Open Access Article
Open Access ArticleTowards site-specific manipulation in cysteine-mediated redox signaling
Jing
Yang
 *abc
*abc
aGuangzhou National Laboratory, Guangzhou International Bio-Island, Guangzhou, China. E-mail: yangjing@ncpsb.org.cn
bSchool of Pharmaceutical Sciences, Guangzhou Medical University, Guangzhou, China
cState Key Laboratory of Medical Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences – Beijing, Beijing Institute of Lifeomics, Beijing, China
First published on 24th April 2025
Abstract
Cysteine sulfenic acid (SOH) modifications are pivotal in redox signaling, yet establishing their causal biological roles remains challenging due to methodological limitations. Traditional approaches often lack precision or disrupt non-redox cysteine functions. This perspective highlights two innovative chemical biology strategies to address these challenges: (1) integrating bioorthogonal cleavage chemistry with genetic code expansion for site-specific SOH incorporation in proteins of interest, enabling controlled activation of redox events, and (2) developing redox-targeted covalent inhibitors (TCIs) to selectively block SOH modifications. By bridging technological innovation with mechanistic inquiry, these strategies not only help elucidate SOH-mediated signaling networks for a better understanding of redox biology, but also hold therapeutic promise for precise redox medicine.
Cells sense and adapt to interior environmental changes or external stresses through intricate signaling mechanisms.1 These processes often begin when signaling molecules bind to receptors on or within the cell, triggering cascades of biological events that amplify and fine-tune cellular responses. A cornerstone of signaling regulation is post-translational modification (PTM), a mechanism first elucidated in the 1950s by Edmond Fischer and Edwin Krebs.2 Their work revealed how adrenaline binding to its receptor initiates a phosphorylation cascade—a reversible enzymatic process mediated by kinases and phosphatases that ensures specificity in this canonical signaling pathway (Fig. 1A).
Over time, our understanding of signaling has expanded to include diverse signal molecules, such as reactive oxygen species (ROS).3 Hydrogen peroxide (H2O2), a non-radical ROS, exemplifies this complexity.4 While H2O2 is recognized as a signaling molecule, its reactive nature sparks debate. Specifically, H2O2 generally leads to non-enzymatic chemical reactions with specific atoms of target proteins, particularly cysteinyl thiols (SH). Among various products of H2O2-mediated oxidation, cysteine sulfenic acid (SOH, also known as sulfenylation) may serve as both a sensor and amplifier of H2O2 signals, playing a pivotal role in redox signaling pathways. Due to its labile nature, SOH is often considered an intermediate en route to additional cysteine redox modifications such as sulfinic acid (SO2H), sulfonic acid (SO3H), and persulfides (SSH). Thus, various chemical approaches have been developed for stabilizing this oxidative intermediate, serving as the method-of-choice for reliable detection of endogenous SOHs. Notably, the last decade has witnessed tremendous progress in the development of chemoselective SOH probes and chemoproteomic technologies to enable global mapping of SOH modifications in intact cells, revealing relative specificity in their formation.5–9 This specificity arises from factors such as cysteine intrinsic reactivity, protein structural motifs, and local microenvironments.10 Seminal studies underscore the biological relevance of SOH. For example, the Toledano group demonstrated in 2002 that yeast cells employ a “redox relay” mechanism (Fig. 1B): the peroxidase Gpx3 senses H2O2, forms SOH, and transfers oxidative signals via disulfide bonds to the transcription factor Yap1, regulating redox homeostasis.11 Similar mechanisms, mediated by peroxidases, operate in plants and mammals.12–16 Beyond these canonical pathways, site-specific SOH modifications are proposed to influence diverse physiological processes. Mitochondrial H2O2, for instance, induces SOH on thermogenic protein UCP1 C253 in brown fat, enhancing energy expenditure.17 In plants, SOH on C51 of the transcription factor CHE boosts salicylic acid production, enabling systemic disease resistance.18 Given that ROS production is thought to be a hallmark of pathogenesis and disease progression in many human diseases, additionally, emerging evidence demonstrates that site-specific SOH modifications play important roles in pathophysiological processes.19–21 For instance, the elevated SOH levels of pyruvate kinase M2 (PKM2) at C358 were observed in disease models of hyperglycemia and diabetes, and this specific cysteine oxidation event was potentially associated with the progression of diabetic glomerular pathology by decreasing PKM2 activity.19
Despite these advances, skepticism persists regarding the causal link between SOH modifications and biological outcomes. Current experimental approaches face limitations (Fig. 2A): gain-of-function methods (e.g., adding exogenous oxidants or expressing H2O2-generating enzymes) lack precision, while loss-of-function strategies (e.g., chemical alkylation or genetic mutagenesis) disrupt non-redox cysteine roles, such as structural disulfide or metal binding. To address these challenges, new methodologies are warranted to selectively induce or block SOH modifications in specific proteins without perturbing other redox processes in living systems. This perspective highlights emerging chemical biology strategies for site-specific manipulation of SOH and discusses their potential to resolve longstanding debates in cysteine-mediated redox signaling. We further explore the translational promise of redox-based, targeted covalent inhibitors (TCIs), which could target dysregulated pathways in diseases linked to oxidative stress. By bridging mechanistic insights with innovative tools, this field is poised to redefine our understanding of redox biology in health and disease.
Gain-of-function manipulation of specific SOH events on target proteins
Bioorthogonal cleavage chemistry, combined with genetic code expansion, has emerged as a transformative tool for precise protein manipulation in living systems.22 This approach enables spatially and temporally controlled activation of proteins by releasing functional groups on demand.23,24 Building on these advances, we propose leveraging bioorthogonal cleavage chemistry and genetic code expansion to achieve site-specific incorporation of SOH within a protein of interest (POI). By replacing a target cysteine residue with chemically “caged” unnatural amino acids (UAAs), the redox-sensitive SOH modification could be masked during protein synthesis and later activated via bioorthogonal cleavage chemistry under controlled conditions (Fig. 2B). Current bioorthogonal cleavage chemistry strategies predominantly focus on releasing amine or hydroxyl groups through transition-metal catalysis or small-molecule triggers, limiting their application to residues like lysine, serine, or tyrosine.25 However, traditional photocaged cysteine derivatives offer a versatile solution.26 For instance, light-controlled activation of catalytic cysteines has been demonstrated in enzymes such as DNA methyltransferase (DNMT), where a photocaged UAA temporarily blocks enzymatic activity until UV irradiation restores function.27 This principle could be adapted to redox biology by developing analogous systems for SOH formation.Despite this potential, photoactivation of Cys-SOH remains underexplored. Pioneering work by the Carroll group introduced photocaged cysteine sulfoxide analogs, where ultraviolet (UV) light cleaves photolabile groups (e.g., ortho-nitrobenzyl, ONB or 4,5-dimethoxy-2-nitrobenzyl, DMNB, Fig. 2B) to generate SOH quantitatively in aqueous environments at neutral pH values.28 Notably, these precursors resist reduction by using methionine sulfoxide reductase, ensuring intracellular stability. While incorporated into peroxidase Gpx3, such photocaged sulfoxides enabled controlled SOH formation, highlighting their potential utility for studying redox signaling. To extend this strategy to genetic encoding, orthogonal synthetase/tRNA pairs must be developed. While EcLeu synthetase—previously used to incorporate DMNB-caged serine29—could theoretically adapt to DMNB-caged cysteine sulfoxide, experimental validation is needed. Alternatively, directed evolution of synthetases may yield systems with enhanced specificity. On the other hand, a key limitation of UV-dependent decaging is its unintended generation of ROS. Future efforts should prioritize bioorthogonal decaging methods that avoid ROS induction, ensuring precise SOH activation without perturbing redox homeostasis.
Loss-of-function manipulation of specific SOH events on target proteins
The success of covalent kinase inhibitors, such as the FDA (US Food and Drug Administration)-approved drugs afatinib and ibrutinib, demonstrates the feasibility of designing covalent small molecules with high target specificity.30 These inhibitors selectively bind to cysteine residues (Cys797 in the epidermal growth factor receptor and EGFR and Cys215 in Bruton's tyrosine kinase, BTK) through a two-step mechanism. First, the inhibitor reversibly associates with its target, positioning a weakly electrophilic group (“warhead”) near a nucleophilic residue (typically cysteine in a reduced form). This proximity enables covalent bond formation, irreversibly blocking protein function. With this in mind, we reasoned that it is also possible for developing TCIs to afford loss-of-function manipulation of SOH events in a site/target-specific manner (Fig. 2C).Progress in bioconjugation methods for chemoselective labeling and detecting SOH modifications has laid a foundation for such efforts.31 However, existing tools are primarily designed for broad profiling of SOH across the proteome, rather than selective targeting. A key challenge lies in balancing reactivity: while state-of-the-art chemoselective SOH warheads or probes prioritize strong labeling efficiency, TCIs require warheads with precisely tuned, moderate reactivity to avoid off-target effects. Early examples of redox-dependent TCIs include dimedone-based compounds, which pair this nucleophilic warhead with binding modules targeting protein tyrosine phosphatases (PTPs).32,33 However, these initial designs lack proteome-wide specificity, and their biological impact on SOH-mediated PTP regulation remains unexplored. Peroxidases—critical sensors of H2O2 as aforementioned—represent another promising yet challenging target class for redox-based TCIs. These enzymes rely on reactive cysteine residues for catalysis, but their structural complexity has hindered small-molecule inhibitor development. Most known peroxidase inhibitors are discovered by serendipity or high-throughput screening rather than de novo structure-based design.34–37 Notably, they typically contain electrophilic groups that covalently modify catalytic/peroxidatic or resolving cysteines. Replacing these groups with nucleophilic warheads reactive toward SOH could offer a novel route to probe peroxidase-mediated redox signaling.
Beyond target-centric approaches, recent advances in combining covalent ligand screening and activity-based protein profiling (ABPP, a chemoproteomic strategy that employs covalent probes to directly interrogate protein function within complex proteomes38) have provided versatile opportunities to discover reactive ligandable hotspots across complex proteomes and to generate small molecules that selectively bind challenging protein targets.39,40 This strategy was pioneered by Cravatt using ABPP to identify proteome-wide targets of a library of electrophilic covalent ligands by using competing individual ligands against a broad-spectrum thiol-reactive probe (i.e., iodoacetamide-alkyne).41 Since its first report in 2016, the strategy has been widely used for ‘high-throughput’ serendipitous discovery of ligandable cysteines, leading to the identification of cysteine-oriented TCIs of proteins previously viewed as “undruggable”.42–48 Moreover, this strategy has also been adapted by targeting other amino acid residues including lysine,46,49–51 tyrosine,52–54 serine,55 aspartate and glutamate,56 and extends beyond cysteine as a source for TCI development. Inspired by these efforts, we reasoned that this compound-centric strategy could also be adapted to expand the ligandability map of the oxidized cysteine proteome and enable discovery of SOH-reactive ligands. Thus, we recently developed a library of 65 SOH-reactive compounds based on cyanoacetamide and nitroacetamide scaffolds (Fig. 2C), which exhibit mild reactivity toward SOHs.57 Screening this library against thousands of cellular SOHs identified 524 liganded sites on 441 proteins. Functional studies revealed that select compounds disrupt protein interactions: one ligand blocked oxidized PRXL2A from suppressing MAPK signaling, while others inhibited nuclear transport of HDGF or DNA repair by BCCIP. Though further optimization (e.g., designing more advanced chemical probes through traditional structure–activity relationship-based methods) is needed to enhance specificity, this global map of ligand–SOH interactions provides a roadmap for developing TCIs that selectively inhibit SOH-driven signaling pathways.
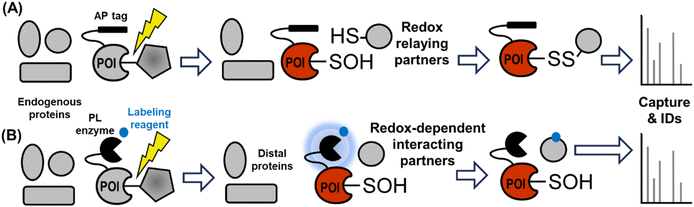
Future outlook
Causal validation of SOH-mediated redox signaling remains a critical frontier in biology. While advances in SOH-specific probes and chemoproteomics have greatly expanded catalogs of SOH sites, their functional consequences in living systems are often obscured by technical limitations inherent to traditional methods. To address this, we propose two complementary chemical biology strategies: (1) integrating bioorthogonal cleavage chemistry with genetic code expansion to enable site-specific SOH incorporation in POIs, and (2) designing redox-dependent TCIs to selectively block site-specific SOH modifications in POIs. These approaches not only establish causal links between SOH events and their biological outcomes but also empower global mapping of SOH-driven protein interaction networks. For instance, POIs engineered with inducible SOHs could serve as baits for affinity purification-mass spectrometry (AP-MS)58 or proximity labeling-mass spectrometry (PL-MS)59 to identify downstream redox-relaying effectors or redox-dependent interacting partners that may empower the identification of ‘erasers’ (e.g., reductases) of specific SOH events and redox-independent ‘readers’ of this type of modification (Fig. 3). In a complementary way, using a redox-dependent TCI allows us to chemically perturb one specific SOH event on a POI, followed by the measurements of resulting changes in the interacting partners of this target.Beyond basic research, redox-based TCIs hold therapeutic promise. For example, oxidized cysteines in oncogenic targets like EGFR or KRAS G12C may drive resistance to conventional electrophilic inhibitors, which fail to engage oxidized residues.60–62 Nucleophilic TCIs targeting these oxidized states could circumvent such resistance. However, challenges remain: the transient nature of SOH demands careful consideration of stoichiometry and irreversible oxidation, while distinguishing functional roles of reduced versus oxidized protein states adds complexity. Conversely, the context-dependent increase in SOH in disease states (e.g., cancers with elevated oxidative stress) offers opportunities for disease-state-specific targeting. Expanding libraries of nucleophilic warheads paired with optimized binding modules will accelerate discovery of druggable SOH sites and refine lead compounds.
Collectively, these strategies bridge technological innovation with biological inquiry, poised to transform our understanding of redox signaling. By engaging chemists and chemical biologists in refining precision tools, this nascent field promises not only mechanistic insights into redox biology but also novel therapeutic avenues for precise redox medicine.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
J. Y. conceived the perspective and wrote the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The work was supported by grants from the National Key R&D Program of China (2022YFA1304700) to J. Y. and the National Natural Science Foundation of China (81973279 and 21922702) to J. Y.References
- M. J. Lee and M. B. Yaffe, Protein Regulation in Signal Transduction, Cold Spring Harbor Perspect. Biol., 2016, 8, a005918 CrossRef PubMed.
- N. Kresge, R. D. Simoni and R. L. Hill, The process of reversible phosphorylation: the work of Edmond H. Fischer, J. Biol. Chem., 2011, 286, e1–e2 CrossRef CAS PubMed.
- B. D'Autreaux and M. B. Toledano, ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis, Nat. Rev. Mol. Cell Biol., 2007, 8, 813–824 CrossRef PubMed.
- T. F. Brewer, F. J. Garcia, C. S. Onak, K. S. Carroll and C. J. Chang, Chemical approaches to discovery and study of sources and targets of hydrogen peroxide redox signaling through NADPH oxidase proteins, Annu. Rev. Biochem., 2015, 84, 765–790 CrossRef CAS PubMed.
- L. Fu, K. Liu, R. B. Ferreira, K. S. Carroll and J. Yang, Proteome-Wide Analysis of Cysteine S-Sulfenylation Using a Benzothiazine-Based Probe, Curr. Protoc. Protein Sci., 2019, 95, e76 CrossRef PubMed.
- Y. Shi, L. Fu, J. Yang and K. S. Carroll, Wittig reagents for chemoselective sulfenic acid ligation enables global site stoichiometry analysis and redox-controlled mitochondrial targeting, Nat. Chem., 2021, 13, 1140–1150 CrossRef CAS PubMed.
- J. Meng, L. Fu, K. Liu, C. Tian, Z. Wu, Y. Jung, R. B. Ferreira, K. S. Carroll, T. K. Blackwell and J. Yang, Global profiling of distinct cysteine redox forms reveals wide-ranging redox regulation in C. elegans, Nat. Commun., 2021, 12, 1415 CrossRef CAS.
- J. Yang, V. Gupta, K. S. Carroll and D. C. Liebler, Site-specific mapping and quantification of protein S-sulphenylation in cells, Nat. Commun., 2014, 5, 4776 CrossRef CAS PubMed.
- J. Yang, V. Gupta, K. A. Tallman, N. A. Porter, K. S. Carroll and D. C. Liebler, Global, in situ, site-specific analysis of protein S-sulfenylation, Nat. Protoc., 2015, 10, 1022–1037 CrossRef CAS PubMed.
- H. Xiao, M. P. Jedrychowski, D. K. Schweppe, E. L. Huttlin, Q. Yu, D. E. Heppner, J. Li, J. Long, E. L. Mills, J. Szpyt, Z. He, G. Du, R. Garrity, A. Reddy, L. P. Vaites, J. A. Paulo, T. Zhang, N. S. Gray, S. P. Gygi and E. T. Chouchani, A Quantitative Tissue-Specific Landscape of Protein Redox Regulation during Aging, Cell, 2020, 180, 968–983 CrossRef CAS PubMed.
- A. Delaunay, D. Pflieger, M. B. Barrault, J. Vinh and M. B. Toledano, A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation, Cell, 2002, 111, 471–481 CrossRef CAS PubMed.
- S. Stocker, M. Maurer, T. Ruppert and T. P. Dick, A role for 2-Cys peroxiredoxins in facilitating cytosolic protein thiol oxidation, Nat. Chem. Biol., 2018, 14, 148–155 CrossRef PubMed.
- M. C. Sobotta, W. Liou, S. Stocker, D. Talwar, M. Oehler, T. Ruppert, A. N. Scharf and T. P. Dick, Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling, Nat. Chem. Biol., 2015, 11, 64–70 CrossRef CAS PubMed.
- G. Bi, M. Hu, L. Fu, X. Zhang, J. Zuo, J. Li, J. Yang and J. M. Zhou, The cytosolic thiol peroxidase PRXIIB is an intracellular sensor for H2O2 that regulates plant immunity through a redox relay, Nat. Plants, 2022, 8, 1160–1175 CrossRef CAS PubMed.
- T. N. Vo, J. Malo Pueyo, K. Wahni, D. Ezerina, J. Bolduc and J. Messens, Prdx1 Interacts with ASK1 upon Exposure to H2O2 and Independently of a Scaffolding Protein, Antioxidants, 2021, 10, 1060 CrossRef CAS.
- P. E. Pace, A. V. Peskin, A. Konigstorfer, C. J. Jasoni, C. C. Winterbourn and M. B. Hampton, Peroxiredoxin interaction with the cytoskeletal-regulatory protein CRMP2: investigation of a putative redox relay, Free Radical Biol. Med., 2018, 129, 383–393 CrossRef CAS.
- E. T. Chouchani, L. Kazak, M. P. Jedrychowski, G. Z. Lu, B. K. Erickson, J. Szpyt, K. A. Pierce, D. Laznik-Bogoslavski, R. Vetrivelan, C. B. Clish, A. J. Robinson, S. P. Gygi and B. M. Spiegelman, Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1, Nature, 2016, 532, 112–116 CrossRef CAS.
- L. Cao, S. Karapetyan, H. Yoo, T. Chen, M. Mwimba, X. Zhang and X. Dong, H2O2 sulfenylates CHE, linking local infection to the establishment of systemic acquired resistance, Science, 2024, 385, 1211–1217 CrossRef CAS.
- W. Qi, H. A. Keenan, Q. Li, A. Ishikado, A. Kannt, T. Sadowski, M. A. Yorek, I. H. Wu, S. Lockhart, L. J. Coppey, A. Pfenninger, C. W. Liew, G. Qiang, A. M. Burkart, S. Hastings, D. Pober, C. Cahill, M. A. Niewczas, W. J. Israelsen, L. Tinsley, I. E. Stillman, P. S. Amenta, E. P. Feener, M. G. Vander Heiden, R. C. Stanton and G. L. King, Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction, Nat. Med., 2017, 23, 753–762 CrossRef CAS PubMed.
- N. Zhou, J. Chen, Z. Ling, C. Zhang, Y. Zhou, D. Wang, L. Zhou, Z. Wang, N. Sun, X. Wang, H. Zhang, K. Tang, J. Ma, J. Lv and B. Huang, Aryl hydrocarbon receptor sulfenylation promotes glycogenolysis and rescues cancer chemoresistance, J. Clin. Invest., 2023, 133, e170753 CrossRef CAS PubMed.
- B. Mu, Y. Zeng, L. Luo and K. Wang, Oxidative stress-mediated protein sulfenylation in human diseases: past, present, and future, Redox Biol., 2024, 76, 103332 CrossRef CAS PubMed.
- J. Wang, X. Wang, X. Fan and P. R. Chen, Unleashing the Power of Bond Cleavage Chemistry in Living Systems, ACS Cent. Sci., 2021, 7, 929–943 CrossRef CAS PubMed.
- J. Wang, Y. Liu, Y. Liu, S. Zheng, X. Wang, J. Zhao, F. Yang, G. Zhang, C. Wang and P. R. Chen, Time-resolved protein activation by proximal decaging in living systems, Nature, 2019, 569, 509–513 CrossRef CAS PubMed.
- S. Tang, A. T. Beattie, L. Kafkova, G. Petris, N. Huguenin-Dezot, M. Fiedler, M. Freeman and J. W. Chin, Mechanism-based traps enable protease and hydrolase substrate discovery, Nature, 2022, 602, 701–707 CrossRef CAS.
- M. Charette, C. Rosenblum, O. Shade and A. Deiters, Optogenetics with Atomic Precision horizontal line A Comprehensive Review of Optical Control of Protein Function through Genetic Code Expansion, Chem. Rev., 2025, 125, 1663–1717 CrossRef CAS.
- C. Y. Chang, B. Niblack, B. Walker and H. Bayley, A photogenerated pore-forming protein, Chem. Biol., 1995, 2, 391–400 CrossRef CAS.
- J. Wolffgramm, B. Buchmuller, S. Palei, A. Munoz-Lopez, J. Kanne, P. Janning, M. R. Schweiger and D. Summerer, Light-Activation of DNA-Methyltransferases, Angew Chem. Int. Ed. Engl., 2021, 60, 13507–13512 CrossRef CAS PubMed.
- J. Pan and K. S. Carroll, Light-Mediated Sulfenic Acid Generation from Photocaged Cysteine Sulfoxide, Org. Lett., 2015, 17, 6014–6017 CrossRef CAS PubMed.
- E. A. Lemke, D. Summerer, B. H. Geierstanger, S. M. Brittain and P. G. Schultz, Control of protein phosphorylation with a genetically encoded photocaged amino acid, Nat. Chem. Biol., 2007, 3, 769–772 CrossRef CAS PubMed.
- L. Boike, N. J. Henning and D. K. Nomura, Advances in covalent drug discovery, Nat. Rev. Drug Discovery, 2022, 21, 881–898 CrossRef CAS.
- L. J. Alcock, M. V. Perkins and J. M. Chalker, Chemical methods for mapping cysteine oxidation, Chem. Soc. Rev., 2018, 47, 231–268 RSC.
- S. E. Leonard, F. J. Garcia, D. S. Goodsell and K. S. Carroll, Redox-based probes for protein tyrosine phosphatases, Angew Chem. Int. Ed. Engl., 2011, 50, 4423–4427 CrossRef CAS.
- F. J. Garcia and K. S. Carroll, Redox-based probes as tools to monitor oxidized protein tyrosine phosphatases in living cells, Eur. J. Med. Chem., 2014, 88, 28–33 CrossRef CAS PubMed.
- C. X. Liu, Q. Q. Yin, H. C. Zhou, Y. L. Wu, J. X. Pu, L. Xia, W. Liu, X. Huang, T. Jiang, M. X. Wu, L. C. He, Y. X. Zhao, X. L. Wang, W. L. Xiao, H. Z. Chen, Q. Zhao, A. W. Zhou, L. S. Wang, H. D. Sun and G. Q. Chen, Adenanthin targets peroxiredoxin I and II to induce differentiation of leukemic cells, Nat. Chem. Biol., 2012, 8, 486–493 CrossRef CAS PubMed.
- J. D. Haraldsen, G. Liu, C. H. Botting, J. G. Walton, J. Storm, T. J. Phalen, L. Y. Kwok, D. Soldati-Favre, N. H. Heintz, S. Muller, N. J. Westwood and G. E. Ward, Identification of Conoidin a as a Covalent Inhibitor of Peroxiredoxin II, Org. Biomol. Chem., 2009, 7, 3040–3048 RSC.
- W. Wei, C. Ma, Y. Cao, L. Yang, Z. Huang, D. Qin, Y. Chen, C. Liu, L. Xia, T. Wang, H. Lei, Y. Yu, M. Huang, Y. Tong, H. Xu, F. Gao, J. Zhang and Y. L. Wu, Identification of H7 as a novel peroxiredoxin I inhibitor to induce differentiation of leukemia cells, Oncotarget, 2016, 7, 3873–3883 CrossRef.
- H. Xu, H. Zhao, C. Ding, D. Jiang, Z. Zhao, Y. Li, X. Ding, J. Gao, H. Zhou, C. Luo, G. Chen, A. Zhang, Y. Xu and H. Zhang, Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1, Signal Transduction Targeted Ther., 2023, 8, 51 CrossRef CAS.
- E. O. J. Porta and P. G. Steel, Activity-based protein profiling: A graphical review, Curr. Res. Pharmacol. Drug Discov., 2023, 5, 100164 CrossRef.
- M. J. Niphakis and B. F. Cravatt, Ligand discovery by activity-based protein profiling, Cell Chem. Biol., 2024, 31, 1636–1651 CrossRef CAS.
- I. Forrest and C. G. Parker, Proteome-Wide Fragment-Based Ligand and Target Discovery, Isr. J. Chem., 2023, 63, e202200098 CrossRef CAS.
- K. M. Backus, B. E. Correia, K. M. Lum, S. Forli, B. D. Horning, G. E. Gonzalez-Paez, S. Chatterjee, B. R. Lanning, J. R. Teijaro, A. J. Olson, D. W. Wolan and B. F. Cravatt, Proteome-wide covalent ligand discovery in native biological systems, Nature, 2016, 534, 570–574 CrossRef CAS PubMed.
- S. G. Kathman, S. J. Koo, G. L. Lindsey, H. L. Her, S. M. Blue, H. Li, S. Jaensch, J. R. Remsberg, K. Ahn, G. W. Yeo, B. Ghosh and B. F. Cravatt, Remodeling oncogenic transcriptomes by small molecules targeting NONO, Nat. Chem. Biol., 2023, 19, 825–836 CrossRef CAS PubMed.
- M. E. Kavanagh, B. D. Horning, R. Khattri, N. Roy, J. P. Lu, L. R. Whitby, E. Ye, J. C. Brannon, A. Parker, J. M. Chick, C. L. Eissler, A. J. Wong, J. L. Rodriguez, S. Rodiles, K. Masuda, J. R. Teijaro, G. M. Simon, M. P. Patricelli and B. F. Cravatt, Selective inhibitors of JAK1 targeting an isoform-restricted allosteric cysteine, Nat. Chem. Biol., 2022, 18, 1388–1398 CrossRef CAS PubMed.
- E. Njomen, R. E. Hayward, K. E. DeMeester, D. Ogasawara, M. M. Dix, T. Nguyen, P. Ashby, G. M. Simon, S. L. Schreiber, B. Melillo and B. F. Cravatt, Multi-tiered chemical proteomic maps of tryptoline acrylamide-protein interactions in cancer cells, Nat. Chem., 2024, 16, 1592–1604 CrossRef CAS PubMed.
- E. V. Vinogradova, X. Zhang, D. Remillard, D. C. Lazar, R. M. Suciu, Y. Wang, G. Bianco, Y. Yamashita, V. M. Crowley, M. A. Schafroth, M. Yokoyama, D. B. Konrad, K. M. Lum, G. M. Simon, E. K. Kemper, M. R. Lazear, S. Yin, M. M. Blewett, M. M. Dix, N. Nguyen, M. N. Shokhirev, E. N. Chin, L. L. Lairson, B. Melillo, S. L. Schreiber, S. Forli, J. R. Teijaro and B. F. Cravatt, An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells, Cell, 2020, 182, 1009–1026 CrossRef CAS PubMed.
- Y. Wang, M. M. Dix, G. Bianco, J. R. Remsberg, H. Y. Lee, M. Kalocsay, S. P. Gygi, S. Forli, G. Vite, R. M. Lawrence, C. G. Parker and B. F. Cravatt, Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs, Nat. Chem., 2019, 11, 1113–1123 CrossRef CAS.
- X. Zhang, V. M. Crowley, T. G. Wucherpfennig, M. M. Dix and B. F. Cravatt, Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16, Nat. Chem. Biol., 2019, 15, 737–746 CrossRef CAS.
- Y. Zhang, Z. Liu, M. Hirschi, O. Brodsky, E. Johnson, S. J. Won, A. Nagata, D. Bezwada, M. D. Petroski, J. D. Majmudar, S. Niessen, T. VanArsdale, A. M. Gilbert, M. M. Hayward, A. E. Stewart, A. R. Nager, B. Melillo and B. F. Cravatt, An allosteric cyclin E-CDK2 site mapped by paralog hopping with covalent probes, Nat. Chem. Biol., 2025, 21, 420–431 CrossRef CAS.
- M. E. Abbasov, M. E. Kavanagh, T. A. Ichu, M. R. Lazear, Y. Tao, V. M. Crowley, C. W. Am Ende, S. M. Hacker, J. Ho, M. M. Dix, R. Suciu, M. M. Hayward, L. L. Kiessling and B. F. Cravatt, A proteome-wide atlas of lysine-reactive chemistry, Nat. Chem., 2021, 13, 1081–1092 CrossRef CAS.
- S. M. Hacker, K. M. Backus, M. R. Lazear, S. Forli, B. E. Correia and B. F. Cravatt, Global profiling of lysine reactivity and ligandability in the human proteome, Nat. Chem., 2017, 9, 1181–1190 CrossRef CAS.
- A. Cuesta and J. Taunton, Lysine-Targeted Inhibitors and Chemoproteomic Probes, Annu. Rev. Biochem., 2019, 88, 365–381 CrossRef CAS.
- H. S. Hahm, E. K. Toroitich, A. L. Borne, J. W. Brulet, A. H. Libby, K. Yuan, T. B. Ware, R. L. McCloud, A. M. Ciancone and K. L. Hsu, Global targeting of functional tyrosines using sulfur-triazole exchange chemistry, Nat. Chem. Biol., 2020, 16, 150–159 CrossRef CAS PubMed.
- J. W. Brulet, A. L. Borne, K. Yuan, A. H. Libby and K. L. Hsu, Liganding Functional Tyrosine Sites on Proteins Using Sulfur-Triazole Exchange Chemistry, J. Am. Chem. Soc., 2020, 142, 8270–8280 CrossRef CAS PubMed.
- A. M. Ciancone, S. Hosseinibarkooie, D. L. Bai, A. L. Borne, H. A. Ferris and K. L. Hsu, Global profiling identifies a stress-responsive tyrosine site on EDC3 regulating biomolecular condensate formation, Cell Chem. Biol., 2022, 29, 1709–1720 CrossRef CAS PubMed.
- H. A. Sharma, M. Bielecki, M. A. Holm, T. M. Thompson, Y. Yin, J. B. Cravatt, T. B. Ware, A. Reed, M. Nassir, T. E. Ewing, B. Melillo, J. F. Bazan, P. S. Baran and B. F. Cravatt, Proteomic Ligandability Maps of Phosphorus(V) Stereoprobes Identify Covalent TLCD1 Inhibitors, bioRxiv, 2025, preprint, DOI:10.1101/2025.01.31.635883.
- K. Bach, B. L. H. Beerkens, P. R. A. Zanon and S. M. Hacker, Light-Activatable, 2,5-Disubstituted Tetrazoles for the Proteome-wide Profiling of Aspartates and Glutamates in Living Bacteria, ACS Cent. Sci., 2020, 6, 546–554 CrossRef CAS.
- L. Fu, Y. Jung, C. Tian, R. B. Ferreira, R. Cheng, F. He, J. Yang and K. S. Carroll, Nucleophilic covalent ligand discovery for the cysteine redoxome, Nat. Chem. Biol., 2023, 19, 1309–1319 CrossRef CAS.
- A. Bauer and B. Kuster, Affinity purification-mass spectrometry. Powerful tools for the characterization of protein complexes, Eur. J. Biochem., 2003, 270, 570–578 CrossRef CAS PubMed.
- W. Qin, K. F. Cho, P. E. Cavanagh and A. Y. Ting, Deciphering molecular interactions by proximity labeling, Nat. Methods, 2021, 18, 133–143 CrossRef CAS PubMed.
- T. H. Truong, P. M. Ung, P. B. Palde, C. E. Paulsen, A. Schlessinger and K. S. Carroll, Molecular Basis for Redox Activation of Epidermal Growth Factor Receptor Kinase, Cell Chem. Biol., 2016, 23, 837–848 CrossRef CAS PubMed.
- M. V. Huynh, D. Parsonage, T. E. Forshaw, V. R. Chirasani, G. A. Hobbs, H. Wu, J. Lee, C. M. Furdui, L. B. Poole and S. L. Campbell, Oncogenic KRAS G12C: kinetic and redox characterization of covalent inhibition, J. Biol. Chem., 2022, 298, 102186 CrossRef CAS PubMed.
- P. A. Schwartz, P. Kuzmic, J. Solowiej, S. Bergqvist, B. Bolanos, C. Almaden, A. Nagata, K. Ryan, J. Feng, D. Dalvie, J. C. Kath, M. Xu, R. Wani and B. W. Murray, Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 173–178 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |