Open Access Article
Open Access ArticleIn situ amplified colorimetric immunoassay coupled with a dual-enzyme-functionalized UiO-66(Ce) framework for quantitative detection of the dengue virus†
Luanfeng
Lin‡
a,
Xiangqun
Fan‡
b,
Yinmei
Yan‡
c,
Ting
Lin‡
a,
Xiao
Han
ac,
Dianping
Tang
 d and
Lifen
Han
*a
d and
Lifen
Han
*a
aThe United Innovation of Mengchao Hepatobiliary Technology Key Laboratory of Fujian Province, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou 350025, China. E-mail: 13655083639@163.com
bFujian Maternity and Child Health Hospital, College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, No.18, Daoshan Road, Fuzhou 350108, China
cCollege of Biological Science and Engineering, Fuzhou University, Fuzhou 350108, China
dKey Laboratory of Analytical Science for Food Safety and Biology (MOE & Fujian Province), Department of Chemistry, Fuzhou University, Fuzhou 350108, China
First published on 29th November 2024
Abstract
Signal amplification and noise reduction are very crucial for the sensitive determination of disease-related biomarkers. The development of rapid and laborious testing is urgently required due to extended incubation time and high mortality. Herein, we developed a Ce-based mesoporous metal–organic framework-based multi-enzyme encapsulation device for the realization of enzyme-linked immunosorbent assay (ELISA) for the dengue virus (DENV). Briefly, a UiO-66(Ce) framework structure with uniform mesopores was synthesized by a one-pot method, which allowed efficient encapsulation of natural enzymes with an encapsulation efficiency of 700%. The developed multi-enzyme reaction probes were able to maintain more than 90% of catalytic activity stable at room temperature for 60 days while ensuring efficient enzyme immobilization. Sensitive evaluation of DENV was achieved by encapsulating glucose oxidase and horseradish peroxidase, combined with the formation of an immune sandwich in the presence of DENV. The developed sensor enabled flexible detection of recombinant dengue virus serotype 2 NS1 protein (DENV2-NS1) from 0.05 to 100 ng mL−1, with a low limit of detection of 39.7 pg mL−1. In addition, there were no significant differences in the test results of the samples obtained through the developed multi-enzyme probes when compared to commercially available ELISA kits. This work provides new horizons for the development of efficient enzyme encapsulation systems.
Introduction
Dengue fever is an acute viral disease caused by the dengue virus (DENV), which is mostly transmitted by Aedes aegypti and Aedes albopictus mosquitos.1,2 Dengue fever has increased substantially in recent years, particularly in tropical and subtropical countries, and has emerged as a major public health problem worldwide.3 The World Health Organization estimates that more than 100 million individuals are infected with the dengue virus each year, with 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases progressing to severe dengue (e.g., dengue hemorrhagic fever or dengue shock syndrome), which has a 20% case fatality rate.4,5 An early diagnosis not only dramatically reduces the danger of severe disease and death, but also allows public health authorities to implement timely isolation and control measures to avoid.6 As a result, the development of quick detection technologies based on molecular biology and nanotechnology, such as isothermal amplification, fluorescence tagging, and electrochemical sensors, has become a priority in dengue studies.6–10 These approaches can produce very sensitive and specific detection results, which are useful for early diagnosis, epidemic prevention, and dengue fever control.
000 cases progressing to severe dengue (e.g., dengue hemorrhagic fever or dengue shock syndrome), which has a 20% case fatality rate.4,5 An early diagnosis not only dramatically reduces the danger of severe disease and death, but also allows public health authorities to implement timely isolation and control measures to avoid.6 As a result, the development of quick detection technologies based on molecular biology and nanotechnology, such as isothermal amplification, fluorescence tagging, and electrochemical sensors, has become a priority in dengue studies.6–10 These approaches can produce very sensitive and specific detection results, which are useful for early diagnosis, epidemic prevention, and dengue fever control.
Currently, the most often used clinical tests for DENV are serologic tests, particularly for distinct antigenic types, which are performed using enzyme-linked immunosorbent assay (ELISA).11–13 However, the immunoassay's sensitivity and specificity are largely determined by the enzyme's activity and stability. These enzymes lose activity during immobilization and long-term storage, resulting in reduced test sensitivity. Traditional enzyme immobilization techniques, such as physical adsorption or covalent binding, are frequently inefficient in preserving enzyme activity, particularly in complex biological samples.14–16 Furthermore, the long-term stability of enzymes poses a significant barrier for immunoassays, particularly under high temperature or extreme pH settings, when enzyme activity is easily lost.17 As a result, improving the performance of immunoassays requires understanding how to efficiently immobilize and stabilize enzyme activity.
A family of highly organized porous materials known as metal–organic frameworks (MOFs) is made up of metal ions or metal clusters joined by ligand connections to organic ligands.18 MOFs have attracted a lot of interest in the domains of gas storage, catalysis, and drug delivery because of their distinctive structural characteristics, which include ultra-high specific surface area, variable pore size, and high structural designability.19 An ideal microenvironment for encapsulating enzymes is provided by the porous structure of MOFs, which can effectively prevent denaturation and inactivation of enzymes under extreme conditions (e.g., high temperature, strong acid or alkaline environment).20 The application of MOFs in biocatalysis has gradually emerged in recent years, particularly in the immobilization and protection of enzymes.21–23 Additionally, the MOFs' large surface area and pore-size adjustability can maximize the enzyme–substrate contact, enhancing the enzyme's catalytic efficiency. According to some research, MOFs can improve the stability of the enzyme by modifying the microenvironment inside the pore in addition to providing physical barriers to preserve the structural integrity of the enzyme.24,25 To further improve the durability and activity of the enzyme, certain MOFs, for instance, can interact with residues on the enzyme's surface through particular functional groups.26 Consequently, MOFs offer a fresh approach to the unsolvable issues with conventional materials in the field of enzyme immobilization and demonstrate a wide range of potential applications in the construction of biosensors and catalysts.
In this work, inspired by previous work, we developed a suitably sized multi-enzyme reactor for rapid and sensitive immunoassays for recombinant dengue virus serotype 2 NS1 protein (DENV2-NS1) (Scheme 1). The synthesized Ce-based MOF structures exhibited suitable pore sizes that could be suitable for the filling of glucose oxidase (GOx) and horseradish peroxidase (HRP). Further, a multistep oxidation of glucose was catalyzed in a cascade in the presence of the target DENV to achieve color change of the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) by the developed multi-enzyme reactor as a signal probe. The developed multi-enzyme reactor enhanced the sensitivity of the sensor on the one hand, and solved the immobilization and stability limitations of natural enzymes on the other. This work provides new ideas for the development of rapid DENV assays, contributing new horizons for the realization of enzyme immobilization and the design of cascade reactions.
Experimental section
Synthesis of Ce-based MOFs
Briefly, probenecid tri-block F127 (PEO106–PPO70–PEO106) was used as a soft-template for the generation of well-ordered macropores in microporous UiO-66(Ce).21 In a typical synthesis sequence, 0.2 g of F127 was dissolved in 12 mL of deionized water, and then acetic acid (0.6 mL) and sodium perchlorate (NaClO4, 1.0 g) were stirred into the solution until the solution became turbid and there was no visible precipitation. Subsequently, cerium precursor (Ce(NH4)2(NO3)6, 1.20 g) and triethylbenzene (0.32 mL) were then added to the mixture. The mixture was transferred to a water bath at 40 °C and agitated for 40 min to promote the self-assembly of UiO-66(Ce). To eliminate the incompletely reacted F127, the obtained product was then centrifuged and washed twice with water and DMF. The DMF from the above steps required extra ethanol washing and prolonged sonication (60 °C, 48 h) to remove. The final product was dried in a vacuum oven (60 °C, 12 h) overnight.Encapsulation of glucose oxidase and horseradish peroxidase
10 mg of UiO-66(Ce) MOF material was dispersed in 5.0 ml of PBS buffer solution (10 mM, pH 7.4) containing 5.0 mg of GOx and 5.0 mg of HRP. The natural enzyme GOx and HRP were confined to the mesopores of the MOFs by shaking. The enzyme-immobilized UiO-66(Ce)@GOx/HRP was collected by centrifugation, washed three times with PBS buffer solution, and then dispersed in 1 mL of PBS buffer solution and set aside.The fabrication protocol for recognizing antibody-functionalized multi-enzyme MOFs was based on previous work. To activate the carboxyl groups on the surface of the nanozyme, 0.6 M 1-(3-(dimethylamino)propyl)-3-ethylcarbodiimide hydrochloride (10 μL) and 0.4 M N-hydroxysuccinimide (10 μL) were added to the previously prepared MOF multienzyme reactor solution (1 mL). Subsequently, incubation was carried out on a shaker at 30 °C for 1 h. The excess active agent was removed by centrifugation and dispersed into an equal volume of PBS buffer solution. 500 μL of diluted DENV2-NS1 antibody solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) was added to the activated MOF solution, followed by incubation at 37 °C for 2 h to obtain UiO-66(Ce)@GOx/HRP-pAb2.
200) was added to the activated MOF solution, followed by incubation at 37 °C for 2 h to obtain UiO-66(Ce)@GOx/HRP-pAb2.
Multi-enzyme catalytic assays for targeting DENV
A 96-microtiter plate capturing the anti-DENV2-NS1 antibody-coupled monoclonal antibody (mAb1) was prepared in advance by adding 50 μL of mAb1 solution (10 μg mL−1) to each microtiter well and placed in the refrigerator at 4 °C overnight. It should be noted that a cling film was required to be placed on the microplate to prevent evaporation of the liquid. The incompletely attached mAb1 was then washed away using PBS. Then, 300 μL of blocking buffer (0.1 M PBS, 1.0% BSA) was added to each well and incubated with oscillation for 1 h (37 °C). The microwells were washed using the same buffer solution. The target response signal of the sensor was obtained by adding different gradient dilutions of DENV2-NS1 samples to it. Briefly, different concentrations of the target were added to each well separately and then incubated with shaking under the best optimized conditions (37 °C, 45 min). Afterwards, the incompletely reacted solution was carefully removed and pre-prepared UiO-66(Ce)@GOx/HRP-pAb2 was added to it. The incompletely reacted secondary antibody was again removed using wash buffer. After that, 100 μL of TMB solution (1 mg mL−1 in PBS buffer with 1% DMSO, v/v) and 100 μL of glucose solution were sequentially added to it, and the incubation was continued at a constant temperature in an oscillator (37 °C, 40 min). At the end of the reaction, the absorbance data at 652 nm were read using a microplate reader, and each set of concentration was tested three times in parallel and the data were processed using the Origin 8.0 software.Results and discussion
Structural and compositional characterization
In this study, we mainly concentrated on the long-term effective usage of natural enzymes in a test technique similar to ELISA. The selection of UiO-66(Ce) metal–organic framework (MOF) as the carrier material for this study was based on several key factors: (i) UiO-66(Ce) showed a uniform mesoporous structure, which provides an ideal space for efficient encapsulation and protection of the enzyme. This structure facilitates the immobilization of the enzyme while maintaining accessibility to its active site; (ii) the pore size of UiO-66(Ce) could be adjusted with the synthesis conditions, which allowed us to optimize the pore size to fit specific enzyme molecules, ensuring efficient encapsulation and release of the enzyme; (iii) UiO-66(Ce) could be synthesized by a simple one-pot method and its surface could be further modified to enhance interaction with enzymes and improve encapsulation efficiency. The theoretical foundation for this work was established by the invention of mesoporous MOFs. Thus, the first thing that must be ascertained is if the MOFs that were guided by this synthesis methodology demonstrated a distinct and sturdy porous structure. High-resolution transmission electron microscopy (HRTEM) was used to confirm the structure of the produced UiO-66(Ce). The results showed that the Ce-based MOFs had complex pore structures on the surface together with homogeneous and stable polyhedral structures (Fig. 1A).21 Because the surface pores were only around 10 nm in size and the specific UiO-66(Ce) particle size was approximately 300 nm, additional surface structure analysis using scanning electron microscopy (SEM) was necessary. The produced UiO-66(Ce), as seen in Fig. 1B, had a pore size of roughly 12 nm and a well-defined dodecahedral shape, suggesting that the majority of small proteins could be enclosed in the precisely designed pore. In addition, the pore size distribution curve of UiO-66(Ce) determined by nitrogen isothermal adsorption and desorption curves and the Barrett–Joyner–Halenda (BJH) method were also found to be in agreement with the SEM results (Fig. S1†). Using elemental mapping spectroscopy, additional elemental level analysis of the synthesized UiO-66(Ce) was performed. As illustrated in Fig. 1C, this analysis revealed that well-defined elements C, N, O, and Ce were uniformly distributed on the structure of Ce-MOFs, and no other hetero-elemental distributions were discovered. The above results indicated that UiO-66(Ce) with mesoporous structure was initially considered to have been synthesized.Structural validation and characterization of enzymatic encapsulation efficiency of UiO-66(Ce)
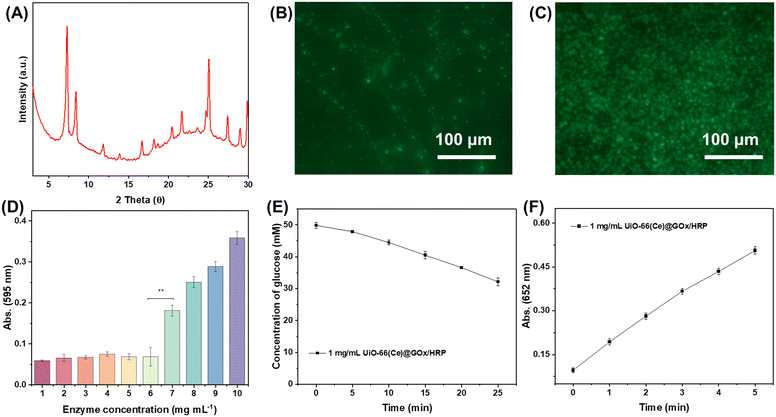
Another question is whether natural enzymes like GOx and HRP might be effectively encapsulated by the synthetic UiO-66(Ce) and if this would have any effect on the enzyme's catalytic qualities. Using X-ray diffraction (XRD), the structure of the synthesized UiO-66(Ce) was first determined. The results indicated the presence of microporous and mesoporous structures in the structure, as shown in Fig. 2A, with significant diffraction signals in the range of low diffraction angles (3–10°). SEM pictures were used as a cross-check for the results above. To ascertain the encapsulation efficiency of GOx and HRP, they were further tagged with fluorescein isothiocyanate (FTIC) and co-incubated with UiO-66(Ce) in an oscillating manner. In theory, FTIC possessed the ability to attach itself to amine groups in proteins (like ε-amino groups on lysine residues) and create covalent bonds.23 Once attached to FITC, glucose oxidase would retain its biological activity and acquire the fluorescence characteristics of FITC, which would allow the natural enzyme to be localized. It was determined that GOx and HRP could be absorbed and immobilized by UiO-66(Ce) based on the ability of the natural enzyme tagged by FTIC to localize to particular particles that are thought to be specific accumulations of UiO-66(Ce), as shown in Fig. 2B and C.To ascertain whether GOx and HRP could be immobilized in the microporous structure of UiO-66(Ce), more measurements of the catalytic efficiency of the enzymes were needed. The natural enzyme lacking FTIC labeling was reconstituted, and the same experimental conditions were followed by uptake and immobilization studies to generate UiO-66(Ce) with both GOx and HRP immobilized, as well as UiO-66(Ce) with both immobilized. The Bradford assay was used to determine the maximum concentrations of implanted GOx and HRP first. In a nutshell, the Thomas Brilliant Blue G-250 dye bound to the basic amino acids (lysine, arginine, etc.) in the proteins, changing the color of the solution from brown to blue. The absorbance was measured at 595 nm. The evaluation of enzyme embedding was carried out using a unit mass concentration (1 mg mL−1, 1 mL) of UiO-66(Ce). As shown in Fig. 2D, the Bradford assay was performed on the supernatant after embedding, and it was found that the absorbance of the substrate corresponding to the absorbance at 595 nm increased dramatically when the concentration of the natural enzyme was increased to 7.0 mg mL−1, indicating that each unit of the synthesized UiO-66(Ce) was able to encapsulate 7 units of the natural enzyme. The catalytic efficiency of the encapsulated glucose oxidase was further explored by means of the actual harvested glucometer. The hydrogen peroxide reactivity of the synthesized UiO-66(Ce)@GOx/HRP was explored following a POD-like enzyme testing protocol.27 Considering the high affinity of HRP for TMB, the determination of HRP activity was carried out using a 1 μg mL−1 multi-enzyme reactor. The results showed that the absorbance at 652 nm showed a linear increase within 5 min of the start of the reaction with a reaction rate of 0.72 μM min−1 μg−1 (Fig. 2F). The above results showed that the rationally designed UiO-66(Ce)@GOx/HRP was able to load the natural enzyme efficiently and maintain an efficient enzymatic reaction activity. Some of the prevailing views are that enzyme molecules could be immobilized within the pores of UiO-66(Ce) by physisorption, in which interaction was usually non-covalent and involves van der Waals forces, π–π stacking and electrostatic interactions. In addition, hydrogen bonds might form between the organic ligands of UiO-66(Ce) and the enzyme molecules. These hydrogen bonds also could help stabilize the enzyme molecule and reduce its movement within the pores, thus protecting its active site from destruction.
Multi-enzyme probe stability and analytical performance of immunosensing platform
Prior to the ELISA-like testing of DENV, the stability of the synthesized multi-enzyme reactors was evaluated. As the results were shown in Fig. 3A, the synthesized UiO-66(Ce)@GOx/HRP stored at room temperature was able to maintain nearly 90% of the catalytic activity for 60 days, implying that the reactor could be suitable for long term stable DENV determination. Further, based on previous experience and reports, capture and bio-signal amplification tests were performed for DENV2-NS1 at low (0.2 ng mL−1) and high concentrations, and it was found that both low and high concentrations could result in a significant absorbance shift of TMB, indicating the feasibility of this strategy, as shown in Fig. 3B. Therefore, using serum gradient dilution standards of DENV2-NS1, the absorbance values at 652 nm were determined at different concentrations as shown in Fig. 3C.28,29 Specific working concentrations were determined in the range of 0.05–100 ng mL−1 where the fitted working concentration curve equation followed Y = 0.3529 + 0.2058![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (n = 11, R2 = 0.992). The limit of detection (LOD) of the developed dengue sensing system was further determined to be as low as 39.7 pg mL−1 by linear equations and error values in blank sample tests (n = 21, 3 s N−1). Last but not least, it was necessary to determine whether the sensing platform developed for the ELISA-like test was capable of sensitively and accurately evaluating the analyte. Therefore, double-blind testing of the target DENV2-NS1 using a commercially available ELISA kit revealed that the two analytical methods maintained a high degree of concordance over a set concentration range, thus demonstrating that the developed multi-enzymatic reactor-based ELISA test platform was expected to achieve a highly sensitive and accurate DENV assay, as shown in Fig. 3D.
C (n = 11, R2 = 0.992). The limit of detection (LOD) of the developed dengue sensing system was further determined to be as low as 39.7 pg mL−1 by linear equations and error values in blank sample tests (n = 21, 3 s N−1). Last but not least, it was necessary to determine whether the sensing platform developed for the ELISA-like test was capable of sensitively and accurately evaluating the analyte. Therefore, double-blind testing of the target DENV2-NS1 using a commercially available ELISA kit revealed that the two analytical methods maintained a high degree of concordance over a set concentration range, thus demonstrating that the developed multi-enzymatic reactor-based ELISA test platform was expected to achieve a highly sensitive and accurate DENV assay, as shown in Fig. 3D.
Additional accuracy experiments on the developed sensor with real samples further validated its performance in detecting DENV in complex biological samples. The experimental results showed that the sensor exhibited excellent sensitivity and specificity in detecting NS1 protein in real clinical samples. Compared with the results of commercially available ELISA kits, the t-value between the two was less than 2.78 (t[0.05,4] = 2.78) (Table 1).30–32 The sensor was not only able to provide reliable results in a shorter period of time, but also effectively avoided the interference of other pathogens and significantly reduced the false-positive and false-negative rates. This showed that the sensor was not only capable of detecting DENV in complex biological samples, but also of detecting the protein in a more sensitive and specific manner, as well as in a more sensitive manner. In addition, the developed sensors were able to perform accurate testing over an ultra-wide detection range, and comparable to those already reported (Table S1†). These results indicated that the multi-enzyme reactor-modified sensor based on the MOF material not only could effectively protect the enzyme activity, but also possessed the ability to rapidly and accurately detect DENV in complex biological samples. Therefore, the sensor has broad applications in the early diagnosis of dengue and outbreak monitoring, especially in resource-limited areas, and provides strong technical support for public health.
| Simple | Methods (mean ± SD) | t value | |
|---|---|---|---|
| D-ELISA | ELISA | ||
| 1 | 5.86 ± 0.42 | 5.73 ± 0.53 | 1.02 |
| 2 | 8.08 ± 0.66 | 8.39 ± 0.31 | 0.86 |
| 3 | 12.87 ± 0.86 | 12.41 ± 0.87 | 1.28 |
| 4 | 2.38 ± 0.09 | 2.52 ± 0.27 | 0.71 |
| 5 | 16.74 ± 1.92 | 18.17 ± 0.90 | 1.08 |
Conclusions
In summary, this work reported a multi-enzyme-assisted metal–organic framework encapsulation system for achieving precise quantification of DENV. The main contributions of this work compared to conventional enzyme-related sensing platforms were summarized as follows: (i) the introduction of block polymers in the synthesis of UiO-66(Ce) enabled the generation of surface mesopores, which facilitated the encapsulation of the natural enzyme; (ii) the natural enzyme was able to efficiently enter and be protected by the framework structure, which exhibited good stability; and (iii) the system encapsulating multiple enzymes could realize multi-step cascade reactions under mild conditions and achieve efficient signal amplification. The strategy of combining multiple natural enzymes with rationally designed metal–organic frameworks opens up new avenues for the precise detection of disease-related biomolecules.Ethical statement
All experiments were performed in accordance with the Guidelines of Fujian Medical University and Fuzhou University (China), and approved by the ethics committee at Fujian Medical University and Fuzhou University (China). Informed consents were obtained from human participants of this study.Data availability
Data will be made available on request.Author contributions
Luanfeng Lin: conceptualization, investigation, methodology, and writing – original draft. Xiangqun Fan: investigation, methodology, writing – review & editing. Yinmei Yan: visualization, investigation, methodology, writing – review & editing. Ting Lin: writing – review & editing, methodology, and project administration. Xiao Han: methodology, writing – review & editing, and project administration. Dianping Tang: methodology, project administration, writing – review & editing. Lifen Han: visualization, investigation, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is funded by the Natural Science Foundation of Fujian Province, China (2021J011287 & 2022J011287), and Integrated Chinese and Western Medicine 'Flagship' Department (Construction Project) of National Health Commission of the People's Republic of China and National Administration of Traditional Chinese Medicine.References
- S. Iqtadar, J. Akram and A. Khan, Vaccines, 2024, 12, 913 CrossRef.
- Y. Lin, D. A. Joubert, S. Kaeser, C. Dowd, J. Germann, A. Khalid, J. A. Denton, K. Retski, A. Tavui, C. Simmons, S. O'Neill and J. R. L. Gilles, Sci. Robot., 2024, 9, eadk7913 CrossRef PubMed.
- M. P. Fallah, T. Raji, A. N. Ngongo, N. Ndembi, A. Ogwell, M. Abdulaziz, M. Aragaw, S. Sembuche, E. Gonese, N. Dereje, P. Materu and J. Kaseya, BMJ Glob. Health, 2024, 9, e014872 CrossRef.
- L. H. Chen, C. Marti, C. Diaz Perez, B. M. Jackson, A. M. Simon and M. Lu, J. Travel Med., 2023, 30, taad127 CrossRef.
- H. Guo, X. Ding, D. Hua, M. Liu, M. Yang, Y. Gong, N. Ye, X. Chen, J. He, Y. Zhang, X. Xu and J. Li, Vaccines, 2024, 12, 563 CrossRef CAS.
- C. Singhal, S. K. Shukla, A. Jain, C. Pundir, M. Khanuja, J. Narang and N. P. Shetti, ACS Biomater. Sci. Eng., 2020, 6, 5886–5894 CrossRef CAS PubMed.
- T. Trakoolwilaiwan, Y. Takeuchi, T. S. Leung, M. Sebek, L. Storozhuk, L. Nguyen, L. D. Tung and N. T. K. Thanh, Nanoscale, 2023, 15, 12915–12925 RSC.
- J. Kim, C. Cho, M. Y. Ryu, J. Kim, S. Lee, T. Park and J. Park, PLoS One, 2019, 14, e0222144 CrossRef CAS PubMed.
- T.-N. Le, W. W. Hsiao, Y. Cheng, C. Lee, T. Huynh, D. M. Pham, M. Chen, M. Jen, H. Chang and W. Chiang, Anal. Chem., 2022, 94, 17819–17826 CrossRef CAS PubMed.
- A. Sangili, T. Kalyani, S. Chen, K. Rajendran and S. Jana, Composites, Part B, 2022, 238, 109876 CrossRef CAS.
- Z. Yu, J. Tang, M. Xu, D. Wu, Y. Gao, Y. Zeng, X. Liu and D. Tang, Anal. Chem., 2024, 96, 11463–11471 CrossRef CAS.
- C. Zhou, Z. Fang, C. Zhao, X. Mai, S. Emami, A. Taha, G. Sun and T. Pan, Anal. Chem., 2021, 93, 11424–11432 CrossRef CAS.
- J. Chi, B. Gao, M. Sun, F. Zhang, E. Su, H. Liu and Z. Gu, Anal. Chem., 2017, 89, 7727–7733 CrossRef CAS.
- Z. Yu, H. Gong, F. Xue, Y. Zeng, X. Liu and D. Tang, Anal. Chem., 2022, 94, 13233–13242 CrossRef CAS PubMed.
- Z. Yu, H. Gong, J. Xu, Y. Li, Y. Zeng, X. Liu and D. Tang, Anal. Chem., 2022, 94, 3418–3426 CrossRef CAS.
- D. Lee, N. Asmare and A. F. Sarioglu, Lab Chip, 2023, 23, 251–260 RSC.
- L. S. Ferraraccio, D. Di Lisa, L. Pastorino and P. Bertoncello, Anal. Chem., 2022, 94, 16122–16131 CrossRef CAS PubMed.
- Y. Xu, Q. Li, H. Xue and H. Pang, Coord. Chem. Rev., 2018, 376, 292–318 CrossRef CAS.
- G. Cai, P. Yan, L. Zhang, H.-C. Zhou and H.-L. Jiang, Chem. Rev., 2021, 121, 12278–12326 CrossRef CAS PubMed.
- Q. Wang, X. Zhang, L. Huang, Z. Zhang and S. Dong, Angew. Chem., Int. Ed., 2017, 56, 16082–16085 CrossRef CAS PubMed.
- Z. Yu, J. Tang, H. Gong, Y. Gao, Y. Zeng, D. Tang and X. Liu, Adv. Funct. Mater., 2023, 33, 2301457 CrossRef CAS.
- W. Xu, Y. Wu, L. Jiao, M. Sha, X. Cai, Y. Wen, Y. Chen, W. Gu and C. Zhu, Nano Res., 2022, 16, 3364–3371 CrossRef.
- Z. Yu, R. Zeng, H. Gong, Y. Gao, S. Chen, Y. Wang and D. Tang, Nano Res., 2023, 17, 2451–2461 CrossRef.
- W. Liu, J. Wu, X. Ji, Y. Ma, L. Liu, X. Zong, H. Yang, J. Dai, X. Chen and W. Xue, Theranostics, 2020, 10, 6245–6260 CrossRef CAS.
- W. Lu, Z. Wei, Z. Gu, T. Liu, J. Park, J. Park, J. Tian, M. Zhang, Q. Zhang, T. Gentle Iii, M. Bosch and H. Zhou, Chem. Soc. Rev., 2014, 43, 5561–5593 RSC.
- Y. Zhang, C. Xing, Z. Mu, Z. Niu, X. Feng, Y. Zhang and B. Wang, J. Am. Chem. Soc., 2023, 145, 13469–13475 CrossRef CAS PubMed.
- B. Jiang, D. Duan, L. Gao, M. Zhou, K. Fan, Y. Tang, J. Xi, Y. Bi, Z. Tong, G. F. Gao, N. Xie, A. Tang, G. Nie, M. Liang and X. Yan, Nat. Protoc., 2018, 13, 1506–1520 CrossRef CAS.
- Z. Yu, Q. Lin, H. Gong, M. Li and D. Tang, Biosens. Bioelectron., 2023, 223, 115028 CrossRef CAS.
- Z. Yu, H. Gong, Y. Gao, L. Li, F. Xue, Y. Zeng, M. Li, X. Liu and D. Tang, Small, 2022, 18, e2202564 CrossRef.
- Z. Yu, C. Qiu, L. Huang, Y. Gao and D. Tang, Anal. Chem., 2023, 95(8), 4212–4219 CrossRef CAS PubMed.
- Z. Yu, H. Gong, M. Li and D. Tang, Biosens. Bioelectron., 2022, 218, 114751 CrossRef CAS PubMed.
- Z. Yu, Z. Xu, R. Zeng, M. Xu, H. Zheng, D. Huang, Z. Weng and D. Tang, Angew. Chem., Int. Ed., 2024, e202414625, DOI:10.1002/anie.202414625.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00335g |
| ‡ These authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |