Open Access Article
Open Access ArticleNHS-ester conjugated gold nanoparticles for spermine detection: a potential tool in meat spoilage monitoring†
Teody
Gumabat
 *a,
Jeanne Phyre Lagare
Oracion
*a,
Jeanne Phyre Lagare
Oracion
 a,
Jolina
Fedelis
a,
Jolina
Fedelis
 a,
Ethel
Keleste
a,
Ethel
Keleste
 a,
Rey
Capangpangan
a,
Rey
Capangpangan
 b,
Noel Lito
Sayson
cd,
Gerard
Dumancas
ef,
Arnold
Alguno
cd and
Felmer
Latayada
b,
Noel Lito
Sayson
cd,
Gerard
Dumancas
ef,
Arnold
Alguno
cd and
Felmer
Latayada
 *ag
*ag
aCenter for Nanoscience and Technology for Research and Entrepreneurship (CeNTRE), Material Science and Polymer Chemistry (MSPC) Laboratory, Caraga State University, 8600 Butuan City, Philippines. E-mail: tlgumabat@carsu.edu.ph
bDepartment of Physical Sciences and Mathematics, College of Marine and Allied Sciences, Mindanao State University at Naawan, Naawan 9023, Misamis Oriental, Philippines
cDepartment of Physics, Mindanao State University-Iligan Institute of Technology, Iligan City 9200, Philippines
dResearch Center for Energy Efficient Materials (RCEEM), Premier Research Institute of Science and Mathematics (PRISM), MSU-Iligan Institute of Technology, Iligan City 9200, Philippines
eHonors College, Henry E. and Shirley T. Frye Hall, Suite 110, North Carolina Agricultural and Technical State University, 1601 East Market Street, Greensboro, NC 27411, USA
fDepartment of Chemistry, New Science Building, North Carolina Agricultural and Technical State University, 1601 E. Market Street, Greensboro, NC 27411, USA
gDepartment of Chemistry, Caraga State University, Butuan City 8600, Philippines. E-mail: fslatayada@carsu.edu.ph
First published on 6th January 2025
Abstract
Spermine, a key biogenic amine indicating food freshness, is typically detected using costly and time-consuming chromatographic methods. This study introduces a more efficient, eco-friendly alternative—a label-free colorimetric sensing platform using gold nanoparticles (AuNPs) functionalized with 11-mercaptoundecanoic acid and capped with an N-hydroxysuccinimide (NHS)-ester. Transmission electron microscopy revealed monodisperse, spherical AuNPs (13 nm), with an increase in size upon functionalization. Fourier transform infrared spectra confirmed successful functionalization. The hydrodynamic size of the AuNPs increased from 17.26 nm to 2167 nm, and the zeta potential shifted from −48.86 mV to −35.59 mV. The platform takes advantage of the selective interaction between spermine and NHS-ester-functionalized AuNPs, inducing nanoparticle aggregation, as shown by a red shift in surface plasmon resonance (SPR). UV-vis spectroscopy demonstrated a robust linear correlation (R2 = 0.958) between spermine concentration (1.0–4.0 μM) and nanoparticle aggregation index, with a limit of detection (LOD) of 0.77 μM. The sensor also exhibited high reproducibility in pork extract matrices (coefficient of variation <5%) and selectivity for spermine amid various interfering analytes. Its eco-friendly design and rapid response time position it as a viable tool for real-time spermine monitoring in food spoilage, offering comparable performance metrics to traditional chromatographic techniques while addressing sustainability concerns.
1 Introduction
Food safety and security issues have inexplicably caused heightened concerns worldwide amidst a looming climate crisis.1,2 Between 1991 and 2021, red meat consumption caused 1729 reported outbreaks globally. These outbreaks resulted in 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 438 illnesses, 10
438 illnesses, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 691 hospitalizations, and 10
691 hospitalizations, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 063 deaths. Many cases went unreported in developing countries.3 In the face of these staggering statistics, there is a crucial need to develop an advanced method for detecting food spoilage. Consuming spoiled meat poses a significant public health risk, contributing to food waste and impacting global food security, due to harmful microorganisms like Salmonella, E. coli, Listeria, molds (Aspergillus), and yeasts (Candida), as well as elevated levels of volatile organic compounds (e.g., sulfides) and biogenic amines (naturally occurring compounds like histamine present in high amounts due to spoilage) which can lead to allergic reactions, severe infections and other adverse health effects. Therefore, early detection of meat spoilage is crucial. It helps in preventing illnesses like food poisoning and bacterial infections.4,5 Regrettably, current methods for detecting meat spoilage heavily rely on subjective visual inspections and manual monitoring. These methods are error-prone, difficult to quantify, lack repeatability, and human judgment shows inconsistencies. Additionally, variations in meat intrinsic properties like fat content, connective tissue content, and skeletal structure differ among meat types. These variations restrict the effectiveness of physical assessment.6,7 To address these challenges and proactively enhance the safety and quality of meat products, researchers are exploring new technologies. These technologies detect specific molecules or compounds produced during meat spoilage, reducing waste and promoting sustainable practices.8–10
063 deaths. Many cases went unreported in developing countries.3 In the face of these staggering statistics, there is a crucial need to develop an advanced method for detecting food spoilage. Consuming spoiled meat poses a significant public health risk, contributing to food waste and impacting global food security, due to harmful microorganisms like Salmonella, E. coli, Listeria, molds (Aspergillus), and yeasts (Candida), as well as elevated levels of volatile organic compounds (e.g., sulfides) and biogenic amines (naturally occurring compounds like histamine present in high amounts due to spoilage) which can lead to allergic reactions, severe infections and other adverse health effects. Therefore, early detection of meat spoilage is crucial. It helps in preventing illnesses like food poisoning and bacterial infections.4,5 Regrettably, current methods for detecting meat spoilage heavily rely on subjective visual inspections and manual monitoring. These methods are error-prone, difficult to quantify, lack repeatability, and human judgment shows inconsistencies. Additionally, variations in meat intrinsic properties like fat content, connective tissue content, and skeletal structure differ among meat types. These variations restrict the effectiveness of physical assessment.6,7 To address these challenges and proactively enhance the safety and quality of meat products, researchers are exploring new technologies. These technologies detect specific molecules or compounds produced during meat spoilage, reducing waste and promoting sustainable practices.8–10
Spermine, a food decomposition product, is recognized as a crucial indicator in specific meat samples. It plays a pivotal role in assessing the freshness and quality of the meat, making it a key focus of the investigation. Spermine belongs to a group of small, low molecular-weight substances produced during the normal metabolism of organisms called biogenic amines. Under room temperature conditions, spermine concentration tends to change during the processing and storage of meat and meat products.11 For instance, one study found that the concentration of spermine in conventional chicken, organic chicken, and duck meat decreased with increasing storage time.12 Additionally, monitoring the spermine concentration of meat can be potentially useful in determining the efficacy of some food preservation methods, like the high hydrostatic pressure (HHP) process13 and gamma irradiation,14 providing valuable insights for both food manufacturers and consumers about the impact of different storage conditions and processing techniques on product quality.
The widely used approach for spermine detection in a complex sample like meat, would be through chromatographic analysis, but has several drawbacks. Numerous reports for detecting spermine and other biogenic amines have already been established using high-performance liquid chromatography-mass spectrometry (HPLC-MS), liquid chromatography-mass spectrometry (LC-MS), or gas chromatography-mass spectrometry (GC-MS). Conversely, these methods are time-consuming, expensive, and not environmentally friendly since the optimization usually requires significant solvent consumption, making it impractical for routine monitoring. Furthermore, the equipment and expertise necessary for these analyses may be inaccessible to everyone.15–17 Hence, an alternative approach18 that is more efficient, cost-effective, and environment friendly is needed to detect spermine in various samples easily.
Gold nanoparticles (AuNPs) have emerged as a sustainable solution to address the existing gap by providing an efficient and cost-effective method to detect biogenic amines such as spermine. In the presence of an analyte, it can display a colorimetric response (color change) when dispersed or aggregated due to its shift in surface plasmon resonance (SPR) wavelength. For instance, citrate-reduced AuNPs can detect biogenic amines at nanomolar concentrations.19 Nevertheless, the selectivity of this method may be questionable, as AuNPs tend to be unstable and would readily respond to other interfering substances causing their aggregation. This could arguably lead to false positives or misleading results. To address this, extensive efforts have been dedicated to functionalizing AuNPs with specific ligands or receptors that will act as a recognition element on the surface of AuNP.9,20–22
In the study by Ling et al.23 on the functionalization of the surface of AuNPs with dithiobis(succinimidylpropionate) (DSP), a notable click reaction was observed between the aliphatic amino group of histamine and the succinimidyl/N-hydroxysuccinimide (NHS)-ester moiety of DSP, demonstrating high sensitivity and selectivity within the range of 0.8 to 2.5 μM. As an alternative to DSP, it might be possible to use a spacer that has a carboxylate group at its end to react with EDC/NHS (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride/N-hydroxysuccinimide) to derivatize an NHS-ester moiety.24 An added benefit of modifying AuNPs with 11-mercaptoundecanoic acid (MUDA) is its tunability and stability. This makes it a valuable component in facilitating specific interactions within the research context. Moreover, its incorporation confers exceptional resistance to small molecules by creating a protective barrier that prevents the aggregation of AuNPs due to nonspecific analyte interactions.24–27 In conclusion, to emulate the study done by Liu and colleagues, carbodiimide chemistry might present a sound method to modify carboxylic groups, converting them into NHS-ester moieties for nanosensor development. There have been reported successes in harnessing this click-reaction as researchers have been able to connect metallic nanoparticles with enzymes,28 tissues,26,29 metal ions,30 and various smaller molecules with EDC/NHS activated MUDA,31–33 demonstrating the versatility and broad applicability of this technique.
In this study, a novel nanosensor was developed for the detection of spermine. This was made possible by MUDA modified AuNPs capped with NHS-ester showcasing advancements in food safety technology for sustainable food production. To demonstrate the superiority of this nanosensor, the performance of MUDA-AuNP (11-mercaptoundecanoic modified AuNP) and NHS-AuNP (NHS ester-capped MUDA-AuNP) was assessed by evaluating their spectral changes in the presence of varying spermine concentrations. Mechanistically, NHS-AuNP was more sensitive. This was due to the formation of stronger covalent bonds with the analyte. As shown in Scheme 1, the NHS-ester moiety in NHS-AuNP links covalently with the aliphatic amino group of spermine through amide formation. Meanwhile, the carboxylic group of MUDA-AuNP links with the aliphatic amino group solely through hydrogen bonding, which is relatively weaker. Furthermore, UV-visible spectroscopy was used to characterize these spectral changes. On the other hand, the successful MUDA functionalization and subsequent verification of NHS-ester capping was conducted using Fourier transform infrared (FTIR) spectroscopy. In addition, the findings of this investigation were juxtaposed with prior research to provide substantiation of the enhancements that were made. Expanding on the findings of this study, future research could explore the development of rapid, on-site testing methods for spermine in meat samples. This could be done using solution-based and paper-based processes.
 | ||
| Scheme 1 Possible interaction of MUDA-AuNP (11-mercaptoundecanoic acid functionalized gold nanoparticles) and NHS-AuNP (NHS-ester capped MUDA-AuNP) towards spermine. | ||
2 Experimental
2.1 Materials
Chloroauric acid trihydrate (HAuCl4·3H2O, 99%), MUDA, histamine, cadaverine, putrescine, spermidine, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide·hydrochloric acid (EDC), L(−)glutamine, lactic acid, D(+)myo-inositol, L(−)asparagine, L(−)histidine, and L(−)lysine were purchased from Sigma-Aldrich (USA). Trisodium citrate dehydrate was purchased from Xilong Science Co., Ltd. (Mainland China). D(+)arabinose, DL-malic acid, sodium hydroxide, sodium chloride, L(−)ascorbic acid, and the copper standard solution were purchased from Scharlau. L(−)cysteine and N-hydroxysuccinimide (NHS) were purchased from Alfa-Aesar (Great Britain). Urea was purchased from Medic Diagnostic Reagents (Philippines). Tetramethylammonium hydroxide pentahydrate was obtained from HiMEDIA Laboratories (India). The cadmium standard was purchased from Buck Scientific (USA). D(+)glucose was obtained from UNI-CHEM Chemical Reagents (India). 2-(N-Morpholino)ethanesulfonic acid (MES) was purchased from Shanghai Aladdin Biochemical Technology (Mainland China). Ammonium sulfide was purchased from Merck (Germany). Ultrapure water purified with a Millipore system (18.2 MΩ) was used as a solvent for aqueous solutions. For dialysis and purification, regenerated cellulose dialysis bag tubing (3500 MWCO) was used which was purchased from Hunan Yibo Biotechnology Co., Ltd., (Mainland China).2.2 Overview of the method
An experimental flowchart is shown in Fig. 1 to aid in the visualization of preparation and characterization procedures. Bare-AuNP was synthesized by the citrate-reduction method, surface functionalized with MUDA, and labeled as MUDA-AuNP. The MUDA-AuNP was divided into three portions with distinct functions: the first for the preparation of NHS-AuNP, the second for MUDA-AuNP-MES preparation, and the third designated as sensor I. Subsequently, NHS-AuNP was synthesized to fulfill the role of sensor II. Following this, MUDA-AuNP-MES was utilized as the precursor for NHS-AuNP-MES, subsequently designated as sensor III. Finally, sensors I, II, and III underwent testing to detect biogenic amines, different spermine concentrations, and potential interferences present in spoiled meat. These were done colorimetrically through a ultraviolet-visible (UV-vis) instrument where their sensitivity and/or selectivity are compared towards various analytes. | ||
| Fig. 1 Experimental flowchart highlighting the processes in this study. Sensor I (MUDA-AuNP), sensor II (NHS-AuNP), and sensor III (NHS-AuNP-MES) were prepared and tested accordingly. | ||
2.3 Preparation of bare-AuNP
The citrate-reduced AuNP was synthesized following the usual Turkevich–Frens method.34 Briefly, a 200 mL Erlenmeyer flask containing 50 mL of 0.667 mM HAuCl4·3H2O was placed in a water bath and heated to 95 °C with constant and vigorous stirring. The solvent loss was prevented by covering the flask with a weighted watch glass. A reflux setup was not chosen due to its uneven temperature gradient tendency. After the desired temperature of the bath was reached, 1.282 mL of 152 mM Na3Ctr·H2O was injected to the HAuCl4 solution, turning the resulting mixture dark after 3 minutes and then burgundy red after about 5 minutes. Finally, after maintaining the desired temperature on the hot plate for 20 minutes, the reaction flask was cooled to room temperature and stored at 4 °C for further characterization and use. This solution was labeled as “bare-AuNP”.2.4 Preparation of MUDA-AuNP and MUDA-AuNP-MES
To increase the selectivity of the as-synthesized AuNPs towards spermine, a functionalization step was necessary. Based on a previous method,31 bare-AuNP was functionalized by first dissolving 0.1 mmol of MUDA in 1 mL of 0.55 M tetramethylammonium hydroxide. Then, this solution was then added to 10 mL of the bare-AuNPs and stirred gently at room temperature for 16 hours. After that, purification was then done by transferring this mixture in a regenerated cellulose dialysis tubing (3500 MWCO) and then dialyzing it against distilled H2O. Through this step, the excess MUDA from inside the dialysis tubing diffused towards the fresh H2O solvent. After 2 hours, the H2O solvent was replaced to prepare for another round of dialysis. This step was repeated twice. Immediately, the remaining solution inside the dialysis tubing was retrieved and this solution was labeled as “MUDA-AuNP”. Finally, 1 mL of the above solution was precipitated via centrifugation (4000 rpm; 30 min) to be used in the preparation of NHS-AuNP-MES. The pelletized MUDA-AuNP was then redispersed in MES buffer (100 mM; pH = 5.5; 500 μL). This solution was labeled as “MUDA-AuNP-MES”.2.5 Preparation of NHS-AuNP and NHS-AuNP-MES
To further increase the sensitivity of the sensor, it was essential that the –COOH groups of MUDA becomes activated via the EDC/NHS coupling reaction, forming NHS-ester capped AuNPs. Firstly, EDC (30 mg mL−1) and NHS (36 mg mL−1) were dissolved in MES buffer (2 mL, 0.1 M, pH = 5.5). Next, activation was then done by incubating 20 μL of the EDC/NHS mixture with 1000 μL of MUDA-AuNP (0.02![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, EDC–NHS
1, EDC–NHS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MUDA-AuNP). This was labeled as “NHS-AuNP”. Another 5 μL of the EDC/NHS mixture was added to 500 μL of the MUDA-AuNP-MES (0.01
MUDA-AuNP). This was labeled as “NHS-AuNP”. Another 5 μL of the EDC/NHS mixture was added to 500 μL of the MUDA-AuNP-MES (0.01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, EDC–NHS
1, EDC–NHS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MUDA-AuNP-MES) solution for 30 min at room temperature. This was labeled as “NHS-AuNP-MES”.
MUDA-AuNP-MES) solution for 30 min at room temperature. This was labeled as “NHS-AuNP-MES”.
2.6 Selectivity and sensitivity assay of AuNP sensors
To conduct a selectivity assay, 6 μL of analyte solution of was introduced into 150 μL of bare-AuNP, MUDA-AuNP, and NHS-AuNP nanosensors in a 96-well polystyrene microplate. For sensitivity, 6 μL of analyte solution was introduced into 150 μL of the MUDA-AuNP, NHS-AuNP, and NHS-AuNP-MES. To predict the performance of the nanosensors in the sample matrix, 6 μL of the common interferences (494 μM) found in spoiled meat extracts was introduced into 150 μL of NHS-AuNP. It must be noted that all analyte and interfering solutions, unless mentioned, are 0.9% saline in ultrapure H2O to simulate salinity in spoiled meat extracts. The resulting mixture was photographed to observe any changes in color after 5 minutes. A wavelength scan within the UV-vis region (400–1000 nm) was then run to evaluate the AuNPs' shift in its SPR wavelength.2.7 Detection of spermine in pork meat
The fresh pork meat was blended then added into 0.9% NaCl solution (w/v = 1/2) and homogenized for 2 min. After that, it was centrifuged (4000 rpm, 25 °C, 30 min), the supernate was obtained and spiked with spermine (to a final concentration of 3575 μM) and stirred thoroughly. To prevent unwanted interferences, 3 mL of this spiked solution was subjected to dialysis by placing in a dialysis membrane (regenerated cellulose membrane; 3500 MWCO). Then, the membrane with sample was dialyzed against 30 mL of dialyzing solvent (0.9% saline). The setup was shaken for homogeneity for 2 h. After that, the dialyzing solvent was collected (assumed to have a total concentration of 325 μM spermine) and added to NHS-AuNP solution sensor at volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 (spermine final concentration with sensor is now at 12.5 μM) and measured by UV-vis spectra to detect the concentration of spermine. Recovery was calculated from found concentration/known concentration × 100%. Precision was calculated from standard deviation/mean × 100%. Accuracy was calculated from [(found concentration − known concentration)/known concentration] × 100%. All measurements were done in triplicates.
25 (spermine final concentration with sensor is now at 12.5 μM) and measured by UV-vis spectra to detect the concentration of spermine. Recovery was calculated from found concentration/known concentration × 100%. Precision was calculated from standard deviation/mean × 100%. Accuracy was calculated from [(found concentration − known concentration)/known concentration] × 100%. All measurements were done in triplicates.
2.8 Measurement and characterization
The sensor's colorimetric response concerning the analyte was acquired using a Microplate reader (EnSight Multi-mode Plate Reader, PerkinElmer, USA). Hydrodynamic size, size distribution, and zeta potential were determined using a Dynamic Light Scattering Instrument (Litesizer 100, Anton-Paar, Austria). The AuNPs' morphology, particle size, and particle aggregation with and without the analyte were conducted using a high-resolution transmission electron microscope (JEM 2100Plus TEM-EDS, JEOL, Japan). Finally, the presence of relevant functional groups in MUDA-AuNP and NHS-AuNP was analyzed using an Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FTIR, Perkin Elmer Spectrum 2, USA) instrument. The samples were air-dried and measured in ambient conditions.3 Results & discussion
3.1 TEM morphology
Transmission electron microscopy (TEM) imaging shown in Fig. S1† reveals the formation of monodisperse and spherical citrate-reduced AuNPs with an average particle size (dave) of 12.89 ± 1.70 nm. With MUDA functionalization, the size, dave = 13.79 ± 1.25 nm (Fig. 2A), has slightly increased, but its spherical morphology is still significantly intact.Clustering of a few key areas was also captured; this might not suggest aggregation since the solution still maintains its original color even with prolonged storage at ambient temperature. This is evidence of the superior stability offered by MUDA. When spermine was added to MUDA-AuNP, the nanoparticles aggregated, as seen in Fig. 2B. However, the level of aggregation was not as pronounced as what was seen when bare-AuNP interacted with histamine (9.5 μM), which can be seen in Fig. S2.† This phenomenon may be attributed to the impact of the large MUDA spacers, which reduce the tendency of particles to aggregate, resulting in the formation of evenly spaced gaps between AuNPs.
3.2 UV-vis characterization
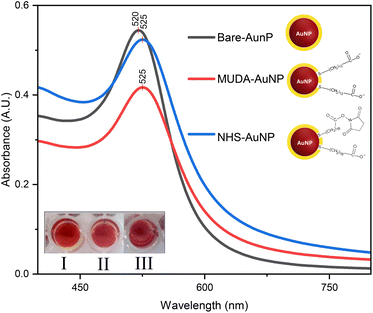
The UV-vis spectra in Fig. 3 exhibit unique peaks for bare-AuNP, MUDA-AuNP, and NHS-AuNP, highlighting variations in optical properties and surface interactions. An evident plasmonic shift from 520 nm to 525 nm was observed when AuNP was ligand exchanged with MUDA. This indicates a change in plasmon resonance35 and size36 consistent with the shifts observed in previous reports. Furthermore, there was a significant increase in absorbance observed at 525 nm when the 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide (EDC/NHS) mixture was added to the MUDA-AuNP solution. This is to be expected since the carboxyl ends of MUDA have a strong affinity for EDC/NHS activation.37The alteration of the SPR peak from 520 nm to 525 nm following NHS/EDC activation of MUDA-AuNP did not yield a plasmonic shift, as the chemical modification predominantly influences the surface chemistry without directly impacting the spatial configuration of the nanoparticles required to induce a shift in the SPR band. The EDC/NHS activation principally enables the binding of biomolecules or analytes through to the MUDA functional groups on the AuNP surface. This improves the sensor's binding capacity for biogenic amines but does not substantially affect the interparticle spacing or the aggregation behavior of the AuNPs.
The increase in absorbance upon the introduction of EDC/NHS to MUDA-AuNP may not exhibit a clear association with the successful alteration of AuNP-bound MUDA carboxyl groups. One possible explanation for the rise in absorbance may stem from the heightened density of absorbing molecules on the surface of MUDA-functionalized AuNP after EDC/NHS treatment; nevertheless, this does not definitively validate or invalidate the efficacy of EDC/NHS activation.
3.3 FTIR characterization
To confirm successful MUDA ligand exchange with AuNP and subsequent activation, bare-AuNP, MUDA-AuNP, and NHS-AuNP solutions were centrifuged for 30 min at 4000 rpm, and their precipitate was analyzed using an ATR-FTIR instrument. It can be seen in Fig. 4 that the characteristic –SH peaks (2548–2680 cm−1) of MUDA have disappeared in MUDA-AuNP and NHS-AuNP. This strongly suggests Au–S bond formation between Au and MUDA.32,38 Strong peaks at 3000–2850 cm−1 corresponded to asymmetric and symmetric C–H stretching characteristics in long-chain groups and were also found in both II and III, further proving ligand exchange.39 Now, we look into the activation of MUDA-AuNP into NHS-AuNP. The EDC/NHS activation of the MUDA-capped AuNP was supported by the disappearance of the carboxyl stretching (1577 cm−1 and 1538 cm−1) found in MUDA-AuNP and the enhancement of 1708 cm−1 and 1770 cm−1 peaks in NHS-AuNP. These enhanced peaks correspond to the symmetric and asymmetric stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups in the succinimidyl cycle of the NHS-ester, proving that the carboxyl group of the MUDA self-assembled monolayer was modified. In addition, the signal at 1816 cm−1 in NHS-AuNP, which corresponds to the C
O groups in the succinimidyl cycle of the NHS-ester, proving that the carboxyl group of the MUDA self-assembled monolayer was modified. In addition, the signal at 1816 cm−1 in NHS-AuNP, which corresponds to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of the succinimidyl ester and the symmetric C
O stretching of the succinimidyl ester and the symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O anhydride, also supports the above assumption.24,40,41
O anhydride, also supports the above assumption.24,40,41
3.4 Particle size distribution and -potential
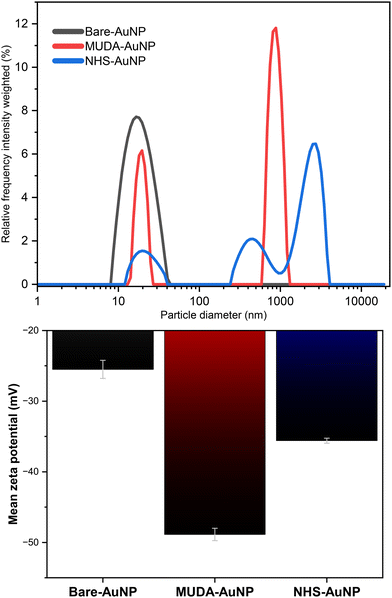
As seen in Fig. 5, the shifting hydrodynamic particle size (dhydro) distribution and zeta (ζ)-potential obtained from dynamic light scattering equipment justifies the previous arguments. We observed larger average dhydro and broader size distributions as further modifications were done. From a dhydro = 17.26 nm in I to a dhydro = 493 nm in II, if we disregard the aggregation-induced peaks in the 100–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm, the increased size can be attributed to the ligand modification of a small citrate molecule to a larger and bulkier MUDA molecule. This is supported by a similar study by colleagues of Ansar,36 who reported an increase in size for the MUDA functionalization of 13 nm AuNPs.
000 nm, the increased size can be attributed to the ligand modification of a small citrate molecule to a larger and bulkier MUDA molecule. This is supported by a similar study by colleagues of Ansar,36 who reported an increase in size for the MUDA functionalization of 13 nm AuNPs.
Aside from particle size, measuring the ζ-potential would also solidify the success of our functionalization and subsequent EDC/NHS activation. The variations in ζ-potential measurements are also shown in Fig. 5. ζ-Potential measurements are a reliable indicator of nanoparticles' surface charge, which can impact their stability and interactions in solution.42 Comparing bare-AuNP with MUDA-AuNP, the ζ-potential became even more negative due to the oxidized carboxyl moiety of the new MUDA monolayer of the AuNP. When this carboxyl moiety got activated via EDC/NHS, the ζ-potential increased due to the formation of an NHS-ester capping.
3.5 Detection mechanism of NHS-AuNP
AuNPs reduced with citrate demonstrate a strong affinity for the amine ends (–NH2) of biogenic amines, enabling their detection through an aggregation mechanism. This process is characterized by a red shift in SPR, leading to the emergence of a peak at 600 nm, while the initial peak at 525 nm diminishes. The intensity of the 600 nm peak increases proportionally to the analyte concentration, as observed in Fig. 6a. Among the biogenic amines tested (histamine, spermine, spermidine, cadaverine, and putrescine at 9.5 μM), histamine caused the most significant SPR shift, while putrescine exhibited negligible response. This variability arises from differences in molecular structure and interaction potential. Upon functionalization with MUDA, this aggregation phenomenon changes, as shown in Fig. 6. Except for spermine and spermidine, no significant SPR shifts were observed, suggesting a selective response attributed to the hydrogen bonding between the amine groups of these polyamines and the carboxyl groups of MUDA. This interaction results in uniform interparticle spacing,43,44 as evidenced by TEM images (Fig. 2b), which display no clumping compared to the random interparticle spacing observed in the bare-AuNP system (Fig. 6a). This uniformity suggests a mechanism distinct from the double-peak aggregation behavior of unmodified AuNPs.The enhanced selectivity of MUDA-AuNP arises from the steric hindrance provided by the bulky MUDA spacer, which limits smaller molecules such as putrescine and cadaverine from accessing the AuNP surface. Larger polyamines like spermine, with multiple reactive amine sites, interact more effectively, inducing aggregation and a corresponding SPR shift. This behavior aligns with similar findings by Liu et al.,27 where carboxyl group hydrogen bonding contributed to selective sensing. To establish a linear response of the MUDA-AuNP colorimetric sensor to spermine, the absorbance ratio at 600 nm to 525 nm (Abs600/Abs525) was plotted against the concentration of the added spermine standard. For MUDA-AuNP, 600 nm was chosen as the final SPR shift wavelength since, at higher concentrations, it becomes more apparent that this is the maximum SPR shift the sensor can respond to. Fig. 7 shows that MUDA-AuNP exhibits a linear response when Abs600/Abs525 is plotted against increasing spermine concentrations ranging from 5–25 μM. The limit of detection (LOD) and limit of quantitation (LOQ) were also calculated45 (see Table 1). An excellent R2 (0.994), which represents the coefficient of determination, was also obtained, further establishing linearity. Conversely, the color change of the solution only becomes noticeable at 15 μM, due to steric hindrance of the MUDA spacer warranting an optimization of this sensor.
| Nanosensor | LOD, μM | LOQ, μM | Linear range, μM |
|---|---|---|---|
| a LOD – limit of detection (3.3 × z). b LOQ – limit of quantitation (10 × z); z = Sy/S where Sy is the standard deviation of the y-intercept and S is the slope of the linear calibration curve. | |||
| MUDA-AuNP | 1.5 | 4.5 | 5–25 |
| NHS-AuNP | 2.2 | 6.6 | 10–25 |
| NHS-AuNP-MES | 0.77 | 2.3 | 1–4 |
To address the sensitivity limitations, MUDA-AuNP was activated with an EDC/NHS mixture, producing NHS-AuNP. This modification introduces an NHS-ester functional group, enabling covalent amide bond formation with biogenic amines. When spermine interacts with NHS-AuNP, the covalent linkage promotes nanoparticle aggregation, resulting in a bathochromic shift of the SPR band from 525 nm to 600 nm. The EDC/NHS combination activates the MUDA-AuNP surface by promoting the development of a reactive NHS ester on the carboxyl groups of the MUDA ligands. EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide) functions as a coupling agent, facilitating the formation of an active ester intermediate through its reaction with the carboxyl groups of MUDA, subsequently transforming them into NHS esters. These reactive esters have a strong reactivity towards primary amines, such as those found in spermine, facilitating effective amide bond formation between the amine groups of spermine and the NHS ester on the MUDA-AuNP surface. This process securely attaches spermine to the nanoparticle, resulting in the noted aggregation and colorimetric response. This aggregation mechanism is visually observable through a color change from red to purple, making the system viable for on-site applications.
The SPR shift induced by NHS-AuNP exhibited a linear response to spermine concentrations from 10 to 25 μM, as shown in Fig. 8. The absorbance ratio (Abs600/Abs525) demonstrated excellent linearity (R2 = 0.990), with a limit of detection (LOD) of 6.6 μM (See Table 1). This improvement over MUDA-AuNP is attributed to the more effective interaction mechanism of NHS-ester functional groups, which favor selective aggregation over non-specific binding.
Further optimization was conducted to enhance sensor sensitivity. The NHS activation process was refined by centrifuging MUDA-AuNP at 4000 rpm for 30 minutes, followed by redispersion in 0.1 M MES buffer (pH 5.5). This single-step precipitation removed excess MUDA ligands, reducing unwanted binding. The EDC/NHS mixture volume was reduced to 5 μL per 100 μL MUDA-AuNP to prevent non-specific aggregation while favoring NHS-ester formation. The MES buffer, lacking free amino and phosphate groups, proved ideal for stabilizing the system and enabling NHS ester formation.24 This produced NHS-AuNP-MES.
3.6 Sensitivity of NHS-AuNP-MES
The observed enhancement in sensor sensitivity using NHS-AuNP-MES, as depicted in Fig. 9, can be attributed to the optimization process, resulting in a better analytical performance. This sensor exhibited the best limit of detection (LOD) and limit of quantification (LOQ) values among the tested configurations, as summarized in Table 1. With an R2 of 0.958, the sensor demonstrated excellent linearity over a range of 1–4 μM, a significant improvement in sensitivity compared to its predecessors. A key feature of NHS-AuNP-MES is its capability for naked-eye detection, with a distinct color change from red to purple observed at a spermine concentration as low as 2.0 μM. This property eliminates the need for sophisticated spectrophotometric equipment, making the nanosensor practical for field-based applications in food spoilage monitoring. The visual detectability enhances user accessibility while retaining high sensitivity, bridging laboratory precision with on-site convenience.The optimization process involved refining the MUDA-AuNP functionalization. Centrifugation effectively removed excess MUDA ligands, which persisted even after dialysis, reducing unwanted NHS-EDC activation on unbound MUDA. This purification step minimized nonspecific reactions that could compromise sensor performance. Subsequent redispersion of the nanoparticles in a 0.1 M MES buffer allowed fine-tuning of the solution to a lower pH, favoring the activation of MUDA carboxyl groups. This adjustment enhanced the efficiency of NHS-ester formation, ensuring more consistent functionalization of the AuNPs and improving the sensor's reliability.
The enhanced sensitivity of the NHS-AuNP-MES sensor is attributed to its optimized binding mechanism. By lowering the pH during the redispersion step, the protonation state of carboxyl groups was adjusted to maximize their reactivity with the EDC/NHS mixture. This facilitated a more uniform distribution of NHS-ester groups on the nanoparticle surface, promoting covalent bonding with spermine and other biogenic amines. The resulting nanoparticle aggregation was more predictable, as evidenced by the linearity of the SPR shift, further validating the sensor's performance.
This systematic optimization not only improved the sensor's sensitivity but also addressed challenges related to sensitivity. The combination of purification, pH adjustment, and an MES buffer media contributed to a stable nanosensor platform, offering a promising tool for real-time, cost-effective, and environment friendly detection of spermine.
3.7 Sensor performance in real samples
Amino acids: L(−)cysteine (HS–CH2–CH(NH2)–COOH), L(−)arginine (HN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NH2)–NH–(CH2)3–CH(NH2)–COOH), L(−)asparagine (H2N–CO–CH2–CH(NH2)–COOH), L(−)histidine (NH–CH
C(NH2)–NH–(CH2)3–CH(NH2)–COOH), L(−)asparagine (H2N–CO–CH2–CH(NH2)–COOH), L(−)histidine (NH–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–CH
N–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2–CH(NH2)–COOH), L(−)glutamine (H2N–CO–(CH2)2–CH(NH2)–COOH), L(−)aspartic acid (HOOC–CH2–CH(NH2)–COOH), and L(−)lysine (H2N–(CH2)4–CH(NH2)–COOH); sugars: D(+)glucose (C6H12O6), D(+)arabinose (C5H10O5), and D(−)myo-inositol (C6H12O6); organic acids: lactic acid (C3H6O3) and malic acid (C4H6O5); other molecules and ions: urea (H2NCONH2), ammonium sulfide ((NH4)2S), copper(II) ion (Cu2+) cadmium(II) ion (Cd2+), chromium(III) ion (Cr3+), inosine (C10H12N4O5), and bovine serum albumin (BSA, a polyamine).
C–CH2–CH(NH2)–COOH), L(−)glutamine (H2N–CO–(CH2)2–CH(NH2)–COOH), L(−)aspartic acid (HOOC–CH2–CH(NH2)–COOH), and L(−)lysine (H2N–(CH2)4–CH(NH2)–COOH); sugars: D(+)glucose (C6H12O6), D(+)arabinose (C5H10O5), and D(−)myo-inositol (C6H12O6); organic acids: lactic acid (C3H6O3) and malic acid (C4H6O5); other molecules and ions: urea (H2NCONH2), ammonium sulfide ((NH4)2S), copper(II) ion (Cu2+) cadmium(II) ion (Cd2+), chromium(III) ion (Cr3+), inosine (C10H12N4O5), and bovine serum albumin (BSA, a polyamine).
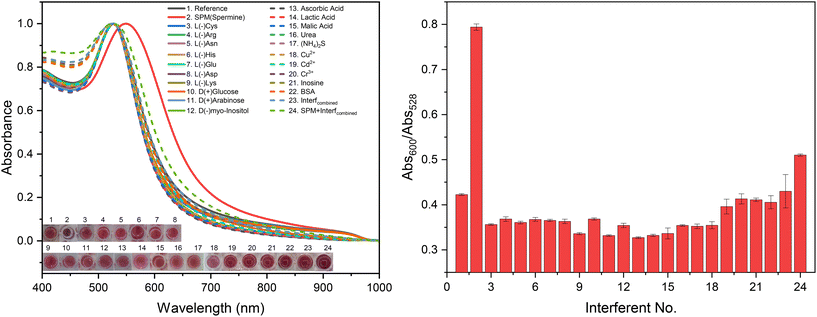
To further assess the sensor's specificity, a combination of these analytes at 50 ppm, with and without spermine (also at 50 ppm), was tested and denoted as SPM + Interf(combined) and Interf(combined), respectively (Fig. 10). The results of these tests provide critical insights into the robustness and selectivity of the nanosensor.
Fig. 10 demonstrates the colorimetric response of the NHS-AuNP nanosensor in the presence of these interfering analytes. Notably, small molecules containing amino or sulfide groups, such as L-cysteine and sulfide ions (S2−) from ammonium sulfide, which are known to aggregate citrate-reduced gold nanoparticles (AuNPs), did not induce significant aggregation in the NHS-AuNP system. The aggregation index (Abs600/Abs528) for these analytes ranged between 0.35 and 0.45, significantly lower than the value of 0.80 observed for spermine only-induced aggregation. This highlights the sensor's enhanced resistance to nonspecific interactions compared to conventional AuNP systems. When spermine was added to a mixture of all interfering analytes (SPM + Interfcombined), a significant aggregation was observed, albeit with a lower spectral ratio (Abs600/Abs528) = 0.51 compared to pure spermine samples (Abs600/Abs528) = 0.79. This reduction can be attributed to the masking effect of such large concentration of interfering molecules, which likely obstructed some of spermine's reactive sites, thereby limiting its interaction with the nanosensor.
It is also crucial to note that the concentrations of interfering analytes tested (50–100 ppm) are substantially higher than those typically encountered in real-world samples, such as meat extracts, where their levels rarely exceed 50 ppm. In practical scenarios, the response of the NHS-AuNP nanosensor to these potential interferents was negligible, ensuring its reliability for detecting spermine in complex mixtures. The observed selectivity of the nanosensor can be attributed to the functionalization with NHS-ester groups, which preferentially react with the multiple amine groups present in spermine, leading to nanoparticle aggregation. The lack of aggregation in the presence of other amino acids and sulfide ions suggests that the steric hindrance provided by bulky MUDA spacers effectively prevents nonspecific interactions, further enhancing the sensor's specificity.
| [Spiked], μM | [Found], μM | Recovery,a % | Precision,b % | Accuracy,c % |
|---|---|---|---|---|
| a Recovery = ([Found]/[Spiked]) × 100%. b Precision = (SD[Found]/[Found]) × 100%. c Accuracy = (|[Found] − [Spiked]|/[Spiked]) × 100% where [Found] is the mean of the found concentration, [Spiked] is the spiked concentration and SD[Found] is the standard deviation of the found concentration. | ||||
| 12.50 | 10.6 ± 0.45 | 84.5 | 4.3 | 15.5 |
The streamlined sample preparation protocol avoids overly complex pre-treatment steps and relies on dialysis to mitigate interference from matrix components. However, the modest recovery rate and reduced accuracy indicate room for optimization in the sample preparation process. This may have been caused by the complexity of the matrix and losses of analytes during sample preparation. Alternative methods to minimize analyte loss during dialysis could be explored, such as shorter dialysis times or alternative separation techniques.
In laboratory conditions, sample preparation is controlled to minimize matrix interference and preserve the integrity of the analyte. Standardized procedures like homogenization, centrifugation, and dialysis are employed to isolate and concentrate the analyte of interest. Conversely, in real-world samples, the matrix is more complex and may contain a variety of interfering substances, which can impact the accuracy of the detection method. For instance, natural variations in the meat's composition and the presence of additional molecules like proteins, lipids, and salts can affect the measurement of analytes. The NHS-AuNP nanosensor, designed to work well in controlled environments, did show good precision and accuracy against actual pork extracts but may experience reduced sensitivity and accuracy due to these complications.
The stability of the AuNP-based nanosensor is crucial during the transition from controlled laboratory environments to practical real-world settings. MUDA-AuNP nanoparticles that have been successfully functionalized for this procedure31 have demonstrated sustained stability for a long period of time. However, N-hydroxysuccinimide (NHS) esters are known for rapid hydrolysis within a short timeframe if not conjugated, potentially leading to functional impairment. Moreover, the long-term storage stability of the nanosensor may not fall under the purview of this research due to the emphasis on immediate functionality and operational effectiveness.
Overall, the results emphasize the robustness of the NHS-AuNP nanosensor in complex environments. Even with proteins, salts, and other potential interferents in the meat extract, the nanosensor maintained high precision and delivered consistent results. These attributes make it a promising tool for applications in food safety and quality control, particularly for detecting biogenic amines in perishable goods.
3.8 Past literature
Comparing the performance of past literature with this work, it can be inferred in Table 3 how NHS-AuNP-MES can compete in terms of its sensitivity to spermine. Its color profile shows potential for development into ready-to-use nanosensors. It exhibited an easily distinguishable color shift from red to blue, whereas others simply shifted from red to colorless. The inclusion of the MUDA spacer in our system significantly enhanced its resistance to handling errors, ensuring extreme stability by providing additional steric hindrance and protection against external influences. We have successfully proposed an alternative functionalization method that is just as promising as using DNA and aptamers, which can often be more challenging to obtain.| Year published | Au-based nanosensor | Technique used | Linear range | LOD | Ref. |
|---|---|---|---|---|---|
| 2013 | Single-stranded | Colorimetric | 13.9–59.2 ng mL−1 | 13.9 ng mL−1 | 46 |
| DNA-AuNPs | (0.0687 μM) | ||||
| 2017 | DNA/aptasensor | Colorimetric | 1–5 μM | 15.25 nM | 47 |
| Capped-AuNPs | (0.01525 μM) | ||||
| 2017 | Calf thymus | Colorimetric | 0.1–2.0 μM | 11.6 nM | 48 |
| DNA-AuNPs | (0.0116 μM) | ||||
| 2017 | Colorimetric | 0.0001–50 μM | 136 pM | 49 | |
| Tyrosine | (0.136 × 103 μM) | ||||
| Protected-AuNPs | Fluorometric | And 10–130 nM | 6.2 nM | ||
| (0.0062 μM) | |||||
| 2018 | Silver–gold/silver | Fluorescence | 2.6–8.0 μM | 0.87 nM | 50 |
| Chloride nanozymes | (0.00087 μM) | ||||
| 2019 | Supramolecular | Colorimetric | 0.1–4.0 μM | 0.034 μM | 51 |
| Pillar[5]arene-AuNPs | |||||
| 2022 | Gluconate-AuNPs | Colorimetric | 0.5–3.5 μM | 0.2511 μM | 52 |
| 2024 | NHS-AuNPs-MES | Colorimetric | 1–4.0 μM | 0.77 μM | This work |
Conclusions
This study shows that using AuNP functionalized with MUDA and conjugated with a succinimidyl/NHS ester is highly effective in detecting spermine. This is due to NHS-esters' interaction with primary amines, emphasizing specificity and concentration-dependent response. TEM imaging shows monodispersed AuNPs with spherical morphology that slightly increased in size with MUDA addition. UV-vis, ATR-FTIR, hydrodynamic size, and zeta potential characterizations suggest successful MUDA ligand exchange and subsequent activation via the EDC/NHS pathway that converts MUDA carboxyl ends to NHS-ester moieties. After a reaction time of 5 min, compared to MUDA-AuNP, NHS-AuNP (the one with activated MUDA) has a better aggregation and visual discrimination at 10 μM of spermine. A linear calibration curve can be obtained by recording the nanosensors' degree of aggregation (Abs600/Abs528) against increasing spermine concentration. Among the three (MUDA-AuNP, NHS-AuNP, and NHS-AuNP-MES) sensors, the linear calibration curve of NHS-AuNP-MES had the lowest range (1.0–4.0 μM) with a detection limit of 0.77 μM in 0.1 M MES buffer (pH = 5.5). The sensor's resistance to interfering analytes found in spoiled meat extracts was also verified, highlighting its potential for practical applications, especially in testing meat products for spoilage.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
The manuscript was written through the contributions of all authors such that conceptualization, investigation, validation, data curation – T. L. G., J. S. F., E. P. K., J. P. O. and F. S. L.; methodology, validation, data curation, and analysis – T. L. G., J. P. O., and F. S. L.; investigation and data curation: T. L. G., F. S. L., and A. C. A; data interpretation and formal analysis: T. L. G., J. P. O., F. S. L. and A. C. A.; writing – original draft preparation and visualization – T. L. G.; writing – review and editing, G. G. D., N. L. S., F. S. L., R. Y. C., A. C. A. All authors have given consent for the publication of this manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project is supported by the Commission on Higher Education – Leading the Advancement of Knowledge in Agriculture and Sciences (CHED-LAKAS) of the Philippines with project code number LAKAS 2022-003. Most of the technical work was done in the Center for Nanoscience and Technology for Research and Entrepreneurship (CeNTRE) and the Chemistry Department of Caraga State University. The Research Center for Energy Efficient Materials (RCEEM) of Mindanao State University (MSU) – Iligan Institute of Technology, the Research on Environment and Nanotechnology Laboratory of MSU – Naawan, and the Central Instrumentation Facility, De La Salle University are also acknowledged.References
- T. King, M. Cole, J. M. Farber, G. Eisenbrand, D. Zabaras, E. M. Fox and J. P. Hill, Trends Food Sci. Technol., 2017, 68, 160–175 CrossRef CAS.
- J. V. Andrei, M. C. Drăgoi, I. G. Rădulescu, V. Erokhin, A. Turek Rahoveanu, J. Subić, M. C. Dragoi, N. Staurskaya and D. Dusmanescu, SHS Web of Conferences, 2021, vol. 95 Search PubMed.
- D. Warmate and B. A. Onarinde, Int. J. Food Microbiol., 2023, 398, 110240 CrossRef CAS.
- B. Ray and A. Bhunia, Fundamental food microbiology, CRC press, 2007 Search PubMed.
- K. Gul, P. Singh and A. A. Wani, in Safety of Meat and Poultry, 2016, pp. 63–77 Search PubMed.
- C. I. E. Del Bosque, B. A. Altmann, M. Ciulu, I. Halle, S. Jansen, T. Nolte, S. Weigend and D. Mörlein, Foods, 2020, 9, 1052 CrossRef CAS.
- M. S. Arshad, M. Sohaib, R. S. Ahmad, M. T. Nadeem, A. Imran, M. U. Arshad, J. H. Kwon and Z. Amjad, Lipids Health Dis., 2018, 17, 223 CrossRef CAS PubMed.
- V. S. Kartika, M. Rivai and D. Purwanto, 2018 International Conference on Information and Communications Technology (ICOIACT), pp. 418–423 Search PubMed.
- P. K. Nanda, D. Bhattacharya, J. K. Das, S. Bandyopadhyay, D. Ekhlas, J. M. Lorenzo, P. Dandapat, L. Alessandroni, A. K. Das and M. Gagaoua, Chemosensors, 2022, 10, 322 CrossRef.
- M. Schirone, L. Esposito, F. D'Onofrio, P. Visciano, M. Martuscelli, D. Mastrocola and A. Paparella, Foods, 2022, 11, 788 CrossRef.
- C. A. L. de la Torre and C. A. Conte-Junior, in Detection of Biogenic Amines: Quality and Toxicity Indicators in Food of Animal Origin, 2018, pp. 225–257 Search PubMed.
- C. A. Lázaro, C. A. Conte-Júnior, A. C. Canto, M. L. G. Monteiro, B. Costa-Lima, A. G. d. Cruz, E. T. Mársico and R. M. Franco, LWT--Food Sci. Technol., 2015, 60, 15–21 CrossRef.
- D. Doeun, H.-S. Shin and M.-S. Chung, Appl. Biol. Chem., 2016, 59, 51–58 CrossRef.
- F. Özogul and O. Özden, Food Bioprocess Technol., 2011, 6, 1343–1349 CrossRef.
- M. Papageorgiou, D. Lambropoulou, C. Morrison, E. Kłodzińska, J. Namieśnik and J. Płotka-Wasylka, TrAC, Trends Anal. Chem., 2018, 98, 128–142 CrossRef.
- I. Guerrero Legarreta and A. N. A. Chavez-Gallardo, J. Muscle Foods, 2007, 2, 263–278 CrossRef.
- D. Rosinská and J. Lehotay, J. Liq. Chromatogr. Relat. Technol., 2014, 37, 609–619 CrossRef.
- M. M. Calabretta, D. Gregucci, R. Desiderio and E. Michelini, Biosensors, 2023, 13, 126 CrossRef.
- A. Lapenna, M. Dell'Aglio, G. Palazzo and A. Mallardi, Colloids Surf., A, 2020, 600, 124903 CrossRef.
- W. A. A. Rosero, A. B. Barbezan, C. D. De Souza and M. E. C. M. Rostelato, Pharmaceutics, 2024, 16, 255 CrossRef.
- F. Zhang and J. Liu, Analysis Sensing, 2020, 1, 30–43 CrossRef.
- Z. Sadiq, S. H. Safiabadi Tali, H. Hajimiri, M. Al-Kassawneh and S. Jahanshahi-Anbuhi, Crit. Rev. Anal. Chem., 2023, 1–36 Search PubMed.
- X. Ling, Y. Liu, D. Zhu, W. An, J. Geng, L. Li, C. Yu and J.-F. Wei, Anal. Methods, 2022, 14, 2698–2702 RSC.
- T. C. Tsai, C. W. Liu, Y. C. Wu, N. A. P. Ondevilla, M. Osawa and H. C. Chang, Colloids Surf., B, 2019, 175, 300–305 CrossRef PubMed.
- A. S. Dileseigres, Y. Prado and O. Pluchery, Nanomaterials, 2022, 12, 292 CrossRef PubMed.
- R. Classen, E. Pouokam, M. Wickleder, M. Diener and A. Mattern, R. Soc. Open Sci., 2022, 9, 220244 CrossRef.
- L. Du, Y. Lao, Y. Sasaki, X. Lyu, P. Gao, S. Wu, T. Minami and Y. Liu, RSC Adv., 2022, 12, 6803–6810 RSC.
- N. R. Nirala, P. S. Saxena and A. Srivastava, Spectrochim. Acta, Part A, 2018, 190, 506–512 CrossRef.
- C.-H. Li, M.-H. Chan, Y.-C. Chang and M. Hsiao, Molecules, 2023, 28, 364 CrossRef.
- Z. Ruiqi, J. Song, Q. Ma, Y. Zhou, J. Yang, S. Shuang and C. Dong, Anal. Methods, 2016, 8, 7232–7236 RSC.
- A. Mattern, F. Machka, M. S. Wickleder, O. S. Ilyaskina, M. Bunemann, M. Diener and E. Pouokam, Org. Biomol. Chem., 2018, 16, 6680–6687 RSC.
- L. Yang, Y. Liao and Z. Zhou, Opt. Mater., 2023, 140, 113867 CrossRef.
- L. Meng, J.-H. Yin, Y. Yuan and N. Xu, RSC Adv., 2018, 8, 9327–9333 RSC.
- J. Kimling, M. Maier, B. Okenve, V. Kotaidis, H. Ballot and A. Plech, J. Phys. Chem. B, 2006, 110, 15700–15707 CrossRef.
- F. Gasiorek, E. Pouokam, M. Diener, S. Schlecht and M. S. Wickleder, Org. Biomol. Chem., 2015, 13, 9984–9992 RSC.
- S. M. Ansar, S. Chakraborty and C. L. Kitchens, Nanomaterials, 2018, 8, 339 CrossRef PubMed.
- G. T. Hermanson, Bioconjugate techniques, Academic press, 2013 Search PubMed.
- Q. Liu, N. Li, M. Wang, L. Wang and X. Su, Anal. Chim. Acta, 2018, 1013, 71–78 CrossRef CAS PubMed.
- M. Timko, P. Kopčanský, M. Antalik, M. Simsikova, E. Valusova, M. Molcan and J. Kováč, Acta Phys. Pol. A, 2012, 121, 1321–1323 CrossRef.
- J. N. Ngunjiri, D. J. Stark, T. Tian, K. A. Briggman and J. C. Garno, Anal. Bioanal. Chem., 2012, 405, 1985–1993 CrossRef.
- B. L. Frey and R. M. Corn, Anal. Chem., 1996, 68, 3187–3193 CrossRef.
- A. Rahdar, N. Amini, F. Askari and Md. A. B. H. Susan, Journal of Nanoanalysis, 2019, 6, 80–89 Search PubMed.
- E. Koushki, RSC Adv., 2021, 11, 23390–23399 RSC.
- I. Isnaeni, I. Irmaniar and Y. Herbani, J. Phys.: Conf. Ser., 2017, 817, 012039 CrossRef.
- S. K. Menon, B. R. Mistry, K. V. Joshi, P. G. Sutariya and R. V. Patel, Spectrochim. Acta, Part A, 2012, 94, 235–242 CrossRef PubMed.
- Z. D. Liu, H. Y. Zhu, H. X. Zhao and C. Z. Huang, Talanta, 2013, 106, 255–260 CrossRef PubMed.
- T. H. Tsoi, Y. J. Gu, W. S. Lo, W. T. Wong, W. T. Wong, C. F. Ng, C. S. Lee and K. L. Wong, ChemPlusChem, 2017, 82, 802–809 CrossRef.
- J. Wang, Z. L. Wu, H. Z. Zhang, Y. F. Li and C. Z. Huang, Talanta, 2017, 167, 193–200 CrossRef.
- K. A. Rawat, J. R. Bhamore, R. K. Singhal and S. K. Kailasa, Biosens. Bioelectron., 2017, 88, 71–77 CrossRef PubMed.
- P.-C. Kuo, C.-W. Lien, J.-Y. Mao, B. Unnikrishnan, H.-T. Chang, H.-J. Lin and C.-C. Huang, Anal. Chim. Acta, 2018, 1009, 89–97 CrossRef PubMed.
- X. Tan, X. Liu, W. Zeng, Z. Zhang, T. Huang, L. Yu and G. Zhao, Spectrochim. Acta, Part A, 2019, 221, 117176 CrossRef CAS.
- S. A. Khan and T. K. Misra, Colloids Surf., A, 2022, 648, 129146 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sd00320a |
| This journal is © The Royal Society of Chemistry 2025 |