Open Access Article
Open Access ArticleA modular scaffold for cellularly-retained fluorogenic probes for sensitive cell-resolved bioactivity imaging
Philipp
Mauker
 ab,
Lucas
Dessen-Weissenhorn
b,
Carmen
Zecha
b,
Nynke A.
Vepřek
ab,
Lucas
Dessen-Weissenhorn
b,
Carmen
Zecha
b,
Nynke A.
Vepřek
 b,
Julia I.
Brandmeier
b,
Daniela
Beckmann
cd,
Annabel
Kitowski
b,
Tobias
Kernmayr
b,
Julia
Thorn-Seshold
b,
Julia I.
Brandmeier
b,
Daniela
Beckmann
cd,
Annabel
Kitowski
b,
Tobias
Kernmayr
b,
Julia
Thorn-Seshold
 ae,
Martin
Kerschensteiner
ae,
Martin
Kerschensteiner
 cdf and
Oliver
Thorn-Seshold
cdf and
Oliver
Thorn-Seshold
 *a
*a
aFaculty of Chemistry and Food Chemistry, Dresden University of Technology, Dresden 01069, Germany. E-mail: oliver.thorn-seshold@tu-dresden.de
bDepartment of Pharmacy, LMU Munich, Munich 81377, Germany
cInstitute of Clinical Neuroimmunology, LMU University Hospital, LMU Munich, Munich 81377, Germany
dBiomedical Center (BMC), Faculty of Medicine, LMU Munich, Martinsried 82152, Germany
eFaculty of Medicine, University Hospital Dresden, Dresden University of Technology, Dresden 01307, Germany
fMunich Cluster for Systems Neurology (SyNergy), Munich 81377, Germany
First published on 16th October 2025
Abstract
Here, we develop a general design for high-quality fluorogenic activity probes to quantify biochemical processes within live cells, via the release of a fully cell-retained, bright fluorescent soluble product upon reaction. Live cell probes must be membrane-permeable to access intracellular biochemistry, but often that means their fluorophore products are similarly permeable resulting in rapid signal loss from the activating cell, which limits their cell-by-cell resolution as well as their sensitivity for quantifying low-turnover processes. Current strategies to retain fluorescent products within cells usually disrupt native biology e.g. by non-specific alkylation or solid precipitation. Here, scanning charge- and polarity-based approaches to swap from permeable to cell-retained states, we developed a bright fluorogenic rhodol-based platform, Trappable Green (TraG), balancing all key requirements for signal integration (rapid probe entry, but effective product retention, across many cell lines) and being modular so it can be adapted to quantify many biochemical target types (examples shown here include probes for GSH, TrxR, and H2O2). The simple and rugged TraG scaffold can now permit straightforward implementation in a range of cell-retained enzyme activity probes, which will enable more accurate cell-resolved imaging as well as higher-sensitivity integration of low-turnover processes, without the drawbacks of alkylation or precipitation-based strategies.
Introduction
Imaging and quantifying biological activity are key challenges in basic and applied research. Focusing on enzyme activity rather than mRNA or protein levels takes post-translational regulation mechanisms into account (PTMs, compartmentalisation, chaperoning), and allows researchers to correctly interpret biochemistry in action. Fluorogenic probes are, ideally, non-fluorescent probes that only generate a fluorescent product after activation by their specific biochemical target or enzyme. They have become crucial tools for sensitive and non-invasive bioactivity imaging, especially in lysates: with probes for peptidases, esterases, phosphatases, glycosidases, and oxidoreductases,1–7 or reactive analytes such as hydrogen peroxide and hydrogen sulfide,8,9 in widespread use.Sensitive, cell-resolved detection is crucial for longitudinally visualising bioactivity during assay time courses, and for understanding the heterogeneity of cell populations. However, fluorogenic probes often encounter a major problem in live cells and tissues: the signal of their fluorescent products becomes diffuse or is lost over time. Apolar, membrane-permeable fluorophores such as coumarins can rapidly exit the cell across the plasma membrane by passive diffusion,10 while negatively charged fluorophores such as fluoresceins are instead excreted from cells by active transport.11,12 This post-activation signal loss sabotages cell-resolved activity imaging, and lowers the sensitivity and reliability of signal quantification (higher and time-dependent background signal). Moreover, the rate of signal loss sets a lower limit on the enzyme activity or analyte concentration that can be detected. For in vivo work, where probe dosage must be low, or for imaging low-turnover processes, or for situations demanding high sensitivity and quantitative reliability, building up and retaining the product signal inside the activating cell in the long term is a crucial challenge for probe design.
The importance of retaining the fluorescence signal within cells has driven three probe designs for signal trapping (full discussion in Fig. S3). (1) Charge/polarity-based product impermeabilization usually suppresses passive membrane transit with ionic motifs, e.g. carboxylates,13 phosphonates,14 sulfonates,15 or tetraalkylammoniums.16,17 To deliver probes into the cell in the first place, intracellularly-cleaved lipophilic masking groups13–15,18 (e.g. carboxylates masked as acetoxymethyl esters,9,13,19,20 or amines as carbamates21), endocytosis (e.g. by cell-penetrating peptides),22 or transporter-mediated uptake16 are often used. None of these approaches has been developed to a state of general applicability; however, typical issues include activation triggers that are not modular; product fluorophore dimness; slow cellular uptake; release of reactive side products; and/or unwanted compartmentalisation. (2) Water-insoluble solid-state fluorophores can be released as reaction products that precipitate as fluorescent cellularly-trapped crystals (e.g. the ELF-97 probe, releasing an HPQ fluorophore).23–25 Yet, crystal deposits cause inflammatory responses and are cytotoxic,26 perturbing biology or preventing longitudinal imaging; moreover, these probes are insensitive at low turnover since the precipitation threshold must be crossed before any signal is seen; also, these probes are rarely well-soluble (mirroring the product insolubility). (3) Products that alkylate cell-impermeable biomolecules can be released: a cell retention strategy pioneered by Urano with e.g. SPiDER probes (non-reactive benzylfluoride probes are enzymatically triggered to give electrophilic quinone methide products that rapidly react with proteins or GSH, enabling long-term signal retention).6,27–30 However, they can also alkylate their target enzyme,30 induce electrophile stress responses, or accumulate toxic effects,28 especially in high-turnover cells.
There are many requirements that must be balanced to deliver a good probe for high-sensitivity enzyme imaging in live cells. Fig. 1 shows the major needs, e.g. (a) good aqueous solubility of the probe, for reproducible handling, high bioavailability, and to avoid aggregation or sequestration; (b) probe robustness, i.e. no occurrence of non-specific (background) product release; (c) reliably effective cell entry across cell lines (e.g. by passive diffusion); (d) linear fluorescence signal (e.g. targets activate fluorescence by just one reaction site per probe molecule); (e) high signal turn-on ratio: the probe is very dark under typical imaging conditions, and the released product is very bright; (f) effective cell retention of the product, allowing long-term signal integration; (g) the fluorophore and probe byproducts must not risk perturbing biology in the long term (unlike precipitating or alkylating probes). Ideally, the probe design would also be modular, i.e. easily chemically adaptable to image enzymes with various reactivity classes. Considering that none of the prior strategies meets all eight of these requirements (discussion in Fig. S3), we set out to develop a probe design that does. We chose O-unmasking of a phenolic fluorophore for activation, a reaction that is applicable for many types of molecular imaging. We now outline the development of a generalised probe design for high-sensitivity enzyme imaging with a cell-retained product that meets all these requirements.
Results and discussion
Design strategy
Facing an eight-factor optimization problem, the challenge was to identify key needs for each factor, choose “good enough” solutions for each (where these already exist) that do not push the other factors out of scope, and focus on the balance of properties that is needed for successful performance overall. For example, ensuring both aqueous solubility and membrane permeability requires a probe with balanced polarity, and sparing use of polar groups e.g. carboxylates or amines. The choice of fluorophore influences the efficiency of fluorescence suppression in its masked probe form (e.g. up to 100% suppression for spirocyclised xanthenes), the signal activation linearity (e.g. 1 or 2 activating reaction sites), and the brightness of the released fluorophore (functionalisation). We prioritised combining known and new strategies to reach a modularly applicable platform that delivers a well-performing probe independent of the chosen biochemical target, and selected the permeable-probe-to-retained-product transition as the key chemical challenge that might need chemical tuning.Design 1: lipidated charged fluoresceins (Fig. S4–S7)
We first focused on using synthetically accessible O-masked fluorogenic probe designs to test which physicochemical features would ensure good cell entry and cell retention and left the other requirements for later. Fluorescein spirolactone O-alkylated O′-esters are a convenient fluorogenic test system with only one reaction site,31 and we had previously noted that some monosulfonated fluorescein diesters were surprisingly capable of cell entry despite their charge.32 We now took these known systems and measured their cell retention after washing, under standard conditions, which was promising for the diester (i2-FS, which has two activation sites which is undesired due to non-linear signal response upon de-acylations (only 10% signal upon the first ester cleavage); Fig. S4) though poor for the monoester (iPS-F, one activation site, desirable linear response). Following the notion that medium-length lipids enhance the cell uptake and retention of natural products,33 we next synthesised a set of more lipid-like O-alkylated sulfonated fluorescein monoester probes (iC4-FS–iC10-FS, for C4–C10 alkyl; for all structures, see Fig. S1). All lipidated designs gave good product retention after washing (Fig. S5bd), but the absolute cellular signal was low, which we attributed to poor cellular uptake due to the sulfonate (Fig. S5c; their signal distribution also varied from uniform C4-FS to membrane-only C10-FS; Fig. S5e). Deleting the sulfonate to raise permeability was a failure (iC4-F: no signal seen in cells (Fig. S6e) or in cell-free esterase assays (Fig. S25)), illustrating the importance of solubility for bioavailability. Using a reversibly ionisable carboxylate in iC4-F![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) (pKa ≈ 4.3) instead of the sulfonate gave 20-fold higher cellular signal intensity than iC4-FS, yet kept its good post-wash retention and uniform signal distribution (Fig. S6). However, we had noted significant cellular distress (rounding and blebbing) with all FS- and FC-ester probes so far. To allow testing the probes in more complete cell media (DMEM with FCS instead of HBSS), we switched the trigger group from an O-isobutyrate ester to a hydrolytically robust, reductively cleavable O-carbamate (GL-C4-FC; Fig. S24 and S26). We then found that the pairing of a strong signal with cell blebbing had been caused by the combination of the lipidated FS/FC probes with the salt buffer solution that had been needed to avoid isobutyrate ester hydrolysis. In more complete buffer (DMEM), cell morphology stayed healthy, and no signal was seen (Fig. S6f and S7). We imagine that the membrane stress of the amphipathic FS/FC probes, plus the lack of nutrients in salt buffer, disrupted membranes enough for the charged probes to enter cells34 (details in Fig. S6), indicating that amphipathic probes are unsuitable for non-invasive cell imaging.
(pKa ≈ 4.3) instead of the sulfonate gave 20-fold higher cellular signal intensity than iC4-FS, yet kept its good post-wash retention and uniform signal distribution (Fig. S6). However, we had noted significant cellular distress (rounding and blebbing) with all FS- and FC-ester probes so far. To allow testing the probes in more complete cell media (DMEM with FCS instead of HBSS), we switched the trigger group from an O-isobutyrate ester to a hydrolytically robust, reductively cleavable O-carbamate (GL-C4-FC; Fig. S24 and S26). We then found that the pairing of a strong signal with cell blebbing had been caused by the combination of the lipidated FS/FC probes with the salt buffer solution that had been needed to avoid isobutyrate ester hydrolysis. In more complete buffer (DMEM), cell morphology stayed healthy, and no signal was seen (Fig. S6f and S7). We imagine that the membrane stress of the amphipathic FS/FC probes, plus the lack of nutrients in salt buffer, disrupted membranes enough for the charged probes to enter cells34 (details in Fig. S6), indicating that amphipathic probes are unsuitable for non-invasive cell imaging.
Design 2: charge-balanced rhodol probes (Fig. 2a)
From our experience with the FS/FC lipidated fluorescein probes, we concluded that amphipathic anionic designs are unsuited for good cell entry and retention. We also prioritised probe solubility, and hydrolytically stable O-carbamate probes that could be studied in complete cell culture media. We thus switched to rhodol scaffolds, since rhodols can be O-acylated to give nonfluorescent (fully spirolactone) probes, but can be much brighter than O′-alkyl fluoresceins (limit: ∼7 × 103 L mol−1 cm−1, Table S1), and their fluorescence is more photostable and is constant over the pH range 4–10.35 We then synthesised a set of six reductively-activated36 “GL-Rho” probes: with (1) either an apolar piperidine (Rho) or a basic piperazine (Rho-A) as an N-substituent; and (2) optionally, a 6-carboxylate (C) or its masked, membrane-permeable acetoxymethyl (AM) ester (Cm) (Fig. 2a). The rhodol fluorophores were accessed from the fluorescein ditriflates by one-sided Buchwald–Hartwig coupling, followed by triflate hydrolysis with LiOH or TBAF. These were transformed into fluorogenic probes by a one-pot sequence, converting their phenolic –OH to a pentafluorophenyl carbonate, then acylating the GSH-labile GL disulfide motif (later deprotecting and optionally masking the 6-carboxylates).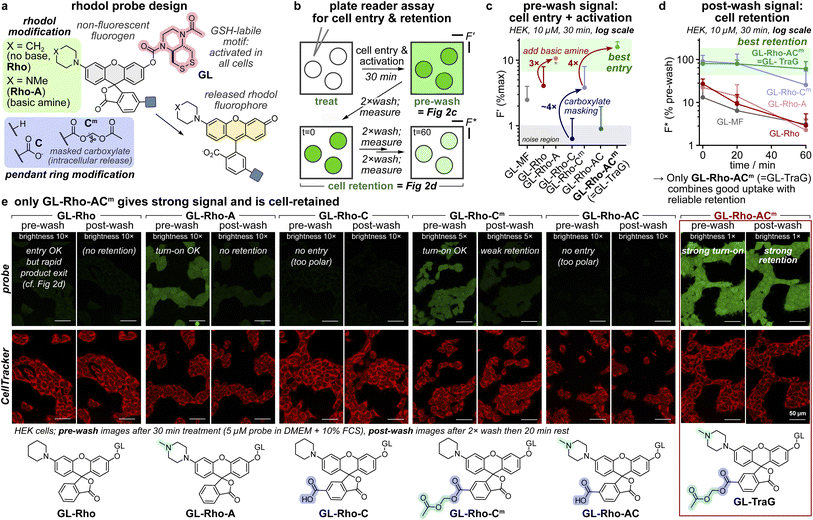 | ||
| Fig. 2 The rhodol probe design Rho-ACm (=Trappable Green: TraG) delivers strong intracellular signal activation and retention. (a) Probe panel overview. (b) Plate reader assay for cell entry, activation, and retention. (c) Entry and activation in HEK293T cells (10 μM probe, DMEM with 10% FCS, 30 min treatment; F′ is fluorescence as a % of full activation of the whole well; error bars: SD; biological replicates normalised to GL-Rho-A (marked with an asterisk); n = 3; error bars: SD; the methyl fluorescein probe GL-MF (ref. 36) is a non-rhodol benchmark for entry and retention, see Fig. S1). (d) Cell retention (conditions as in c; F* is fluorescence as a % of pre-wash value; error bars = SD). (e) Cell turn-on and retention assayed by confocal microscopy (conditions as in c except with a 5 μM probe; scale bars: 50 μm; full data in Fig. S9a). | ||
The optical properties of the rhodol products varied somewhat, with excitation maxima at 490–530 nm, Stokes shifts of ∼25 nm (emission maxima at 515–560 nm), and extinction coefficients of 3–6 × 104 L mol−1 cm−1. Fluorescence quantum yields varied from 4–64%, with piperazinyl products H-Rho-A and H-Rho-AC being the brightest, as expected37 (∼3 × 104 L mol−1 cm−1; Fig. S21 and Table S1). Importantly for high sensitivity, all probes were non-fluorescent, with outstanding probe/product signal turn-on ratios of up to ∼550 (Fig. S21); the piperazinyl probes were particularly efficiently activated by their target GSH (Fig. S26); and the carbamates of all probes were hydrolytically stable for hours in FCS-supplemented DMEM, for long term cell experimentation (Fig. S24).
Probe selection: disulfide reduction sensing (Fig. 2b–e)
We then tested cell entry, activation, and signal retention in HEK cells (Fig. 2b). Cell entry and activation was moderate for piperidinyl GL-Rho, but 3× higher for the basic piperazinyl (GL-Rho-A; Fig. 2c). Adding a carboxylate blocked cellular uptake (GL-Rho-C/GL-Rho-AC); masking it as an acetoxymethyl ester (GL-Rho-Cm/GL-Rho-ACm) let the probes reach the same signal as that of GL-Rho/GL-Rho-A. Importantly, probe treatment does not impair cell morphology, and uptake occurs homogeneously in healthy cells with uniform cellular distribution of the product signal (Fig. S9a and 10). We then subjected cells to three cycles of “wash, measure, and wait”, to monitor intracellular signal retention. The rhodol product from GL-Rho/GL-Rho-A leaked out rapidly from cells, but an added carboxylate (GL-Rho-Cm/GL-Rho-ACm) greatly enhanced cell retention, particularly for GL-Rho-ACm where cells stayed bright despite three medium exchanges over 1 h (Fig. 2d). We used confocal microscopy to complement these plate reader assays. GL-Rho-A, GL-Rho-Cm, and GL-Rho-ACm indeed show cell entry and activation, while GL-Rho-C and GL-Rho-AC do not (Fig. 2e). Only GL-Rho-ACm had strong post-wash signal retention (Fig. 2e and S9ab) and gave good performance in HeLa, MEF, and A549 cell lines (Fig. S11, strong retention in HeLa cells, weaker in A549 cells).The Rho-ACm scaffold features fast cell entry and signal generation, plus good post-wash intracellular signal retention, by combining a basic amine with a masked, intracellularly-revealed carboxylate. The combination of high fluorogenicity with good cell retention allows sensitive cell-resolved imaging either without washing, or with washing (even after a significant delay). Its aqueous solubility avoids aggregation effects; the full spirocyclisation of the probe state (before reaction) plus its biochemical robustness allow zero-background imaging; and with the high brightness of the released fluorophore, its signal turn-on ratio is strong (170×). The probe rapidly enters different cell lines where its phenolic single activation site is efficiently activated and provides linear signal turn-on for reliable signal quantification to give a uniform cellular signal. Crucially, neither the probe nor the fluorophore causes apparent cellular harm (and toxic crystal formation or non-specific bioconjugation are avoided), supporting that the data acquired during longitudinal imaging or enzyme activity integration can be reliably interpreted. We thus considered that this design combines all eight desirable design features (see Introduction) within one probe scaffold. Since the Rho-ACm scaffold showed the best performance, we renamed it  (TraG), and to test the versatility of TraG as a modular platform, we now evaluated two additional types of TraG activity probes.
(TraG), and to test the versatility of TraG as a modular platform, we now evaluated two additional types of TraG activity probes.
Modularity test 2: hydrogen peroxide sensing (Fig. 3)
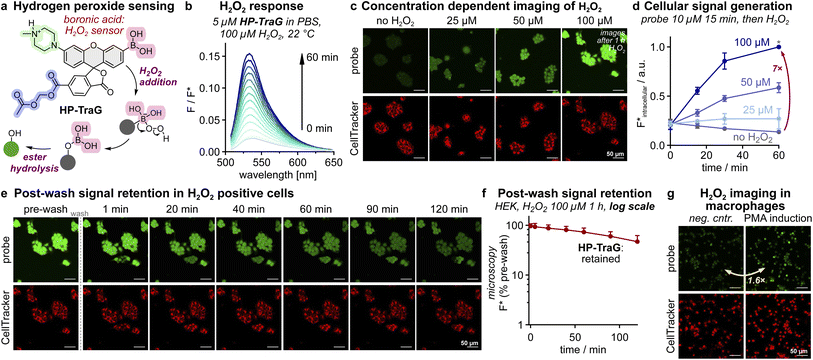
Hydrogen peroxide (H2O2) is a major physiological messenger with baseline levels that fuel cell signalling and metabolic function,38 but which can also be created as an unwanted metabolic byproduct at harmful levels that have been correlated to neurodegeneration, cancer, or autoimmune disorders.39–41 Sensitive and linearly-responsive tools are needed to resolve and study its multiple roles. The most common small molecule probes for sensing H2O2 exposure use arylboronic acids that H2O2 converts to phenols.8 Signal integration is crucial for sensitively detecting low H2O2 concentrations, so cell retained probes (e.g. SPiDER: intracellular quinone-methide trapping) have been utilised despite their moderate cell-toxicity.19,28,42We hoped that a TraG-based design could deliver a more biocompatible cell-retained H2O2 sensor (Fig. 3a), and considered that a boronate's oxidation-hydrolysis mechanism (instead of the previous carbamate cyclisation mechanism) would be a good test of the modularity of the TraG platform. As the common pinacol boronate diester was hydrolytically unstable during purification (as reported elsewhere43), we applied the probe as a free boronic acid (membrane permeable, pKa ≈ 8–9 (ref. 44)). This HP-TraG probe gave linear signal generation with H2O2 (Fig. 3b), with up to 48-fold turn-on (Fig. S22). Loading it into HEK cells (15 min), then washing and extracellularly administering 25–100 μM H2O2 gave H2O2-dependent intracellular fluorescence signals with a high turn-on index (up to a 7-fold increase, Fig. 3c and d) that were cell-retained for >2 h after washing off the extracellular medium (Fig. 3e and f). Finally, we used HP-TraG for imaging endogenous H2O2 in Hoxb8-derived macrophages45 after activation with phorbol 12-myristate 13-acetate (PMA).46 The HP-TraG signal increases by 60% upon PMA treatment, i.e. sensitively detecting both the low endogenous baseline and the slightly increased H2O2 concentrations upon activation (1–4 μM in macrophages47), again with strong post-wash signal retention (Fig. 3g and S14). Thus, the TraG cell-retained fluorogenic design adds a useful new hydrogen peroxide sensor to the toolbox of chemical biology that gives strong performance (rapid, H2O2-dependent intracellular signal) while overcoming the drawbacks of cell-reactive quinone methides as trapping agents.
Modularity test 3: TrxR enzyme imaging (Fig. 4)
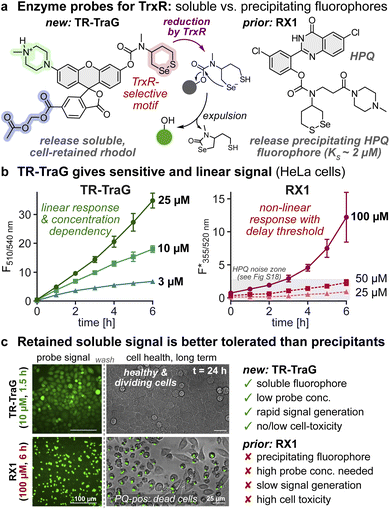
Mammalian thioredoxin reductase (TrxR) is a key enzyme that uses NADPH to reduce thioredoxins, which drive hundreds of redox reactions involved in metabolism, protein folding, and signaling.48,49 TrxR is also one of just 25 selenoproteins in the human proteome; its selenium is needed so that its activity is resilient against biochemical damage.50 TrxR is a difficult target for molecular imaging due to its low expression level (ca. ≤20 nM) vs. high levels of chemically similar thiol off-targets (>10 μM).51 Only activity imaging can map TrxR function, since its activity is decoupled from mRNA levels (Se incorporation is regulated post-transcriptionally), and antibodies do not distinguish non-functional or non-Se forms. The first and only TrxR-selective probe for live cell activity imaging, RX1, was published in 2022,52 and is used for redox biology studies and high-throughput screening.53 RX1's target specificity stems from its cyclic selenenylsulfide, a substrate that is selectively reduced by TrxR then cyclises to release its phenolic cargo HPQ (Fig. 4a). The precipitating and thus cellularly retained solid-state fluorophore HPQ was chosen for signal accumulation to overcome low TrxR levels. However, high probe dosage and long incubation times were needed to surpass the precipitation threshold (KS), and the crystalline HPQ precipitates that generate signal also stimulate inflammatory responses and are toxic to cells. A soluble cell-retained fluorophore product could solve both drawbacks, giving a more biocompatible probe (lower dosage that is less cellularly damaging but still a quantifiable signal) that is also faster to quantify (since KS need no longer be overcome). Thus, we patched RX1's selenenylsulfide onto our rhodol scaffold, hoping the resulting probe TR-TraG would keep TrxR selectivity while accessing the advantages of cell-retained soluble fluorophores.In cell-free experiments, TR-TraG was activated by even 20 nM TrxR1 (vicinal selenolthiol); cell-free selectivity was decent over vicinal dithiols (resisting thioredoxin 1 up to 300 nM, though resistance to glutaredoxin 1 was lower), and outstanding over monothiol GSH (1000 mol eq. of GSH reach only ∼15% activation after 4 h, i.e. the level reached by 0.002 mol eq. of TrxR after 0.5 h; Fig. S15). Pleasingly, in cellular assays, the TR-TraG signal mainly depended on TrxR activity: inhibition with electrophile TRi-1 (ref. 54) (HeLa and A549 cells, Fig. S16) or genetic knockout (in MEF cells,55 Fig. S17) largely controlled its signal. Thus, the selenenylsulfide substrate does set the probe's target-selectivity. We next examined some systematic benefits of the soluble cell-retained design.
A major technical drawback of precipitating fluorophores is their non-linear fluorescence response. In each cell, the released fluorophore concentration has to surpass KS (HPQ: ∼2 μM) before the true signal starts to be observable, whereas soluble fluorophores are theoretically detectable with linear activation response from the first molecule released. Plate reader assays with precipitating fluorophores also suffer from inter-cell variability since turnover must reach ca. 2KS in the majority of cells before the overall signal becomes linear, again, an issue that does not affect soluble probes. Finally, probe quenching in precipitation-based systems is often incomplete: even quenching one fluorescence channel (e.g. HPQ: ESIPT quenching by O-masking) does not suppress all channels (weak long-wavelength tail of normal emission, Fig. S18b, 45 min), whereas xanthene spirocyclisation quenching can be complete. All these advantages were evident when comparing TR-TraG and RX1 in cellular assays. TR-TraG builds up signal linearly from time zero, proportional to its dosage, reaching a usefully quantifiable signal even at ≤1 h at 3 μM (Fig. 4b); while the RX1 signal starts only at >3 h at 100 μM, with no true signal at lower times or doses. Such high RX1 exposure is incompatible with assays in tissues due to limited, variable biodistribution, a limitation that TR-TraG escapes. The rhodol's reproducible signal (Fig. S16) also contrasts to the highly variable signal of RX152 which results from its sensitivity to precipitation effects.
A major biological problem with precipitating fluorophores is that they cause cellular stress and cytotoxicity, which limit or prevent long-term experiments and in vivo assays.26 Typical ways to run high-powered assays, e.g. first imaging and sorting by FACS to stratify cell populations, then further cultivation or parameter testing, are thus impossible. The rhodol TR-TraG instead allowed high-quality cell-resolved imaging at order(s) of magnitude lower probe exposure (10 μM, 90 min) than RX1 (100 μM, 6 h; see Fig. S18), which should already result in far lower biological stress from the probe. Yet, we attribute the major difference to product exposure. Cells treated with TR-TraG were healthy and continued dividing to confluency over 24 h (as did untreated controls), whereas RX1-treated cells that formed PQ crystals were essentially dead (with no escape even after probe removal and culture for 24 h in fresh media; Fig. 4c, S18 and 19). Thus, TraG type probes will likely enable long-term cell tracking by bioactivity, e.g. using FACS to resolve and study cell subpopulations: which at least in the context of TrxR probes is a novel and urgently needed advancement.
Conclusions
We designed a novel, modularly applicable fluorogenic probe scaffold for flexible use in sensitive biochemical activity imaging at low probe doses, which results in the linear generation of a biocompatible, cell-retained, bright fluorescence signal from the released fluorophore H-TraG (=H-Rho-AC) (λex 504 nm, λem 531 nm, ελex = 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1). The combination of a basic amine and an intracellularly unmasked carboxylate on the spirocyclised rhodol precursor allows rapid cell loading and retention of the probe, as well as excellent post-wash retention of the fluorescent open-form rhodol product generated by O-unmasking, across different cell lines (hours in HEK and HeLa cells, up to 1 h in MEF, A549, and Hoxb8-derived macrophages). That the piperazine-rhodol seems to escape significant signal loss by passive diffusion or by active transport contributes to the reliability of signal detection and confidence in signal quantification, even after long “post-wash” incubation times as would be encountered in multi-step cell biology assays (such as cell population sorting) or in situations with wash-in/wash-out (such as ADME kinetics during in vivo enzyme activity imaging). The signal is uniformly distributed across the whole cell with no compartmental accumulation, which is a further advantage for in vivo imaging in 3D environments.
000 M−1 cm−1). The combination of a basic amine and an intracellularly unmasked carboxylate on the spirocyclised rhodol precursor allows rapid cell loading and retention of the probe, as well as excellent post-wash retention of the fluorescent open-form rhodol product generated by O-unmasking, across different cell lines (hours in HEK and HeLa cells, up to 1 h in MEF, A549, and Hoxb8-derived macrophages). That the piperazine-rhodol seems to escape significant signal loss by passive diffusion or by active transport contributes to the reliability of signal detection and confidence in signal quantification, even after long “post-wash” incubation times as would be encountered in multi-step cell biology assays (such as cell population sorting) or in situations with wash-in/wash-out (such as ADME kinetics during in vivo enzyme activity imaging). The signal is uniformly distributed across the whole cell with no compartmental accumulation, which is a further advantage for in vivo imaging in 3D environments.
Previous approaches to ensure cellular signal retention and thus biochemical activity integration have greatly relied on releasing precipitating fluorophore or intracellular alkylator products, which have biological as well as technical disadvantages. Here, we combine known and new strategies to design the modular, broadly applicable TraG probe platform, which distinguishes itself from known retention strategies since it has good performance with respect to all eight features required for live cell probes (including but not only the degree of cell retention) in one scaffold.
We applied our modular scaffold to generate two activity sensor probes showing the superior performance that a soluble fluorophore probe can achieve with a well-tempered cell-entry/exit profile. (1) The hydrogen peroxide sensor HP-TraG senses exogenous and endogenous hydrogen peroxide in cells, adding a novel and milder cell-retained H2O2 probe to the probe toolbox. (2) TR-TraG images the cellular enzyme activity of thioredoxin reductase 1 (TrxR1) and outperforms the current probe RX1 in multiple respects: it more rapidly reaches higher sensitivity quantification, even at vastly lower probe loading, and delivers beneficial linear signal development, as well as allowing long-term cellular viability. Nevertheless, the modular performance of this system, with two probes of rather different overall polarity that are taken up efficiently and perform strongly across multiple cell lines, promises the straightforward design and generation of a variety of other phenol-releasing probes centred on this scaffold (e.g. for O-unmasking by glycosidases or phosphatases, or by peptidases via self-immolative spacers), which can improve the sensitivity and biocompatibility of long-term-compatible cellular and in vivo molecular imaging.
Source of biological samples
Human cervical cancer cell line (HeLa): ACC 57 (German Collection of Microorganisms and Cell Cultures DSMZ); human embryonic kidney cell line (HEK293): ACC 305 (German Collection of Microorganisms and Cell Cultures DSMZ); human lung cancer cell line (A549): CCL-185 (American Type Culture Collection ATCC).Author contributions
P. M. performed synthesis, chemical analysis, enzymatic cell-free studies, cell biology, and coordinated data assembly. L. D.-W., C. Z., N. A. V., D. B. and A. K. performed cell biology. L. D.-W., N. A. V. and D. B. performed confocal microscopy, image analysis and quantification. J. I. B. and T. K. performed synthesis. J. T.-S. and M. K. supervised cell biology and confocal microscopy. P. M. and O. T.-S. designed the concept and experiments. O. T. -S. supervised all other experiments. P. M. and O. T.-S. co-wrote the manuscript with input from all authors.Conflicts of interest
The authors declare no competing financial interests.Abbreviations
| A549 | human lung cancer cell line |
| ADME | absorption, distribution, metabolism, and excretion |
| AM | acetoxymethyl ester |
| DMEM | Dulbecco's modified Eagle's medium (cell culture media) |
| ESIPT | excited state intramolecular proton transfer |
| FACS | fluorescence-activated cell sorting (by flow cytometry) |
| FCS | foetal calf serum |
| Hoxb8 | macrophage precursor cell line |
| HPQ | 2-(2′-hydroxyphenyl)-4(3H)-quinazolinone (fluorophore) |
| GSH | glutathione |
| HEK | human embryonic kidney cell line HEK293T |
| HeLa | human cervical cancer cell line |
| K S | solubility limit |
| MEF | mouse embryonic fibroblast cell line |
| PBS | phosphate buffered saline (buffer) |
| PMA | phorbol 12-myristate 13-acetate |
| RX1 | molecular probe for TrxR1 activity with a cell-retained signal based on HPQ release and precipitation |
| SPiDER | molecular probe scaffold with cell retention of a signal based on enzymatic unfurling of an alkylating ortho-quinone methide |
| TBAF | tetranbutylammonium fluoride |
| TrxR | the mammalian selenoenzyme thioredoxin reductase 1 |
Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: synthesis, analysis, biochemistry, and cell biology, incl. design and performance of lipidated fluorescein probes (Fig. S4–S8), rhodol probes (Fig. S9–S18), photocharacterisations (Fig. S20–S22 and Table S1), and cell-free assays (Fig. S23–S28) (PDF). See DOI: https://doi.org/10.1039/d5sc05388a.Acknowledgements
The authors acknowledge support from the German Research Foundation (DFG; Emmy Noether grant 400324123 to O.T.-S.); the Joachim Herz Foundation (Research Fellowships to P. M. and J. T.-S.); the Studienstiftung des Deutschen Volkes (PhD scholarships to P. M. and D. B.); and the Munich Graduate School of Systemic Neurosciences (D. B.). We thank Lukas Zeisel and Jan Felber (TUDD, LMU) for synthesising the dithiane bioreductive activation triggers for GL- and TR-probes. We thank Markus Conrad (TUM) for the kind gift of the validated TrxR knockout MEF cell line (generation and validation reported in ref. 55). We are grateful to Dr Johannes Morstein (Caltech) for collegial discussions around cellular uptake, retention, and localisation.References
- W. Chyan and R. T. Raines, ACS Chem. Biol., 2018, 13, 1810–1823 CrossRef CAS PubMed.
- A. R. Lippert, G. C. Van de Bittner and C. J. Chang, Acc. Chem. Res., 2011, 44, 793–804 CrossRef CAS PubMed.
- A. E. Albers, B. C. Dickinson, E. W. Miller and C. J. Chang, Bioorg. Med. Chem. Lett., 2008, 18, 5948–5950 CrossRef CAS PubMed.
- T. D. Gruber, C. Krishnamurthy, J. B. Grimm, M. R. Tadross, L. M. Wysocki, Z. J. Gartner and L. D. Lavis, ACS Chem. Biol., 2018, 13, 2888–2896 CrossRef CAS PubMed.
- L. Li, J. Ge, H. Wu, Q.-H. Xu and S. Q. Yao, J. Am. Chem. Soc., 2012, 134, 12157–12167 CrossRef CAS PubMed.
- H. Ito, Y. Kawamata, M. Kamiya, K. Tsuda-Sakurai, S. Tanaka, T. Ueno, T. Komatsu, K. Hanaoka, S. Okabe, M. Miura and Y. Urano, Angew. Chem., Int. Ed., 2018, 57, 15702–15706 CrossRef CAS PubMed.
- L. Tian, Y. Yang, L. M. Wysocki, A. C. Arnold, A. Hu, B. Ravichandran, S. M. Sternson, L. L. Looger and L. D. Lavis, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4756–4761 CrossRef CAS PubMed.
- M. S. Messina, G. Quargnali and C. J. Chang, ACS Bio Med Chem Au, 2022, 2, 548–564 CrossRef CAS PubMed.
- K. G. Fosnacht, M. D. Hammers, M. S. Earp, A. K. Gilbert and M. D. Pluth, Chem.–Asian J., 2022, 17, e202200426 CrossRef CAS PubMed.
- B. G. Rosser, S. P. Powers and G. J. Gores, J. Biol. Chem., 1993, 268, 23593–23600 CrossRef CAS PubMed.
- H. Sun, D. R. Johnson, R. A. Finch, A. C. Sartorelli, D. W. Miller and W. F. Elmquist, Biochem. Biophys. Res. Commun., 2001, 284, 863–869 CrossRef CAS PubMed.
- S. E. Bresler, V. M. Bresler, E. N. Kazbekov, A. A. Nikiforov and N. N. Vasilieva, Biochim. Biophys. Acta, Biomembr., 1979, 550, 110–119 CrossRef CAS PubMed.
- S. Izumi, Y. Urano, K. Hanaoka, T. Terai and T. Nagano, J. Am. Chem. Soc., 2009, 131, 10189–10200 CrossRef CAS PubMed.
- J. L. Turnbull, B. R. Benlian, R. P. Golden and E. W. Miller, J. Am. Chem. Soc., 2021, 143, 6194–6201 CrossRef CAS PubMed.
- S. T. Caldwell, S. N. O'Byrne, C. Wilson, F. Cvetko, M. P. Murphy, J. G. McCarron and R. C. Hartley, Chem. Commun., 2021, 57, 3917–3920 RSC.
- T. Fukuda, S. Yokomizo, S. Casa, H. Monaco, S. Manganiello, H. Wang, X. Lv, A. D. Ulumben, C. Yang, M.-W. Kang, K. Inoue, M. Fukushi, T. Sumi, C. Wang, H. Kang, K. Bao, M. Henary, S. Kashiwagi and H. Soo Choi, Angew. Chem., Int. Ed., 2022, 61, e202117330 CrossRef CAS PubMed.
- S. Jia, E. Y. Lin, E. B. Mobley, I. Lim, L. Guo, S. Kallepu, P. S. Low and E. M. Sletten, Chem, 2023, 9, 3648–3665 CAS.
- L. Rusha and S. C. Miller, Chem. Commun., 2011, 47, 2038–2040 RSC.
- B. C. Dickinson, J. Peltier, D. Stone, D. V. Schaffer and C. J. Chang, Nat. Chem. Biol., 2011, 7, 106–112 CrossRef CAS PubMed.
- H. Ye, L. Sun, Z. Pang, X. Ji, Y. Jiao, X. Tu, H. Huang, X. Tang, Z. Xi and L. Yi, Anal. Chem., 2022, 94, 1733–1741 CrossRef CAS PubMed.
- G. Jiang, X.-F. Lou, S. Zuo, X. Liu, T.-B. Ren, L. Wang, X.-B. Zhang and L. Yuan, Angew. Chem., Int. Ed., 2023, e202218613 CAS.
- X. Li, R. Higashikubo and J.-S. Taylor, Bioconjugate Chem., 2008, 19, 50–56 CrossRef CAS PubMed.
- Y. Liu, C. Xu, H.-W. Liu, L. Teng, S. Huan, L. Yuan and X.-B. Zhang, Anal. Chem., 2021, 93, 6463–6471 CrossRef CAS PubMed.
- Z. Huang, E. Terpetschnig, W. You and R. P. Haugland, Anal. Biochem., 1992, 207, 32–39 CrossRef CAS.
- H.-W. Liu, K. Li, X.-X. Hu, L. Zhu, Q. Rong, Y. Liu, X.-B. Zhang, J. Hasserodt, F.-L. Qu and W. Tan, Angew. Chem., Int. Ed., 2017, 56, 11788–11792 CrossRef CAS PubMed.
- S. R. Mulay, J. Desai, S. V. Kumar, J. N. Eberhard, D. Thomasova, S. Romoli, M. Grigorescu, O. P. Kulkarni, B. Popper, V. Vielhauer, G. Zuchtriegel, C. Reichel, J. H. Bräsen, P. Romagnani, R. Bilyy, L. E. Munoz, M. Herrmann, H. Liapis, S. Krautwald, A. Linkermann and H.-J. Anders, Nat. Commun., 2016, 7, 10274 CrossRef CAS PubMed.
- R. Obara, M. Kamiya, Y. Tanaka, A. Abe, R. Kojima, T. Kawaguchi, M. Sugawara, A. Takahashi, T. Noda and Y. Urano, Angew. Chem., Int. Ed., 2021, 133, 2153–2157 CrossRef.
- H. Iwashita, E. Castillo, M. S. Messina, R. A. Swanson and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2018513118 CrossRef CAS PubMed.
- H. Kashima, M. Kamiya, F. Obata, R. Kojima, S. Nakano, M. Miura and Y. Urano, Chem. Commun., 2021, 57, 5802–5805 RSC.
- S. Wang, W. Tan, W. Lang, H. Qian, S. Guo, L. Zhu and J. Ge, Anal. Chem., 2022, 94, 7272–7277 CrossRef CAS PubMed.
- W. Chyan, H. R. Kilgore, B. Gold and R. T. Raines, J. Org. Chem., 2017, 82, 4297–4304 CrossRef CAS PubMed.
- P. Mauker, D. Beckmann, A. Kitowski, C. Heise, C. Wientjens, A. J. Davidson, S. Wanderoy, G. Fabre, A. B. Harbauer, W. Wood, C. Wilhelm, J. Thorn-Seshold, T. Misgeld, M. Kerschensteiner and O. Thorn-Seshold, J. Am. Chem. Soc., 2024, 146, 11072–11082 CAS.
- J. Morstein, A. Capecchi, K. Hinnah, B. Park, J. Petit-Jacques, R. C. Van Lehn, J.-L. Reymond and D. Trauner, J. Am. Chem. Soc., 2022, 144, 18532–18544 CrossRef CAS PubMed.
- J. Pfeffermann, R. Yadav, T. Glasnov, O. Thorn-Seshold and P. Pohl, bioRxiv preprint, 2025 DOI:10.1101/2025.01.13.632814.
- M. D. Hammers, M. J. Taormina, M. M. Cerda, L. A. Montoya, D. T. Seidenkranz, R. Parthasarathy and M. D. Pluth, J. Am. Chem. Soc., 2015, 137, 10216–10223 CrossRef CAS PubMed.
- L. Zeisel, J. G. Felber, K. C. Scholzen, C. Schmitt, A. J. Wiegand, L. Komissarov, E. S. J. Arnér and O. Thorn-Seshold, J. Am. Chem. Soc., 2024, 146, 5204–5214 CrossRef CAS.
- Z. Ye, W. Yang, C. Wang, Y. Zheng, W. Chi, X. Liu, Z. Huang, X. Li and Y. Xiao, J. Am. Chem. Soc., 2019, 141, 14491–14495 CrossRef CAS PubMed.
- H. Sies, V. V. Belousov, N. S. Chandel, M. J. Davies, D. P. Jones, G. E. Mann, M. P. Murphy, M. Yamamoto and C. Winterbourn, Nat. Rev. Mol. Cell Biol., 2022, 23, 499–515 CrossRef CAS PubMed.
- M. T. Lin and M. F. Beal, Nature, 2006, 443, 787–795 CrossRef CAS PubMed.
- H. J. Forman and H. Zhang, Nat. Rev. Drug Discovery, 2021, 20, 689–709 CrossRef CAS PubMed.
- E. C. Cheung and K. H. Vousden, Nat. Rev. Cancer, 2022, 22, 280–297 CrossRef CAS PubMed.
- E. W. Miller, B. C. Dickinson and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15681–15686 CrossRef CAS PubMed.
- S. E. Caldwell, I. R. Demyan, G. N. Falcone, A. Parikh, J. Lohmueller and A. Deiters, Bioconjugate Chem., 2025, 36, 540–548 CrossRef PubMed.
- K. A. Kurnia, W. Setyaningsih, N. Darmawan and B. Yuliarto, J. Mol. Liq., 2021, 326, 115321 CrossRef CAS.
- V. Redecke, R. Wu, J. Zhou, D. Finkelstein, V. Chaturvedi, A. A. High and H. Häcker, Nat. Methods, 2013, 10, 795–803 CrossRef CAS PubMed.
- R. Huang, L. Zhao, H. Chen, R.-H. Yin, C.-Y. Li, Y.-Q. Zhan, J.-H. Zhang, C. Ge, M. Yu and X.-M. Yang, PLoS One, 2014, 9, e96246 CrossRef PubMed.
- J. M. Slauch, Mol. Microbiol., 2011, 80, 580–583 CrossRef CAS PubMed.
- C. G. Miller, A. Holmgren, E. S. J. Arnér and E. E. Schmidt, Free Radical Biol. Med., 2018, 127, 248–261 CrossRef CAS PubMed.
- E. S. J. Arnér and A. Holmgren, Eur. J. Biochem., 2000, 267, 6102–6109 CrossRef PubMed.
- A. P. Lothrop, G. W. Snider, E. L. Ruggles and R. J. Hondal, Biochemistry, 2014, 53, 554–565 CrossRef CAS PubMed.
- L. Zeisel, M. S. Maier and O. Thorn-Seshold, Synthesis, 2023, 55, 1385–1393 CrossRef CAS.
- L. Zeisel, J. G. Felber, K. C. Scholzen, L. Poczka, D. Cheff, M. S. Maier, Q. Cheng, M. Shen, M. D. Hall, E. S. J. Arnér, J. Thorn-Seshold and O. Thorn-Seshold, Chem, 2022, 8, 1493–1517 CAS.
- F. H. Abdalbari and C. M. Telleria, Discover Oncol., 2021, 12, 1–18 CrossRef PubMed.
- W. C. Stafford, X. Peng, M. H. Olofsson, X. Zhang, D. K. Luci, L. Lu, Q. Cheng, L. Trésaugues, T. S. Dexheimer, N. P. Coussens, M. Augsten, H.-S. M. Ahlzén, O. Orwar, A. Östman, S. Stone-Elander, D. J. Maloney, A. Jadhav, A. Simeonov, S. Linder and E. S. J. Arnér, Sci. Transl. Med., 2018, 10, eaaf7444 CrossRef PubMed.
- M. Conrad, Biochim. Biophys. Acta Gen. Subj., 2009, 1790, 1575–1585 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |