. This variability is proportional to the physical diversity within the sample. High SIV values indicate significant fluctuations in scattering intensity, suggesting a heterogeneous bacterial composition. Conversely, low SIV values suggest a more uniform scattering response, characteristic of a homogeneous composition. Thus, SIV is a crucial metric for assessing the underlying physical properties of a sample, making it valuable for differentiating between bacterial species and analyzing sample composition. | (6) |
After standardization, we applied principal component analysis (PCA) to reduce the dimensionality of the dataset. PCA identifies the most important patterns in the data by generating components that capture the maximum variance. We retained the two most significant components, enabling us to represent the data in a simplified two-dimensional space. This transformation can be understood as finding the optimal linear combinations of the original features that best capture the variance in the data.
| PC = Dscaled × W | (7) |
Here, PC represents the principal components, Dscaled is the standardized data matrix, and W is the matrix of weights (or loadings) that define the linear combinations of the original features used to form the principal components.
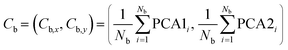 | (8) |
To assess the clustering of data points around their respective centroids, we calculated the Euclidean distance between each point and its corresponding centroid. This distance serves as a metric for quantifying how far each point deviates from the central tendency of its group. This computation, as shown in eqn (9), computes the measurement of the dispersion of data points within each bacterial group in the PCA-transformed space.
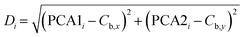 | (9) |
To ensure that only representative data points were retained, we established a threshold based on the distribution of distances. Specifically, we excluded any data point that exceeded the average distance plus one standard deviation from the centroid.
| T = D + σd | (10) |
This filtering criterion allowed us to focus on data points that were more closely aligned with the central trends of their respective bacterial groups, thereby enhancing the reliability of our final analysis.
Each trial produced a set of feature vectors as described in characterization metrics section:
X = {x1, x2, …, xN}, xk ∈ ![[Doublestruck R]](https://www.rsc.org/images/entities/char_e175.gif) d∀k ∈ {1, …, N} d∀k ∈ {1, …, N} | (11) |
To evaluate separability of bacterial data from background/noise, K-means32 clustering was applied independently on each trial dataset X. The K-means objective minimizes within cluster variance:
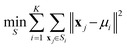 | (12) |
Cluster quality was quantified using the silhouette score s(i)33 for each data point xi:
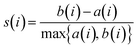 | (13) |
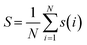 | (14) |
Higher values of S indicate more compact and well-separated clusters, suggesting stronger bacterial signal detection.
For each bacteria type, silhouette scores were aggregated across slides and trials:
 | (15) |
![[S with combining macron]](https://www.rsc.org/images/entities/i_char_0053_0304.gif) was considered the optimal condition for bacterial detection.
was considered the optimal condition for bacterial detection.
Finally, for each bacterial species, the optimal conditions were identified as
 | (16) |
4.3 Classification models
The filtered dataset was then utilized for training machine learning algorithms aimed at predicting class labels within the optimal clusters, thereby enabling the identification of distinguishing features for the identified bacterial groups. We applied four advanced machine learning models that include support vector classifier (SVC),34 gradient boosting,35 LightGBM,36 and XGBoost37 to classify different bacterial types based on the extracted features. These models were selected for their ability to handle various aspects of the classification task, contributing to a comprehensive and accurate data analysis.Support vector classifier (SVC)34 works by identifying the optimal hyperplane that maximizes the margin between the nearest points of different classes, effectively separating the data into distinct groups. This makes SVC particularly effective in binary classification tasks with a clear boundary between classes.
Gradient boosting, on the other hand, is a powerful ensemble learning technique that builds models sequentially, each one attempting to correct the errors of the previous models by optimizing the residuals. This approach excels in complex, non-linear relationships between input features and the target variable. LightGBM,36 XGBoost37 a widely adopted implementation of gradient boosting known for its scalability and precision. XGBoost was selected for its ability to handle complex data patterns and provide a robust framework for bacterial classification in this study.
Each of these models was strategically chosen to take advantage of their strengths, ensuring a comprehensive and accurate analysis of the data. For instance, SVM has been chosen based on the literature38,39 which uses similar kind of techniques that are based on optics.
In general, combining these four models provided a broad exploration of machine learning techniques, with each model contributing valuable information to the bacterial classification process. The results demonstrated the feasibility of using machine learning for this purpose while also highlighting opportunities for further refinement and optimization in future studies.
The first metric used in our analysis is accuracy, which is a measure of the correctness of a model's predictions. It is calculated as the ratio of correctly predicted instances to the total instances and computed according to eqn (17)
 | (17) |
F1 score, another important metric, is particularly useful in handling imbalanced datasets. It balances precision and recall, making it more effective in applications where both are critical. The F1 score is calculated as the harmonic mean of precision and recall as shown in eqn (18)
 | (18) |
Here, precision measures the accuracy of positive predictions, calculated as the ratio of true positives (TP) to the sum of true positives and false positives (FP), as shown in eqn (19).
 | (19) |
 | (20) |
Here, false negatives (FN) are instances incorrectly predicted as negative despite being positive, while true negatives (TN) are instances correctly identified as negative.
5 Results and discussion
We collected optical scatter data metrics for multiple Gram-positive and Gram-negative bacteria, as well as their combinations, across three different concentrations (0.05, 0.1, and 0.2) and three different volumes (5 mL, 10 mL, and 20 mL). Our observations indicate that data collected from slides prepared with lower volumes and concentrations are more likely influenced by the sterile phosphate-buffered saline (PBS) used during slide preparation. Microscopic analysis further confirmed that lower volumes and concentrations resulted in a reduced bacterial cell count within the test region. Also, a lower refractive index contrast relative to PBS will diminish elastic-scatter amplitude at the detector.Based on these findings, we determined that a concentration of 0.2 and a volume of 20 mL provided the most representative data for capturing accurate scatter signatures. Therefore, this combination was used for the subsequent analysis.
5.1 Optical scatter measurements and PCA analysis
Optical scatter data plots for beam deflection efficiency (BDE) and direction (DIR) of various Gram-positive and Gram-negative bacteria are presented in Fig. 5. The scatter data signatures exhibit distinct patterns across different bacterial species. However, some cluster overlap is observed, particularly between Klebsiella pneumoniae (KP) and Pseudomonas aeruginosa (PA), as well as between PA + SE and SM + KP. This overlap can be attributed to the similarity in scatter signatures resulting from their comparable rod-shaped morphologies, as detailed in Table 1. | ||
| Fig. 5 Plot of beam deflection efficiency (BDE) versus directionality (dir) computed from the original features Xpos, Ypos, and power. | ||
Building on the insights from existing literature,16,18,19,27,40–43 which highlight significant variations in optical signatures across different bacteria, we further explored these patterns by integrating beam deflection efficiency (BDE) and direction metrics with scattering intensity variation (SIV). By applying principal component analysis (PCA), we uncovered distinct and well-separated clusters for each bacteria, as depicted in Fig. 6. This analysis clearly demonstrates that our portable device effectively captures and differentiates multiple bacteria based on their scatter data signatures.
The ability of the device to accurately distinguish between bacteria, even those with similar shapes and sizes, emphasizes its efficacy in microbial analysis. This not only validates the sensitivity and precision of the optical scatter metrics employed but also highlights the potential of our device for rapid and reliable bacterial characterization in various practical applications.
5.2 Repeatability of experimental data
The test was carried out on Escherichia coli (EC) and Staphylococcus aureus (SA) to assess repeatability, with the resulting scatter data presented in Fig. 7. Multiple trials were performed on the same slide, starting with two trials using SA bacteria, followed by three trials using EC bacteria. As shown in Fig. 7, the scatter data is consistently clustered for each bacterium, demonstrating a high degree of repeatability in the trials. This consistency indicates that the data are both reliable and reproducible under the experimental conditions. The minimal variation observed between trials allowed us to confidently use the results from a single trial for further quantitative analysis, as the consistency across trials ensured that the overall data integrity remained unaffected. | ||
| Fig. 7 Results from repeatability tests conducted, demonstrating the consistency and reliability of the scatter data across different trials. | ||
5.3 Effects of bacteria solution concentration and volume
In order to evaluate the best combination of bacteria concentrations and volumes considered, a cluster analysis was carried out using K-means algorithm.32 The results of silhouette score obtained from the cluster analysis are presented in Table 2. Results show that a concentration of 0.2 and a volume of 20 mL are representative for better capture of scatter data, and this data was used for further analysis for quantification. Also, in all the experiments with lower volumes and concentrations, it is the same volume and concentration of PBS, just less bacteria. We also observed under microscopic study that lower volumes and concentrations of bacteria have lower cell count within the test region. This is consistent with reduced cell counts in the illuminated region and a lower refractive index contrast relative to PBS, which diminishes elastic scatter amplitude at the detector.| Bacteria | Concentration | Volume | Silhouette score |
|---|---|---|---|
| Staphylococcus aureus (SA) | 0.2 | 20 | 0.911 |
| Escherichia coli (EC) | 0.2 | 20 | 0.964 |
| Pseudomonas aeruginosa (PA) | 0.2 | 5 | 0.933 |
| Staphylococcus epidermidis (SE) | 0.2 | 10 | 0.842 |
| Klebsiella pneumoniae (KP) | 0.05 | 20 | 0.971 |
| Streptococcus mutans (SM) | 0.2 | 20 | 0.952 |
| EC + SA | 0.1 | 20 | 0.876 |
| PA + SE | 0.1 | 20 | 0.889 |
| SM + KP | 0.1 | 10 | 0.871 |
5.4 Performance evaluation and comparison of the models
The performance of four machine learning models, gradient boosting (GB), LightGBM, support vector machine (SVM), and XGBoost was evaluated for classifying Gram-positive and Gram-negative bacteria, as shown in Table 3. The evaluation was conducted using different combinations of training and test data derived from three samples, referred to as S1, S2, and S3. Test accuracy and F1 scores were used to assess each model's effectiveness across various combinations of training and test data.| Model | Data group | Training data | Test data | Test accuracy | F1 score |
|---|---|---|---|---|---|
| Gradient boosting (GB) | Gram-positive | (S1, S2) | S3 | 0.8550 | 0.8300 |
| (S1, S3) | S2 | 0.7027 | 0.6300 | ||
| Gram-negative | (S1, S2) | S3 | 0.4937 | 0.4900 | |
| (S1, S3) | S2 | 0.8195 | 0.7900 | ||
| Combinations | (S1, S2) | S3 | 0.9247 | 0.9200 | |
| (S1, S3) | S2 | 1.0000 | 1.0000 | ||
| LightGBM | Gram-positive | (S1, S2) | S3 | 0.9998 | 1.0000 |
| (S1, S3) | S2 | 0.7036 | 0.6400 | ||
| Gram-negative | (S1, S2) | S3 | 0.7365 | 0.7300 | |
| (S1, S3) | S2 | 0.8795 | 0.8700 | ||
| Combinations | (S1, S2) | S3 | 0.8958 | 0.8900 | |
| (S1, S3) | S2 | 1.0000 | 1.0000 | ||
| SVM | Gram-positive | (S1, S2) | S3 | 1.0000 | 1.0000 |
| (S1, S3) | S2 | 0.7128 | 0.6400 | ||
| Gram-negative | (S1, S2) | S3 | 1.0000 | 1.0000 | |
| (S1, S3) | S2 | 1.0000 | 1.0000 | ||
| Combinations | (S1, S2) | S3 | 1.0000 | 1.0000 | |
| (S1, S3) | S2 | 1.0000 | 1.0000 | ||
| XGBoost | Gram-positive | (S1, S2) | S3 | 1.0000 | 1.0000 |
| (S1, S3) | S2 | 0.6857 | 0.6300 | ||
| Gram-negative | (S1, S2) | S3 | 0.7302 | 0.7300 | |
| (S1, S3) | S2 | 0.7661 | 0.7400 | ||
| Combinations | (S1, S2) | S3 | 0.9097 | 0.9100 | |
| (S1, S3) | S2 | 1.0000 | 1.0000 |
Gradient boosting showed varying performance depending on the data split. For Gram-positive bacteria, it performed well when trained on S1 and S2 and tested on S3, achieving a test accuracy of 0.8550 and an F1 score of 0.8300. However, its performance dropped when the training data was switched to (S1, S3) and tested on S2, with accuracy and F1 scores decreasing to 0.7027 and 0.6300, respectively. For Gram-negative bacteria, GB struggled, especially when trained on (S1, S2) and tested on S3, where it managed only 0.4937 accuracy and 0.4900 F1 score. Interestingly, the model's performance improved significantly for Gram-negative bacteria when the training data was (S1, S3), with accuracy and F1 scores rising to 0.8195 and 0.7900, respectively. Notably, GB performed exceptionally well on the combination of bacteria data, achieving perfect accuracy when trained on (S1, S3) and tested on S2.
LightGBM on the other hand, consistently delivered strong results across both Gram-positive and Gram-negative bacteria classifications. For Gram-positive bacteria, it nearly achieved a very high accuracy of 0.9998 and an F1 score of 1.000 when trained on (S1, S2) and tested on S3. The model remained robust across different data splits, showing its reliability. For Gram-negative bacteria, LightGBM performed solidly, with accuracy ranging from 0.7036 to 0.8795 and corresponding F1 scores. Its performance on combination data was similarly strong, achieving perfect accuracy and F1 scores when trained on (S1, S3) and tested on S2, indicating its generalization capability.
SVM also performed exceptionally well, particularly with Gram-positive bacteria. It achieved perfect accuracy and F1 scores (1.000) when trained on (S1, S2) and tested on S3. However, its performance was somewhat sensitive to different data splits, as seen when accuracy dropped to 0.7128 and the F1 score to 0.6400 with the training and test combination of (S1, S3) and S2, respectively. Despite this, SVM maintained high performance across Gram-negative bacteria and combination data, consistently achieving perfect accuracy and F1 scores in several scenarios. These results highlight SVM's effectiveness in classifying bacterial data when trained with the appropriate dataset.
XGBoost displayed strong performance, particularly with Gram-positive bacteria, where it achieved perfect accuracy and F1 scores across multiple data splits. However, its performance was slightly less consistent with Gram-negative bacteria, as evidenced by an accuracy of 0.7302 and an F1 score of 0.7300 when trained on (S1, S2) combination and tested on S3. The model improved significantly when trained on (S1, S3) and tested on S2, achieving perfect accuracy. XGBoost's ability to deliver strong results across different data combinations demonstrates its robustness, though it showed some sensitivity to specific training-test configurations.
In comparing the models, LightGBM and SVM emerged as the most consistent and reliable classifiers, delivering high accuracy and F1 scores across various data splits. Their performance was particularly impressive with Gram-positive bacteria, where they often achieved perfect or near-perfect results. XGBoost also performed well, particularly for Gram-positive bacteria, though it showed some sensitivity to different training and test data combinations. Gradient Boosting displayed more variability, especially with Gram-negative bacteria, indicating that it may require further tuning or an alternative approach to optimize its performance.
The sample decision boundary visualizations in Fig. 8 illustrate how well the SVM models classify both Gram-positive and Gram-negative bacteria. The clear decision boundaries and accurate test data predictions confirm the model's effectiveness. Additionally, the shared decision boundary observed between PA + SE and SM + KP suggests that non-linear patterns in the data may be present, which could potentially be addressed using a non-linear kernel in SVM.
Nonpathogenic organisms can be detected optically; however, classification is restricted to trained pathogenic genera. If a new sample is kept under test and the probability of prediction of the model falls below an adjustable threshold, the sample is treated as ‘unknown’. These signatures are stored for future model updates. The planned future work includes expanding the reference libraries and implementing advanced anomaly detection approaches.
6 Limitations
An earlier aerosol study27 used a device based on backscatter geometry and dynamic aerosols, which provided richer temporal features that allowed bacteria discrimination. In contrast, the current forward-scatter bacteria device uses static-slide configuration that integrates over a quasi-static field, reducing fine-grained cues. It is very difficult to directly compare static slide and aerosol configurations due to different experimental conditions and protocols. In addition, genus-level predictions can inform early clinical assessment even if species/strain confirmation requires downstream methods. We now explicitly state that the present study demonstrates robust genus-level discrimination and species-level separation only within Staphylococcus. Future work will explore time-resolved scattering for finer resolution. In addition, the current approach and the model developed are optimized for bacteria. Fungi are outside the model training set, and the present model will not be able to classify, and hence future work needs to include dedicated training data for fungi in the model for characterization.7 Conclusion
In this study a portable photonics-based point-of-care device designed for rapid and accurate characterization of bacteria is investigated. Experiments were conducted on multiple Gram-positive and Gram-negative bacteria and their combinations that can capture optical scatter data. The results of bacteria characterization are obtained using the optical scatter data and adopting clustering and machine learning algorithms. Based on the results obtained, it was demonstrated that the current portable device can be used for robust genus-level bacteria discrimination, and not bacterial strains. More work is needed to investigate the device for bacterial strains and fungi.Author contributions
Ramana Pidaparti conceived the research idea, supervised the research, and contributed to the writing and review of the manuscript. Sanjay Oruganti led the device development and to the writing and review of the manuscript. Naveen Kurra conducted the experiments and data analysis, and to the writing and review of the manuscript. Patrick Maffe and Arnab Mondal prepared the bacterial solutions, Everett Grizzle and Rebecca Johnson conducted some experiments. Hitesh Handa and Rao Tatavarti reviewed the manuscript.Conflicts of interest
The authors state that there are no conflicts to declare.Data availability
The data supporting this article will be made available upon request.Acknowledgements
The first author thanks the Georgia Research Alliance (GRA) and the UGA Start-up funds for supporting this research. Also, thanks to students Alek Bruckman and Nathaniel Leblanc for their help with CAD, fabrication and assembly of the portable device.Notes and references
- T. Ferkol and D. Schraufnagel, Ann. Am. Thorac. Soc., 2014, 11, 404–406 CrossRef PubMed.
- M. Hao, J. Clin. Res., 2024, 8, 233 Search PubMed.
- O. I. Guliy, O. A. Karavaeva, A. V. Smirnov, S. A. Eremin and V. D. Bunin, Sensors, 2023, 23, 9391 CrossRef CAS PubMed.
- L. Lin, H. Yang and X. Xu, Front. Environ. Sci., 2022, 10, 880246 CrossRef.
- M. H.-U. Rahman, R. Sikder, M. Tripathi, M. Zahan, T. Ye, Z. E. Gnimpieba, B. K. Jasthi, A. B. Dalton and V. Gadhamshetty, Chemosensors, 2024, 12, 140 CrossRef CAS.
- A. Bari, S. Aslam, H. Khan, S. Shakil, M. Yaseen, S. Shahid, A. Yusaf, N. Afshan, S. Shafqat and M. Zafar, Plasmonics, 2025, 1–27 Search PubMed.
- A. Sloan, G. Wang and K. Cheng, Clin. Chim. Acta, 2017, 473, 180–185 CrossRef CAS PubMed.
- M. Vouga and G. Greub, Clin. Microbiol. Infect., 2016, 22, 12–21 CrossRef PubMed.
- R. Kshikhundo and S. Itumhelo, World News Nat. Sci., 2016, 26–38 Search PubMed.
- B. Obara, M. A. Roberts, J. P. Armitage and V. Grau, BMC Bioinf., 2013, 14, 1–13 CrossRef PubMed.
- R. A. Alharbi, Saudi J. Biol. Sci., 2020, 27, 968–974 CrossRef CAS PubMed.
- K. B. Mullis, Sci. Am., 1990, 262, 56–65 CrossRef CAS PubMed.
- F. S. Costa, C. C. Bezerra, R. M. Neto, C. L. Morais and K. M. Lima, Sci. Rep., 2020, 10, 12994 CrossRef CAS PubMed.
- E. Rubio, Y. Zboromyrska, J. Bosch, M. J. Fernandez-Pittol, B. I. Fidalgo, A. Fasanella, A. Mons, A. Román, C. Casals-Pascual and J. Vila, PLoS One, 2019, 14, e0220307 CrossRef CAS PubMed.
- D. A. Buzatu, T. J. Moskal, A. J. Williams, W. M. Cooper, W. B. Mattes and J. G. Wilkes, PLoS One, 2014, 9, e94254 CrossRef PubMed.
- Y. Xu, M. M. Hassan, A. S. Sharma, H. Li and Q. Chen, Crit. Rev. Food Sci. Nutr., 2023, 63, 486–504 CrossRef CAS PubMed.
- R. Amann and B. M. Fuchs, Nat. Rev. Microbiol., 2008, 6, 339–348 CrossRef CAS PubMed.
- H. Wang, H. Ceylan Koydemir, Y. Qiu, B. Bai, Y. Zhang, Y. Jin, S. Tok, E. C. Yilmaz, E. Gumustekin and Y. Rivenson, et al. , Light: Sci. Appl., 2020, 9, 118 CrossRef PubMed.
- M. E. Berry, H. Kearns, D. Graham and K. Faulds, Analyst, 2021, 146, 6084–6101 RSC.
- C. Cuntín-Abal, et al. , TrAC, Trends Anal. Chem., 2024, 172, 117565 CrossRef.
- X. Wang, T. Yang and J.-H. Wang, Sens. Diagn., 2024, 3, 1590–1612 RSC.
- V.-M. Tsitou, D. Rallis, M. Tsekova and N. Yanev, Biotechnol. Biotechnol. Equip., 2024, 38, 2349587 CrossRef.
- S. Romphosri, et al. , Sci. Rep., 2024, 14, 20498 CrossRef CAS PubMed.
- T. Lehnert and M. A. M. Gijs, Lab Chip, 2024, 24, 1441–1493 RSC.
- N. K. S. Amitabha Acharya, Nanosensors for Point-of-Care Diagnostics of Pathogenic Bacteria, Springer, Singapore, 2023 Search PubMed.
- Y. Wang, K. Jia and J. Lin, TrAC, Trends Anal. Chem., 2024, 177, 117785 CrossRef CAS.
- R. Tatavarti, S. Nadimpalli, G. V. K. Mangina, N. Kiran Machiraju, A. Pachiyappan, S. Hiremath, V. Jagannathan and P. Viswanathan, Front. Phys., 2023, 11, 261 Search PubMed.
- V. S. N. R. Tatavarti, R. M. Pidaparti and S. S. O. Venkata, Systems and methods of use thereof for determining aerosol particle characteristics, U.S. Pat., 11957447B2, 2024 Search PubMed.
- P. Thangaraju and S. B. Varthya, Medical Device Guidelines and Regulations Handbook, Springer, 2022, pp. 163–187 Search PubMed.
- V. S. Pugachev, Probability Theory and Mathematical Statistics for Engineers, Elsevier, 2014 Search PubMed.
- S. Sullivant, Algebraic Statistics, American Mathematical Society, 2023, vol. 194 Search PubMed.
- J. MacQueen, Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 1967, pp. 281–297 Search PubMed.
- P. J. Rousseeuw, J. Comput. Appl. Math., 1987, 20, 53–65 CrossRef.
- B. E. Boser, I. M. Guyon and V. N. Vapnik, Proceedings of the Fifth Annual Workshop on Computational Learning Theory, 1992, pp. 144–152 Search PubMed.
- J. H. Friedman, Ann. Stat., 2001, 1189–1232 Search PubMed.
- G. Ke, Q. Meng, T. Finley, T. Wang, W. Chen, W. Ma, Q. Ye and T.-Y. Liu, Adv. Neural Inf. Process. Syst., 2017, 30, 3146–3154 Search PubMed.
- T. Chen and C. Guestrin, Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, pp. 785–794 Search PubMed.
- K.-X. Mu, Y.-Z. Feng, W. Chen and W. Yu, Chemom. Intell. Lab. Syst., 2018, 179, 46–53 CrossRef CAS.
- K. De Bruyne, B. Slabbinck, W. Waegeman, P. Vauterin, B. De Baets and P. Vandamme, Syst. Appl. Microbiol., 2011, 34, 20–29 CrossRef CAS PubMed.
- Y.-L. Pan, K. Aptowicz, J. Arnold, S. Cheng, A. Kalume, P. Piedra, C. Wang, J. Santarpia and G. Videen, J. Quant. Spectrosc. Radiat. Transfer, 2022, 279, 108067 CrossRef CAS.
- S. Banik, S. K. Melanthota, Arbaaz, J. M. Vaz, V. M. Kadambalithaya, I. Hussain, S. Dutta and N. Mazumder, Anal. Bioanal. Chem., 2021, 413, 2389–2406 CrossRef CAS PubMed.
- S.-Z. Yang, Q.-A. Liu, Y.-L. Liu, G.-J. Weng, J. Zhu and J.-J. Li, Microchim. Acta, 2021, 188, 1–23 CrossRef PubMed.
- J. Robinson, B. Rajwa, E. Bae, V. Patsekin, A. Roumani, A. Bhunia, J. Dietz, V. Davisson, M. Dundar and J. Thomas, et al. , Opt. Photonics News, 2011, 22, 20–27 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |





