 Open Access Article
Open Access ArticleBiosynthesis of reveromycin derivatives by altering the regioselectivity of cytochrome P450revI†
Ya Fen
Yong
 ab,
Song
Liu
ac,
Katsuyuki
Sakai
ab,
Song
Liu
ac,
Katsuyuki
Sakai
 a,
Keisuke
Fujiyama
a,
Keisuke
Fujiyama
 a,
Hiroshi
Takagi
a,
Yushi
Futamura
a,
Hiroshi
Takagi
a,
Yushi
Futamura
 de,
Takeshi
Shimizu
d,
Hiroyuki
Osada
de,
Takeshi
Shimizu
d,
Hiroyuki
Osada
 de,
Eugene Boon Beng
Ong
de,
Eugene Boon Beng
Ong
 b and
Shunji
Takahashi
b and
Shunji
Takahashi
 *a
*a
aNatural Product Biosynthesis Research Unit, RIKEN Center for Sustainable Resource Science, Wako, Saitama 351-0198, Japan. E-mail: shunjitaka@riken.jp
bInstitute for Research in Molecular Medicine, Universiti Sains Malaysia, 11800 USM, Penang, Malaysia
cScience Center for Future Foods, Jiangnan University, Lihu Ave. 1800, Wuxi, Jiangsu 214122, China
dChemical Resource Development Research Unit and Drug Discovery Chemical Bank Unit, RIKEN Center for Sustainable Resource Science, Wako, Saitama 351-0198, Japan
eInstitute of Microbial Chemistry, Shinagawa, Tokyo 141-0021, Japan
First published on 23rd June 2025
Abstract
Reveromycin A (RM-A) (1) has a 6,6-spiroacetal core structure that is important for its biological activity. However, 1 undergoes a spiroacetal rearrangement to form RM-B (2) with a 5,6-spiroacetal core, which exhibits reduced bioactivity. This undesired rearrangement is partly due to the hemisuccinate moiety at the C18 position of 1. In 1 biosynthesis, P450revI catalyses the C18-hydroxylation of RM-T (3), which is essential for its subsequent hemisuccinylation to generate 1. In this study, we aimed to alter the P450revI regioselectivity to improve the stability of the 6,6-spiroacetal core and expand the structural diversity of RMs. Candidate amino acid residues for mutagenesis studies were selected by comparing the co-crystal structure of P450revI with the docking models of the P450revI mutant-3 complexes. Notably, the P450revI-A241L mutant selectively produced novel RM derivatives. Nuclear magnetic resonance analysis revealed that P450revI-A241L catalysed the C17-hydroxylation of 3 to produce 17-hydroxy-RM-T (6). Co-crystal structure analysis of the P450revI-A241L-3 complex revealed that the pro-R hydrogen at the C17 position faces toward the haem iron. Introduction of the P450revI-A241L mutant gene into the Actinacidiphila reveromycinica SN-593-ΔrevI strain led to the production of 17-hemisuccinyloxy-RM-T (7). After the successful bioproduction of RM derivatives, we evaluated their structural stabilities and biological activities. Compounds 6 and 7 exhibited better stabilities than 18-hydroxylated-3 (RM-T1; 4) and 1, respectively. Biological activity analysis revealed that 6 and 7 exhibited anti-malarial and anti-multiple myeloma activities, respectively, comparable to those of 1 and 3, while showing low cytotoxicity against human cell lines. Overall, this study highlights the potential of RM derivatives as pharmaceuticals.
Introduction
Natural products with diverse chemical structures and properties are of great significance for drug discovery. To date, natural products and their derivatives account for over one-third of the therapeutic agents approved for human use.1 Modifications of natural product scaffolds facilitate structure–activity relationship studies to enhance their biological activity and selectivity,2–4 optimise their drug-like properties,5–7 and elucidate their mechanisms of action.8 These pave the way to the development of new drugs to address unmet medical needs.Reveromycins (RMs) are a class of polyketide compounds isolated from Streptomyces sp. SN-593,9 which was recently classified as Actinacidiphila reveromycinica SN-593.10 The RM structure contains a 6,6- or 5,6-spiroacetal core, two polyene carboxylic acids, two alkyl groups, and a hemisuccinate moiety (Fig. 1a).9,11–14 RM-A (1) exhibits various biological activities, including anti-osteoclastic,15 anti-periodontitis,16–18 anti-tumour,19,20 and anti-fungal effects.21,22 The molecular target of 1 is eukaryotic isoleucyl-tRNA synthetase (IleRS),23,24 a housekeeping enzyme that charges tRNA with isoleucine at the initial phase of protein synthesis. Notably, an acidic environment is closely related to the efficacy of 1.15,21,22 Under acidic conditions, proton dissociation of tricarboxylic acid-containing 1 is suppressed, thereby enhancing its cell permeability. Considering the selectivity for eukaryotic IleRSs, it is intriguing to diversify the chemical scaffold of 1 for creating drug-like compounds.
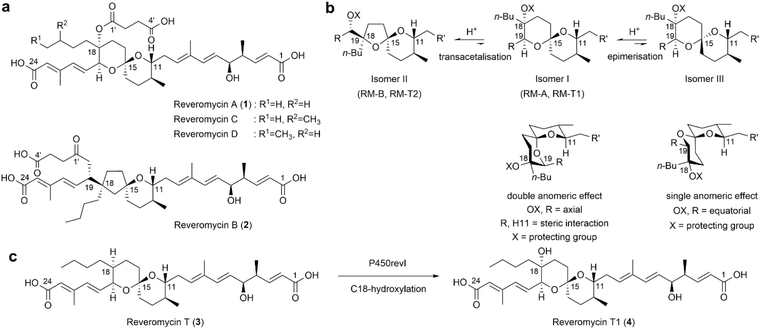 | ||
| Fig. 1 Chemical structures of RMs. (a) RMs isolated from Actinacidiphila reveromycinica SN-593. (b) Transacetalisation of RM from 6,6-spiroacetal to the 5,6-spiroacetal structure. Chemical synthetic studies revealed that the presence of axially oriented C18 and C19 substituted groups introduced steric hindrance into the 6,6-spiroacetal core (Isomer I). This strain was reduced via structure rearrangement to the 5,6-spiroacetal core (Isomer II)26,28,29 or epimerisation to form Isomer III26,27 under acidic conditions.12,26–30,35 (c) P450revI catalyses hydroxylation of RM-T (3) to form a C18-hydroxylated product, RM-T1 (4). | ||
RM-B (2), which possesses a 5,6-spiroacetal core (Fig. 1a), exhibits diminished biological activity compared to 6,6-spiroacetal RMs.21,25 Compound 1 has a thermodynamically preferred 6,6-spiroacetal core (isomer I) with a double anomeric effect. However, axial orientation of the C18 hemisuccinate moiety and C19 diene acid side chain14 and steric hindrance between the H11 and C19 substituents exerts strain12 on the 6,6-spiroacetal core of 1, leading to its spiroacetal epimerisation (from the isomer I core to the isomer III core; Fig. 1b).26,27 When the C18 tertiary hydroxyl group is free, transacetalisation of the 6,6-spiroacetal core to the 5,6-spiroacetal core occurs. Inherent instability of the 6,6-spiroacetal core28 has been shown in synthetic studies of 1 and 2, indicating a greater preference for the formation of the 5,6-spiroacetal core under acidic conditions (Fig. 1b).26,28–34
Cytochrome P450s are ubiquitously distributed haem-containing biocatalysts.36 They play crucial roles in bioactive natural product biosynthesis by catalysing precise stereo- or regioselective C–H activation,37–39 enabling the modifications of the building blocks40,41 and chemical scaffolds,42,43 thereby achieving structural diversification.42–48 As selective C–H functionalisation remains a challenge in synthetic chemistry,49,50 various P450 engineering strategies, such as rational design,39,51–54 directed evolution,55–57 and semi-rational design,58,59 have been established to create valuable compounds.60 We previously elucidated the biosynthetic pathway of 1 in A. reveromycinica SN-593.33,34 P450revI (CYP107E6) catalyses the C18-hydroxylation of RM-T (3), which is essential for hemisuccinate formation for the biosynthesis of 1 (Fig. 1c and S1†).34
Axial orientation of the substituent at the C18 position accelerates undesired transacetalisation. In this study, we hypothesised that modifying the hydroxylation site in compound 3 enhances the stability of the 6,6-spiroacetal core while introducing new biological activity. To verify this, we engineered P450revI mutants to catalyse the hydroxylation of compound 3 at an alternative carbon position. Here, we report the successful generation of a P450revI-A241L mutant efficiently catalysing hydroxylation at the C17 position of compound 3. Our synthetic biology approach enabled the substantial production of novel RM derivatives. We also evaluated the stabilities, biological activities, and potential applications of the synthesised compounds.
Results and discussion
Alteration of the regiospecific hydroxylation of 3 by the P450revI mutant
The crystal structure of P450revI in complex with 3 (PDB entry: 3WVS)34 served as the basis for mutagenesis analysis. As the haem cofactor is a central player in P450 oxidative reactions,36 we first examined the distance between the haem iron and carbon or hydrogen atoms of 3. Although P450revI catalyses C18-hydroxylation of 3, the carbon or hydrogen atom that is most proximal to the haem iron is C16/H16 (4.5/3.6 Å), followed by C17/H17 (4.9/4.3 Å) and C18/H18 (5.2/4.5 Å; Fig. S2†).34 This suggests that 3 relocates upon oxygen binding to the haem cofactor, leading to a short distance between C18/H18 and haem iron, reflecting the catalytic conformation for selective C18-hydroxylation. Given the stringent substrate recognition of P450revI through the formation of salt bridges and hydrogen bonds involving residues R81 and R190 (Fig. 2b),34 the orientation of 3 in the catalytic site of mutants is expected to remain largely similar to that in the wild-type. Therefore, to create P450revI mutants with altered regioselectivity, our strategy was to restrict the movement of the C18 of 3 toward the haem cofactor. | ||
| Fig. 2 P450revI engineering based on crystal structure and docking simulation studies. (a) Overall structure of P450revI in complex with 3 (protein data bank [PDB] entry: 3WVS). Compound 3 is indicated in yellow, and the haem cofactor is shown in grey. Two amino acid residues (R81 and R190) involved in salt bridge formation are shown as cyan sticks, and the G237 residue forming a water-mediated hydrogen bond with 3 is shown as a wheat stick. Red sphere denotes the water molecule. (b) Close-up view of the active site. Target residues (S89, G237, A241, L287, and V391) for site-directed mutagenesis are shown as wheat sticks. (c) Product profiles of wild-type P450revI and its mutants determined via ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) analysis. Absorbance at 238 nm was used to monitor the target compounds. RM-T1 (4) (m/z 559), non-enzymatically isomerised 5,6-spiroacetal product (5) (m/z 559)12,27,28 from 4, and novel hydroxylated product peak 6 (m/z 559) from substrate 3 (m/z 543) were detected in the negative-ion mode. Kinetic analyses of (d) wild-type P450revI and (e) P450revI-A241L for hydroxylation of 3. | ||
To identify the amino acid residues influencing the orientation of 3 in the active site of P450revI, we searched for the residues within 5 Å from 3 and identified 25 residues (Fig. S2c†). As R81 and R190 are essential for substrate recognition, these two residues and their proximal residues were excluded from the mutation. Owing to its role in dioxygen activation during the P450 reaction,34,61 T245 was omitted from the target residues. Furthermore, residues G290–A293 in the flexible loop were not considered for mutations based on a previous report that introducing a mutation in the flexible loop significantly reduces the P450revI activity.34
To select candidate amino acid residues for mutagenesis, the co-crystal structure of P450revI-3 and docking models of P450revI mutant-3 complexes were compared. Based on the reported distance between the haem iron and substrate in P450 s (Table S1†), P450revI mutant docking models predicted to catalyse C16- or C17- hydroxylation, but not C18-hydroxylation, were collected (Fig. S3†). First, distance from the haem iron to C16 or C17 in compound 3 must be shorter than that to C18. Second, in the mutant docking models of the P450revI mutant, the distance from the haem iron to C18 must be greater than 5.2 Å to reduce the likelihood of C18-hydroxylation (Table S1†). Based on these criteria, five amino acid residues–S89 (B′′ helix), G237, A241 (I helix), L287 (loop following the K helix), and V391 (loop following the L helix) (Fig. 2a)–were selected for site-directed mutagenesis (Fig. S3†).
To investigate whether the P450revI mutants catalyse the novel regiospecific hydroxylation of 3, wild-type and mutants were heterologously expressed in Escherichia coli and purified until homogeneity. Enzymatic assays were performed using 3 as a substrate. Samples were prepared as described in the ESI† and analysed by UPLC-MS. The product amount was quantified using a calibration curve constructed with 3 as the standard. Wild-type P450revI catalysed C18-hydroxylation of 3 to form RM-T1 (4). S89M, L287V, and V391I mutants produced a novel hydroxylated product peak 6 with m/z 559 [M–H]−, although only trace amounts were detected (Fig. 2c and S4†). The C18-hydroxylase activity of the S89M, L287V, and V391I mutants was comparable to that of wild-type P450revI (Fig. 2c and S4†). G237I, G237V, and L287A mutants produced trace amounts of peak 6 and showed significantly reduced C18-hydroxylase activity compared to the wild-type P450revI (Fig. 2c and S4†). Interestingly, A241I, A241L, A241T, and A241V mutants (Fig. 2c) showed capacity to produce peak 6. Notably, the A241L mutant yielded peak 6 as the main product with a relative activity of 25.6 ± 1.6% (Fig. 2c and S4†). A241I and A241V mutants produced peak 6 with relative activities of 40.0 ± 2.0 and 41.3 ± 4.2%, respectively, and the mutants also retained the ability to catalyse C18-hydroxylation of 3 (Fig. S4†). Product ratios of 4 to 6 for A241I and A241V were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.5 and 1
4.5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.9, respectively. Replacement of A241 with other amino acid residues abolished the activities of all mutant enzymes (Fig. S5†).
1.9, respectively. Replacement of A241 with other amino acid residues abolished the activities of all mutant enzymes (Fig. S5†).
Next, kinetic parameters of the P450revI-A241L mutant were analysed. The Km value (0.90 ± 0.10 μM) of the mutant for 3 was comparable to that of the wild-type enzyme (Fig. 2d and e). Moreover, the kcat (25.7 ± 2.5 min−1) value of the mutant was 3.9-fold lower than that of the wild-type enzyme. The kcat/Km value (28.9 min−1 μM−1) of the mutant enzyme was approximately 3.3-times lower than that of wild-type. Based on the Km value of 0.9 μM, the A241L mutant was considered suitable for the in vivo production of a novel hydroxylated product from compound 3 using A. reveromycinica SN-593.
Production of novel RM derivatives using A. reveromycinica SN-593
Obtaining substantial amounts of RM derivatives is essential to evaluate their stabilities and biological activities. We used synthetic biology approaches with the A. reveromycinica SN-593-ΔrevI strain.34The wild-type revI gene or the P450revI-A241L mutant gene was introduced into the A. reveromycinica SN-593-ΔrevI strain.34 The resulting wild-type revI transformant produced 1.13 mg L−1 of hemisuccinylated product 1. Interestingly, the P450revI-A241L mutant transformant generated the novel hydroxylated peak 6 (m/z 559 [M–H]−) and hemisuccinylated peak 7 (m/z 659 [M–H]−), Fig. 3a iii. Yields of 6 and 7 were 0.34 and 0.47 mg L−1, respectively. These results indicate that the novel hydroxylated peak 6 underwent hemisuccinylation, which is similar to the biosynthetic process of 1. Here, the revQ gene,33 which acts as a Streptomyces antibiotic regulatory protein, was introduced into the P450revI-A241L mutant-transformed strain to enhance its productivity. The yield (5.18 mg L−1) of peak 7 was increased by 11-fold in the resulting revQ transformant (Fig. 3a iv; Tables S2 and S3†). Interestingly, a small amount of compound 1 was also detected in the revQ transformant (Fig. 3a iv). This is likely due to the imperfect product specificity of the A241L mutant, leading to the production of a small amount of compound 4, which was subsequently converted to 1in vivo.
Isolation and structure determination of novel RMs
Ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) revealed peaks 6 and 7 as a novel hydroxylated compound of 3 and hemisuccinylated compound of 6, respectively. To determine the chemical structures and physicochemical properties of 6 and 7, A. reveromycinica SN-593 transformants harbouring revI-A241L-revQ gene (Table S2†) were cultivated on a large scale to obtain sufficient amounts of 6 and 7 for subsequent analyses. Compound 7 was obtained as a white amorphous powder. As evidenced by HR-ESI-ToF-MS analysis, 7 exhibited the same molecular formula as 1,12 which was C36H52O11 ([M–H]−, m/z 659.3429, calculated for C36H51O11, 659.3431; Fig. S6†). Similar to 1,12 the UV-vis spectrum of 7 showed absorption maxima at 238 and 263 nm (Fig. S7†). The infrared spectrum indicated characteristic absorption at 1697 cm−1 (Fig. S8†), indicating the presence of carbonyl groups. The 1D and 2D nuclear magnetic resonance (NMR) spectra of 7 (Fig. S9–S15†) indicated the presence of five methyls, nine methylenes, fifteen methines, eight sp2 methines, and seven non-protonated carbons, including four carbonyl carbons. Chemical shift values obtained via NMR analysis are presented in Table S4.† The 2D NMR data revealed that the structures of the 5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid (C1–C10) and diene carboxylic acid (C19–C24) side chains of 7 were consistent with those of 1. 1H–1H correlation spectroscopy of H216 (δH 1.55, 2.09)/H17 (δH 5.26), H17/H18, and H18/H19 (δH 4.50) revealed connections from C16 (δC 42.6) to C19. Heteronuclear multiple bond coherence (HMBC) correlation from H216/H19 to C15 (δC 99.4) suggested a 6,6-spiroacetal core in 7.26 The remaining side chain of 7 with molecular formula C4H5O4 was suggested to be a hemisuccinate ester. According to heteronuclear single quantum coherence and DEPT135 data, the 1H NMR signal at 2.59 ppm indicated two methylenes, which showed HMBC correlation with two carbonyl carbons (δC 173.8 and 176.2). Chemical shift values in the 13C NMR spectrum for 1 at C17 and C18 were 25.4 and 82.2 ppm, respectively. However, 7 showed the chemical shifts of 70.8 and 44.6 ppm at C17 and C18, respectively, indicating that the hemisuccinate ester was attached to C17 instead of C18. Therefore, compound 7 was named 17-hemisuccinyloxy-RM-T, and its planar structure is shown in Fig. 3b. The configuration at C17 was determined to be R* by the rotating-frame nuclear Overhauser effect spectroscopy (ROESY) correlations between H17 (δH 5.26)/H20 (δH 6.44), H16 (δH 1.55)/H18 (δH 1.87), and H19 (δH 4.50)/H21 (δH 6.38) in 7 (Fig. 3b and S15†).Compound 6 exhibited physicochemical properties similar to 1 (Fig. S16–S18†). The molecular formula of 6 was determined as C32H48O8 ([M–H]−, m/z 559.3274, calculated for C32H47O8, 559.3271), indicating that the hemisuccinate ester of 1 was substituted with a hydroxyl group. As summarised in Table S5,† NMR data of 6 (Fig. S19–S25†) were consistent with those of 7, except that the hemisuccinate ester signal observed for 7 was not detected in 6. The 13C NMR chemical shift value at C17 was 66.0 ppm, indicating that a hydroxyl group was bound to the C17 of 6. Compound 6 was named 17-hydroxy-RM-T, and its chemical structure is shown in Fig. 3c. The configuration at C17 was determined to be R* by the ROESY correlations between H17 (δH 3.92)/H20 (δH 6.42), H17/H25 (δH 1.02 and 1.77), and H19 (δH 4.45)/H21 (δH 6.35) in 6 (Fig. 3c and S25†). Based on the structures of compounds 6 and 7, their predicted biosynthetic pathways are shown in Fig. 3d.
X-ray crystallography of P450revI-A241L in complex with 3
To elucidate the mechanisms of the regioselective hydroxylation of 3, we crystallised the P450revI-A241L mutant in complex with 3 and successfully determined its co-crystal structure at 2.13 Å. All crystallographic data and refinement statistics are summarised in Table S6.† The overall structure of P450revI-A241L closely resembled that of wild-type P450revI34 (root-mean-square deviation = 0.264; Fig. 4a and b). However, substrate-binding conformation of 3 in P450revI-A241L was partially different from that in wild-type P450revI (Fig. 4c). In the A241L mutant, the C24 carboxyl group of 3 formed (1) a water-mediated hydrogen bond with R81, which was different from that in the wild-type, which formed a direct hydrogen bond with R81, and (2) a hydrogen bond with the main chain of A80 through a water-mediated network, which was absent in the wild-type. C1 carboxyl and C5 hydroxyl groups of 3 exhibited the same interactions as those observed in the wild-type (Fig. 2b and 4c). Mutated residue A241L established hydrophobic interactions with the spiroacetal ring of 3, resulting in a shift in the binding coordinates of 3 on the haem plane (Fig. 4d). While the distances of C17/H17 of 3 to haem iron were not significantly changed, the distances of Fe–C18 and Fe–H18 were increased up to 0.5 Å compared to that in the wild-type (Fig. S26†). This poses a challenge for hydroxylation at the C18 position, leading to selective hydroxylation at the C17 position. At the C17 position, pro-R hydrogen was oriented toward, whereas pro-S hydrogen was oriented away from the haem iron (Fig. 4d). Binding conformation of 3 in the A241L mutant permitted the facile hydroxylation of the pro-R hydrogen at its C17 position. Additionally, ROESY correlations observed between H17 and H20 of 6 and 7, respectively (Fig. S15 and S25†), suggested the likelihood of the R-configuration at C17 of both 6 and 7 (Fig. 4d).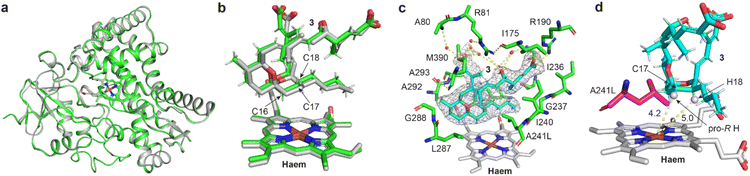 | ||
| Fig. 4 Co-crystal structure of P450revI-A241L with 3. (a and b) Superimposition of the co-crystal structure of the P450revI-A241L mutant (PDB entry: 9LN5; green) with wild-type P450revI (PDB entry: 3WVS; grey). (c and d) Close-up views of the substrate binding site in the P450revI-A241L mutant. Red and white spherical models indicate the ordered water molecules and hydrogen atoms, respectively. Yellow dashed lines indicate the hydrogen bonds or distances to the haem iron atom (Å). (c) Mesh model shows the polder map of 3 contoured at 3.0σ. | ||
Stabilities of novel RMs
As depicted in Fig. 1b, the strain on the 6,6-spiroacetal core of 1 due to the axial orientations of the C18 hemisuccinate moiety and C19 diene acid side chain was relieved upon exposure to acidic conditions.12,26–28 Consistent with previous reports on synthetic 4 (18-hydroxylated-3),12,26,27,30in vitro enzyme conversion product 4 was isomerised from 6,6-spiroacetal to 5,6-spiroacetal (5) under acidic conditions (Fig. 2c and S27†). However, 6 (17-hydroxylated-3), produced using the P450revI-A241L mutant, was more stable than 4 as no equivalent 5,6-spiroacetal structures were detected (Fig. 2c and S27†). Moreover, 1 (18-hemisuccinylated-3) isomerised from the 6,6-spiroacetal to 5,6-spiroacetal form (2) after incubation at pH 3 for 20 h. Notably, 7 (17-hemisuccinylated-3) was more stable than 1 under the same conditions (Fig. 5). Compound 3 without any substituent at the C18 position was the most stable RM among the tested four derivatives (Fig. 5), suggesting that axially oriented C18 substituents are among the key factors for RM spiroacetal rearrangement. These results confirm our hypothesis that the stability of the 6,6-spiroacetal core is enhanced by changing the reaction positions of hydroxylation and hemisuccinylation.Biological activity evaluation
Biological activities of compounds 6 and 7 against bacteria, fungi, malarial parasites, and human cancer cell lines were investigated (Table 1). Consistent with previous reports,9,21 no RM derivative exhibited anti-bacterial activity. Their low susceptibility was due to the absence of key residues in bacterial IleRS that are responsible for interactions with compounds.24 Moreover, no RM derivative exhibited anti-fungal activity against Aspergillus fumigatus and Candida albicans but inhibited the growth of Pyricularia oryzae and Aspergillus oryzae. Additionally, inhibitory activities of compounds 6 and 7 against Pyricularia oryzae were lower than those of compounds 1 and 3. Plasmodium cytoplasmic IleRS is a potential target to develop new multistage anti-malarial agents.62 Here, we evaluated the anti-malarial activities of RMs. Compounds 1, 3, and 6 inhibited the growth of Plasmodium falciparum at sub-micromolar concentrations. Notably, compound 6 exhibited lower cytotoxicity in human cell lines than compounds 1 and 3, indicating its potential for drug development. A recent study reported that compound 1 exhibits anti-multiple myeloma (MM) activity.20 Therefore, we evaluated the cytotoxic effects of RM derivatives on PCM6 cell lines. Compounds 3 and 7 exhibited anti-MM activity comparable to that of compound 1, whereas compound 6 showed no activity at concentrations up to 10 μM. Interestingly, anti-MM activities of compounds 1, 3, and 7 were high at low pH conditions. MM is a haematological malignancy associated with osteolytic bone disease, in which in vivo interactions between MM cells and bone-resorbing osteoclasts create an acidic microenvironment.20 This environment plays a crucial role in mediating the cytotoxic effects of compounds 1, 3, and 7 on MM cells and osteoclasts by enhancing their cellular uptake. Under acidic conditions, suppression of proton dissociation of the carboxylic groups facilitates cellular uptake, while the neutral intracellular pH contributes to the stability of RMs. Although such acidic conditions promote formation of the inactive 5,6-spiroacetal core, the pH gradient between the acidic extracellular environment and the neutral cytoplasm—as observed in osteoclasts63—is likely an important factor underlying the bioactivity of RMs. Nonetheless, the intracellular dynamics of RMs under physiologically relevant conditions remain to be elucidated. Future studies should investigate the interactions between the IleRSs of target organisms and RM derivatives to elucidate the potential of RM derivatives for drug development. Considering the challenges in establishing in vitro cell assays under acidic conditions, future in vivo studies should address these limitations to elucidate the biological potency and stability of RM derivatives.| Cell lines | IC50 values (μM) | |||||
|---|---|---|---|---|---|---|
| 17-Hemisuccinyloxy-RM-T (7) | 17-Hydroxy-RM-T (6) | RM-T (3) | RM-A (1) | Positive controls | ||
| S.a., Staphylococcus aureus 209; E.c., Escherichia coli HO141; A.f., Aspergillus fumigatus Af293; C.a., Candida albicans JCM154; P.o., Pyricularia oryzae kita-1; A.o., Aspergillus oryzae; P.f., Plasmodium falciparum 3D7; HeLa, human cervical cancer cell line; HL-60, human promyelocytic leukaemia cell line; PCM6, human multiple myeloma cell line. Positive controls:a Chloramphenicol.b Amphotericin B.c Artemisinin.d Paclitaxel.e 1 was use as a control.20 Data are represented as the mean ± standard deviation of three replicates. Numbers in parentheses indicate the standard deviations. Values < 0.0001 μM are indicated as 0. | ||||||
| Bacteria | S. aureus | >30 | >30 | >30 | >30 | 7.8 (0.3) |
| E. coli | >30 | >30 | >30 | >30 | 3.0 (0.08) | |
| Fungi | A. fumigatus | >30 | >30 | >30 | >30 | 0.44 (0.05) |
| C. albicans | >30 | >30 | >30 | >30 | 0.057 (0.03) | |
| P. oryzae | 3.6 (0.5) | 22 (8) | 0.62 (0.2) | 0.21 (0.04) | 0.17 (0.02) | |
| A. oryzae | 0.37 (0.09) | 0.95 (0.2) | 0.21 (0.05) | 0.67 (0.05) | 0.68 (0.2) | |
| Malaria | P. falciparum | 4.2 (0.7) | 0.090 (0.003) | 0.22 (0.03) | 0.22 (0.004) | 0.017 (0.0004) |
| Mammalian | HeLa | >30 | >30 | 2.1 (0.4) | 6.9 (0.7) | 0.0020 (0.0001) |
| HL-60 | >30 | 13.6 (1) | 0.60 (0.01) | 4.2 (0.4) | 0.0059 (0) | |
| PCM6 (pH 7) | 3.6 (0.2) | >10 | 0.75 (0.08) | 2.1 (0.2) | NA | |
| PCM6 (pH 6) | 0.61 (0.1) | >10 | 0.18 (0.03) | 0.47 (0.03) | NA | |
Conclusions
Based on its selective C–H activation properties, P450revI was engineered to alter the regioselectivity, resulting in a novel hydroxylated compound, 6. Introduction of the P450revI-A241L gene into the ΔrevI strain facilitated the in vivo production of 17-hydroxylated-3 (6) and 17-hemisuccinylated 3 (7), whose structures were determined via NMR analyses. Together with co-crystal structure analysis of the P450revI-A241L mutant in complex with 3, the combined data support the likelihood that compounds 6 and 7 possess an R-configuration at the C17 position. However, further analyses are required to definitively assign the stereochemistry. Overall, this study outlines an integrated approach to expand the structural diversity of RMs via P450 engineering and synthetic biology approaches using Actinacidiphila as a biosynthetic platform. Based on their chemical properties, novel RM derivatives with improved stability under acidic conditions were successfully synthesised in this study. Furthermore, biological evaluation revealed that these derivatives retained activities comparable to those of the original compounds with varying sensitivities to human cells. Future studies should examine RM-IleRS interactions in target organisms and assess their activities in acidic tissues in vivo to determine their drug development potential.Data availability
Structural data files have been deposited into the Protein Data Bank (PDB) under accession code 9LN5.Author contributions
S. T., Y. F. Y., and S. L. designed the experiments. Y. F. Y., K. S., K. F., H. T., and Y. F. performed the experiments and analysed the data. Y. F. Y. drafted the original manuscript. S. T., E. B. B. O., T. S., K. S., K. F., and H. O. edited and revised the manuscript. All authors have read and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by funds from JSPS KAKENHI for Scientific Research (A; 20H00416 and 25H00924 [S. T.]) and Transformative Research Areas (A; 23H04564 [S. T.], 25H01597 [S. T.], and 21J01340 [K. F.]). Additionally, this work was supported by the Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) of AMED under grant number JP22ama121001 (support number 5499). Y. F. Y. was supported by RIKEN IPA and the Universiti Sains Malaysia Short-Term Grant 000723. We would like to thank the beamline staff for BL1A at the Photon Factory (Ibaraki, Japan) for their assistance and advice regarding the experiments. We would also like to thank Dr Toshihiko Nogawa (RIKEN CSRS) for assistance with HR-ESI-ToF-MS analysis. We are grateful to Ms. Harumi Aono and Ms. Mari Shime (RIKEN CSRS) for assistance with biological activity evaluations. Aspergillus oryzae was kindly provided by Noda Institute for Scientific Research.Notes and references
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS.
- S. Groß, F. Panter, D. Pogorevc, C. E. Seyfert, S. Deckarm, C. D. Bader, J. Herrmann and R. Müller, Chem. Sci., 2021, 12, 11882–11893 RSC.
- J.-W. Peng, X.-D. Yin, H. Li, K.-Y. Ma, Z.-J. Zhang, R. Zhou, Y.-L. Wang, G.-F. Hu and Y.-Q. Liu, J. Agric. Food Chem., 2021, 69, 4604–4614 CrossRef CAS PubMed.
- L.-W. Zou, Y.-G. Li, P. Wang, K. Zhou, J. Hou, Q. Jin, D.-C. Hao, G.-B. Ge and L. Yang, Eur. J. Med. Chem., 2016, 112, 280–288 CrossRef CAS PubMed.
- C. M. F. Ancajas, A. S. Oyedele, C. M. Butt and A. S. Walker, Nat. Prod. Rep., 2024, 41, 1543–1578 RSC.
- J. C. J. M. D. S. Menezes, RSC Adv., 2017, 7, 9357–9372 RSC.
- N. J. White, Science, 2008, 320, 330–334 CrossRef CAS PubMed.
- Y. Phang, X. Wang, Y. Lu, W. Fu, C. Zheng and H. Xu, Eur. J. Med. Chem., 2020, 205, 112646 CrossRef CAS PubMed.
- H. Osada, H. Koshino, K. Isono, H. Takahashi and G. Kawanishi, J. Antibiot., 1991, 44, 259–261 CrossRef CAS PubMed.
- H. Komaki, H. Takagi, S. Takahashi and H. Osada, Biosci., Biotechnol., Biochem., 2024, 88, 689–695 Search PubMed.
- H. Takahashi, H. Osada, H. Koshino, T. Kudo, S. Amano, S. Shimizu, M. Yoshihama and K. Isono, J. Antibiot., 1992, 45, 1409–1413 CrossRef CAS.
- H. Koshino, H. Takahashi, H. Osada and K. Isono, J. Antibiot., 1992, 45, 1420–1427 CrossRef CAS.
- L. Fremlin, M. Farrugia, A. M. Piggott, Z. Khalil, E. Lacey and R. J. Capon, Org. Biomol. Chem., 2011, 9, 1201–1211 RSC.
- M. Ubukata, H. Koshino, H. Osada and K. Isono, J. Chem. Soc., Chem. Commun., 1994,(16), 1877–1878 RSC.
- J. T. Woo, M. Kawatani, M. Kato, T. Shinki, T. Yonezawa, N. Kanoh, H. Nakagawa, M. Takami, K. H. Lee, P. H. Stern, K. Nagai and H. Osada, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4729–4734 CrossRef CAS PubMed.
- M. Tanaka, K. Miyazawa, M. Tabuchi, T. Yabumoto, M. Kadota, M. Yoshizako, C. Yamane, M. Kawatani, H. Osada, H. Maeda and S. Goto, J. Dent. Res., 2012, 91, 771–776 CrossRef CAS PubMed.
- T. Yabumoto, K. Miyazawa, M. Tabuchi, S. Shoji, M. Tanaka, M. Kadota, M. Yoshizako, M. Kawatani, H. Osada, H. Maeda and S. Goto, Am. J. Orthod., 2013, 144, 368–380 CrossRef.
- M. Mizuno, K. Miyazawa, M. Tabuchi, M. Tanaka, M. Yoshizako, C. Minamoto, Y. Torii, Y. Tamaoka, M. Kawatani, H. Osada, H. Maeda and S. Goto, Sci. Rep., 2015, 5, 16510 CrossRef CAS PubMed.
- A. Yano, S. Tsutsumi, S. Soga, M. J. Lee, J. Trepel, H. Osada and L. Neckers, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15541–15546 CrossRef CAS PubMed.
- W. Keiichiro, B.-E. Ariunzaya, T. Hirofumi, C. Qu, T. Jumpei, H. Masahiro, O. Asuka, H. Takeshi, M. Hirokazu, S. Kimiko, O. Masahiro, S. Ryohei, M. Yukari, E. Itsuro, T. Eiji, K. Makoto, O. Hiroyuki, M. Toshio and A. Masahiro, Haematologica, 2021, 106, 1172–1177 CrossRef.
- H. Takahashi, H. Osada, H. Koshino, M. Sasaki, R. Onose, M. Nakakoshi, M. Yoshihama and K. Isono, J. Antibiot., 1992, 45, 1414–1419 CrossRef CAS.
- A. Lyu, H. Liu, H. Che, L. Yang, J. Zhang, M. Wu, W. Chen and G. Li, Front. Microbiol., 2017, 8, 550 Search PubMed.
- Y. Miyamoto, K. Machida, M. Mizunuma, Y. Emoto, N. Sato, K. Miyahara, D. Hirata, T. Usui, H. Takahashi, H. Osada and T. Miyakawa, J. Biol. Chem., 2002, 277, 28810–28814 CrossRef CAS PubMed.
- B. Chen, S. Luo, S. Zhang, Y. Ju, Q. Gu, J. Xu, X. L. Yang and H. Zhou, Nat. Commun., 2021, 12, 1616 CrossRef CAS.
- T. Shimizu, T. Usui, K. Machida, K. Furuya, H. Osada and T. Nakata, Bioorg. Med. Chem. Lett., 2002, 12, 3363–3366 CrossRef CAS.
- K. E. Drouet, T. Ling, H. V. Tran and E. A. Theodorakis, Org. Lett., 2000, 2, 207–210 CrossRef CAS PubMed.
- T. Shimizu, T. Masuda, K. Hiramoto and T. Nakata, Org. Lett., 2000, 2, 2153–2156 CrossRef CAS.
- T. Shimizu, R. Kobayashi, K. Osako, H. Osada and T.-i. Nakata, Tetrahedron Lett., 1996, 37, 6755–6758 CrossRef CAS.
- M. El Sous and M. A. Rizzacasa, Tetrahedron Lett., 2000, 41, 8591–8594 CrossRef CAS.
- M. El Sous, D. Ganame, P. Tregloan and M. A. Rizzacasa, Synthesis, 2010, 2010, 3954–3966 CrossRef.
- K. J. McRae and M. A. Rizzacasa, J. Org. Chem., 1997, 62, 1196–1197 CrossRef CAS.
- A. N. Cuzzupe, C. A. Hutton, M. J. Lilly, R. K. Mann, K. J. McRae, S. C. Zammit and M. A. Rizzacasa, J. Org. Chem., 2001, 66, 2382–2393 CrossRef CAS.
- S. Takahashi, A. Toyoda, Y. Sekiyama, H. Takagi, T. Nogawa, M. Uramoto, R. Suzuki, H. Koshino, T. Kumano, S. Panthee, T. Dairi, J. Ishikawa, H. Ikeda, Y. Sakaki and H. Osada, Nat. Chem. Biol., 2011, 7, 461–468 CrossRef CAS PubMed.
- S. Takahashi, S. Nagano, T. Nogawa, N. Kanoh, M. Uramoto, M. Kawatani, T. Shimizu, T. Miyazawa, Y. Shiro and H. Osada, J. Biol. Chem., 2014, 289, 32446–32458 CrossRef CAS.
- M. El Sous, D. Ganame, P. A. Tregloan and M. A. Rizzacasa, Org. Lett., 2004, 6, 3001–3004 CrossRef CAS PubMed.
- Z. Li, Y. Jiang, F. P. Guengerich, L. Ma, S. Li and W. Zhang, J. Biol. Chem., 2020, 295, 833–849 CrossRef.
- K. Zhang, B. M. Shafer, M. D. Demars II, H. A. Stern and R. Fasan, J. Am. Chem. Soc., 2012, 134, 18695–18704 CrossRef CAS.
- G.-D. Roiban, R. Agudo and M. T. Reetz, Angew. Chem., Int. Ed., 2014, 53, 8659–8663 CrossRef CAS PubMed.
- X. Zhang, P. Shen, J. Zhao, Y. Chen, X. Li, J. W. Huang, L. Zhang, Q. Li, C. Gao, Q. Xing, C. C. Chen, R. T. Guo and A. Li, ACS Catal., 2023, 13, 1280–1289 CrossRef CAS.
- M. J. Cryle, A. Meinhart and I. Schlichting, J. Biol. Chem., 2010, 285, 24562–24574 CrossRef CAS PubMed.
- S. Uhlmann, R. D. Süssmuth and M. J. Cryle, ACS Chem. Biol., 2013, 8, 2586–2596 CrossRef CAS PubMed.
- J. Staunton and B. Wilkinson, Chem. Rev., 1997, 97, 2611–2630 CrossRef CAS.
- Y. Anzai, S. Li, M. R. Chaulagain, K. Kinoshita, F. Kato, J. Montgomery and D. H. Sherman, Cell Chem. Biol., 2008, 15, 950–959 CAS.
- Y. Xue, D. Wilson, L. Zhao, H. Liu and D. H. Sherman, Cell Chem. Biol., 1998, 5, 661–667 CAS.
- O. Volokhan, H. Sletta, T. E. Ellingsen and S. B. Zotchev, Appl. Environ. Microbiol., 2006, 72, 2514–2519 CrossRef CAS.
- B. Zhao, L. Lei, D. G. Vassylyev, X. Lin, D. E. Cane, S. L. Kelly, H. Yuan, D. C. Lamb and M. R. Waterman, J. Biol. Chem., 2009, 284, 36711–36719 CrossRef CAS PubMed.
- S. Huang, S. S. Elsayed, M. Lv, J. Tabudravu, M. E. Rateb, R. Gyampoh, K. Kyeremeh, R. Ebel, M. Jaspars, Z. Deng, Y. Yu and H. Deng, Cell Chem. Biol., 2015, 22, 1633–1642 CAS.
- M. Daum, H.-J. Schnell, S. Herrmann, A. Günther, R. Murillo, R. Müller, P. Bisel, M. Müller and A. Bechthold, ChemBioChem, 2010, 11, 1383–1391 CrossRef CAS.
- M. Milan, M. Bietti and M. Costas, Chem. Commun., 2018, 54, 9559–9570 RSC.
- C. N. Stout, N. M. Wasfy, F. Chen and H. Renata, J. Am. Chem. Soc., 2023, 145, 18161–18181 CrossRef CAS PubMed.
- C. Calvó-Tusell, Z. Liu, K. Chen, F. H. Arnold and M. Garcia-Borràs, Angew. Chem., Int. Ed., 2023, 62, e202303879 CrossRef PubMed.
- Y. Luo, Z. Du, C. Jiang, Z. Yu, J.-J. Zhong, T. Shi and H. Xiao, ACS Catal., 2023, 13, 15673–15681 CrossRef CAS.
- Q. Chen, Z. Chao, K. Wang, X. Wang, H. Meng, X. Liu, X. Shan and J. Zhou, ACS Catal., 2024, 14, 4117–4129 CrossRef CAS.
- P. Zhao, J. Chen, N. Ma, J. Chen, X. Qin, C. Liu, F. Yao, L. Yao, L. Jin and Z. Cong, Chem. Sci., 2021, 12, 6307–6314 RSC.
- O. F. Brandenberg, K. Chen and F. H. Arnold, J. Am. Chem. Soc., 2019, 141, 8989–8995 CrossRef CAS PubMed.
- C. G. Acevedo-Rocha, C. G. Gamble, R. Lonsdale, A. Li, N. Nett, S. Hoebenreich, J. B. Lingnau, C. Wirtz, C. Fares, H. Hinrichs, A. Deege, A. J. Mulholland, Y. Nov, D. Leys, K. J. McLean, A. W. Munro and M. T. Reetz, ACS Catal., 2018, 8, 3395–3410 CrossRef CAS.
- K. Zhang, A. Yu, X. Chu, F. Li, J. Liu, L. Liu, W.-J. Bai, C. He and X. Wang, Angew. Chem., Int. Ed., 2022, 61, e202204290 CrossRef CAS.
- Y. Pan, G. Li, R. Liu, J. Guo, Y. Liu, M. Liu, X. Zhang, L. Chi, K. Xu, R. Wu, Y. Zhang, Y. Li, X. Gao and S. Li, Nat. Commun., 2023, 14, 1669 CrossRef CAS PubMed.
- H. Li, W. Dai, S. Qin, S. Li, Y. Yu and L. Zhang, Biotechnol. Bioeng., 2023, 120, 2230–2241 CrossRef CAS.
- S. T. Jung, R. Lauchli and F. H. Arnold, Curr. Opin. Biotechnol., 2011, 22, 809–817 CrossRef CAS.
- S. Nagano and T. L. Poulos, J. Biol. Chem., 2005, 280, 31659–31663 CrossRef CAS PubMed.
- E. S. Istvan, F. Guerra, M. Abraham, K. S. Huang, F. Rocamora, H. Zhao, L. Xu, C. Pasaje, K. Kumpornsin, M. R. Luth, H. Cui, T. Yang, S. Palomo Diaz, M. G. Gomez Lorenzo, T. Qahash, N. Mittal, S. Ottilie, J. Niles, M. C. S. Lee, M. Llinas, N. Kato, J. Okombo, D. A. Fidock, P. Schimmel, F. J. Gamo, D. E. Goldberg and E. A. Winzeler, Sci. Transl. Med., 2023, 15, eadc9249 CrossRef CAS PubMed.
- A. Teti, H. C. Blair, S. L. Teitelbaum, A. J. Kahn, C. Koziol, J. Konsek, A. Zambonin-Zallone and P. H. Schlesinger, J. Clin. Invest., 1989, 83, 227–233 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc01355k |
| This journal is © The Royal Society of Chemistry 2025 |


