Open Access Article
Open Access ArticleRedirecting electron flows in glutamate oxidases by selective anchoring of osmium complexes†
Minjung
Han‡
a,
Sun-heui
Yoon‡
 a,
Jaehee
Lee
a,
Taek Dong
Chung
a,
Jaehee
Lee
a,
Taek Dong
Chung
 *ab and
Woon Ju
Song
*ab and
Woon Ju
Song
 *a
*a
aDepartment of Chemistry, Seoul National University, Seoul 08826, Republic of Korea. E-mail: tdchung@snu.ac.kr; woonjusong@snu.ac.kr
bAdvanced Institute of Convergence Technology, Suwon-Si, Gyeonggi-do 16229, Republic of Korea
First published on 27th March 2025
Abstract
L-Glutamate is the most abundant and essential excitatory neurotransmitter in the nervous system. However, its direct electrochemical detection is challenging due to its inherently non-electroactive nature. In this study, we redesigned L-glutamate oxidase (GlutOx) by covalently attaching osmium polypyridyl complexes as electron mediators at selected sites. Most engineered enzymes retained their native catalytic activity, while exhibiting significantly altered catalytic currents during L-glutamate oxidation, depending on the proximity, orientation, and microenvironments of the osmium complexes relative to the FAD cofactors. Notably, two mutants significantly enhanced catalytic currents, revealing selectively and efficiently rerouted electron transfer pathways from the enzyme active site to Os complexes. These findings provide an effective strategy for designing redox-active enzymes for electrochemical biosensors.
Introduction
Protein environments regulate electron transfer with remarkable efficiency and precision.1,2 The protein matrix not only tunes the redox potential of cofactors3,4 but also determines the distance between electron donor and acceptor molecules, as well as reorganization energy, thereby governing the thermodynamics and kinetics of electron transfers.5 Additionally, surrounding redox-active amino acids, such as tryptophan and tyrosine, often play direct roles in proton-coupled electron transfer.6,7 Hydrophobic residues, such as leucine, and other chemical signals can gate conformational changes8 essential for facilitating electron transfer.9 Therefore, controlling electron flow within protein environments remains a critical yet challenging aspect of enzyme redesign and engineering.L-Glutamate, the primary excitatory neurotransmitter in the mammalian central nervous system, plays a key role in cognitive function, memory, and learning.10,11 Accurate and rapid detection of extracellular glutamate is essential, as dysregulation is associated with numerous neurological and psychiatric disorders.12 However, its redox-inactive nature presents significant challenges for monitoring dynamic biological processes under physiological conditions.
Enzyme-based electrochemical biosensors are powerful tools for detecting local concentrations of redox-innocent small molecules.13 Flavin adenine dinucleotide (FAD)-dependent enzymes are among the most commonly utilized for this purpose due to the redox properties of their native cofactor, as exemplified by FAD-dependent glucose oxidases.14,15 During substrate oxidation, FAD is reduced to FADH2, which can be electrochemically detected directly, indirectly via redox mediators, or through hydrogen peroxide (H2O2) generated by the reaction of FADH2 with dioxygen. Accordingly, FAD-dependent enzymes that utilize glutamate as a substrate represent a promising strategy for developing enzyme-based glutamate-specific biosensors.16
L-Glutamate oxidase (GlutOx), a member of the L-amino acid oxidase family (EC 1.4.3.2), catalyzes the oxidative deamination of L-amino acids, producing α-ketoglutarate and ammonia17,18 (Fig. 1a). By converting electrochemically undetectable L-glutamate into measurable electrochemical signals, GlutOx could enable high-resolution temporal and spatial monitoring of glutamate levels, making it highly suitable for biosensor applications. Specifically, GlutOx from Streptomyces sp. X-119-6 is an ideal candidate due to its efficient preparation through E. coli heterologous overexpression or commercial availability. This enzyme also shows high catalytic activities with a turnover number (kcat) of 35 s−1 and a Michaelis constant (KM) of 5.0 mM, significantly lower than the glutamate level in synaptic vesicles (>100 mM).19 These properties satisfy the biochemical, kinetic, and thermodynamic requirements for effective biosensor design (Fig. 1b).
However, GlutOx encounters physical constraints. The FAD cofactor is deeply embedded within the protein matrix, approximately 15–30 Å from the protein surface, despite multiple voids connecting to the active sites.20,21 Consequently, direct electron transfer from FADH2 to the electrode was barely detectable (Fig. 1c). Previous studies22 have addressed such constraints by detecting H2O2,23–25 introducing excess electron mediators, or conjugating gold nanoparticles.26,27
To achieve high sensitivity of the chemical signals associated with L-glutamate and optimize electron transfer systematically, we prepared osmium (Os) polypyridyl complexes designed for introduction at specific sites of GlutOx as electron mediators. Notably, two mutants mediated electron transfer in a more controlled and pronounced manner, indicating that the integration of inorganic and biochemical modifications allowed for precise construction and control of electron transfer pathways. This work highlights the potential for developing versatile enzyme-based biosensors for L-glutamate detection by effectively translating chemical signals into electric outputs.
Results and discussion
Design and synthesis of osmium complexes for bioconjugation
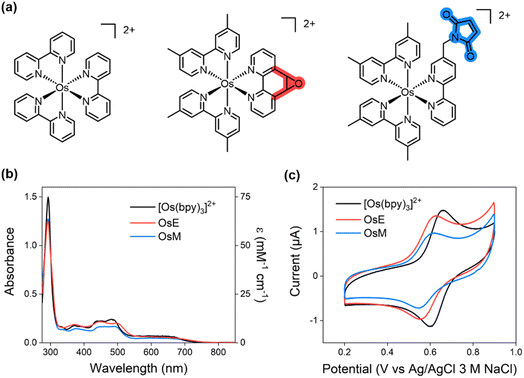
Various inorganic and organic molecules, such as ferrocene, osmium polypyridyl complexes, and benzoquinone, have been employed as redox mediators to facilitate electron transfer between enzymes and electrodes.28–31 In this study, we selected Os polypyridyl complexes owing to their suitable redox potential (E1/2 = 111−730 mV vs. Ag/AgCl) for FADH2 oxidation under biochemically compatible conditions, high stability in both oxidized and reduced states in aqueous media, adequate solubility, and most importantly, large electron transfer rate constants (105 to 106 M−1 s−1).29,32 While tris(2,2′-bipyridine)osmium or [Os(bpy)3]2+ (ca. 0.1 mM) in solution can diffusively facilitate intermolecular electron transfer (ca. 5 μA) from FAD cofactor in glucose oxidase (8 μM),32 GlutOx exhibited a low catalytic current (0.66 μA) under similar conditions (Fig. S1a in the ESI†). Even at a 10-fold higher concentration (1.07 mM) of [Os(bpy)3]2+, only a modest increase—approximately 4.6-fold—in catalytic current was observed (Fig. S1b in the ESI†). This discrepancy is attributed to the deeper embedding of the FAD cofactor within the GlutOx protein matrix.To achieve more efficient and controlled electron transfer, we hypothesized that covalent anchoring of an Os complex in close proximity to the FAD cofactor is essential. To this end, we designed two Os polypyridyl complexes for selective bioconjugation to GlutOx (Fig. 2a): [Os(dmbpy)2(phen-epoxide)]2+ (dmbpy = 4,4′-dimethyl-2,2′-bipyridine, phen-epoxide = 5,6-epoxy-5,6-dihydro-[1,10]phenanthroline, OsE) and [Os(dmbpy)2(bpy-maleimide)]2+ (bpy-maleimide = 1-([2,2′-bipyridin]-5-ylmethyl)-1H-pyrrole-2,5-dione, OsM). Both phen-epoxide33,34 and bpy-maleimide35 serve as moieties capable of forming covalent bonds with cysteine residues, facilitating selective attachment. These Os complexes are designed to mediate electron transfer from the FAD cofactor to the electrode, either directly or with additional assistance from free Os(bpy)3 in solution (Fig. 1b). Specifically, both Os complexes feature two dimethyl-substituted bpy (dmbpy) ligands to lower oxidation potentials, while phen-epoxide or bpy-maleimide ligands provide a relatively short, rigid, and discrete orientation of the complex on the protein surface.
These complexes were synthesized in two steps with overall yields of 30%, purified by preparative HPLC, and characterized by 1H NMR and ESI-MS (Fig. S2 in the ESI†). The UV-Vis spectra of the Os complexes (Fig. 2b) were similar to those of other analogous complexes with some variations (molar absorption coefficient, ε450 nm = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 597 cm−1 M−1 for OsE and 8871 cm−1 M−1 for OsM).
597 cm−1 M−1 for OsE and 8871 cm−1 M−1 for OsM).
Cyclic voltammetry (CV) of the two Os complexes in DPBS (pH 7.4) (Fig. 2c) demonstrated that OsE and OsM exhibit reversible behavior with comparable redox potentials: 0.590 V and 0.573 V vs. Ag/AgCl (3 M NaCl), respectively. The peak current of OsM was slightly lower than that of OsE, possibly due to differences in their molecular structure and diffusion coefficients. Using the Randles–Sevcik equation36 (Fig. S3 in the ESI†), the diffusion coefficients were determined to be 2.06 × 10−6, 1.26 × 10−6, and 3.68 × 10−6 cm2 s−1 for OsE, OsM, and [Os(bpy)3]2+, respectively. More importantly, both oxidation potentials of OsE and OsM are significantly more positive than that of FADH2/FAD couple (−0.416 V vs. Ag/AgCl, 3 M NaCl),37 indicating their capacity to mediate electron transfer from FADH2 to the electrode. Additionally, the redox potential of [Os(bpy)3]2+ (0.630 V) is higher than those of the two Os complexes, suggesting that [Os(bpy)3]2+ can serve as a secondary redox mediator, further facilitating electron transfer from enzyme-tethered OsE or OsM to the electrode, if necessary (Fig. 1b).
Redesign of GlutOx for anchoring inorganic electron mediators
We inspected the X-ray crystal structure of the homodimer GlutOx20 to identify suitable sites for the selective, covalent attachment of Os complexes (Fig. 3a and Table S3 in the ESI†). The wild-type protein contains two cysteine residues: C615, which is located at the dimeric interface and thus inaccessible for bioconjugation, and C224, which is surface-exposed but remote from the active site, with a 35.6 Å separation between the Cβ atom of C224 and the C4a position in FAD. Therefore, we used the wild-type protein as a negative control, attaching one equivalent of the Os complex per monomer at a site unlikely to mediate electron transfer. | ||
| Fig. 3 Biochemical characterization of GlutOx. (a) X-ray crystal structure of L-glutamate oxidase (PDB 2E1M). One protomer of the homodimer is shown in surface representation, while the other is depicted in cartoon representation. The FAD cofactor is illustrated as yellow sticks. Native cysteine residues (C224 and C615) are shown with black spheres, and the selected sites for cysteine mutation near the FAD cofactor are indicated with colored spheres. (b) UV-Vis spectra of FAD and the wild-type protein (20 μM) before and after Os complexes conjugation. (c) Cyclic voltammograms of Os complexes-bound wild-type protein and [Os(bpy)3]2+ (100 μM) in DPBS pH 7.0 (scan rate: 5 mV s−1). (d) Initial rates of H2O2 formation (e) The ratio of the two reaction products, H2O2 and α-ketoglutarate, from a 15 minute enzymatic reaction. A value of unity is represented by a dotted line. | ||
The optimal location for attaching Os complex is midway between FADH2 and the protein surface, ensuring that it does not interfere with key steps in native enzyme catalysis, such as substrate access and binding near the FAD cofactor, and product release. While predicting the exact spatial orientation at atomic resolution is challenging, we measured the distances between the Cβ atoms of candidate cysteine residues and the C4a position in the FAD cofactor, assuming that cysteine modification would not induce significant structural changes. Based on the criterion of an interatomic distance of less than approximately 20 Å, we selected five residues (K99, K104, P110, R334, and P612) on a flexible surface loop and individually mutated them to cysteine. To prevent competitive side reactions, we replaced both pre-existing cysteine residues with serine for all cysteine mutants.
The wild-type protein and all mutants were prepared through heterologous overexpression in E. coli (Fig. S4 in the ESI†). The UV-Vis spectra of the purified proteins showed three peaks at 280, 393, and 472 nm, regardless of mutations, and were similar to those of the wild-type protein (Fig. 3b and S6 in the ESI†). Notably, the latter two bands are characteristic features of the protein-bound FAD cofactor, redshifted relative to free FAD (λmax = 375 and 450 nm with ε450 nm = 9820 cm−1 M−1). To accurately determine the FAD content of GlutOx variants, we extracted the FAD cofactor and quantified the holo-form fraction by HPLC and UV-Vis spectrophotometry, as reported previously.38 The molar absorptivity of the FAD bound to GlutOx was determined to be ε450 nm = 7687 cm−1 M−1 (Fig. S5 in the ESI†).
Next, we attached two Os complexes to the wild-type protein or cysteine mutants by incubating GlutOx (10 μM) with OsE or OsM (3 equiv.) and tris(2-carboxyethyl)phosphine (TCEP, 10 equiv.) in 100 mM Tris buffer pH 8.2 at 4 °C overnight. Following purification using ultrafiltration, the samples were characterized by ICP-OES (Table S5†), HPLC, UV-Vis (Fig. 3b and S6 in the ESI†), and LC-MS (Fig. S7 in the ESI†). Although the fraction of Os complex-bound proteins was higher as determined by ICP-OES and UV-Vis experiments compared to LC-MS, the effects of mutations and linker variations within the Os complexes were consistently observed across the proteins, with OsM generally achieving comparable to or higher yields than OsE (Table S5a in the ESI†).
The wild-type protein, which contains the C224 residue, exhibited conjugation yields of up to 60% and 96% per monomer for OsE and OsM, respectively. The K104C and P110C mutants demonstrated nearly quantitative conjugation yields with OsM, likely due to the high surface exposure of their introduced cysteine residues. In contrast, these mutants exhibited lower yields with OsE (47% and 72%, respectively), suggesting that OsE and OsM may require different spatial orientations for efficient Michael addition or exhibit distinct reactivity towards specific cysteine residues. The P612C mutant showed moderate conjugation levels for both Os complexes (87% for OsE and 65% for OsM). Conversely, K99C and R334C exhibited considerably lower conjugation yield with both Os complexes (26% and 27% for OsE and 43% and 22% for OsM, respectively). K99C is positioned in a region less exposed to the bulk solvent, while R334C is located in a more rigid region at the terminus of an α-helix. Consequently, their limited accessibility or structural rigidity likely hinder their ability to accommodate sterically bulky complexes.
We also assessed the electrochemical properties of the proteins (Fig. 3c and S8 in the ESI†). GlutOx without any Os complex conjugation (Non) exhibited residual current at 0.672 V, likely due to the involvement of redox-active amino acids on the protein surface at high oxidation potential, as reported previously.39,40 OsE- or OsM-bound proteins exhibited higher currents than the residual current, corresponding to the oxidation of the anchored Os complexes. However, both displayed nearly irreversible behavior at a low scan rate (2–5 mV s−1). This feature was consistent across multiple scans and comparable across all six GlutOx mutants and both Os complexes. These results suggest that the observed perturbations in electrochemical redox behavior are primarily induced by covalent anchoring rather than variations in the surrounding chemical microenvironments. The loss of reversibility may result from the sacrificial oxidation of redox-active amino acid residues, as observed with GlutOx prior to conjugation. Then, the height of oxidation current may vary with the number of amino acids oxidized at the electrode. Nonetheless, faster scan rates (>20 mV s−1) significantly recovered the reduction currents of the Os complex (Fig. S9 in the ESI†), demonstrating that the anchored redox mediators on the protein surface remain functional and viable for the intended application.
Biochemical characterization of redesigned GlutOx
We investigated whether cysteine mutations or the introduction of Os complexes interfered with the native enzymatic reactions with L-glutamate. Two activity assays were employed to independently detect the reaction products, H2O2 and α-ketoglutarate (Scheme S1 in the ESI†). Notably, the bound Os complexes remained in their reduced states and would not participate in the steady-state catalysis.First, continuous coupled activity assays with horseradish peroxidase (HRP) and o-dianisidine (o-DNS) were conducted to measure the formation rate of H2O2 generated by the reaction between FADH2 and dioxygen, as reported previously.41 Time-dependent absorption changes were monitored at 440 nm (oxidized o-DNS, ε440 nm = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1) upon the addition of 20 mM L-glutamate. The initial catalytic activity, normalized to protein-bound FAD concentrations (Fig. S5 in the ESI†), revealed that all cysteine mutants, prior to conjugation, exhibited kcat values ranging from 21.0 to 34.0 s−1, comparable to that of the wild-type protein (Fig. 3d, grey bars). These results suggest that the mutations caused no substantial perturbations in enzyme catalysis and some may have even slightly enhanced the overall catalytic activities.
000 M−1 cm−1) upon the addition of 20 mM L-glutamate. The initial catalytic activity, normalized to protein-bound FAD concentrations (Fig. S5 in the ESI†), revealed that all cysteine mutants, prior to conjugation, exhibited kcat values ranging from 21.0 to 34.0 s−1, comparable to that of the wild-type protein (Fig. 3d, grey bars). These results suggest that the mutations caused no substantial perturbations in enzyme catalysis and some may have even slightly enhanced the overall catalytic activities.
We measured H2O2 formation rates following the conjugation of Os complex mediators (Fig. 3d, red or blue bars). The wild-type proteins covalently attached with OsE or OsM exhibited kcat values of 26.3 s−1 or 28.8 s−1, respectively, comparable to their activity prior to conjugation. These results indicate that the Os complex, when introduced remotely in their reduced states, retain dormant during steady-state catalysis. The specific activities of all cysteine mutants, except for P612C, were comparable to the wild-type protein, indicating that covalently bound Os complexes at these positions do not kinetically interfere with critical steps of enzyme catalysis, including glutamate access and binding to the active site, multi-electron/proton-associated steps, and product release. In contrast, P612C exhibited significantly reduced activity with both Os complexes, producing only 24.4% and 13.5% of the wild-type activity for OsE and OsM, respectively. As P612C position is located at the dimer interface, the introduction of sterically bulky Os complexes likely obstructs substrate access or product release from the active site of the adjacent subunit, thereby reducing overall steady-state catalytic activities.
Next, we performed an alternative coupled activity assay using 3-methyl-2-benzothiazolinone hydrazone (MBTH) to detect the other reaction product, α-ketoglutarate, as reported previously.42 Following a 15 min enzymatic reaction, the formation of MBTH–azine–α-ketoglutaric acid was quantified by UV-Vis spectrophotometry (ε368 nm = 4000 cm−1 M−1) (Fig. S10a in the ESI†). The concentrations of α-ketoglutarate produced from the multiple turnover reactions of the cysteine mutants were comparable to those of the wild-type proteins (Fig. S10b and Table S5 in the ESI†). The catalytic activity of all mutants, except P612C, were retained even after the conjugation with OsE or OsM, consistent with the HRP assay results described above. The catalytic activities of the P612C mutant were approximately 50% of those observed in the mutant without Os complexes, which is substantially higher than the levels detected in the HRP assay. This discrepancy suggests that Os complex conjugation at the P612C position reduces coupling efficiency for H2O2 formation.
Consequently, the concentration of H2O2 generated during the reaction was re-measured using the HRP assay to calculate the ratio of the two reaction products, [H2O2]/[α-ketoglutarate], as an indicator of electron coupling efficiency (Fig. 3e). This ratio was determined to be 0.84 for the wild-type protein, indicating that FADH2 rapidly reacts with dioxygen, facilitating the transfer of nearly all electrons from glutamate to dioxygen. All mutants, except for P612C, exhibited ratios between 0.6 and 0.8, similar to that of the wild type enzyme, both before and after the conjugation of the Os complexes. These results suggest that the native enzymatic reactions are preserved upon attaching Os complexes near the FAD cofactor. The P612C mutant showed significantly lower ratios of 0.32 and 0.31 with OsE and OsM, respectively, further supporting the hypothesis that not all electrons produced from glutamate oxidation are efficiently coupled with H2O2 formation. These results also suggest that the P612C mutant remains viable for electrochemical studies, as the primary object is to detect electrons from FADH2 rather than H2O2.
Electrochemical detection with the assistance of the secondary redox mediator
We applied linear gradients of oxidation potentials to the OsE- or OsM-attached wild-type protein and cysteine mutants in the presence of L-glutamate (200 mM) (Fig. 4). To minimize dependence on dioxygen concentration and competition with H2O2 formation, all CV experiments were conducted under anaerobic conditions. Additionally, electrochemical measurements were initiated with an excess of [Os(bpy)3]2+ as a secondary electron mediator in the solution to maximize catalytic currents. The addition of the wild-type protein to the free [Os(bpy)3]2+ complex produced a low steady-state catalytic current (Fig. 4a). Similarly, OsM or OsE attached to positions irrelevant to the FAD resulted in no net increase in steady-state catalytic current, likely due to the passive and slow electron transfers from the active sites to the free [Os(bpy)3]2+.Prior to Os complex conjugation, K104C (Fig. 4c) and R334C (Fig. 4e) showed voltammetric behavior similar to the wild-type protein in the presence of excess [Os(bpy)3]2+. In contrast, other cysteine mutants—K99C (Fig. 4b), P110C (Fig. 4d), and P612C (Fig. 4f)—induced a noticeable increase in current even before Os complex conjugation. These data suggest that these cysteine mutations may facilitate electron transfer from FADH2 to free [Os(bpy)3]2+, potentially by reducing steric or electrostatic hindrance near the active site pocket and promoting intermolecular electron transfer in a diffusive manner.
Next, we measured the CV data of the OsE-bound proteins in the presence of an excess secondary mediator, [Os(bpy)3]2+ (Fig. 4). To our disappointment, none of the cysteine mutants exhibited an increase in catalytic current compared to their non-conjugated counterparts. These data indicate that OsE, despite being selectively anchored near the active site pocket, do not effectively facilitate electron transfer from FADH2 to the secondary mediator or the electrode. It is possible that the negative results observed with OsE-conjugated proteins are partially attributable to relatively low conjugation yields. The increased catalytic current due to the K99C or P110C mutations was retained even after OsE conjugation, but to the significantly less extent with P612C, suggesting that the introduction of OsE to the latter position might compensate for the increased accessibility of free [Os(bpy)3]2+ complex.
Similarly, the OsM-bound K99C (Fig. 4b) and P612C (Fig. 4f) displayed higher catalytic current than the OsM-bound wild-type protein, but not relative to their corresponding mutants without OsM. These results indicate that the covalently bound Os complexes do not directly accept electrons from FADH2, and free [Os(bpy)3]2+ instead diffusively mediate electron transfer. R334C (Fig. 4e) behaved similarly to the wild-type protein upon OsM conjugation. However, K104C (Fig. 4c) and P110C (Fig. 4d) gratifyingly exhibited significant increases in catalytic currents upon OsM conjugation. These results indicate that the OsM covalently attached to the K104 or P110 positions can selectively reroute electron from FADH2 to the protein surface. Given that all mutants demonstrated substantially high conjugation yields with OsM, their distinct electrochemical behaviors are likely attributable to their specific locations within the protein.
Several electron transfer pathways may occur, as depicted in Fig. 1b. However, assuming that the intermolecular electron transfer between the enzyme and the secondary redox mediator is the rate-determining step allows us to estimate the second-order electron transfer rate constants (Table S6†). Even single cysteine mutations altered the electron transfer rate constants by up to 4.7-fold, whereas OsE-conjugation resulted in negligible changes across all proteins. The effects of OsM-conjugation were also minimal, except for the K104C and P110C mutants. While the OsM-conjugated wild-type protein exhibited an electron transfer rate constant of 1.4 × 103 M−1 s−1, the K104C and P110C mutants facilitated intermolecular electron transfer at enhanced rate constants of 7.9 × 103 M−1 s−1 and 11 × 103 M−1 s−1, respectively.
Electrochemical detection without the secondary redox mediator
To evaluate whether the OsM-bound mutants can mediate electron transfer directly to the electrode, we conducted CV experiments without adding the secondary electron mediator, [Os(bpy)3]2+ (Fig. 5 and S11 in the ESI†). For comparison, we also measured the catalytic currents of OsE-anchored proteins. None of the OsE-conjugated proteins exhibited a noticeable increase in currents upon the addition of L-glutamate. In contrast, the OsM-bound K104C and P110C mutants (Fig. 5b and c), but not others (Fig. 5a and S11 in the ESI†), exhibited approximately two- and three-fold increases in catalytic current, respectively, compared to the diffusion-limited current of OsM in the protein. These data support that OsM conjugation at these positions selectively and effectively mediates electron transfer to the electrode, eliminating the need for a secondary mediator. This condition establishes a more streamlined and directly mediated electron transfer pathway, advancing enzyme-based glutamate biosensor designs. The lack of catalytic currents in OsE-bound mutants suggest that the distinct conjugation linker alters the spatial orientation and distance of Os complexes relative to the FAD, as well as their interactions with adjacent amino acid residues.Our manual docking simulation supports that OsM attached to the K104C or P110C positions may be situated between the isoalloxazine ring of FAD and the bulk solvent, enabling efficient electron transfer (Fig. S12 in the ESI†). The interatomic distance between the FAD cofactor and the OsM complex is estimated to be approximately 15 Å, which is shorter than that measured with other cysteine mutants, such as R334C and P612C (20−28 Å) (Fig. S13 in the ESI†), but too far for a single electron hopping process to occur efficiently.
Therefore, we investigated whether redox-active amino acids could participate in electron transfer, as reported previously.43 Notably, GlutOx contains a higher proportion of redox-active amino acids, such as tryptophan (Trp) and tyrosine (Tyr), compared to other proteins: Trp accounts for 4.5% and Tyr for 2.3% of the total amino acid sequence in GlutOx, surpassing the average proportions of 2.86% and 1.5%, respectively.44 More specifically, W608 is positioned between the protein-bound OsM and the FAD cofactor, within a 7–15 Å distance, suggesting its potential roles in facilitating efficient electron transfer via the introduced Os complexes. Thus, we replaced W608 with phenylalanine, generating two double mutants: K104C/W608F and P110C/W608F. However, both proteins exhibited significant spectral perturbation of the FAD cofactor, causing a distinct blue shift in the maximum wavelength (λmax) from 472 nm to 456 nm relative to the wild-type protein and other single mutants (Fig. S14 in the ESI†). The altered spectra closely resemble that of free FAD, likely due to changes in the confined environments caused by the removal of tryptophan residues.45 Regardless of OsM conjugation, these mutants exhibited a marked decrease in enzymatic activity with L-glutamate (0.13 and 0.27 s−1 for K104C/W608F and P110C/W608F, respectively) compared to the wild-type enzyme (24.3 s−1), reporting significant impairment of the native enzymatic reaction. Consequently, the OsM-bound K104C/W608F and P110C/W608F mutants showed no increase in catalytic current upon the addition of L-glutamate (200 mM), preventing further investigation into the specific role of the tryptophan in electron transfer. Therefore, additional mutational and structural studies are required to elucidate which residues contribute to the newly created electron transfer pathway and how these residues interact with the Os complexes to facilitate electron flow.
Electrochemical analysis of OsM–GlutOx
If the protein-bound OsM mediates electron transfer in a reversible manner, the steady-state catalytic current (icat), derived from the electron transfer between FADH2 and the protein-bound Os complex, can be determined using eqn (1).46 Here, nFAD represents the number of electrons transferred per FAD cofactor; F is the Faraday constant; A is the electrode area; DGlutOx is the diffusion coefficient; kFAD–Os is the electron transfer rate constant; and [GlutOx] is the enzyme concentration. | (1) |
However, all Os-bound GlutOx exhibited nearly irreversible redox behavior at the scan rate at which we observed the catalytic current (Fig. 3c and S8 in the ESI†), suggesting the involvement of additional redox-active amino acid residues in electron transfer. Thus, we measured the current (iprotein) in the absence of L-glutamate using eqn (2), where nprotein represents the number of electron transfers associated with the protein matrix, and kprotein–Os is the electron transfer rate constant between the protein and the Os complex.
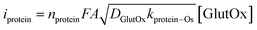 | (2) |
Finally, icat was estimated by subtracting the observed currents in the presence and absence of L-glutamate (itotal and iprotein, respectively), as shown in eqn (3) and Fig. S16 in the ESI.†
| icat = itotal − iprotein | (3) |
For simplicity, we independently determined the diffusion coefficients (D) of Os-bound GlutOx protein using dynamic light scattering measurements (Fig. S15 in the ESI†). For the wild-type GlutOx protein, the hydrodynamic diameter was determined to 16.1 nm, resulting in D = 3.06 × 10−7 cm2 s−1 according to the Stokes–Einstein equation.47 Upon attachment of OsM, the hydrodynamic diameters of the wild-type protein, K104C and P110C mutants remained similar. These values indicate that single mutation or OsM conjugation does not cause significant perturbation in the diffusion coefficients of GlutOx proteins.
Consequently, intramolecular electron transfer rate constants from FADH2 to OsM in the OsM-conjugated K104C and P110C mutants were estimated at 3.5 s−1 and 13 s−1, respectively. These values are significantly faster than those observed with glucose oxidase, where multiple ferrocene or osmium complexes are covalently but randomly attached, exhibiting electron transfer rate constants of 0.0005–0.90 s−1 or 0.2–0.4 s−1, respectively.46,48 These data indicate that the site-selective conjugation of osmium complexes near the active site pocket optimizes electron transfer, highlighting its potential for enhancing enzyme-based biosensor performance.
Using the best-forming OsM-bound K104C and P110C single mutants, we measured the catalytic current with L-aspartate or D-glutamate instead of L-glutamate to assess the specific sensing capabilities of these GlutOx mutants. HRP assays revealed that neither of the single mutants with OsM exhibited detectible steady-state activity with L-aspartate or D-glutamate. Consistently, the OsM-anchored K104C and P110C mutant showed no catalytic current with these analogous amino acids (Fig. 5b and c). These results demonstrate that this enzyme-based biosensor selectively detects L-glutamate, making it particularly suitable for accurately monitoring L-glutamate concentrations under biological conditions.
Conclusions
We demonstrated that site-specific conjugation of osmium polypyridyl complexes to redesigned L-glutamate oxidase enables efficient electron transfer, endowing the molecular basis for the development of L-glutamate biosensors. Two novel osmium complexes were designed and synthesized for selective bioconjugation, offering versatility for use with various redox-active enzymes. Among the engineered mutants, K104C and P110C emerged as optimal sites for OsM attachment, facilitating direct electron transfer from the FADH2 cofactor to the electrode without requiring secondary mediators. The selective introduction of OsM retained the native catalytic activity of GlutOx while enhancing electron transfer at a faster rate compared to other engineered FAD-dependent enzymes. Notably, only one Os complex per FAD site was introduced in this study, enabling selective rerouting of electron flows in a more controllable manner. Although further optimization, including tuning the redox potentials of Os complexes or modifying nearby amino acid residues, is necessary for practical applications, these findings offer a versatile and promising approach for engineering redox-active enzymes and developing efficient electrochemical biosensors useful in neuroscience and biomedical diagnostics.Data availability
The data supporting this article have been included as part of the ESI.† Biochemical and synthetic procedures, supplementary schemes, figures and tables are available in the ESI.†Author contributions
M. Han and S.-h. Yoon: conceptualization, formal analysis, investigation, methodology, visualization, writing – original draft and review & editing. J. Lee: investigation, writing – review & editing. T. D. Chung and W. J. Song: conceptualization, funding acquisition, project administration, supervision, validation, writing – original draft, writing – review & editing.Conflicts of interest
The authors declare no competing interests.Acknowledgements
We appreciate Minjung Keum for assisting the synthesis and purification of Os complexes, and Jaewon Lee and Hyunuk Eom for their efforts in identifying alternative GlutOx enzymes. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS-2021-NR060082 and No. RS-2022-NR070547) and the Creative-Pioneering Researchers Program from Seoul National University.Notes and references
- H. B. Gray and J. R. Winkler, Annu. Rev. Biochem., 1996, 65, 537–561 CrossRef CAS PubMed.
- J. Stubbe, D. G. Nocera, C. S. Yee and M. C. Y. Chang, Chem. Rev., 2003, 103, 2167–2202 CAS.
- N. M. Marshall, D. K. Garner, T. D. Wilson, Y. G. Gao, H. Robinson, M. J. Nilges and Y. Lu, Nature, 2009, 462, 113–116 CAS.
- J. G. Kleingardner, B. D. Levin, G. Zoppellaro, K. K. Andersson, S. J. Elliott and K. L. Bren, J. Biol. Inorg. Chem., 2018, 23, 1073–1083 CAS.
- X. Chen, M. Chen, P. G. Wolynes, P. Wittung-Stafshede and H. B. Gray, J. Am. Chem. Soc., 2022, 144, 4178–4185 CAS.
- S. Y. Reece and D. G. Nocera, Annu. Rev. Biochem., 2009, 78, 673–699 CAS.
- A. Migliore, N. F. Polizzi, M. J. Therien and D. N. Beratan, Chem. Rev., 2014, 114, 3381–3465 CAS.
- S. Fatima and L. Olshansky, Nat. Rev. Chem, 2024, 8, 762–775 CAS.
- T. Min, C. E. Ergenekan, M. K. Eidsness, T. Ichiye and C. Kang, Protein Sci., 2001, 10, 613–621 CAS.
- T. V. Bliss and G. L. Collingridge, Nature, 1993, 361, 31–39 CrossRef CAS PubMed.
- C. A. Tamminga, S. Southcott, C. Sacco, A. D. Wagner and S. Ghose, Schizophr. Bull., 2012, 38, 927–935 CrossRef PubMed.
- W. J. McEntee and T. H. Crook, Psychopharmacology, 1993, 111, 391–401 CrossRef CAS PubMed.
- J. Wang, Chem. Rev., 2008, 108, 814–825 CrossRef CAS PubMed.
- L. C. Clark Jr and C. Lyons, Ann. N. Y. Acad. Sci., 1962, 102, 29–45 CAS.
- A. E. Cass, G. Davis, G. D. Francis, H. A. Hill, W. J. Aston, I. J. Higgins, E. V. Plotkin, L. D. Scott and A. P. Turner, Anal. Chem., 1984, 56, 667–671 CrossRef CAS PubMed.
- Q. Wang, C. Yang, S. Chen and J. Li, Angew. Chem., Int. Ed. Engl., 2024, 63, e202406867 CAS.
- P. D. Pawelek, J. Cheah, R. Coulombe, P. Macheroux, S. Ghisla and A. Vrielink, EMBO J., 2000, 19, 4204–4215 CAS.
- A. Faust, K. Niefind, W. Hummel and D. Schomburg, J. Mol. Biol., 2007, 367, 234–248 CAS.
- J. Arima, T. Tamura, H. Kusakabe, M. Ashiuchi, T. Yagi, H. Tanaka and K. Inagaki, J. Biochem., 2003, 134, 805–812 CAS.
- J. Arima, C. Sasaki, C. Sakaguchi, H. Mizuno, T. Tamura, A. Kashima, H. Kusakabe, S. Sugio and K. Inagaki, FEBS J., 2009, 276, 3894–3903 CAS.
- T. Utsumi, J. Arima, C. Sakaguchi, T. Tamura, C. Sasaki, H. Kusakabe, S. Sugio and K. Inagaki, Biochem. Biophys. Res. Commun., 2012, 417, 951–955 CAS.
- F. Wu, P. Yu, X. Yang, Z. Han, M. Wang and L. Mao, J. Am. Chem. Soc., 2018, 140, 12700–12704 CAS.
- L. Yang, R. Bai, B. Xie, N. Zhuang, Z. Lv, M. Chen, W. Dong, J. Zhou and M. Jiang, Food Chem., 2023, 405, 134792 CAS.
- M. Yuan, S. Sahin, R. Cai, S. Abdellaoui, D. P. Hickey, S. D. Minteer and R. D. Milton, Angew. Chem., Int. Ed. Engl., 2018, 57, 6582–6586 CAS.
- Y. Wang, H. Fathali, D. Mishra, T. Olsson, J. D. Keighron, K. P. Skibicka and A.-S. Cans, J. Am. Chem. Soc., 2019, 141, 17507–17511 CAS.
- J. T. Holland, C. Lau, S. Brozik, P. Atanassov and S. Banta, J. Am. Chem. Soc., 2011, 133, 19262–19265 CAS.
- Y. Xiao, F. Patolsky, E. Katz, J. F. Hainfeld and I. Willner, Science, 2003, 299, 1877–1881 CrossRef CAS PubMed.
- Y. Degani and A. Heller, J. Phys. Chem. A, 1987, 91, 1285–1289 CAS.
- S. M. Zakeeruddin, D. M. Fraser, M. K. Nazeeruddin and M. Grätzel, J. Electroanal. Chem., 1992, 337, 253–283 CAS.
- P.-C. Nien, J.-Y. Wang, P.-Y. Chen, L.-C. Chen and K.-C. Ho, Bioresour. Technol., 2010, 101, 5480–5486 CrossRef CAS PubMed.
- D. P. Hickey, R. C. Reid, R. D. Milton and S. D. Minteer, Biosens. Bioelectron., 2016, 77, 26–31 CrossRef CAS PubMed.
- Y. Nakabayashi, K. Nakamura, M. Kawachi, T. Motoyama and O. Yamauchi, J. Biol. Inorg. Chem., 2003, 8, 45–52 CrossRef CAS PubMed.
- S. Dwaraknath, N.-H. Tran, T. Dao, A. Colbert, S. Mullen, A. Nguyen, A. Cortez and L. Cheruzel, J. Inorg. Biochem., 2014, 136, 154–160 CAS.
- A. E. Wertz, S. C. Marguet, C. Turro and H. S. Shafaat, Inorg. Chem., 2024, 63, 20438–20447 CAS.
- J. Lee and W. J. Song, J. Am. Chem. Soc., 2023, 145, 5211–5221 CAS.
- A. J. Bard and L. R. Faulkner, Electrochemical Methods: Fundamentals and Applications, Wiley, 2000 Search PubMed.
- C. Walsh, J. Fisher, R. Spencer, D. W. Graham, W. T. Ashton, J. E. Brown, R. D. Brown and E. F. Rogers, Biochemistry, 1978, 17, 1942–1951 CAS.
- V. Cafaro, R. Scognamiglio, A. Viggiani, V. Izzo, I. Passaro, E. Notomista, F. D. Piaz, A. Amoresano, A. Casbarra, P. Pucci and A. Di Donato, Eur. J. Biochem., 2002, 269, 5689–5699 CAS.
- E. V. Suprun, Electrochem. Commun., 2021, 125, 106983 CAS.
- E. V. Suprun, M. S. Zharkova, G. E. Morozevich, A. V. Veselovsky, V. V. Shumyantseva and A. I. Archakov, Electroanalysis, 2013, 25, 2109–2116 CAS.
- E. Rosini, L. Caldinelli and L. Piubelli, Front. Mol. Biosci., 2018, 4, 102 Search PubMed.
- Y. Yano, S. Matsuo, N. Ito, T. Tamura, H. Kusakabe, K. Inagaki and K. Imada, Protein Sci., 2021, 30, 1044–1055 CAS.
- C. Shih, A. K. Museth, M. Abrahamsson, A. M. Blanco-Rodriguez, A. J. Di Bilio, J. Sudhamsu, B. R. Crane, K. L. Ronayne, M. Towrie, A. Vlcek Jr, J. H. Richards, J. R. Winkler and H. B. Gray, Science, 2008, 320, 1760–1762 CAS.
- L. P. Kozlowski, Nucleic Acids Res., 2017, 45, D1112–D1116 CAS.
- K. Schwinn, N. Ferré and M. Huix-Rotllant, Phys. Chem. Chem. Phys., 2020, 22, 12447–12455 RSC.
- A. Badia, R. Carlini, A. Fernandez, F. Battaglini, S. R. Mikkelsen and A. M. English, J. Am. Chem. Soc., 1993, 115, 7053–7060 CrossRef CAS.
- P. Wiltzius, Phys. Rev. Lett., 1987, 58, 710–713 CrossRef CAS PubMed.
- F. Battaglini, P. N. Bartlett and J. H. Wang, Anal. Chem., 2000, 72, 502–509 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Biochemical and synthetic procedures, supplementary schemes, figures and tables. See DOI: https://doi.org/10.1039/d5sc00166h |
| ‡ M. Han and S.-h. Yoon contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |