Open Access Article
Open Access ArticleThermal proteome profiling of itaconate interactome in macrophages†
Yunzhu
Meng‡
ab,
Tiantian
Wei‡
acd,
Chenlin
Zhang
ab,
Anqi
Yu
ab,
Yuan
Liu
ab,
Junyu
Xiao
ad and
Chu
Wang
 *abe
*abe
aPeking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China. E-mail: chuwang@pku.edu.cn
bSynthetic and Functional Biomolecules Center, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, China
cState Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, Beijing 100191, China
dThe State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, Beijing 100871, China
ePeking University Chengdu Academy for Advanced Interdisciplinary Biotechnologies, Chengdu 610231, China
First published on 2nd July 2025
Abstract
Itaconate (ITA) is an upregulated immunometabolite in macrophages during pathogen infection. It is known to influence oxidation stress, cellular metabolism, programmed cell death and many other biological processes to regulate the immune response via interaction with proteins. Previous studies capture covalently ITA-modified proteins by activity-based proteome profiling with bioorthogonal chemical probes; however, how itaconate interacts non-covalently with other proteins at the proteome level remains unexplored. Here we applied thermal proteome profiling (TPP) to globally identify a large number of ITA-interacting proteins in macrophage proteomes. Among these targets, we verified mitochondrial branched-chain aminotransferase (BCAT2) as a novel non-covalent binding target of itaconate via biochemical and structural experiments. The binding of itaconate could inhibit transamination activity of BCAT and regulate the metabolism of branched-chain amino acids (BCAAs) in lipopolysaccharide (LPS)-activated inflammatory macrophages. This study offers a valuable resource that helps decipher novel and comprehensive functions of ITA in macrophages during the immune response and other related biological processes.
Introduction
Macrophages are one of the key immune cell types that defend against pathogen invasion. Upon pathogenic signal stimulation, they undergo a series of changes such as upregulating the transcription of particular genes and reprogramming metabolic processes.1,2 Itaconate (ITA) is a by-product of the tricarboxylic acid (TCA) cycle catalyzed by aconitate decarboxylase 1 (ACOD1),3 which is significantly up-regulated and plays a crucial regulatory role during pathogen infection (Fig. 1a).2,4,5 This immunoregulatory metabolite has been reported to help hosts defend against pathogen infection,6–8 and affect cell metabolism,9,10 redox homeostasis,11 and signal transduction.12,13 Recent studies have also highlighted the versatile roles of itaconate in moderating effects on epigenetics14 and obesity.15–17Itaconate accomplishes these regulatory roles by interacting with important proteins primarily through covalent modification.5 As itaconate has an electrophilic double bond, it can modify the cysteine residues in proteins via Michael addition, which is a process known as “itaconation”.18 Since the discovery of itaconation on KEAP1 in 2018,13 a series of chemical probes and proteomic methods have been developed to identify itaconation proteins and sites in macrophages18,19 and pathogens,20,21 which have greatly promoted functional investigation of itaconate.12,22 More recently, an ITA-mediated lysine acylation was discovered whose functional impact is yet to be investigated.23
In addition to covalent modification, itaconate has also been shown to non-covalently interact with certain proteins and affect their functions. For example, succinate dehydrogenase (SDH)9 and Tet methylcytosine dioxygenase 2 (TET2)14 are two non-covalent interacting proteins of itaconate. In both cases, itaconate binds to the proteins' substrate-binding pockets mainly through dicarboxylic groups, mimicking the action of their native substrates of succinate24 and α-ketoglutarate (AKG),14 respectively. These non-covalent interactions between itaconate and the target proteins resulted in the suppression of protein activity and regulation of downstream metabolic9 or transcriptional pathways.14 However, since the current chemical probes and proteomic methods can only profile the covalent targets of itaconate, the non-covalent interactome landscape of itaconate in macrophages is yet to be explored.
Thermal proteome profiling (TPP) is a powerful technology that enables interactome profiling of metabolites or ligands in their native forms.25–29 Operating based on the rationale that ligand binding or modification to a protein may alter its thermal stability, TPP, as enabled by the multiplex quantitative proteomics (such as TMT), can be used to draw and compare melting curves for thousands of proteins in proteomes simultaneously in the absence vs. presence of ligand cotreatment, which can be analysed to identify targets of a ligand molecule at the proteome level.27 The power of this chemoproteomic technology has been highlighted in numerous applications such as detecting targets of drugs,25,28,30,31 endogenous metabolites32–35 and even metal cofactors36 without derivatization. In addition, it also helps profile targets of post-translational modification such as phosphorylation,37–39O-GlcNAcylation,40 and methylation.41
Considering that itaconate can both covalently modify and non-covalently interact with proteins, we reasoned that TPP can aid in identification of more ITA-interacting proteins in proteomes, especially those non-covalent binding targets that were previously neglected by probe-based chemoproteomic methods. We therefore applied TPP to globally map itaconate's interactome in macrophage cells. By comparing with the published data for covalently itaconated proteins, we obtained a list of non-covalent targets of itaconate, among which the mitochondrial branched-chain amino acid aminotransferase 2 (BCAT2) was structurally and functionally characterized. The results showed that itaconate can block transamination activity of BCAT and regulate catabolism of branched-chain amino acids in inflammatory macrophages.
Results
Profiling of ITA-interacting proteins by TPP
To profile ITA-interacting proteins by TPP (Fig. 1b), RAW 264.7 cells were suspended in PBS buffer and lysed by freezing–thawing with liquid nitrogen. After incubation with 1 mM itaconate or PBS buffer at room temperature, the resulting proteomes were split into 8 aliquots and subjected to a series of thermal stimulations at the indicated temperatures for 3 minutes. The soluble contents were extracted using centrifugation and analyzed using either gel-based or mass spectrometry (MS)-based methods. In the gel-based analysis, relative immunoblotting intensities were employed for quantification of individual proteins, while in the MS-based analysis, the samples were further digested by trypsin, labeled by using the IBTpro-16plex42 labeling reagents and analyzed by LC-MS/MS to quantify protein abundance in each aliquot for a large number of proteins simultaneously. In both cases, thermal shift curves were fitted using the quantitative data and the Tm values with or without itaconate treatment were determined (Fig. 1b).We performed the MS-based TPP experiments in three biological replicates. A total of 4912 proteins were quantified in at least one replicate and 3214 proteins were quantified across all three biological replicates. Based on these raw proteomic data, we obtained thermal shift curves for 2450 proteins under both conditions with high confidence after a strict filter was applied (Fig. 1c), and their Tm values demonstrated good correlation across the three biological replicates (Fig. S1†). To identify potential ITA-interacting targets, a volcano plot was created to display the ΔTm value for each protein along with its statistical significance (Fig. 1c). Given that the global thermal stability of the RAW 264.7 proteome was not dramatically changed upon itaconate treatment as visualized by gel-based staining (Fig. S2†), we defined proteins with ΔTm values exceeding 1 °C in all three biological replicates and a relatively significant P value (|ΔTm × (−log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P)| > 1.5) as ITA-interacting targets. Finally, 190 proteins (8%) were found to meet these criteria and interestingly, all of them exhibited increasing thermal stability after ITA treatment (Fig. 1c and Table S1†).
P)| > 1.5) as ITA-interacting targets. Finally, 190 proteins (8%) were found to meet these criteria and interestingly, all of them exhibited increasing thermal stability after ITA treatment (Fig. 1c and Table S1†).
As protein thermal stability can be affected by both covalent modification and non-covalent interactions of itaconate, we cross-checked with a list of 862 covalently itaconated proteins (with 1131 itaconated cysteine sites) obtained by the bioorthogonal ITalk probe18 to further distinguish the covalent and non-covalent targets in the TPP data (Fig. 1c and Table S1†). A total of 35 proteins were found in both ITalk and TPP data, including a prominent example of peroxiredoxin 1 (PRDX1) that is essential for cellular redox control. In particular, C173 of PRDX1 was previously identified to undergo itaconation, and mutation of this cysteine inhibited the formation of the PRX1 homodimer, which is associated with suppression of TNF-α-induced release.43 The gel-based TPP showed that after itaconate treatment, the ΔTm value of PRDX1 was changed by 2.0 ± 1.0 °C (Fig. 1d), which was consistent with the MS-based analysis (Fig. S3a†), further confirming the interaction between itaconate and PRDX1.
Aside from the covalent targets of itaconate, 155 proteins were identified specifically in the TPP profiling (Fig. 1c and Table S1†), which could be considered in principle as non-covalent binding targets of itaconate. Satisfyingly, this group included the known non-covalent target of itaconate, SDHA, which showed a ΔTm of 1.8 ± 0.9 °C from the MS-based analysis and a ΔTm of 2.0 ± 1.0 °C in the gel-based validation (Fig. 1e and S3b†). We also validated one novel target, lysine methyltransferase 2 (EFMT2), by overexpressing the protein, and the gel-based result demonstrated a similar change in thermal stability after itaconate treatment (Fig. 1f and S3c†).
Characterization of BCAT2 as a direct non-covalent target of ITA
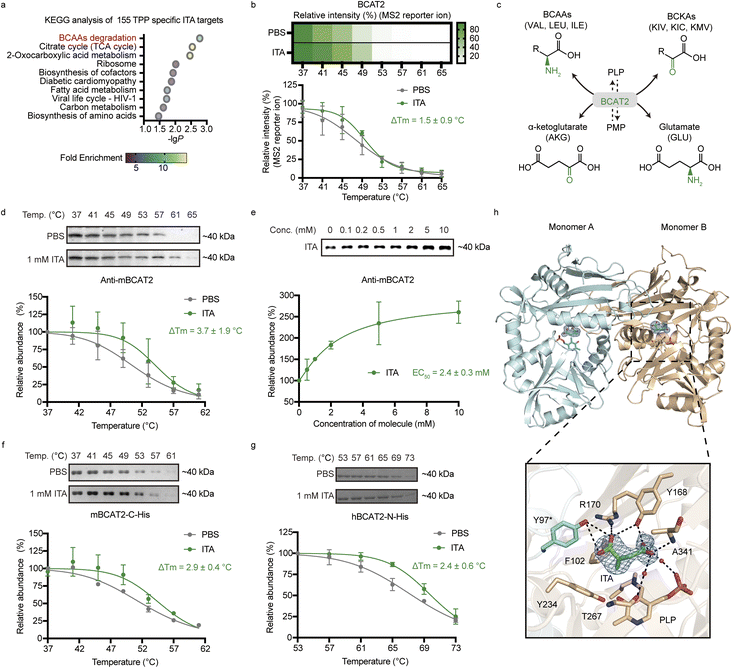
We further performed the KEGG pathway enrichment analysis for these 155 TPP-specific ITA-interacting targets (Fig. 2a). The results showed that they were mainly involved in the citrate cycle, fatty acid metabolism and the pathway related to virus infection, all of which were associated with known functions of itaconate.5 Additionally, there were two novel pathways discovered by TPP, including branched chain amino acids (BCAA) degradation and 2-oxocarboxylic acid metabolism, indicating that itaconate might play a regulatory role in these pathways. In particular, the mitochondrial branched-chain-amino-acid aminotransferase (BCAT2) drew our attention as the enzyme works in the initial phase of BCAA degradation and in the MS-based analysis, BCAT2 showed well-fitted melting curves with an ITA-induced ΔTm of 1.5 ± 0.9 °C (Fig. 2b).BCAT2 catalyzes the conversion from BCAAs, namely valine (VAL), leucine (LEU) and isoleucine (ILE), to their corresponding branched-chain-keto-acids (BCKAs), including 3-methyl-2-oxopentanoic acid (KMV), 3-methyl-2-oxobutanoic acid (KIV), and 4-methyl-2-oxopentanoic acid (KIC), and the reaction occurs with the concomitant conversion from α-ketoglutarate (AKG) to glutamate (GLU) (Fig. 2c).44 The protein is known to regulate cancer proliferation45,46 and chemokine release,47 as well as cellular metabolism such as lipid metabolism.48 Since the relevance of BCAT2 and itaconate has never been reported, we were therefore motivated to characterize their interaction and investigate the functional impact.
We first validated the thermal shifting result by the gel-based TPP assay in RAW 264.7 cell lysates. In both temperature- and concentration-based TPP experiments, itaconate could significantly increase thermal stability of BCAT2 (Fig. 2d and e). We further purified mouse BCAT2 (mBCAT2) and human BCAT2 (hBCAT2), and found that itaconate could also enhance thermal stability of these purified proteins (Fig. 2f and g), suggesting that this interaction between itaconate and BCAT2 is direct and conserved in mammalian cells.
To elucidate how itaconate specifically binds to BCAT2, we attempted to crystallize mBCAT2 or hBCAT2 in the presence of itaconate and successfully determined the structure of hBCAT2 in a complex with itaconate at 2.0 Å (Fig. 2h and Table S2†) (Protein Data Bank (PDB): 9LEP). The co-crystal structure of hBCAT2–ITA clearly showed that hBCAT2 forms a dimer and in each monomer, one molecule of itaconate was observed to directly bind to the pocket which was normally occupied by its native substrate, α-ketoglutarate. Specifically, itaconate formed several strong hydrogen bonds with hBCAT2 residues, including the sidechain phenol hydroxyl groups of Tyr168 from one monomer and Tyr97 from the other monomer, the sidechain guanidino group of Arg170, as well as the backbone amino group of Ala341. In addition, Phe102, Tyr234 and the cofactor pyridoxal phosphate (PLP) were involved in hydrophobic/van der Waals interactions with itaconate. Two water molecules were observed in the pocket, which also mediated a network of hydrogen bonds between itaconate and hBCAT2 residues. Together, all these data unambiguously supported that itaconate can non-covalently bind to hBCAT2.
Inhibition of the transaminase activity of BCAT2 by ITA
Given that itaconate and α-ketoglutarate shares the same binding pocket in BCAT2, we next investigated how the non-covalent binding of itaconate could regulate BCAT2's activity. Theoretically, BCAT2 is able to catalyze both the forward reaction and reverse reaction of the BCAAs–BCKAs conversion. To evaluate the activity of BCAT2, we used VAL and AKG as the substrates for measuring the forward reaction, and KIV and GLU as the substrates for measuring the reverse reaction. 10 mM itaconate was incubated with the purified mBCAT2 and corresponding substrates at 37 °C for 1 hour. The relative activity of mBCAT2 was determined by quantifying the amount of glutamate or α-ketoglutarate generated by the forward or reverse reaction using LC-MS. The results showed that itaconate could inhibit the forward activity of BCAT2 but did not affect that of the reverse reaction (Fig. 3a). With regard to the forward reaction, we also observed that a higher ITA/AKG ratio could result in more obvious inhibition (Fig. 3b), which indicated a model where itaconate could compete with α-ketoglutarate to inhibit activity of BCAT2. Detailed Michaelis–Menten kinetics analysis in vitro revealed a Ki value of 15.32 mM and a competitive inhibition mechanism (Fig. S4†). Considering that the local concentration of ITA might be higher in mitochondria, it is possible that BCAT2 will be inhibited in macrophages upon activation.Interestingly, itaconate hardly changed the transamination activity of BCAT1 (Fig. S5a†), which is a cytosolic isoform of BCAT2, highlighting the unique isoform selectivity of itaconate against these two functionally similar enzymes. Consistent with this observation, purified BCAT1 did not show a thermal shift upon ITA incubation either (Fig. S5b†). To clarify this selectivity, the published structure of hBCAT1 (PDB: 7NWA) and our hBCAT2 were aligned for comparison. We noticed a different loop region outside the binding pocket (Pro192-Tyr193 in hBCAT1 and Ala199-Tyr200 in hBCAT2) (Fig. S6†) that may cause the selectivity of itaconate; however, a detailed structural study is necessary to fully explain this isoform selectivity.
We then investigated if the inhibition of BCAT2 by itaconate could also occur in living cells. RAW 264.7 cells were treated with 10 mM itaconate for 12 hours as in a previous study,49 and the cellular metabolome was extracted using 80% methanol. The relative abundance of BCAAs, BCKAs, glutamate and α-ketoglutarate was quantified by LC-MS and the ratios of the corresponding BCKAs/BCAAs were calculated to reflect the transamination activity of BCAT. As expected, we observed a decreased level of BCKAs and an increased level of BCAAs after itaconate treatment, suggesting that itaconate could indeed inhibit BCAT's activity and interfere with BCAA metabolism in living cells (Fig. 3c and d). Surprisingly, we noticed that the level of AKG was also decreased after itaconate treatment (Fig. S7†). This change might be caused by other influenced proteins in the AKG metabolism such as IDH3a, which is an AKG synthase and was identified as an ITA target in our TPP data.
As itaconate is significantly up-regulated during pathogen inflection and inflammation, we next examined the alteration of BCAT activity and BCAA metabolism in lipopolysaccharide (LPS) activated macrophages. When macrophage cells were treated with 100 ng per mL LPS for 12 hours, the expression of Acod1 was promoted, resulting in the elevated production of endogenous itaconate as expected. To confirm that the inhibitory effect was solely dependent on itaconate, we also knocked out Acod1 in macrophages (“sgACOD1”) by using CRISPR-Cas9,21 which effectively eliminated the cellular production of itaconate under LPS induction in comparison with the control cells (“sgNC”) (Fig. S8†). Along with the accumulation of itaconate, the forward activity of BCAT in the sgNC cells dropped, resulting in the reduction of KIV and accumulation of VAL (Fig. 3e and f). Satisfyingly, the change in BCAAs and BCKAs as well as the inhibited activity of BCAT was no longer observed in the sgACOD1 cells upon stimulation by LPS and the trend was reversed by adding exogenous itaconate to inhibit the activity of BCAT again (Fig. 3e and f). Collectively, these results indicated that itaconate generated by Acod1 was the key metabolite for regulating BCAT's activity and BCAA metabolism in LPS-activated inflammatory macrophages.
Conclusions
As one of the key immunoregulatory metabolites in macrophages, itaconate can bind to pivotal proteins to regulate their functions and associated biological processes. Here, we successfully identified itaconate-interacting proteins in macrophages by thermal proteome profiling, which provided more hints for deciphering the versatile functions of itaconate.As chemical derivation would alter the natural reactivity of itaconate7,49 and might prevent the formation of hydrogen bonds with amino acids in specific binding pockets of targets,14,24 it is crucial to identify its targets using the unmodified molecules. While we have identified both non-covalent and covalent targets of itaconate with using TPP in this study, other advanced proteomic strategies such as LIP-MS,50 TRAP,51 ipHSA52 and PELSA34 would also be valuable for the identification and cross-validation of more itaconate targets, which will help overcome the problem of both false-positive and false-negative targets by the inherent limitation of TPP. In addition, while we attempted to distinguish non-covalent targets by comparing with the ITalk-captured itaconation targets18 for simplicity, highly specific antibodies or probes are still highly desired to directly capture endogenous itaconation proteins at the proteome level. Given that itaconate binds to the BCAT2's pocket and is unlikely to be converted due to the lack of the α-keto acid moiety, it might be interesting to perform comparative profiling of itaconate with similar dicarboxylate metabolites to characterize their overlapping and unique interactome landscapes in living cells.
Subsequent functional research is also needed to focus on investigating in depth the biological impact of the BCAT2–ITA interaction. Non-covalent binding mode is often associated with reversible inhibition, which means that the inhibition on BCAT might vary depending on the level of itaconate during the inflammation or infection process. Additionally, recent studies also showed that BCAAs and BCAKs have different immune regulatory roles.44,53,54 Therefore, a systematic investigation might be required to better understand the relation between BCAT2–ITA interaction and inflammation. Furthermore, BCAA metabolism is also reported to be linked to fatty acid metabolism and β-oxidation.48 These two crucial biological processes have been shown to be regulated by itaconate, which results in a weight-reducing effect in the obese mouse model.17 Hence, it is reasonable to speculate that there is a link between this BCAT2–ITA interaction and the weight-reducing effect. We envisage that the chemoproteomic study presented here will offer valuable resources and novel clues to study itaconate-interacting proteins to better understand the versatile functions of this intriguing immunoregulatory metabolite.
Data availability
The MS proteomics data were deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD060673. Source data are provided with this paper.Author contributions
Y. M. and C. W. conceived the project. Y. M. conducted most of the chemoproteomic and biochemical experiments. J. X. guided and supervised T. W and C. Z. to solve the structure of hBCAT2–ITA. A. Y. helped purify mBCAT2 and hBCAT2 mutants. Y. L. developed computational algorithms to analyze the iBT proteomic data. Y. M. and C. W. analyzed the data and wrote the manuscript with input from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Computing Platform of the Center for Life Science for supporting the proteomics data analysis. We thank the National Center for Protein Sciences at Peking University for assistance with protein crystallization and the staff of the Shanghai Synchrotron Radiation Facility for assistance with X-ray data collection. C. W. acknowledges the support from the National Natural Science Foundation of China (22525071, 22361142837, and 22321005), the Ministry of Science and Technology (2022YFA1304700), the Beijing National Laboratory for Molecular Sciences (BNLMS-CXTD-202401) and the PKUSZ-AI4S program.References
- K. Willmann and L. F. Moita, Physiologic disruption and metabolic reprogramming in infection and sepsis, Cell Metab., 2024, 36, 927–946 CrossRef CAS PubMed.
- G. Rosenberg, S. Riquelme, A. Prince and R. Avraham, Immunometabolic crosstalk during bacterial infection, Nat. Microbiol., 2022, 7, 497–507 CrossRef CAS PubMed.
- A. Michelucci, T. Cordes, J. Ghelfi, A. Pailot, N. Reiling, O. Goldmann, T. Binz, A. Wegner, A. Tallam, A. Rausell, M. Buttini, C. L. Linster, E. Medina, R. Balling and K. Hiller, Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7820–7825 CrossRef CAS PubMed.
- C. L. Strelko, W. Lu, F. J. Dufort, T. N. Seyfried, T. C. Chiles, J. D. Rabinowitz and M. F. Roberts, Itaconic Acid Is a Mammalian Metabolite Induced during Macrophage Activation, J. Am. Chem. Soc., 2011, 133, 16386–16389 CrossRef CAS PubMed.
- D. Ye, P. Wang, L.-L. Chen, K.-L. Guan and Y. Xiong, Itaconate in host inflammation and defense, Trends Endocrinol. Metab., 2024, 35, 586–606 CrossRef CAS PubMed.
- E. A. Day and L. A. J. O'Neill, Protein targeting by the itaconate family in immunity and inflammation, Biochem. J., 2022, 479, 2499–2510 CrossRef CAS PubMed.
- A. F. McGettrick and L. A. J. O'Neill, Two for the price of one: itaconate and its derivatives as an anti-infective and anti-inflammatory immunometabolite, Curr. Opin. Immunol., 2023, 80, 102268 CrossRef CAS PubMed.
- S. A. Riquelme, K. Liimatta, T. Wong Fok Lung, B. Fields, D. Ahn, D. Chen, C. Lozano, Y. Sáenz, A.-C. Uhlemann, B. C. Kahl, C. J. Britto, E. DiMango and A. Prince, Pseudomonas aeruginosa Utilizes Host-Derived Itaconate to Redirect Its Metabolism to Promote Biofilm Formation, Cell Metab., 2020, 31, 1091–1106 CrossRef CAS PubMed.
- V. Lampropoulou, A. Sergushichev, M. Bambouskova, S. Nair, E. E. Vincent, E. Loginicheva, L. Cervantes-Barragan, X. Ma, S. C. Huang, T. Griss, C. J. Weinheimer, S. Khader, G. J. Randolph, E. J. Pearce, R. G. Jones, A. Diwan, M. S. Diamond and M. N. Artyomov, Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation, Cell Metab., 2016, 24, 158–166 CrossRef CAS PubMed.
- S. T. Liao, C. Han, D. Q. Xu, X. W. Fu, J. S. Wang and L. Y. Kong, 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects, Nat. Commun., 2019, 10, 5091 CrossRef PubMed.
- M. Bambouskova, L. Gorvel, V. Lampropoulou, A. Sergushichev, E. Loginicheva, K. Johnson, D. Korenfeld, M. E. Mathyer, H. Kim, L.-H. Huang, D. Duncan, H. Bregman, A. Keskin, A. Santeford, R. S. Apte, R. Sehgal, B. Johnson, G. K. Amarasinghe, M. P. Soares, T. Satoh, S. Akira, T. Hai, C. de Guzman Strong, K. Auclair, T. P. Roddy, S. A. Biller, M. Jovanovic, E. Klechevsky, K. M. Stewart, G. J. Randolph and M. N. Artyomov, Electrophilic properties of itaconate and derivatives regulate the IκBζ–ATF3 inflammatory axis, Nature, 2018, 556, 501–504 CrossRef CAS PubMed.
- M. C. Runtsch, S. Angiari, A. Hooftman, R. Wadhwa, Y. Zhang, Y. Zheng, J. S. Spina, M. C. Ruzek, M. A. Argiriadi, A. F. McGettrick, R. S. Mendez, A. Zotta, C. G. Peace, A. Walsh, R. Chirillo, E. Hams, P. G. Fallon, R. Jayaraman, K. Dua, A. C. Brown, R. Y. Kim, J. C. Horvat, P. M. Hansbro, C. Wang and L. A. J. O'Neill, Itaconate and itaconate derivatives target JAK1 to suppress alternative activation of macrophages, Cell Metab., 2022, 34, 487–501 CrossRef CAS PubMed.
- E. L. Mills, D. G. Ryan, H. A. Prag, D. Dikovskaya, D. Menon, Z. Zaslona, M. P. Jedrychowski, A. S. H. Costa, M. Higgins, E. Hams, J. Szpyt, M. C. Runtsch, M. S. King, J. F. McGouran, R. Fischer, B. M. Kessler, A. F. McGettrick, M. M. Hughes, R. G. Carroll, L. M. Booty, E. V. Knatko, P. J. Meakin, M. L. J. Ashford, L. K. Modis, G. Brunori, D. C. Sévin, P. G. Fallon, S. T. Caldwell, E. R. S. Kunji, E. T. Chouchani, C. Frezza, A. T. Dinkova-Kostova, R. C. Hartley, M. P. Murphy and L. A. O'Neill, Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1, Nature, 2018, 556, 113–117 CrossRef CAS PubMed.
- L. L. Chen, C. Morcelle, Z. L. Cheng, X. Chen, Y. Xu, Y. Gao, J. Song, Z. Li, M. D. Smith, M. Shi, Y. Zhu, N. Zhou, M. Cheng, C. He, K. Y. Liu, G. Lu, L. Zhang, C. Zhang, J. Zhang, Y. Sun, T. Qi, Y. Lyu, Z. Z. Ren, X. M. Tan, J. Yin, F. Lan, Y. Liu, H. Yang, M. Qian, C. Duan, X. Chang, Y. Zhou, L. Shen, A. S. Baldwin, K. L. Guan, Y. Xiong and D. Ye, Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses, Nat. Cell Biol., 2022, 24, 353–363 CrossRef CAS PubMed.
- R. A. Frieler, T. M. Vigil, J. Song, C. Leung, D. R. Goldstein, C. N. Lumeng and R. M. Mortensen, Aconitate decarboxylase 1 regulates glucose homeostasis and obesity in mice, Obesity, 2022, 30, 1818–1830 CrossRef CAS PubMed.
- X. Zhang, Y. Zhi, X. Zan, K. Fan, K. Chen, S. Zhao, X. Dai, L. Li, Y. Yang, K. Hu, X. Gong and L. Zhang, Immune response gene 1 deficiency aggravates high fat diet-induced nonalcoholic fatty liver disease via promotion of redox-sensitive AKT suppression, Biochim. Biophys. Acta, Mol. Basis Dis., 2023, 1869, 166656 CrossRef CAS PubMed.
- Z. Yu, X. Li, Y. Quan, J. Chen, J. Liu, N. Zheng, S. Liu, Y. Wang, W. Liu, C. Qiu, Y. Wang, R. Zheng and J. Qin, Itaconate alleviates diet-induced obesity via activation of brown adipocyte thermogenesis, Cell Rep., 2024, 43, 114142 CrossRef CAS PubMed.
- W. Qin, Y. Zhang, H. Tang, D. Liu, Y. Chen, Y. Liu and C. Wang, Chemoproteomic Profiling of Itaconation by Bioorthogonal Probes in Inflammatory Macrophages, J. Am. Chem. Soc., 2020, 142, 10894–10898 CrossRef CAS PubMed.
- W. Qin, K. Qin, Y. Zhang, W. Jia, Y. Chen, B. Cheng, L. Peng, N. Chen, Y. Liu, W. Zhou, Y.-L. Wang, X. Chen and C. Wang, S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate, Nat. Chem. Biol., 2019, 15, 983–991 CrossRef CAS PubMed.
- Y. Zhang, W. Qin, D. Liu, Y. Liu and C. Wang, Chemoproteomic profiling of itaconations in Salmonella, Chem. Sci., 2021, 12, 6059–6063 RSC.
- Z. Liu, D. Liu and C. Wang, In situ chemoproteomic profiling reveals itaconate inhibits de novo purine biosynthesis in pathogens, Cell Rep., 2024, 43, 114737 CrossRef CAS PubMed.
- C. Wei, Z. Xiao, Y. Zhang, Z. Luo, D. Liu, L. Hu, D. Shen, M. Liu, L. Shi, X. Wang, T. Lan, Q. Dai, J. Liu, W. Chen, Y. Zhang, Q. Sun, W. Wu, P. Wang, C. Zhang, J. Hu, C. Wang, F. Yang and Q. Li, Itaconate protects ferroptotic neurons by alkylating GPx4 post stroke, Cell Death Differ., 2024, 31, 983–998 CrossRef CAS PubMed.
- D. Liu, W. Xiao, H. Li, Y. Zhang, S. Yuan, C. Li, S. Dong and C. Wang, Discovery of Itaconate-Mediated Lysine Acylation, J. Am. Chem. Soc., 2023, 145, 12673–12681 CrossRef CAS PubMed.
- F. Chen, W. A. M. Elgaher, M. Winterhoff, K. Büssow, F. H. Waqas, E. Graner, Y. Pires-Afonso, L. Casares Perez, L. de la Vega, N. Sahini, L. Czichon, W. Zobl, T. Zillinger, M. Shehata, S. Pleschka, H. Bähre, C. Falk, A. Michelucci, S. Schuchardt, W. Blankenfeldt, A. K. H. Hirsch and F. Pessler, Citraconate inhibits ACOD1 (IRG1) catalysis, reduces interferon responses and oxidative stress, and modulates inflammation and cell metabolism, Nat. Metab., 2022, 4, 534–546 CrossRef CAS PubMed.
- M. M. Savitski, F. B. Reinhard, H. Franken, T. Werner, M. F. Savitski, D. Eberhard, D. Martinez Molina, R. Jafari, R. B. Dovega, S. Klaeger, B. Kuster, P. Nordlund, M. Bantscheff and G. Drewes, Tracking cancer drugs in living cells by thermal profiling of the proteome, Science, 2014, 346, 1255784 CrossRef PubMed.
- H. Franken, T. Mathieson, D. Childs, G. M. A. Sweetman, T. Werner, I. Tögel, C. Doce, S. Gade, M. Bantscheff, G. Drewes, F. B. M. Reinhard, W. Huber and M. M. Savitski, Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry, Nat. Protoc., 2015, 10, 1567–1593 CrossRef CAS PubMed.
- K. V. M. Huber, K. M. Olek, A. C. Müller, C. S. H. Tan, K. L. Bennett, J. Colinge and G. Superti-Furga, Proteome-wide drug and metabolite interaction mapping by thermal-stability profiling, Nat. Methods, 2015, 12, 1055–1057 CrossRef CAS PubMed.
- I. Becher, T. Werner, C. Doce, E. A. Zaal, I. Tögel, C. A. Khan, A. Rueger, M. Muelbaier, E. Salzer, C. R. Berkers, P. F. Fitzpatrick, M. Bantscheff and M. M. Savitski, Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat, Nat. Chem. Biol., 2016, 12, 908–910 CrossRef CAS PubMed.
- Y. Tu, L. Tan, H. Tao, Y. Li and H. Liu, CETSA and thermal proteome profiling strategies for target identification and drug discovery of natural products, Phytomedicine, 2023, 116, 154862 CrossRef CAS PubMed.
- F. B. M. Reinhard, D. Eberhard, T. Werner, H. Franken, D. Childs, C. Doce, M. F. Savitski, W. Huber, M. Bantscheff, M. M. Savitski and G. Drewes, Thermal proteome profiling monitors ligand interactions with cellular membrane proteins, Nat. Methods, 2015, 12, 1129–1131 CrossRef CAS PubMed.
- M. Kalxdorf, I. Günthner, I. Becher, N. Kurzawa, S. Knecht, M. M. Savitski, H. C. Eberl and M. Bantscheff, Cell surface thermal proteome profiling tracks perturbations and drug targets on the plasma membrane, Nat. Methods, 2021, 18, 84–91 CrossRef CAS PubMed.
- S. Sridharan, N. Kurzawa, T. Werner, I. Günthner, D. Helm, W. Huber, M. Bantscheff and M. M. Savitski, Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP, Nat. Commun., 2019, 10, 1155 CrossRef PubMed.
- Y. T. Lim, N. Prabhu, L. Dai, K. D. Go, D. Chen, L. Sreekumar, L. Egeblad, S. Eriksson, L. Chen, S. Veerappan, H. L. Teo, C. S. H. Tan, J. Lengqvist, A. Larsson, R. M. Sobota and P. Nordlund, An efficient proteome-wide strategy for discovery and characterization of cellular nucleotide-protein interactions, PLoS One, 2018, 13, e0208273 CrossRef PubMed.
- K. Li, S. Chen, K. Wang, Y. Wang, L. Xue, Y. Ye, Z. Fang, J. Lyu, H. Zhu, Y. Li, T. Yu, F. Yang, X. Zhang, S. Guo, C. Ruan, J. Zhou, Q. Wang, M. Dong, C. Luo and M. Ye, A peptide-centric local stability assay enables proteome-scale identification of the protein targets and binding regions of diverse ligands, Nat. Methods, 2025, 22, 278–282 CrossRef CAS PubMed.
- K. J. Li, Y. Y. Ye, X. L. Zhang, J. H. Zhou, Y. N. Li and M. L. Ye, Identification of the binding proteins of organic acid metabolites by matrix thermal shift assay, Sepu, 2024, 42, 702–710 CAS.
- X. Zeng, T. Wei, X. Wang, Y. Liu, Z. Tan, Y. Zhang, T. Feng, Y. Cheng, F. Wang, B. Ma, W. Qin, C. Gao, J. Xiao and C. Wang, Discovery of metal-binding proteins by thermal proteome profiling, Nat. Chem. Biol., 2024, 20, 770–778 CrossRef CAS PubMed.
- J. X. Huang, G. Lee, K. E. Cavanaugh, J. W. Chang, M. L. Gardel and R. E. Moellering, High throughput discovery of functional protein modifications by Hotspot Thermal Profiling, Nat. Methods, 2019, 16, 894–901 CrossRef CAS PubMed.
- C. M. Potel, N. Kurzawa, I. Becher, A. Typas, A. Mateus and M. M. Savitski, Impact of phosphorylation on thermal stability of proteins, Nat. Methods, 2021, 18, 757–759 CrossRef CAS PubMed.
- I. R. Smith, K. N. Hess, A. A. Bakhtina, A. S. Valente, R. A. Rodríguez-Mias and J. Villén, Identification of phosphosites that alter protein thermal stability, Nat. Methods, 2021, 18, 760–762 CrossRef CAS PubMed.
- D. T. King, J. E. Serrano-Negrón, Y. Zhu, C. L. Moore, M. D. Shoulders, L. J. Foster and D. J. Vocadlo, Thermal Proteome Profiling Reveals the O-GlcNAc-Dependent Meltome, J. Am. Chem. Soc., 2022, 144, 3833–3842 CrossRef CAS PubMed.
- C. Sayago, J. Sánchez-Wandelmer, F. García, B. Hurtado, V. Lafarga, P. Prieto, E. Zarzuela, P. Ximénez-Embún, S. Ortega, D. Megías, O. Fernández-Capetillo, M. Malumbres and J. Munoz, Decoding protein methylation function with thermal stability analysis, Nat. Commun., 2023, 14, 3016 CrossRef CAS PubMed.
- Y. Ren, Y. He, Z. Lin, J. Zi, H. Yang, S. Zhang, X. Lou, Q. Wang, S. Li and S. Liu, Reagents for Isobaric Labeling Peptides in Quantitative Proteomics, Anal. Chem., 2018, 90, 12366–12371 CrossRef CAS PubMed.
- L. Mullen, E. M. Hanschmann, C. H. Lillig, L. A. Herzenberg and P. Ghezzi, Cysteine Oxidation Targets Peroxiredoxins 1 and 2 for Exosomal Release through a Novel Mechanism of Redox-Dependent Secretion, Mol. Med., 2015, 21, 98–108 CAS.
- A. Dimou, V. Tsimihodimos and E. Bairaktari, The Critical Role of the Branched Chain Amino Acids (BCAAs) Catabolism-Regulating Enzymes, Branched-Chain Aminotransferase (BCAT) and Branched-Chain α-Keto Acid Dehydrogenase (BCKD), in Human Pathophysiology, Int. J. Mol. Sci., 2022, 23, 4022 CrossRef CAS PubMed.
- J. Li, M. Yin, D. Wang, J. Wang, M.-Z. Lei, Y. Zhang, Y. Liu, L. Zhang, S.-W. Zou, L.-P. Hu, Z.-G. Zhang, Y.-P. Wang, W.-Y. Wen, H.-J. Lu, Z.-J. Chen, D. Su and Q.-Y. Lei, BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma, Nat. Cell Biol., 2020, 22, 167–174 CrossRef CAS PubMed.
- K. Wang, Z. Zhang, H.-I. Tsai, Y. Liu, J. Gao, M. Wang, L. Song, X. Cao, Z. Xu, H. Chen, A. Gong, D. Wang, F. Cheng and H. Zhu, Branched-chain amino acid aminotransferase 2 regulates ferroptotic cell death in cancer cells, Cell Death Differ., 2021, 28, 1222–1236 CrossRef CAS PubMed.
- Z. Cai, J. Chen, Z. Yu, H. Li, Z. Liu, D. Deng, J. Liu, C. Chen, C. Zhang, Z. Ou, M. Chen, J. Hu and X. Zu, BCAT2 Shapes a Noninflamed Tumor Microenvironment and Induces Resistance to Anti-PD-1/PD-L1 Immunotherapy by Negatively Regulating Proinflammatory Chemokines and Anticancer Immunity, Adv. Sci., 2023, 10, e2207155 CrossRef PubMed.
- Q. X. Ma, W. Y. Zhu, X. C. Lu, D. Jiang, F. Xu, J. T. Li, L. Zhang, Y. L. Wu, Z. J. Chen, M. Yin, H. Y. Huang and Q. Y. Lei, BCAA–BCKA axis regulates WAT browning through acetylation of PRDM16, Nat. Metab., 2022, 4, 106–122 CrossRef CAS PubMed.
- A. Swain, M. Bambouskova, H. Kim, P. S. Andhey, D. Duncan, K. Auclair, V. Chubukov, D. M. Simons, T. P. Roddy, K. M. Stewart and M. N. Artyomov, Comparative evaluation of itaconate and its derivatives reveals divergent inflammasome and type I interferon regulation in macrophages, Nat. Metab., 2020, 2, 594–602 CrossRef CAS PubMed.
- I. Piazza, K. Kochanowski, V. Cappelletti, T. Fuhrer, E. Noor, U. Sauer and P. Picotti, A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication, Cell, 2018, 172, 358–372 CrossRef CAS PubMed.
- Y. Tian, N. Wan, H. Zhang, C. Shao, M. Ding, Q. Bao, H. Hu, H. Sun, C. Liu, K. Zhou, S. Chen, G. Wang, H. Ye and H. Hao, Chemoproteomic mapping of the glycolytic targetome in cancer cells, Nat. Chem. Biol., 2023, 19, 1480–1491 CrossRef CAS PubMed.
- X. Zhang, C. Ruan, Y. Wang, K. Wang, X. Liu, J. Lyu and M. Ye, Integrated Protein Solubility Shift Assays for Comprehensive Drug Target Identification on a Proteome-Wide Scale, Anal. Chem., 2023, 95, 13779–13787 CrossRef CAS PubMed.
- Y. Dong, X. Zhang, R. Miao, W. Cao, H. Wei, W. Jiang, R. Gao, Y. Yang, H. Sun and J. Qiu, Branched-chain amino acids promotes the repair of exercise-induced muscle damage via enhancing macrophage polarization, Front Physiol, 2022, 13, 1037090 CrossRef PubMed.
- L. S. Silva, G. Poschet, Y. Nonnenmacher, H. M. Becker, S. Sapcariu, A. C. Gaupel, M. Schlotter, Y. Wu, N. Kneisel, M. Seiffert, R. Hell, K. Hiller, P. Lichter and B. Radlwimmer, Branched-chain ketoacids secreted by glioblastoma cells via MCT1 modulate macrophage phenotype, EMBO Rep., 2017, 18, 2172–2185 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and supplemental figures and tables. See DOI: https://doi.org/10.1039/d5sc02378e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |