Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Headspace paper-based analytical device for ammonia quantification in human biological samples
Kawin
Khachornsakkul
 *ab,
Darrien
Johnsen
ab and
Sameer
Sonkusale
*ab,
Darrien
Johnsen
ab and
Sameer
Sonkusale
 *ab
*ab
aDepartment of Electrical and Computer Engineering, Tufts University, Medford, MA 02155, USA. E-mail: Kawin.khachornsakkul@tufts.edu; sameer.sonkusale@tufts.edu
bSonkusale Research Labs, Halligan Hall, Tufts University, Medford, MA 02155, USA
First published on 5th March 2025
Abstract
This article presents a simple and cost-effective headspace paper-based analytical device (hPAD) for the quantification of ammonia in human biological samples. The aim of this approach is to enhance the detection selectivity for ammonia in complex samples. The detection principle leverages basic chemistry, wherein ammonia reacts with copper sulfate (CuSO4) to form the complex ion tetraamminecopper(II) sulfate ([Cu(NH3)4]SO4), resulting in a colour change from pale blue to dark blue on a paper substrate. The quantitative analysis of ammonia is straightforward through placement of the sensor on the inside lid of the sample vial, and the resulting colour change is measured using a smartphone and image processing software. Upon optimization, the developed assay demonstrated a linear range between 2.5 and 40.0 μM (R2 = 0.9955) with a detection limit (LOD) of 0.90 μM. The sensor also exhibited high precision, with the highest relative standard deviation (RSD) recorded at 6.17%. Moreover, the method showed remarkable selectivity, as the sensor showed no response to common interfering molecules in a complex biological matrix. The technique is fast, requiring only 4 min for the reaction, and does not necessitate any heating procedures. Furthermore, the developed method provides excellent accuracy for detecting ammonia levels in both human serum and urine samples, with recovery rates ranging from 93.4% to 107.6%. Therefore, the hPAD offers a simple and affordable solution by sensing in the headspace that overcomes the limitations of direct measurement in the sample, which may be affected by the colour, pH, other existing ions and molecules in the sample solution. Overall, this approach is suitable for various applications in both medical and environmental analysis.
Introduction
Infectious diseases caused by bacteria pose a consistent threat to public health worldwide, with high incidence and mortality rates.1–3 Among bacterial pathogens, Helicobacter pylori (H. pylori) is a spiral-shaped bacterium that infects the stomach and duodenum, the first part of the small intestine.4–6 The prevalence of H. pylori infection is estimated at around 50% of the global population, with significant variation across regions and socioeconomic groups.7,8 This bacterium is uniquely adapted to survive in the stomach's harsh acidic environment by producing the enzyme urease, which converts urea into ammonia. Likewise, infection with H. pylori is associated with several conditions, including gastritis (inflammation of the stomach lining) and peptic ulcer disease.9–11 Given these risks, early detection of H. pylori infection is critical for initiating timely medical intervention, which can significantly reduce the morbidity and mortality associated with this pathogen.Ammonia serves as a key biomarker for the presence of H. pylori infection due to its excessive production from urea by the urease enzyme generated by this bacterium.12–14 Elevated ammonia levels in humans can therefore be used to diagnose and monitor H. pylori infection. Similarly, abnormal ammonia levels in human samples, including blood, urine, serum, saliva, and sweat, can indicate various medical conditions, such as chronic kidney disease (CKD), urea cycle disorders, and sepsis.15,16 In healthy individuals, ammonia levels typically range from 11.0 to 32.0 μM in blood serum and from 5.0 to 45.0 μM in urine.17,18 Elevated levels of ammonia in blood serum or urine may result from kidney disease, liver failure, salicylate poisoning, parenteral nutrition, gastrointestinal (GI) bleeding, or infection.19 Therefore, the quantitative detection of ammonia levels in humans is crucial for assessing health conditions. Current commercially available ammonia sensors primarily rely on electrochemical and optical detection methods.20 Electrochemical assays determine ammonia by measuring changes in current on the surface of the electrode, while optical detection monitors the intensity of colour or fluorescence change in proportion to ammonia levels in the sample. These methods show great promise as sensitive techniques for ammonia quantification, offering advantages such as cost-effectiveness and accessibility. However, a key limitation of these assays is their susceptibility to interference present in complex sample matrices, such as human blood, serum, and urine, which can reduce measurement accuracy. For instance, electrochemical sensors may suffer from interference by ions such as sodium and potassium that are abundant in all biological fluids.21,22 Optical sensors can be affected by the background colour, cloudiness, and the pH of the samples, leading to erroneous readings.23,24 These highlight the need for improvement of the selectivity for ammonia detection in complex biological samples. Hence, the development of higher selective method for ammonia monitoring in human samples is desirable.
Paper-based analytical devices (PADs) represent a revolutionary approach for conducting inexpensive and rapid on-site analysis in various medical and environmental applications.25–27 They offer numerous advantages, including simplicity, cost-effectiveness, user-friendliness, low reagent consumption, and portability. One of the most widely used techniques in PAD development is colorimetric detection, as the paper substrate provides excellent optical contrast.28,29 The colour change produced on PADs can be easily measured using low-cost optical devices such as smartphones, cameras, or scanners. The detection principle of colorimetric PADs begins with the introduction of a sample solution onto the paper sensor, where pre-deposited chemical reagents react with the target analyte. The resulting colour change is proportional to the analyte concentration, and its intensity or shade can be quantitatively analysed.30,31 This straightforward process allows PADs to be used for on-site monitoring in both medical and environmental settings, making them accessible even to untrained personnel. With their simplicity and versatility, PADs hold great potential for widespread deployment in real-world applications. Recently, simple colorimetric PADs for ammonia monitoring have been developed based on the acid–base colour assay principle, using numerous organic dyes as chromogenic agents.20,32–34 While these existing assays show great promise for ammonia quantification, they have limitations in selectivity. The variation in pH solution and the presence of some metal ions can interfere with the detection method. Additionally, colour measurement errors may occur when using this assay in complex sample matrices, such as human samples, due to their natural colour. To address these challenges, developing a more selective colorimetric PAD sensor for quantitative ammonia monitoring is essential for accurate detection in human samples.
Headspace extraction techniques have been extensively used to enhance the sensitivity and selectivity of analytical methods.35–37 These techniques offer the key advantage of being non-destructive, allowing for the preservation of the sample solution while effectively isolating volatile compounds. By concentrating analytes in the vapor phase above the sample, headspace extraction minimizes interference from the sample matrix and improves the detection limits of target analytes.38,39 This makes it a valuable tool in various applications, including environmental monitoring, food analysis, and clinical diagnostics. To date, headspace techniques have been integrated with colorimetric PADs for detecting various analytes such as cyanide (CN−),40 sulfide ion (S2−),41 bromate (BrO3−) and bromide (Br−),42 selenium,43 arsenic,44 formaldehyde45 in different sample matrices. For example, Thepchuay et al.46 achieved a cost-effective headspace paper-based colorimetric method for detecting ethanol in whole blood using an enzymatic reaction. In their technique, ethanol spontaneously diffuses from the sample layer to the detection layer without requiring a heating process. The ethanol level is quantified by measuring the colour change using a digital camera. Similarly, Conrado et al.47 demonstrated a straightforward fabrication of a headspace microextraction system integrated with a PAD sensor for CN− quantification. Using analyte volatilization, CN− is easily extracted into a specific reagent via a single-drop microextraction (SDME) procedure and quantified with a colorimetric PAD sensor. This approach enhances the selective and sensitive detection of CN−. These above studies have successfully demonstrated improved selectivity and sensitivity for target analyte monitoring. Combining headspace techniques with colorimetric PAD sensors could accordingly enhance detection selectivity for ammonia quantification in human sample matrices.
In this article, we present a simple headspace paper-based analytical device (hPAD) for the quantification of ammonia in human urine and serum samples, aimed at rapidly monitoring infectious diseases such as H. pylori infection and their related symptoms. The goal of this work is to improve the selectivity of the detection method in urine and serum samples. By employing the complex reaction between ammonia and copper sulfate (CuSO4), quantitative detection of ammonia levels is easily achieved by measuring the colour intensity in a digital image using ImageJ software. The assay procedure begins with placing the hPAD sensor onto the lid of the sample bottle. At this stage, ammonia evaporates and reacts with the pre-deposited reagent in the hPAD, resulting in a colour change. After a few minutes, the sensor is removed from the lid, and the colour intensity is measured in a custom black box environment to eliminate ambient light during colour readout. Moreover, the proposed technique doesn't require the use of heating process to generate gaseous phase of the analyte as ammonia is naturally volatile at room temperature. With this feature, the selective detection of ammonia is obtained without interference from existing ions, pH variations, or coloured samples. Overall, our sensor offers a simple, cost-efficient, and disposable tool for the selective detection of ammonia levels in complex samples, and it also facilitates to operate without requiring a skilled technician. Additionally, the technique is applicable to further applications in developing selective methods for other molecules that can evaporate into the gas phase.
Experimental section
Materials and instruments
Ammonia solution, copper(II) sulfate (CuSO4), bovine serum albumin (BSA), C-reactive protein (CRP), creatinine, cortisol, dopamine, glucose, lactic acid, and uric acid were bought from Sigma-Aldrich (USA). Human interleukin-6 (IL-6) was purchased from Abcam company (UK). Urea was obtained from Fisher Scientific Company (USA). Sodium hydroxide (NaOH), hydrochloric acid (HCl), dibasic sodium phosphate (Na2HPO4), monobasic sodium phosphate (NaH2PO4), potassium dihydrogen phosphate (KH2PO4), potassium bicarbonate (KHCO3), and sodium chloride (NaCl), were obtained from Sigma-Aldrich (USA). All chemicals used in the experiment were of analytical reagent (AR grade) and the solutions were prepared using PBS (50.0 mmol L−1, pH 7.4). Human control serum (heat inactivated from human male AB plasma, USA origin, sterile-filtered) and human control urine (Surine™ negative urine control samples) were obtained from Sigma-Aldrich (USA). Whatman No.1 filter paper was obtained from Cole-Parmer (VernonHills, IL). UV-visible absorption spectra were recorded with a UV-visible spectrometer (Lambda 35, Perkin15 Elmer Instruments, USA) using quartz cuvettes (1.0 cm path length).Fabrication of the proposed hPAD sensor
The procedure of the fabrication of the paper sensor for ammonia quantification is illustrated in Scheme 1(a). The design for the paper sensors was created using Adobe Illustrator software program with a circle hydrophilic zone (diameter 5.0 mm) and the hydrophobic barrier. Next, the designed paper was printed on Whatman No. 1 filter paper using a wax printer and then was braked at 120 °C for 2 min. Afterwards, the back side of the paper was taped with a masking tape to prevent solution leakage through the device. Subsequently, 5.0 μL of CuSO4 at 12.5 mM was deposited into the circle hydrophilic zone and allowed to dry at room temperature. Finally, the prepared paper device was placed into the inside of the lid using double size tape. | ||
| Scheme 1 The illustration of (a) the fabrication process of the developed hPAD sensor and (b) its general procedure of the detection method. | ||
General procedure and analytical characteristics
To optimize the parameter conditions of the developed hPAD sensor for ammonia quantification, 5.0 mL of the ammonia solution at 20.0 μM was introduced into a 20.0 mL vial, which was then sealed with a lid containing the paper sensor and left to stand at temperature for 4 min. The paper sensor was then removed from the lid and placed in a 3D black box to take a picture. The colour signal was determined using the ImageJ program in red-green-blue (RGB) mode. The analytical signal was calculated as the ratio between the B value and the total RGB value (Scheme 1(b)). All experiments were performed in triplicate (n = 3).A linear range of the proposed method was performed by introducing 5.0 mL of ammonia solution at different concentration into the vial. After that, the procedure was conducted as described earlier. The detection limit (LOD) of the method was determined by calculating from average blank signal ± three-fold standard deviation (3S.D.). Reproducibility was investigated though the measurement of the analytical signal of the three-different ammonia concentrations at 10.0, 20.0, and 30.0 μM for ten times (n = 10). Afterwards, the relative standard deviation (RSD) was calculated to validate the precision of the method. Moreover, the selectivity was also tested by measuring the analytical signals of ammonia at 20.0 μM compared to other substrates expected to have in real samples. Similarly, the interference was evaluated through the detection of the ammonia in the mixture with other potential interfering substrates (n = 3).
Real sample analysis
To validate the practical application of the proposed assay, 5.0 mL of human control serum and urine samples were introduced into the vial, and then the procedure was carried out as described earlier. Subsequently, the percentage recovery was calculated. This research did not involve human or animal samples.Results and discussion
Working principle of the proposed hPAD sensor
In this study, the main principle underlying the development of the proposed sensor is associated with the chemical reaction between ammonia and CuSO4 to form a soluble complex ion, tetraamminecopper(II) sulfate ([Cu(NH3)4]SO4), which produces a dark blue colour, as shown in eqn (1). This complex ion is considerably stable, with a high formation constant (Kf) as 2.1 × 1013.48–50| NH3(aq) + CuSO4(aq) → [Cu(NH3)4]SO4(aq) | (1) |
Furthermore, to reduce the interference effect from background colour and existing ions or molecules in solution, we monitor aqueous ammonia level with the headspace technique using the proposed paper sensor. Generally, the aqueous ammonia can immediately evaporate to be a gaseous form at room temperature,20 as shown in eqn (2).
| NH3(aq) ⇌ NH3(g) + H2O(aq) | (2) |
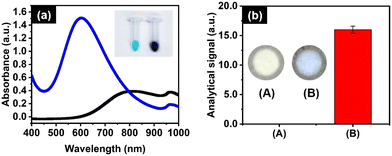
We accordingly evaluated whether the proposed sensor can be applied to monitor the gaseous ammonia generated from the aqueous ammonia in the close vial. The sensor was placed into the inside lid of the vial. As shown in Fig. 1(b), the analytical signal was 0.0 a.u. for a blank signal, DI water, while it was 16.0 a.u. when detecting ammonia at 20.0 μM. This result indicates that the developed sensor can be applied for the headspace detection of ammonia, although there is no need for external heating procedure. Therefore, the developed hPAD sensor provides a great feasibility to quantitatively monitor ammonia concentrations in human samples, while remaining the basic chemistry.
Parameter optimization
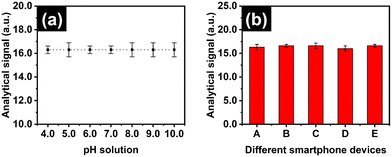
We optimized the parameter that can affect the assay efficiency through the detection of the analytical signal of ammonia at 20.0 μM. First, the impact of sample volume of the aqueous ammonia in the vial was investigated, as it is associated with the generated ammonia gas in the vial which can react with the sensor. As shown in Fig. 2(a), the average analytical signal increased from 2.5 to 5.0 mL, after that, it gradually decreased with the volume. This perhaps can be explained by the condensation of these generated gases to regenerate the aqueous ammonia at gas equilibrium.51 So, it reduces the mass of gaseous ammonia in the system, leading to less reaction with the sensor. We accordingly selected sample volume at 5.0 mL for our further experiment. Subsequently, the reaction time of the proposed method was examined by counting the time after placing the sensor and closing the vial, as shown in Fig. 2(b). The outcome demonstrated the slight increase in the analytical signal from 67.0 to 71.0 a.u. for 2 and 4 min, respectively. Beyond this threshold, the signal remained constant. Thus, the reaction time of 4 min was chosen as a sufficient value to reduce the total analysis time. Following that, the concentration of CuSO4 pre-deposited onto the sensor was varied between 12.5 and 100.0 mM, as this parameter plays a crucial role for colorimetric detection of ammonia in this assay, affecting both sensitivity and linearity. Below this level, we could not observe the pale blue colour of CuSO4 for its deposition on the paper sensor which may subsequently influence the colour measurement. In Fig. 2(c), we observed a progressive increase in the analytical signal with the CuSO4 proportionally. However, we further detected ammonia at 10.0, 20.0, and 30.0 μM employing the sensor pre-deposited with each different CuSO4 concentration. In Fig. 2(d), we found that the change in the analytical signal towards ammonia concentrations was less when the sensor with a higher CuSO4 concentration was used. This phenomenon can be attributed with the fast saturation of the reaction between ammonia and CuSO4 on paper sensors. To clarify, when the higher CuSO4 is applied, the precipitation reaction with ammonia immediately occurs and leads to the saturation of the colour product. Thereby, the colour signal does not significantly change when ammonia concentrations increase, being able to provide less sensitivity and linearity. Consequently, we chose the lowest concentration of CuSO4 at 12.5 mM for fabricating the paper sensor in the proposed method.Likewise, we validated whether pH variations influence the proposed system by measuring ammonia concentrations at 20.0 μM in solutions with pH values ranging from 4.0 to 10.0. As shown in Fig. 3(a), the results indicated that the analytical signals remained consistent across the different pH values. This phenomenon is perhaps due to the lack of contact between hPAD and sample solution. Therefore, our technique is unaffected by pH changes, making it suitable for ammonia quantification in a wide range of pH environments. Notably, given that the typical pH range of human samples is between 5.5 and 8.0, the developed sensor is well-suited for determining ammonia levels in human samples. Additionally, the impact of using different smartphones was investigated by detecting ammonia levels at 20.0 μM with five different devices. In Fig. 3(b), the results indicated no significant differences in analytical measurements across the smartphones. This therefore suggests that the assay can be performed without requiring a specific smartphone, making it extensively adaptable in real world applications.
Analytical characteristics
The analytical performance of the colorimetric hPAD sensor for ammonia determination was evaluated. The test was run with both a blank solution and a solution containing various ammonia concentrations. The average analytical signals were 0.0, 13.7, and 14.3 a.u. for blank and ammonia detection at 2.5 and 5.0 μM. This indicates the direct rise in colour signal with ammonia concentration proportionally. The linear range established between 2.5 and 40.0 μM (R2 = 0.9955), as shown in Fig. 4. This merit falls within the clinically relevant range for ammonia detection in human serum and urine samples. Additionally, the LOD of the proposed assay was found to be 0.90 μM calculated from average blank signal ± three-fold standard deviation (3S.D.). Furthermore, the reproducibility of the method was investigated through the detection of ammonia at various concentrations for ten times (n = 10), and then the relative standard deviation (RSD) was calculated for this validation. The RSD of 5.41%, 5.05%, and 6.17% for ammonia detection at 10.0, 20.0, and 30.0 μM, respectively, was obtained, suggesting the acceptable precision of the developed method. | ||
| Fig. 4 Linear range of ammonia detection using our proposed hPAD sensor, including the images (a) blank, (b) 2.5, (c) 5.0, (d) 10.0, (e) 20.0, (f) 30.0, and (g) 40.0 μM (n = 3). | ||
Investigation of selectivity and interferences
The selectivity of the method was investigated by measuring the analytical signal of other interfering substrates that expect to present in real samples, including BSA, CRP, creatinine, cortisol, dopamine, glucose, IL-6, lactic acid, urea, and uric acid, compared to that of ammonia. The results in Fig. 5(a) indicated that only ammonia could provide the colour change, while other substrates exhibited the analytical signal of 0.0, similar to blank signal. The method consequently shows exceptional specificity for ammonia monitoring. Similarly, the effect of the interferences was assessed through the detection of the analytical signal of the mixture solution between ammonia and other interfering substrates. In Fig. 5(b), the average analytical signals for both only ammonia detection and mixed solution with interfering substrates were not significantly different over 5.0%. This outcome hence concludes that our method can be used to detect ammonia without the effect of interferences from other substrates. Based on the above performance, the proposed colorimetric hPAD sensor for ammonia quantification enables for POC testing in human biological samples, addressing the interference effect from existing ions in the solution and pH variation and colour of the solution.Real sample application
To validate the practical use for biomedical application, the method was employed to determine ammonia levels in both human control serum and urine samples. The assay was run using a spiking technique. Prior spiking standard solution, the analytical signal was 0.0 a.u., similar to blank signal, implying no ammonia content in these samples. Upon spiking standard solution, the recovery percentage was found in the range of 93.4–107.6% with the highest RSD of 6.3%, as indicated in Table 1. Furthermore, the obtained results were compared with a reference method using the Berthelot reaction.52 We found that there were no significant differences between both methods for detecting ammonia levels in human urine and serum samples with T-test critical at 95% confident level. This demonstrates the high accuracy of the developed method for ammonia detection in human samples. Notably, our technique is safer and more cost-effective than the reference method, as it does not require harmful chemicals such as phenol and alkaline hypochlorite or expensive instruments like spectrophotometer. Additionally, by utilizing paper as a substrate, the analytical procedure is simpler and more accessible for unskilled users, eliminating the need for solution mixing. Moreover, our assay is unaffected by physical matrix interferences such as pH, colour, turbidity, and viscosity, which may impact the performance of the reference method. With these characteristics, the developed hPAD sensor can be an alternative analytical technique for detecting ammonia in the human biological fluids with the advantages of simplicity, cost-effectiveness, portability, rapidity, and disposability.| Sample type | Ammonia standard added (μM) | Proposed method found (μM) | Berthelot reaction (μM) | % recovery | % RSD |
|---|---|---|---|---|---|
| Human urine | 0.0 | 0.0 | 0.0 | — | — |
| 7.5 | 7.6 | 7.9 | 101.4 | 4.0 | |
| 12.5 | 11.9 | 12.4 | 95.0 | 3.8 | |
| 15.0 | 16.1 | 15.5 | 107.6 | 6.3 | |
| Human serum | 0.0 | 0.0 | 0.0 | — | — |
| 7.5 | 7.6 | 7.5 | 101.4 | 4.0 | |
| 12.5 | 11.9 | 12.2 | 95.0 | 3.8 | |
| 15.0 | 14.1 | 13.6 | 93.8 | 3.7 |
Comparison of the proposed method and other PADs for ammonia quantification
The analytical performance of the hPAD sensor developed in this study was compared with other colorimetric PAD sensors for ammonia detection, as shown in Table 2.32–34,53,54 By comparing, the developed hPAD sensor offers safer, more selective, sensitive, and accessible than previous method that used bromothymol blue, anthocyanin, 3-nitrophenol, methyl ammonium lead iodide, and Nessler's reagent as colorimetric substrates. Notably, these methods employ the direct introduction of sample solution into the paper substrate and then produce the colour change. Still, they have limitations regarding the chemical substrate used for chemical interactions, relating to their specificity and selectivity. For example, acid–base indicators like bromothymol blue and 3-nitrophenol can somehow cause a false positive measurement due to the interference from pH variation in the solution. While the green extraction of anthocyanin offers sustainable chemistry, it employs organic solvents, is time-consuming, and demands skilled labour, which can lead to reduced precision and batch-to-batch variability during sensor fabrication. Furthermore, although methyl ammonium lead iodide deposited on the PAD sensor shows good potential for ammonia detection, its stability is reasonably low, as the substrate is prone to degradation when exposed to light. The assay of Nessler's reagent also has limitations in less determining ammonia levels in complex samples, like human samples, as it interferes with the electrolyte and protein in the sample solution. In our method, the combination of the headspace technique and the use of CuSO4 on PAD sensor provides better selectivity and safety compared to previous methods. It thus enables the determination of ammonia levels in complex samples, resulting in improved analytical performance for ammonia quantification.Conclusion
In this article, a simple and inexpensive hPAD for the selective and sensitive detection of ammonia in human samples has been successfully developed. By leveraging basic chemistry, the colorimetric detection is based on the precipitation reaction between CuSO4, pre-deposited onto the paper substrate, and evaporated ammonia in the vial. Quantitative analysis is easily determined by measuring the colour change on the paper substrate. With the headspace method, selective monitoring of ammonia levels is achieved without interference from either physical or chemical factors. Moreover, this assay does not require any heating procedure to generate gaseous ammonia, making it highly practical for unskilled labour. Our method exhibited a linear working range within the clinically relevant range for ammonia monitoring, requiring no additional sample preparation or dilution. Furthermore, this assay provided remarkable accuracy and precision in detecting ammonia in human urine and serum samples. Based on these characteristics, the developed hPAD offers a great solution for enhancing the selectivity of ammonia assays without the need for expensive instruments and chemicals. This proposed concept can thus serve as an effective alternative method for ammonia quantification for rapid diagnosis of the infection and related diseases and can be expanded for future advancements in developing selective methods for other molecules.Data availability
The data that supports the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Kawin Khachornsakkul: conceptualization, methodology, investigation, validation, project administration, supervision, data curation, writing – original draft, writing – reviewing & editing. Darrien Johnsen: conceptualization, methodology, investigation, validation, data curation, writing – original draft. Sameer Sonkusale: resources, supervision, project administration, writing – review & editing.Conflicts of interest
The author declares no conflict of interest.References
- V. Sekar, A. Santhanam and P. Arunkumar, J. Water Process Eng., 2024, 59, 105102 CrossRef.
- M. Asif, F. R. Awan, Q. M. Khan, B. Ngamsom and N. Pamme, Analyst, 2020, 145, 7320–7329 RSC.
- K. Khachornsakkul, R. Del-Rio-Ruiz, H. Creasey, G. Widmer and S. R. Sonkusale, ACS Sens., 2023, 8, 4364–4373 CrossRef CAS.
- P. Malfertheiner, M. C. Camargo, E. El-Omar, J. M. Liou, R. Peek, C. Schulz, S. I. Smith and S. Suerbaum, Nat. Rev. Dis. Primers, 2023, 9, 1–24 Search PubMed.
- Y. Fei, R. Fang, L. Xiao, Y. Zhang, K. Fan, Y. Jiang, S. Lei, R. Xu, D. Yang, Y. Ye, S. Xiang, P. Wang, C. Zhou and T. Tang, Anal. Biochem., 2022, 651, 114737 Search PubMed.
- M. Li, Y. Sun, J. Yang, C. de Martel, H. Charvat, G. M. Clifford, S. Vaccarella and L. Wang, Helicobacter, 2020, 25, e12729 CrossRef.
- C. Celik, G. Can Sezgin, U. G. Kocabas, S. Gursoy, N. Ildiz, W. Tan and I. Ocsoy, Anal. Chem., 2021, 93, 6246–6253 CrossRef CAS.
- M. Leja, I. Grinberga-Derica, C. Bilgilier and C. Steininger, Helicobacter, 2019, 24, e12635 CrossRef.
- J. E. Sykes and S. L. Marks, Canine Feline Infect. Dis., 2014, pp. 465–473 Search PubMed.
- G. Y. Pih, J. H. Noh, J. Y. Ahn, G. S. Han, H. S. Jung, H. Y. Jung and J. M. Kim, J. Korean Med. Sci., 2022, 37, e227 CrossRef CAS PubMed.
- A. Koumi, T. Filippidis, V. Leontara, L. Makri and M. Z. Panos, World J. Gastroenterol., 2011, 17, 349 CrossRef.
- D. Y. Graham and M. Miftahussurur, J. Adv. Res., 2018, 13, 51 CrossRef CAS PubMed.
- E. F. Miller and R. J. Maier, J. Bacteriol., 2014, 196, 3074 CrossRef PubMed.
- P. P. Ricci and O. J. Gregory, Sci. Rep., 2021, 11, 1–7 CrossRef.
- M. J. Chan, Y. J. Li, C. C. Wu, Y. C. Lee, H. W. Zan, H. F. Meng, M. H. Hsieh, C. S. Lai and Y. C. Tian, Biomedicines, 2020, 8, 468 CrossRef CAS PubMed.
- L. Zhao, S. Hou, R. Na, B. Liu, Z. Wang, Y. Li and K. Xie, Front. Public Health, 2023, 10, 1016931 CrossRef.
- Henry's Clinical Diagnosis and Management by Laboratory Methods - 24th Edition | Elsevier Shop, https://shop.elsevier.com/books/henrys-clinical-diagnosis-and-management-by-laboratory-methods/mcpherson/978-0-323-67320-4, (accessed 2 January 2025).
- Ammonia Levels: Causes, Symptoms & Treatment, https://my.clevelandclinic.org/health/articles/22686-ammonia-levels, (accessed 2 January 2025).
- Sleisenger and Fordtran's Gastrointestinal and Liver Disease- 2 Volume Set - 11th Edition | Elsevier Shop, https://shop.elsevier.com/books/sleisenger-and-fordtrans-gastrointestinal-and-liver-disease-2-volume-set/feldman/978-0-323-60962-3, (accessed 2 January 2025).
- K. Khachornsakkul, K. H. Hung, J. J. Chang, W. Dungchai and C. H. Chen, Analyst, 2021, 146, 2919–2927 RSC.
- Y. C. Chang, H. Bai, S. N. Li and C. N. Kuo, Sensors, 2011, 11, 4060–4072 CrossRef CAS.
- W. N. Opdycke, S. J. Parks and M. E. Meyerhoff, Anal. Chim. Acta, 1983, 155, 11–20 CrossRef CAS.
- J. Courbat, D. Briand, J. Damon-Lacoste, J. Wöllenstein and N. F. de Rooij, Sens. Actuators, B, 2009, 143, 62–70 CrossRef.
- Á. Markovics, G. Nagy and B. Kovács, Sens. Actuators, B, 2009, 139, 252–257 CrossRef.
- W. Dungchai, O. Chailapakul and C. S. Henry, Anal. Chim. Acta, 2010, 674, 227–233 CrossRef CAS.
- C. Chen, L. Zhao, H. Zhang, X. Shen, Y. Zhu and H. Chen, Anal. Chem., 2019, 91, 5169–5175 CrossRef CAS PubMed.
- K. Khachornsakkul, F. J. Rybicki and S. Sonkusale, Talanta, 2023, 260, 124538 CrossRef CAS PubMed.
- J. Noiphung, K. Talalak, I. Hongwarittorrn, N. Pupinyo, P. Thirabowonkitphithan and W. Laiwattanapaisal, Biosens. Bioelectron., 2015, 67, 485–489 CrossRef CAS.
- W. Wonsawat, S. Limvongjaroen, S. Supromma, W. Panphut, N. Ruecha, N. Ratnarathorn and W. Dungchai, Analyst, 2020, 145, 4637–4645 RSC.
- K. Khachornsakkul, R. Del-Rio-Ruiz, C. Asci and S. Sonkusale, Analyst, 2024, 149, 3756–3764 RSC.
- D. M. Cate, S. D. Noblitt, J. Volckens and C. S. Henry, Lab Chip, 2015, 15, 2808–2818 RSC.
- M. A. Vargas-Muñoz, J. Morales, V. Cerdà, L. Ferrer and E. Palacio, Microchem. J., 2023, 188, 108469 CrossRef.
- S. U. Haq, M. Aghajamali and H. Hassanzadeh, RSC Adv., 2021, 11, 24387–24397 RSC.
- B. M. Jayawardane, I. D. McKelvie and S. D. Kolev, Anal. Chem., 2015, 87, 4621–4626 CrossRef CAS PubMed.
- C. Bicchi, C. Iori, P. Rubiolo and P. Sandra, J. Agric. Food Chem., 2002, 50, 449–459 CrossRef CAS PubMed.
- J. E. Ruvalcaba, E. Durán-Guerrero, C. G. Barroso and R. Castro, Foods, 2020, 9, 255 CrossRef CAS PubMed.
- N. Fukana, T. Sonsa-ard, N. Chantipmanee, P. C. Hauser, P. Wilairat and D. Nacapricha, Sens. Actuators, B, 2021, 339, 129838 CrossRef CAS.
- A. Shahvar, M. Saraji, H. Gordan and D. Shamsaei, Talanta, 2019, 197, 578–583 CrossRef CAS.
- M. Saraji and B. Farajmand, J. Chromatogr. A, 2013, 1314, 24–30 CrossRef CAS.
- M. Saraji and N. Bagheri, Sens. Actuators, B, 2018, 270, 28–34 CrossRef CAS.
- Y. Pan, Z. Fang, H. Chen, Z. Long and X. Hou, Luminescence, 2021, 36, 1525–1530 CrossRef CAS PubMed.
- K. Phoonsawat and W. Dungchai, Talanta, 2021, 221, 121590 CrossRef CAS PubMed.
- K. Huang, K. Xu, W. Zhu, L. Yang, X. Hou and C. Zheng, Anal. Chem., 2016, 88, 789–795 CrossRef CAS.
- F. Pena-Pereira, L. Villar-Blanco, I. Lavilla and C. Bendicho, Anal. Chim. Acta, 2018, 1011, 1–10 CrossRef CAS.
- V. Mohammadi and M. Saraji, Microchim. Acta, 2024, 191, 1–7 CrossRef PubMed.
- Y. Thepchuay, T. Sonsa-ard, N. Ratanawimarnwong, S. Auparakkitanon, J. Sitanurak and D. Nacapricha, Anal. Chim. Acta, 2020, 1103, 115–121 CrossRef CAS.
- J. A. M. Conrado, D. A. G. Araújo and J. F. da S. Petruci, Anal. Chim. Acta, 2023, 1281, 341882 CrossRef CAS.
- J. A. Dean and N. A. Lange, Lange's Handbook of Chemistry, Google Books, 1999, https://books.google.com/books/about/Lange_s_Handbook_of_Chemistry.html?id=56KPMQEACAAJ, (accessed 2 January 2025) Search PubMed.
- 174: The Formation of Complex Ions - Chemistry LibreTexts, https://chem.libretexts.org/Bookshelves/General_Chemistry/Book%3A_General_Chemistry%3A_Principles_Patterns_and_Applications_(Averill)/17%3A_Solubility_and_Complexation_Equilibria/17.04%3A_The_Formation_of_Complex_Ions, (accessed 2 January 2025).
- An equilibrium using copper(II) and ammonia | Experiment | RSC Education, https://edu.rsc.org/experiments/an-equilibrium-using-copperii-and-ammonia/1711.article, (accessed 18 October 2024).
- 14: Reaction Equilibrium in the Gas Phase - Chemistry LibreTexts, https://chem.libretexts.org/Bookshelves/General_Chemistry/Book%3A_Concept_Development_Studies_in_Chemistry_(Hutchinson)/14%3A_Reaction_Equilibrium_in_the_Gas_Phase, (accessed 28 January 2021).
- APHA (1989) Standard Methods for Examination of Water and Wastewater, American Public Health Association, Washington DC, − References - Scientific Research Publishing, 17th edn, https://www.scirp.org/reference/referencespapers?referenceid=1518549, (accessed 2 January 2025).
- A. Maity and B. Ghosh, Sci. Rep., 2018, 8, 1–10 CAS.
- A. Sheini, Microchim. Acta, 2020, 187, 1–11 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |