Open Access Article
Open Access ArticleA dual-enzyme activated fluorescent probe for precise identification of tumor senescence†
Xianzhu
Luo
ab,
Erzhuo
Hu
a,
Fei
Deng
c,
Cuiling
Zhang
 *a and
Yuezhong
Xian
*a and
Yuezhong
Xian
 *a
*a
aShanghai Engineering Research Center of Molecular Therapeutics and New Drug Development, Department of Chemistry, School of Chemistry and Molecular Engineering, East China Normal University, Shanghai 200241, China. E-mail: clzhang@chem.ecnu.edu.cn; yzxian@chem.ecnu.edu.cn
bKey Laboratory of Haikou Trauma, Key Laboratory of Hainan Trauma and Disaster Rescue, Key Laboratory of Emergency and Trauma, Ministry of Education, The First Affiliated Hospital of Hainan Medical University, Hainan Medical University, Haikou 571199, China
cARC Centre of Excellence for Nanoscale Biophotonics, Graduate School of Biomedical Engineering, Faculty of Engineering, University of New South Wales, Sydney 2052, Australia
First published on 11th March 2025
Abstract
Precise recognition of senescent cells is essential owing to their key role in various diseases, including aging and tumor suppression. Although senescence-associated β-galactosidase (SA-β-Gal) is widely used as a senescence biomarker, it is not remarkably accurate due to its overexpression in some non-senescent cells. Herein, we developed a dual-channel fluorescent probe to improve the identification accuracy of senescent cells through simultaneous detection of β-gal and α-L-fucosidase (AFU) because the two markers are upregulated in senescent cells. The dual-channel fluorescent probe named HDQ-NA-AFU-Gal was employed to detect β-gal and AFU and identify senescence in living cells and tumor-bearing mice. When the two are present, the dual-enzyme activated probe emits strong red and green fluorescence at 740 nm and 550 nm, respectively, enabling independent detection of β-gal and AFU. This dual-enzyme detection approach allows for the precise differentiation between normal and senescent cells, particularly in ovarian cancer cells overexpressing β-gal. Furthermore, the probe can be applied as an effective tool for tracing β-gal and AFU during tumor senescence in mice. Thus, the dual-enzyme-responsive fluorescent probe has promising applications in biological research and clinical medicine.
Introduction
Cellular senescence has traditionally been defined as a durable arrest of the cell cycle, occurring in response to oxidative stress or exogenous stimuli. The process serves to promote tissue repair and prevent the proliferation of stressed or damaged cells.1,2 Cellular senescence can be induced by various factors, including DNA damage, telomere dysfunction, oncogene activation, and oxidative stress.1,3 Recent studies have indicated its involvement in tissue repair and tumor suppression.4,5 However, accumulating evidence suggests that an abnormal buildup of senescent cells may contribute to the development of age-related diseases, including inflammation, tissue aging, fibrosis, and neurodegenerative diseases.6,7 For instance, cellular senescence plays a key role in cancer development. On the one hand, it contributes to curbing tumor progression by inhibiting the proliferation of cancer cells. On the other hand, excessive senescence in cancer cells can lead to a persistent inflammatory environment, which can create a “breeding ground” for tumor development.8 Therefore, specific monitoring of cancer cells senescence is of great significance.Several biochemical indicators may be associated with cellular senescence, including the lack of proliferation markers (such as Ki67), elevated levels of β-galactosidase (β-gal) in lysosomes, and the up-regulation or activation of tumor suppressors, e.g., p53, p21, and p16.9–13 Among them, β-gal is not only easy to detect in cells and tissues but has also become one of the most widely used biomarkers for assessing senescence.14 However, overexpression of β-gal has also been found in some non-senescent cells, such as ovarian cancer cells (Ovcar-3 cells).15,16 In some cases, a decrease in β-gal expression does not prevent the occurrence of senescence, as observed in human fibroblasts and cervical cancer cells. These indicated that only the β-gal biomarker is insufficient for the precise detection of cellular senescence.17,18 As a kind of glycosidase, α-L-fucosidase (AFU) plays a role in the metabolism of specific glycolipids and glycoproteins, and it has been identified as a promising biomarker for cellular senescence.19,20 Therefore, the simultaneous detection of dual enzymes (β-gal and AFU) could improve the precision of cell senescence diagnosis.
Compared to co-staining and optical microscopy, fluorescent probes for monitoring biomarkers (such as β-gal) offer some advantages, including non-invasiveness, high sensitivity, and real-time visualization.21–26 Although it is possible to use multiple fluorescent probes to simultaneously detect multiple targets, the differences in probe uptake and localization in the cell might lead to variability in experimental results. The development of one probe with multi-response functions can address these issues.27
In recent years, multi-analyte fluorescent probes have received attention for their higher accuracy in related research, including reversible probes,28,29 AND logic probes,30,31 sequential detection probes,32,33 and non-interference fluorescent probes.34–38 These systems address the challenges of using multiple independent fluorescent probes to understand the relationships between various biological species. However, many fluorescent probes are limited to detect multiple targets individually or simultaneously. And probes may be consumed after reacting with one target, resulting in inaccurate detection of other targets. AND logic systems and sequential detection only provide information about the synergistic effects of two species and do not do so for the independent study of each species. Dual-channel emission should be able to simultaneously evaluate each species' role and study their interactions.36–38 For example, Wu et al. developed a dual-channel fluorescent probe (ATP-LW) that can simultaneously detect peroxynitrite (ONOO−) and adenosine-5′-triphosphate (ATP), revealing that an increase in ONOO− and ATP depletion are believed to be the causes of liver necrosis.36 Ren et al. monitored endogenous hypochlorous acid (HClO) and hydrogen sulfide (H2S) in lysosomes of living cells using a single fluorescent probe with a dual-channel, providing a powerful molecular tool for studying the relationship between redox balance and lysosomal function.37 Wang et al. developed a fluorescent probe for monitoring the dynamic changes of hydrogen peroxide (H2O2) and glutathione (GSH), elucidating the biochemical mechanism of ferroptosis in different disease models.38
Recently, some multi-target recognition fluorescent probes have been developed for the detection of cellular senescence.39–42 For example, Gao et al. developed a fluorescent probe for tracking cellular senescence, relying on detecting senescence-related β-gal activity and monitoring lysosomal pH.41 Liu et al. developed an excellent fluorescent probe based on β-gal, HClO, and lysosomal pH for tracking of cellular senescence.39 Wang et al. reported a dual-locked enzyme-activatable bioorthogonal fluorescence (DEBOF) imaging approach for the detection of senescent cancer cells, allowing tracking of the population fluctuation of senescent cancer cells in tumor-bearing mice.40 These fluorescent probes are mainly based on AND logical or sequential response, which cannot be used for independent research on each species and evaluation of their interactions. Therefore, it is urgently necessary to develop a multi-target independent detection system for multiple analytes in cellular senescence.
Herein, we successfully designed a dual-target activated fluorescent probe (HDQ-NA-AFU-Gal) for precise imaging of cellular senescence based on β-gal and AFU enzymes. In the presence of β-gal and AFU, the dual-response fluorescent probe emits strong fluorescence signals at 740 nm and 550 nm, respectively, enabling the simultaneous and non-interfering detection of both enzymes. This approach not only effectively avoids spectral overlap but also studies the roles and interactions between each species. The biocompatible HDQ-NA-AFU-Gal exhibits high specificity and sensitivity for β-gal and AFU. Moreover, the probe can be effectively localized in lysosomes. This probe can distinguish senescent cells from non-senescent cells based on the high expression of β-gal or AFU, such as human hepatoma cells (HepG2 cells), Ovcar-3 cells, and mouse breast 4T1 tumor cells. Furthermore, HDQ-NA-AFU-Gal has been successfully applied in tracing the activity of β-gal and AFU during tumor senescence in mice.
Results and discussion
Design and synthesis of the fluorescent HDQ-NA-AFU-Gal probe
A fluorescent probe (HDQ-NA-AFU-Gal) was rationally designed to detect β-gal and AFU separately or simultaneously. The synthesis route is illustrated in Scheme S1.† The naphthylimide scaffold was modified by introducing α-L-fucose as a specific recognition group for AFU. To enable simultaneous detection of β-gal, we incorporated β-D-galactopyranoside and hemicarbocyanine moieties as the responsive and fluorescent moieties of β-gal, respectively. When β-gal and AFU are present, the probe exhibits significant red and green fluorescence at 740 nm and 550 nm, respectively, enabling independent detection of β-gal and AFU. When both AFU and β-gal are present, the probe emits strong fluorescence at 550 nm and 740 nm, respectively. The dual-enzyme activation probe enhances the accuracy of tracking cell senescence. The HDQ-NA-AFU-Gal probe was characterized by nuclear magnetic resonance (NMR) and mass spectrometry (MS). The NMR and MS results (m/z [C53H54N3O15+], calcd: 972.3545; found [M + Na]+: 995.3380) indicated the successful synthesis of the probe.Absorption spectra of the HDQ-NA-AFU-Gal probe towards β-gal and AFU
UV-vis absorption spectra of the HDQ-NA-AFU-Gal probe were recorded in PBS (10 mM, pH = 6) containing 5% DMSO as a co-solvent. As shown in Fig. S1,† the probe exhibited a strong absorption peak at 470 nm. Upon the addition of AFU, a new absorption peak around 420 nm appeared, which was attributed to the hydrolysis of α-L-fucopyranoside by AFU, which resulted in the release of the naphthalimide fluorophore (Scheme 1). Subsequently, the addition of β-gal led to the cleavage of β-D-pyranose lactoside and release of the phthalocyanine fluorophore. This caused a reduction of the absorption at 470 nm and an increase of the absorption at 620 nm. When both enzymes were present, the absorption peak at 470 nm completely disappeared, and strong absorption peaks at 420 nm and 620 nm appeared. These results indicate that the probe can be used for detecting β-gal and AFU separately or simultaneously.Fluorescence spectra of HDQ-NA-AFU-Gal toward β-gal and AFU
We further investigated the fluorescence of HDQ-NA-AFU-Gal towards β-gal and AFU. As shown in Fig. 1A and D, the probe exhibited minimal fluorescence at 550 nm and 740 nm under excitation at 450 nm and 640 nm, respectively. Upon the addition of AFU and β-gal, we observed the fluorescence enhancement of the probe at 550 nm (λex = 450 nm) and 740 nm (λex = 640 nm), respectively, which indicated that the probe could be used for the detection of AFU and β-gal. Moreover, a linear relationship was observed between the fluorescence intensity of the probe and the concentrations of AFU (2–100 U per L) and β-gal (5–500 U per L) (Fig. 1B and E). The corresponding linear equations were Y1 = 10.2356 [AFU] + 34.4198 (U per L) (R12 = 0.9985) and Y2 = 0.6033 [β-gal] – 0.1197 (U per L) (R22 = 0.9983), respectively. The detection limits for AFU and β-gal, based on 3σ/k (where σ is the standard deviation and k is the slope), were about 0.32 U per L and 2.80 U per L, respectively. In addition, we investigated the stability of the probe in different physiological environments. The probe remained stable in PBS buffer or FBS containing solution at 37 °C, and almost no decomposition was observed (Fig. S2 and S3†).Subsequently, we investigated whether the probe could be used for the simultaneous detection of β-gal and AFU. As shown in Fig. S4A,† in the presence of 500 U per L β-gal, the fluorescence intensity was enhanced with the increase of AFU concentration, indicating that the detection of AFU was not affected by β-gal. As shown in Fig. S4B,† the presence of AFU did not interfere with the detection of β-gal. These results indicate that the probe enables separate or simultaneous detection of β-gal and AFU through dual channels without cross-interference.
Selectivity of the HDQ-NA-AFU-Gal probe for β-gal and AFU
We investigated the selectivity of HDQ-NA-AFU-Gal for β-gal and AFU by examining the probe's responsiveness to various biological species, including carboxylic esterase, BSA (100 μg mL−1), alkaline phosphatase (ALP, 1 U mL−1), common amino acids (cysteine (Cys), glutathione (GSH), tryptophan (Try), methionine (Met), leucine (Leu), threonine (Thr), glycine (Gly), and alanine (Ala)), several active small molecular species (H2O2, ClO−, and NO2−), and common ions (Fe3+, Ca2+, Mg2+, Zn2+, SO42−, CO32−, and NO3−). The results showed that only AFU induced a strong fluorescence signal change at 550 nm, while the other substances did not result in significant fluorescence changes (Fig. 1C and S5A†). Furthermore, only β-gal caused a noticeable fluorescence signal change at 740 nm (Fig. 1F and S5B†). These results suggest that the probe exhibits excellent specificity for β-gal and AFU.Reaction kinetics of HDQ-NA-AFU-Gal towards β-gal and AFU
We then investigated the fluorescence kinetics of the HDQ-NA-AFU-Gal probe toward targets. As shown in Fig. S6A,† in the presence of AFU, the fluorescence intensity of HDQ-NA-AFU-Gal at 550 nm increased with prolonged reaction time and then reached a maximum around 45 min. We also tested the change in the fluorescence signal of the HDQ-NA-AFU-Gal probe in the presence of β-gal with reaction time. The fluorescence intensity at 740 nm showed an increase with reaction time and then remained stable after approximately 25 min (Fig. S6B†).Effect of pH on the performance of HDQ-NA-AFU-Gal
We further assessed the effect of pH (ranging from 4 to 9) on the fluorescence signal of the HDQ-NA-AFU-Gal probe for detecting AFU and β-gal. As shown in Fig. S7,† in the absence of AFU and β-gal, the probe exhibited negligible fluorescence at both 550 nm and 740 nm. Upon the addition of AFU, the fluorescence signal at 550 nm gradually increased with the increase of pH and reached a maximum at pH 6 before the plateau (Fig. S7A†). Conversely, in the presence of 500 U per L β-gal, the fluorescence signal at 740 nm decreased slowly with increasing pH, remaining relatively stable at pH 4–6 (Fig. S7B†). Given that β-gal and AFU are primarily localized in weak acidic lysosomes, this probe exhibits great potential for monitoring their activity.HDQ-NA-AFU-Gal probe for the detection of β-gal and AFU in living cells
We assessed the feasibility of this probe for the detection of β-gal and AFU in biological systems. Firstly, we tested its cytotoxicity using the CCK-8 assay. As shown in Fig. S8,† when the concentration of HDQ-NA-AFU-Gal was at 50 μM, all cell lines (LO2, 4T1, Ovcar-3 and HepG2 cells) exhibited high viability, indicating that the HDQ-NA-AFU-Gal probe has low cytotoxicity.We further investigated whether HDQ-NA-AFU-Gal could be used to monitor β-gal and AFU in living cells using laser confocal scanning microscopy. Previous studies have demonstrated that β-gal and AFU were overexpressed in Ovcar-3 and HepG2, respectively. Thus, we selected these two cell lines as the models. LO2 cells were used as a negative control with relatively low levels of β-gal and AFU. First, we tested the incubation time of the probe to detect β-gal and AFU in different cell lines. As shown in Fig. S9 and S10,† both the green and red fluorescence signals gradually increased with time and then remained stable after around 60 min. Therefore, we selected 60 min as the optimal incubation time. As shown in Fig. 2A, the probe exhibited weak green and red fluorescence in LO2 cells. In contrast, a significant green fluorescence signal was observed in HepG2 cells with overexpressed AFU, indicating that the probe can be used for the detection of AFU in living cells. Subsequently, we investigated the performance of the probe for monitoring β-gal. A strong red and a slightly enhanced green fluorescent signal were detected in Ovcar-3 cells compared to LO2 cells, confirming that the probe can effectively detect β-gal in Ovcar-3 cells. These results demonstrate that the probe can simultaneously detect β-gal and AFU in living cells.
Lysosomal co-localization studies
Given that AFU and β-gal are primarily localized in the lysosomes of cells, we further explored whether the probe could be localized in the lysosomes of living cells. The pyridine nitrogen atom in the probe with excellent acidophilic properties makes it a potential candidate for lysosome targeting.43 We then used the commercial lysosomal dye Lyso-Tracker Green as a control in co-localization experiments. We chose Ovcar-3 cells as the model and co-incubated with Lyso-Tracker Green and HDQ-NA-AFU-Gal probe. As shown in Fig. 3A and B, a strong correlation was observed between the green fluorescence channel of Lyso-Tracker Green and the red channel of HDQ-NA-AFU-Gal, with a Pearson's correlation coefficient of 0.86. Furthermore, a high degree of fluorescence synchronization was evident between the green channel of Lyso-Tracker Green and the red channel of HDQ-NA-AFU-Gal, as analyzed at the site indicated by the white box and the yellow arrow. These results indicate that the HDQ-NA-AFU-Gal probe can be efficiently localized in the lysosomes of cells.HDQ-NA-AFU-Gal probe for detection of β-gal and AFU in senescent cells
The ability of HDQ-NA-AFU-Gal to monitor AFU and β-gal in senescent cells was further investigated using laser confocal scanning microscopy. We selected two cell lines with low expression of AFU and β-gal: a normal cell line (LO2 cells) and a breast cancer cell line (4T1 cells). In addition, we also studied other cell lines, such as HepG2 cells overexpressing AFU and Ovcar-3 cells overexpressing β-gal. Different methods were employed to induce cellular senescence in these cell lines. We pretreated LO2 cells and HepG2 with H2O2 for 3 days to induce senescence.20 As for 4T1 cells, they were treated with palbociclib,13 a common breast cancer kinase inhibitor, for one week.44,45 For Ovcar-3 cells, the cells were co-incubated with doxorubicin (DOX, 0.5 μM) for 3 days to induce senescence. To validate the successful induction of cellular senescence, we used a commercially available senescence kit (X-gal) to test (Fig. 4A). A strong blue signal is observed in senescent cells after using X-gal, while almost no signal was detected in non-senescent cells.46 After treatment with H2O2 or palbociclib, we observed a distinct blue signal in both LO2 and 4T1 cells, confirming the successful establishment of the senescence models. In addition, we tested the levels of cell cycle-related factors (such as p21 and p16) and some typical senescence-associated secretory phenotypes (IL-6, IL-1β) (Fig. 4D, S11 and S12†). The results showed that the levels of p21, P16, IL-6 and IL-1β were upregulated, further demonstrating the successful establishment of the cell senescence model.As shown in Fig. 4B, untreated LO2 and 4T1 cells displayed weak fluorescence in green and red channels. After H2O2 or palbociclib treatment, both cell lines exhibited obvious fluorescence in the green and red channels (Fig. 4B and C), suggesting the elevation of AFU and β-gal in senescence normal and cancer cells. In addition, both HepG2 cells overexpressing AFU and Ovcar-3 cells overexpressing β-gal showed strong red and green fluorescence signals after induction of cellular senescence. These results further implied that the HDQ-NA-AFU-Gal probe could be used for monitoring cellular senescence. Overall, the probe can be used to differentiate different senescent cells by detecting the levels of AFU and β-gal, thereby enhancing the accuracy of senescent cell detection, avoiding the inaccuracy of identifying cellular aging solely through β-gal detection.
HDQ-NA-AFU-Gal probe for imaging of AFU and β-gal during tumor senescence
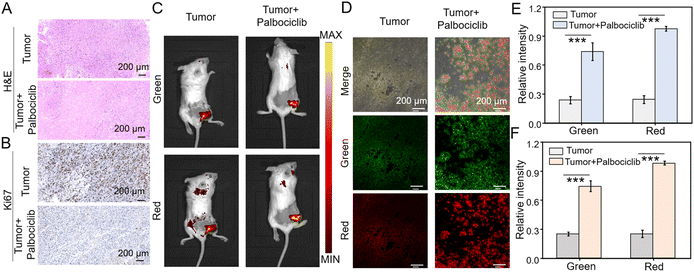
Based on the excellent performance of the dual-channel fluorescence HDQ-NA-AFU-Gal probe in living cells, we then investigated the feasibility of the probe in a tumor senescence mouse model. Balb/c mice were divided into two groups: the control group (4T1 tumor mice) and the treatment group (4T1 tumor mice treated with palbociclib). A tumor-bearing mouse model was first established by subcutaneously injecting 4T1 cells. For the treatment group, mice were treated with palbociclib daily for 7 days to induce senescence and inhibit tumor growth.The successful establishment of the tumor model was initially validated through H&E staining (Fig. 5A). Additionally, the absence of the proliferation marker Ki67 further confirmed tumor senescence (Fig. 5B), suggesting the effective induction of senescence in the tumor. Subsequently, in vivo fluorescence imaging based on the probe was performed. In the control group, weak fluorescence signals were observed in the tumor region, whereas significantly enhanced fluorescent signals in green and red fluorescent channels were observed in the treatment group. These results suggest that the probe can effectively detect AFU and β-gal in tumor senescence (Fig. 5C), and can be used to assess tumor treatment effects through monitoring AFU and β-gal levels. Fresh tumor sections were then imaged and analyzed using confocal microscopy. As shown in Fig. 5D, the control group displayed only weak fluorescence in both channels. In contrast, the treatment group exhibited strong fluorescent signals in both green and red channels. These results indicate that HDQ-NA-AFU-Gal can be employed for precise detection of tumor senescence, providing an effective tool for monitoring senescence in vivo.
Conclusions
In summary, we have successfully developed a dual-enzyme activated fluorescent probe, HDQ-NA-AFU-Gal, for tracking senescence in vivo. HDQ-NA-AFU-Gal shows the features of separate or simultaneous detection of AFU and β-gal without cross-talking. In the presence of AFU and β-gal, the probe emits strong fluorescence at 550 nm and 740 nm, respectively, demonstrating high sensitivity and specificity towards AFU and β-gal. Based on dual response design, HDQ-NA-AFU-Gal can effectively distinguish β-gal or AFU overexpressed cells and senescent cells, thereby improving the accuracy and precision of aging diagnosis. In addition, the probe has been successfully applied in imaging tumor senescence, providing a new strategy for the evaluation of senescence and assessment of potential tumor treatment.Data availability
Additional experimental data supporting this article are included in the ESI.† Reasonable requests for additional information can be made to the corresponding authors.Author contributions
Xianzhu Luo: conceptualization, investigation, writing – original draft. Erzhuo Hu: investigation, visualization. Cuiling Zhang: supervision, conceptualization, writing – review & editing, funding acquisition. Yuezhong Xian: supervision, writing – review & editing, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (22274054, 21974050, and 11727810), the Natural Science Foundation of Shanghai (20ZR1418000), and the Fundamental Research Funds for the Central Universities. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of East China Normal University and approved by the Animal Ethics Committee of East China Normal University (No. m20240204).References
- M. Collado, M. A. Blasco and M. Serrano, Cell, 2007, 130, 223–233 CrossRef CAS PubMed.
- D. Muñoz-Espín and M. Serrano, Nat. Rev. Mol. Cell Biol., 2014, 15, 482–496 CrossRef PubMed.
- S. He and N. E. Sharpless, Senescence in Health and Disease, Cell, 2017, 169, 1000–1011 CrossRef CAS PubMed.
- M. Rhinn, B. Ritschka and W. M. Keyes, Development, 2019, 146, dev151837 CrossRef CAS PubMed.
- L. Yang, G. Liu, Q. Chen, Y. Wan, Z. Liu, J. Zhang, C. Huang, Z. Xu, S. Li, C. Lee, L. Zhang and H. Sun, Anal. Chem., 2022, 94, 5425–5431 CrossRef CAS PubMed.
- J. L. Kirkland, T. Tchkonia, Y. Zhu, L. J. Niedernhofer and P. D. Robbins, J. Am. Geriatr. Soc., 2017, 65, 2297–2301 CrossRef PubMed.
- L. J. Niedernhofer and P. D. Robbins, Nat. Rev. Drug Discovery, 2018, 17, 377 CrossRef CAS PubMed.
- L. Wang, L. Lankhorst and R. Bernards, Nat. Rev. Cancer, 2022, 22, 340–355 CrossRef CAS PubMed.
- B. Lozano-Torres, J. F. Blandez, I. Galiana, J. A. Lopez-Dominguez, M. Rovira, M. Paez-Ribes, E. Gonzalez-Gualda, D. Munoz-Espín, M. Serrano, F. Sancenon and R. Martinez-Manez, Anal. Chem., 2021, 93, 3052–3060 CrossRef CAS PubMed.
- M. Serrano, A. W. Lin, M. E. McCurrach, D. Beach and S. W. Lowe, Cell, 1997, 88, 593–602 CrossRef CAS PubMed.
- N. E. Sharpless and C. J. Sherr, Nat. Rev. Cancer, 2015, 15, 397–408 CrossRef CAS PubMed.
- A. Takahashi, N. Ohtani and E. Hara, Cell Div., 2007, 2, 10–15 CrossRef PubMed.
- B. Lozano-Torres, J. F. Blandez, I. Galiana, A. Garca-Fernandez, M. Alfonso, M. D. Marcos, M. Orzaez, F. Sancenon and R. Martinez-Manez, Angew. Chem., Int. Ed., 2020, 59, 15152–15156 CrossRef CAS PubMed.
- G. P. Dimri, X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj and O. Pereira-Smith, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 9363–9367 CrossRef CAS PubMed.
- J. Wang, M. Liu, X. Zhang, X. Wang, M. Xiong and D. Luo, Exploration, 2024, 4, 20230027 CrossRef CAS PubMed.
- T. Su, R. Shen, D. Tu, X. Han, X. Luo and F. Yu, Smart Mol., 2025, e20240062 CrossRef.
- S. Chatterjee, M. Bhattacharya and J. Barlow, J. Cancer Res., 1979, 39, 1943–1951 CAS.
- H.-G. Kopp, A. T. Hooper, S. V. Shmelkov and S. Rafii, Histol. Histopathol., 2007, 22, 971–976 CAS.
- S. Koo, M. Won, H. Li, W. Y. Kim, M. Li, C. Yan, A. Sharma, Z. Guo, W. H. Zhu, J. L. Sessler, J. Y. Lee and J. S. Kim, Chem. Sci., 2021, 12, 10054–10062 RSC.
- D. Hildebrand, S. Lehle, A. Borst, S. Haferkamp, F. Essmann and K. Schulze-Osthoff, Cell Cycle, 2013, 12, 1922–1927 CrossRef CAS PubMed.
- B. Lozano-Torres, I. Galiana, M. Rovira, E. Garrido, S. Chaib, A. Bernardos, D. Munoz-Espin, M. Serrano, R. Martinez-Manez and F. Sancenon, J. Am. Chem. Soc., 2017, 139, 8808–8811 CrossRef CAS PubMed.
- X. Chai, H. H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X. L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X. P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 18005–18013 CrossRef CAS PubMed.
- X. Li, W. Qiu, J. Li, X. Chen, Y. Hu, Y. Gao, D. Shi, X. Li, H. Lin, Z. Hu, G. Dong, C. Sheng, B. Jiang, C. Xia, C.-Y. Kim, Y. Guo and J. Li, Chem. Sci., 2020, 11, 7292–7301 RSC.
- J. Li, L. Wang, X. Luo, Y. Xia, Y. Xie, Y. Liu and W. Tan, Anal. Chem., 2023, 95, 3996–4004 CrossRef CAS PubMed.
- D. Liu, G. Fang, Y. Wang, C. Meng, Z. Liu, Q. Chen and X. Shao, Exploration, 2024, 4, 20230145 CrossRef CAS PubMed.
- X. Luo, S. Cheng, W. Zhang, K. Dou, R. Wang, R. Wang and F. Yu, ACS Sens., 2024, 9, 810–819 CrossRef CAS PubMed.
- L. Zhou, X. Zhang, Y. Dong, Y. Pan, J. Li, Y. Zang and X. Li, ACS Sens., 2022, 7, 1958–1966 CrossRef CAS PubMed.
- F. Yu, P. Li, B. Wang and K. Han, J. Am. Chem. Soc., 2013, 135, 7674–7680 CrossRef CAS PubMed.
- L. He, L. He, S. He, T. Xu, X. Ren, Z. Zhang, Z. Qin, X. Zhang and L. Yuan, Angew. Chem., Int. Ed., 2022, 61, e202211409 CrossRef CAS PubMed.
- A. C. Sedgwick, H. H. Han, J. E. Gardiner, S. D. Bull, X. He and T. D. James, Chem. Sci., 2018, 9, 3672–3676 RSC.
- J. Guo, J. Sun, D. Liu, J. Liu, L. Gui, M. Luo, D. Kong, S. Wusiman, C. Yang, T. Liu, Z. Yuan and R. Li, Anal. Chem., 2023, 95, 16868–16876 CrossRef CAS PubMed.
- S. Yang, J. Jiang, A. Zhou, Y. Zhou, W. Ye, D. Cao and R. Yang, Anal. Chem., 2020, 92, 7194–7199 CrossRef CAS PubMed.
- X. Xie, Y. Liu, G. Liu, Y. Zhao, J. Bian, Y. Li, J. Zhang, X. Wang and B. Tang, Anal. Chem., 2022, 94, 10213–10220 CrossRef CAS PubMed.
- Y. Chen, Z. Hu, M. Yang, J. Gao, J. Luo, H. Li and Z. Yuan, Sens. Actuators, B, 2022, 362, 131742 CrossRef CAS.
- Y. Zheng, X. Li, W. Tang, L. Xie, F. Dai and B. Zhou, Sens. Actuators, B, 2022, 368, 132169 CrossRef CAS.
- L. Wu, J. Liu, X. Tian, R. R. Groleau, B. Feng, Y. Yang, A. C. Sedgwick, H. Han, Y. Wang, H. Wang, F. Huang, S. D. Bull, H. Zhang, C. Huang, Y. Zang, J. Li, X. He, P. Li, B. Tang, T. D. James and J. L. Sessler, J. Am. Chem. Soc., 2022, 144, 174–183 CrossRef CAS PubMed.
- Y. Wang, H. Yan, Y. Yue, Y. Zhang, F. Huo, F. Cheng and C. Yin, Chem. Eng. J., 2023, 464, 496 Search PubMed.
- M. Ren, Z. Li, B. Deng, L. Wang and W. Lin, Anal. Chem., 2019, 91(4), 2932–2938 CrossRef CAS PubMed.
- H. Liu, R. Lv, F. Song, Y. Yang, F. Zhang, L. Xin, P. Zhang, Q. Zhang and C. Ding, Chem. Sci., 2024, 15, 5681–5693 RSC.
- X. Wang, S. S. Liew, J. Huang, Y. Hu, X. Wei and K. Pu, J. Am. Chem. Soc., 2024, 146, 22689–22698 Search PubMed.
- Y. Gao, Y. Hu, Q. Liu, X. Li, X. Li, C. Y. Kim, T. D. James, J. Li, X. Chen and Y. Guo, Angew. Chem., Int. Ed., 2021, 60, 10756–10765 CrossRef CAS PubMed.
- L. Wang, J. Li, Z. Zhao, Y. Xia, Y. Xie, D. Hong, Y. Liu and W. Tan, Anal.Chem., 2024, 96, 154–162 CrossRef PubMed.
- Y. Wang, R. Zhao, X. Zhu, H. Gao, C. Gong, X. Liu and H. Zhang, Anal. Chem., 2022, 94, 13413–13421 CrossRef CAS PubMed.
- F. Wu, J. Liu, M. Tao, M. Wang, X. Ren and Z. Hai, Anal. Chem., 2023, 95, 10481–10485 CrossRef CAS PubMed.
- S. R. Whittaker, A. Mallinger, P. Workman and P. A. Clarke, Pharmacol. Ther., 2017, 173, 83–105 CrossRef CAS PubMed.
- J. Gao, F. Li, J. Chen, Y. Gao, C. Fan, Y. Huang, H. Yu, X. Yang and X. Wang, Sens. Actuators, B, 2024, 398, 134696 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc00103j |
| This journal is © The Royal Society of Chemistry 2025 |



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: