 Open Access Article
Open Access ArticleRational design of gold nanoparticle-based chemosensors for detection of the tumor marker 3-methoxytyramine†
Sebastian
Franco-Ulloa‡
ab,
Andrea
Cesari‡
 cd,
Giordano
Zanoni‡
cd,
Giordano
Zanoni‡
 c,
Laura
Riccardi
c,
Laura
Riccardi
 a,
Joseph
Wallace
a,
Beatrice Bernadette
Mascitti
a,
Joseph
Wallace
a,
Beatrice Bernadette
Mascitti
 c,
Federico
Rastrelli
c,
Federico
Rastrelli
 c,
Fabrizio
Mancin
c,
Fabrizio
Mancin
 *c and
Marco
De Vivo
*a
*c and
Marco
De Vivo
*a
aMolecular Modeling and Drug Discovery, Istituto Italiano di Tecnologia, Via Morego 30, 16163 Genova, Italy. E-mail: marco.devivo@iit.it
bExpert Analytics, Møllergata 8, 0179 Oslo, Norway
cDepartment of Chemical Science, University of Padova, Via Marzolo 1, 35131 Padova, Italy. E-mail: fabrizio.mancin@unipd.it
dDepartment of Chemistry and Industrial Chemistry, University of Pisa, Via Moruzzi 13, 56124 Pisa, Italy
First published on 4th March 2025
Abstract
In this study, we combined computational modeling, simulations, and experiments to design gold nanoparticle-based receptors specifically tailored for the NMR detection of 3-methoxytyramine (3-MT), a prognostic marker for asymptomatic neuroblastoma. We used short steered MD simulations to rank a library of 100 newly functionalized, tripeptide-coated AuNPs for their ability to recognize 3-MT. Validation of the computational analysis was performed on a subset of synthesized tripeptide-coated nanoparticles, showing a strong correlation between the predicted and experimental affinities. Eventually, we tested the sensing performance using nanoparticle-assisted NMR chemosensing, a technique which relies on magnetization transfer within a nanoparticle-host/analyte-guest complex to isolate the sole NMR signals of the analyte. This approach led to the identification of novel chemosensors that exhibited better performance compared to existing ones, lowering the limit of detection below 25 μM and demonstrating the utility of this integrated strategy.
Introduction
After decades of research in supramolecular and biomimetic chemistry, the realization of synthetic hosts with programmed affinity and selectivity remains a challenging task.1 Various approaches have been explored to date, starting from synthetic macrocyclic hosts and moving to dynamic combinatorial libraries,2 molecularly imprinted polymers,3,4 stimulated evolution of antibodies and protein-based receptors5 and aptamers.6 In this progression, the original concepts of complementarity and pre-organization have been integrated with self-assembly, library screening and directed evolution, which can provide alternative solutions to the full rational design.Ligand-protected gold nanoparticles (AuNPs) have also demonstrated their wide potential as self-assembled macromolecular hosts.7–9 AuNPs are formed by a monolayer of “ligand” molecules bound to the surface of a metallic gold core.10 Such structure provides multivalent and adjustable binding sites located in the coating monolayer. In addition, the use of nanoparticles allows conjugation of the molecular recognition abilities of the monolayer with the properties of the core, enabling widespread applications in sensing,11–13 catalysis14,15 and plasmonics.16
Accordingly, we recently demonstrated that nanoparticle-based hosts are the key element in a sensing protocol named nanoparticle-assisted NMR chemosensing.17 This method exploits the molecular recognition abilities of nanoparticle receptors to detect and identify small molecules in complex mixtures by means of magnetization or saturation transfer. Indeed, these processes allow editing the NMR spectrum of the mixture analyzed in such a way to isolate the sole signals of the target analytes.18–20 The efficiency of the magnetization/saturation transfer increases with the size of the receptor, and this justifies the superior performance of nanoparticles with respect to “molecular” hosts.21,22 The main advantage of NMR chemosensing, besides the possibility to perform direct analysis with almost no sample preparation, is the unambiguous identification even of unknown target species.23
However, as is common in sensing systems, the affinity of the host for the target guest is also a crucial parameter. In the case of NMR chemosensing, its influence on the sensing performance is even more complex than usual. Not only does a low affinity (<103 M−1) obviously lead to poor performances,20 but also a high affinity (>105 M−1) can be detrimental, as the prolonged association with the slow-tumbling host can broaden the guest signals beyond detection.24
Designing nanoparticle hosts with tailored affinity is hence a crucial but non-trivial task. The binding of the guest molecules to AuNPs is controlled by the three-dimensional arrangement of the monolayer. The correct configuration of the ligands promotes the formation of well-organized complementary interactions at the host–guest interface.25–29 However, as with proteins and polymers,30 the flexibility and dynamics of the monolayer make it difficult to unravel the relationship between the structure of the ligands and the affinity of the nanoparticles for a specific guest molecule. As a matter of fact, the design of AuNP-based supramolecular hosts remains largely a trial-and-error process.
In this context, we and others have shown that extensive atomistic molecular simulations can overcome this design bottleneck, providing guidance for rational design.27–29,31–33 Our group used molecular dynamic (MD) simulations to demonstrate that the monolayer reaches the binding configuration via the formation of dynamic and transient pockets, whose structure dictates affinity and selectivity.34,35 Subsequently, we identified and characterized these binding pockets through a computational tool that returns their exact location in ligand-coated AuNPs.36 This approach demonstrated the relevance of the energetics of pocket formation, beside the formation of complementary interactions, to the overall affinity for the guest.36 More recently, we used MD simulations to direct the design of ligands capable of improving the performance of salicylate-binding AuNPs, reaching a 10-fold increase in affinity (and 1000-fold increase in NMR chemosensing limit of detection) when compared to early candidates.35 These studies provided information on the factors controlling the host–guest interaction in the nanoparticle coating monolayer. Nonetheless, they required microsecond-long MD simulations which were computationally expensive and time-consuming, making them impractical for the fast and efficient rational design of novel ligands engineered as specific chemosensors.
Here, we report on a new combined synthetic and computational approach for the fast screening of nanoparticle-based hosts. We use short, steered MD simulations to computationally evaluate a library of 100 newly functionalized tripeptide-coated AuNPs for their affinity to 3-methoxytyramine (3-MT, Fig. 1a). This monoamine is one of the main prognostic biomarkers for asymptomatic neuroblastoma, an extracranial solid tumor responsible for 15% of pediatric cancer-related deaths.37,38 Accordingly, there is strong interest in developing methods for its detection. After the computational ranking of potential receptors for 3-MT, we could easily synthesize a subset of the library to experimentally validate the outcome of the screening and to demonstrate the superior performance of the newly functionalized AuNPs in the detection of 3-MT by nanoparticle assisted NMR chemosensing.
Results and discussion
Rational design of a 3-MT binding AuNP library
With the aim of creating a library of AuNPs that covers a wide range of chemical variations while ensuring accessibility and straightforward modification, we focused our attention on peptides. Indeed, these provide a wide range of intermolecular interactions while straightforward, high-throughput methods are available for their synthesis.Starting with these considerations, we defined a peptide-based general scaffold for the coating ligands of our initial AuNP library. All the considered ligands were methyl ester-terminated tripeptides conjugated at the N-terminus with 8-thiooctanoic acid (Fig. 1b). This structure grants several relevant features: (i) it permits synthetic accessibility via standard solid-phase protocols; (ii) the thiooctanoic residue ensures the formation of a stable ligand monolayer, as well as the presence of a hydrophobic region; and (iii) the tripeptide moiety provides a variable portion incorporating a large number of unique sequences (even considering only canonical amino acids).
To reduce the total number of possible ligands to a manageable size for initial screening, we focused on the characteristics of 3-MT. This molecule belongs to the phenethylamine class and features a hydrophobic aromatic portion and, at neutral pH, a positively charged, H-bond donor headgroup (the protonated amine). We have already demonstrated that molecules belonging to the phenethylamine class can be recognized by amphiphilic AuNPs coated with alkyl/aryl sulfonates (which combine hydrophobic interactions and ion pairing), with affinities in the order of 105 M−1,23 or by alkyl-crown ethers (which combine hydrophobic interactions and cooperative H-bonding), with affinities in the order of 102 M−1.19 This provided some preliminary indication on the features to incorporate into the ligands.
Moreover, 3-MT is a metabolite of dopamine and many of its native protein receptors are known.
Analysis of the amino acidic composition of the catecholamine binding pockets found in the Protein Data Bank (see Fig. 1c and Table S1† for details) provided hence additional information on the relevant possible interactions, emphasizing the importance of ion pairing, hydrophobic and aromatic interactions.
We opted to use anionic nanoparticles fixing the total charge of each ligand to −1, to avoid excessive ion-pairing interactions with any positively charged interfering molecules. Consequently, Asp, which is the negatively charged amino acid that most often populates the protein pockets, was fixed to the terminal position 3, or A3. The charge of the ligand, being located on the surface of the monolayer, should promote solubility in polar solvents and induce all ligands to adopt an elongated, comparable configuration. Regarding positions 1 and 2, namely A1 and A2, we short-listed the candidate amino acids excluding those that: (1) are charged at physiological pH (Asp, Glu, Arg, His, and Lys); (2) have the potential to interact with the gold core (Cys); (3) are chemically redundant with respect to the rest of the amino acids included (Asn with respect to Gln, Thr with respect to Ser, Val and Met with respect to Ile and Leu). The outcome of this selection was a group of nine amino acids (Ala, Gln, Gly, Ile, Leu, Phe, Pro, Ser, and Tyr) which were all among the most prominent catecholamine binding proteins (Fig. 1c). Eventually, Trp was added to this list, notwithstanding its low abundance in catecholamine binding proteins, since it could potentially provide relevant π-interactions. In the end, positions A1 and A2 iterated over ten amino acids while position A3 was assigned to Asp. This selection resulted in a library of 100 coating molecules (i.e., 100 AuNPs).
Computational screening of the AuNP library
Having defined the first screening library, we devised a fast computational approach that ranked the hosts based on short molecular dynamics (MD) simulations (see Fig. 2a and ESI† for details). Briefly, the NanoModeler webserver39,40 was used to produce a 3D geometry for each AuNP from the Au144(SR)60 template. Nanoparticles with a gold core diameter of 1.6 nm, which correspond to the Au144SR60 cluster, have been demonstrated to have optimal features for NMR chemosensing. In turn, as we already demonstrated, computational simulation performed with Au144SR60 satisfactorily models the structural and molecular recognition features of gold nanoparticles with a metal core diameter of about 1.6 nm, notwithstanding the intrinsic inhomogeneity of the real samples.35 Then, 10 guest molecules were placed randomly inside the monolayer at a distance 1/3 of the monolayer thickness away from the gold core. After a short MD equilibration run to relax the system, all the guest molecules were simultaneously pulled away from the AuNP during a 10 ns-long steered MD simulation. Lastly, a score parameter W (in kcal mol−1) was computed from the median force required to dissociate the guest molecules from the monolayer.To validate and calibrate the results of the computational analysis, we scanned the literature and our unpublished data and found 31 AuNP/guest dyads with known experimental binding affinity, spanning 11 different ligands and 17 different analytes (Table S2†). Their experimental binding free energies (ΔGexp) ranged between −2.8 ± 0.1 and −8.7 ± 0.1 kcal mol−1 in 22 of the 31 dyads, while the remaining 9 had a binding affinity below the experimental detection limit. Notably, most dyads resembled our library of 3-MT hosts; 21 out of 31 dyads had ligands and guests of opposite charges as well as the ligands' charge on the surface of the monolayer. Applying the protocol to this dataset resulted in a raw affinity score, W, spanning from −3.6 ± 0.5 to −20.9 ± 1.5 kcal mol−1, that correlated with the experimental affinity (r2 = 0.49, Fig. S2†). The experimental data were then used to calibrate the method and compute a definitive adjusted score, Wadj (eqn S2†).
The library of tripeptide ligand-coated AuNPs (named according to the three amino acids in the coating ligands) was then scored with our computational approach employing 3-MT as the guest. The calculated raw scores, W, ranged from −8.7 ± 1.4 kcal mol−1 (IleIleAsp) to −19.3 ± 1.9 kcal mol−1 (TrpPheAsp). The adjusted binding scores, Wadj, ranged from −6.0 ± 0.7 kcal mol−1 (IleIleAsp) to −8.7 ± 0.5 kcal mol−1 (TrpPheAsp). The 20 best predicted binders had an adjusted score Wadj below −7.94 kcal mol−1 (Fig. 2b).
Synthesis and testing of the AuNP library
The quality of the predictions resulting from the above computational screening was assessed by selecting a group of 14 tripeptidic ligands from the library and testing them experimentally (Table 1).| Rank | A1–A2 | W adj (kcal mol−1)a | ΔGexp (kcal mol−1)b |
|---|---|---|---|
| a Errors in Wadj are the standard deviations of the W values obtained for each guest pulling-out trajectory. b Errors in ΔGexp are obtained by propagation from the errors on the diffusion rates measured by DOSY experiments. | |||
| 1 | TrpPhe | −8.72 ± 0.48 | −6.27 ± 0.44 |
| 3 | GlnPro | −8.49 ± 0.51 | −5.45 ± 0.41 |
| 4 | TyrPro | −8.42 ± 0.66 | −6.35. ± 0.32 |
| 5 | TrpPro | −8.41 ± 0.47 | −5.51 ± 0.33 |
| 6 | ProLeu | −8.39 ± 0.28 | −5.54 ± 0.14 |
| 12 | GlyIle | −8.10 ± 0.35 | −4.95 ± 0.79 |
| 14 | GlnGln | −8.06 ± 0.66 | −4.80 ± 0.53 |
| 21 | PheSer | −7.93 ± 0.35 | −5.11 ± 0.79 |
| 35 | ProPro | −7.80 ± 0.37 | −4.73 ± 0.60 |
| 39 | TrpGln | −7.76 ± 0.50 | −4.64 ± 0.86 |
| 40 | LeuLeu | −7.73 ± 0.62 | −4.52 ± 0.96 |
| 43 | ProAla | −7.64 ± 0.26 | −4.64 ± 0.76 |
| 47 | GlnAla | −7.60 ± 0.37 | −4.64 ± 0.69 |
| 62 | PheLeu | −7.39 ± 0.53 | −5.02 ± 0.38 |
The tripeptidic ligands were prepared with a solid phase procedure based on standard Fmoc protection and 2-chlorotrityl resin (see ESI†). Detachment from the resin was performed with simultaneous removal of the protecting groups (except for the thiol-protecting acetyl) using 1,1,1,3,3,3-hexafluoro-2-propanol. The thiol residue was eventually deprotected only before nanoparticle functionalization using sodium methoxide. The AuNPs were prepared by NaBH4 reduction of tetrachloroauric acid according to a two-step protocol developed by Scrimin.41 All the AuNPs had average core diameters between 1.4 and 2.0 nm (TEM micrographs and size distribution reported in ESI†) and featured a clear negative ζ-potential (Fig. S45†), confirming the presence of the negative charge provided by the terminal Asp-OMe residue. With this procedure, AuNPs can be produced by a single person at a rate of three AuNPs per week, using solid-phase synthesis, or at a higher pace using automated peptide synthesis.
The affinity of AuNPs for the target 3-MT was determined with 1H-DOSY (Diffusion Ordered SpectroscopY),42 measuring the diffusion coefficients (D, m2s−1) of 3-MT in the presence and in the absence of the AuNPs. When a small guest molecule binds to monolayer-protected AuNPs in fast exchange with respect to the NMR time scale, the apparent diffusion coefficient of the guest is the average between the diffusion coefficients of the free and the AuNP-bound states (weighted for the relative populations). These parameters allow calculating the bound and unbound fractions of the guest and consequently the binding constant (K). Samples containing 3-MT (0.5 mM) and AuNPs (2 mM, note that the AuNP concentration is expressed in terms of the overall concentration of coating ligands) in buffered deuterated water ([HEPES buffer] = 10 mM, pD = 7) were analyzed. Previous studies showed that the number of guest molecules that can bind a single AuNP generally sums to 30–40% of the amount of coating ligands (i.e., the binding site is formed by a cluster of 2–4 coating molecules).23 The conditions chosen hence ensured that binding sites in the AuNPs could not be saturated even in the case of very high affinity. The average values of the affinity constants (converted to ΔG values) for 3-MT measured with DOSY experiments are shown in Fig. 3, where they are compared with the values predicted by the calculations. Remarkably, the correlation between the computed and experimental association constants is good (r2 = 0.68).
In addition, four systems from Fig. 3 attracted our attention, namely TrpPheAsp (rank 1), TyrProAsp (rank 4), PheSerAsp (rank 21) and PheLeuAsp (rank 62), which are predicted to be worse binders than found experimentally (if their affinity values are removed from the plot, the r2 value raises to 0.90). The fact that all these AuNPs have aromatic amino acids in position A1 suggests that this typology might influence the predicted score. We suppose that the π-interactions offered by these amino acids displace the position of the free energy minimum of the monolayer-captured guest inwards, closer to the gold core and below the starting position of the simulations. This would imply that the steered simulations in our protocol might not capture the entire dissociation process. Future iterations of this methodology should explore embedding the guest molecules deeper into the monolayer.
Experimental testing of the nanoparticles' affinity and sensing performance
Having ranked peptide-based AuNPs for their ability to bind to 3-MT, we turned our attention to their exploitation as macromolecular guests for NMR-chemosensing. We analyzed samples containing 3-MT (100 μM) and AuNPs (500 μM) in H2O/D2O 90% buffered with HEPES (1 mM) at pD = 7.0 with the best performing high-power water-STD protocol (HPwSTD, Fig. 4).24,29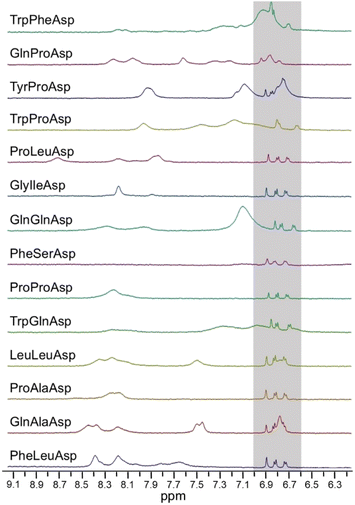 | ||
| Fig. 4 HPwSTD subspectra of 3-MT (100 μM) and AuNPs (at the overall ligand concentration of 500 μM). Conditions: 500 MHz, 25 °C, [HEPES] = 1 mM, pD = 7.0, H2O/D2O 90:10, 512 scans. Full spectra are reported in Fig. S46.† | ||
All the nanoparticles were capable of analyte detection under these conditions. In a few cases (TrpProAsp, TrpGlnAsp, LeuLeuAsp, and GlnAlaAsp), we found that the nanoparticles' NH signals overlapped with the signals of 3-MT, preventing their accurate integration. With the other nanoparticles we could determine S/N ratios for the analyte signals ranging from 20 to 40, which substantially exceeded the limit of detection, LOD, conventionally set to S/N values greater than 3.
Earlier, we demonstrated that AuNPs with affinity (K) for the target below 1 × 103 M−1 (ΔG ≈ −4.1 kcal mol−1) are unable to allow its detection at submillimolar concentrations in HPwSTD experiments.29 On the other hand, as we'll discuss in detail later, AuNPs with affinity above 1 × 104 M−1 (ΔG ≈ −5.5 kcal mol−1) can produce strong signal broadening also affecting the target detection.24 In line with these previous results, nanoparticles with higher affinities (TrpPheAsp, GlnProAsp, TrpProAsp, PheSerAsp, K > 5 × 104 M−1) produced broadened signals, with the exceptions of ProLeuAsp and possibly of TyrProAsp. Such different behaviors of AuNPs with similar affinity confirm, as we previously reported,29 that other factors beside the sole affinity contribute to fine-tuning the sensing performance.
Remarkably, while the experimental affinities correlated well both with computational results and NMR performance, we found no correlations with the average size or even with the size dispersion of the single batches. This observation confirms that, provided that the variations of the sample's dimensions are small (in our case nanoparticles studied featured core sizes between 1.4 and 2.0 nm), the performances of the nanoparticles are not influenced by their size (and consequently by their size dispersion).
Comparison with state-of-the-art AuNPs for NMR chemosensing
Building on the results of this experiment, we identified ProAlaAsp for further investigation. The selection criteria used were: sharp 3-MT signals in the spectra of Fig. 4, a ΔGexp value for 3-MT binding lower than −5.0 kcal mol−1 (K < 4.3 × 103 M−1), and the absence of interfering AuNP signals. Among the AuNP candidates matching these criteria, namely ProLeuAsp, GlyIleAsp, ProProAsp and ProAlaAsp, we selected ProAlaAsp since it produced the largest analyte signals in Fig. 4 (S/N = 30). We first evaluated the ability of ProAlaAsp to isolate the signals of 3-MT from those of other molecules that the nanoparticles do not recognize. Fig. 5 shows an HPwSTD experiment on a sample containing ProAlaAsp (500 μM) and a mixture of 3-MT, hippuric acid, vanillic acid, and homovanillic acid, each at 50 μM concentration. In particular, hippuric acid was included since it is one of the most relevant species observed in the aromatic portion of the NMR spectra of urine samples, while vanillic and homovanillic acids are other two relevant markers in patients affected by neuroblastoma.37,38 The HPwSTD experiment performed in the presence of ProAlaAsp isolates only the 3-MT signals in the difference spectrum, notwithstanding the presence of fully overlapped signals which were completely removed. | ||
| Fig. 5 (a) HPwSTD subspectra of 3-MT (25–100 μM) and SO3–AuNP or ProAlaAsp (500 μM, ligand concentration). Conditions: 500 MHz, 25 °C, [HEPES] = 1 mM, pD = 7.0, H2O/D2O 90:10, 512 scans. (b) Signal-to-noise ratio of the 3-MT signal at 6.81 ppm ([3-MT] = 25–100 μM) with SO3–AuNP or ProAlaAsp at 25 μM or 500 μM. Full spectra are reported in Fig. S47.† | ||
The performance of ProAlaAsp in terms of sensitivity, LOD, and detection range was then compared with that of the high affinity sulfonate functionalized nanoparticles SO3–AuNPs (Fig. 6), which were previously demonstrated to allow the detection of phenethylamine derivatives down to 50 μM in HPwSTD experiments.24 The affinity (K) of ProAlaAsp for 3-MT in HEPES buffer at 25 °C is 2.4 × 103 M−1, while that of SO3–AuNP is >105 M−1 under the same conditions (above the limit that can be measured by DOSY). The HPwSTD spectra of samples containing either ProAlaAsp (500 μM) or SO3–AuNP (500 μM) and 3-MT, at concentrations ranging from 100 μM down to the challenging concentration of 25 μM are reported in Fig. 6a.
 | ||
| Fig. 6 1H NMR subspectra of 3-MT (50 μM), hippuric acid (50 μM), homovanillic acid (50 μM), vanillic acid (50 μM) and ProAlaAsp (500 μM, ligand concentration). Conditions: 500 MHz, 25 °C, [HEPES] = 1 mM, pD = 7.0, H2O/D2O 90:10). (a) 1H spectrum (128 scans); (b) HPwSTD spectrum (512 scans). Signals at 8.2 ppm belong to amide protons that receive saturation via chemical exchange with H2O. The asterisk denotes a subtraction artifact. The full HPwSTD spectrum is reported in Fig. S48.† | ||
Fig. 6a reveals that the SO3–AuNP's performance was poor at all analyte concentrations, since the 3-MT STD signals were substantially broadened and became even undetectable at 25 μM. Fig. 6b shows that only when the 3-MT concentration reaches 100 μM does the signal-to-noise ratio exceed the LOD threshold, but it significantly drops at lower concentrations. As mentioned, this poor performance is the consequence of the high affinity of SO3–AuNP for the analyte. Under the experimental conditions used (excess of nanoparticle binding sites with respect to the analyte), the high affinity results in a large fraction of analyte bound to the nanoparticle and, therefore, in the substantial broadening of the analyte signals because of the reduced tumbling rate. This detrimental effect is at play at any analyte concentrations, but it is more dramatic at low concentration since, under these conditions, the analyte is fully bound to the nanoparticles, and its signals are broadened beyond detection. Noticeably, also nanoparticles GlnProAsp, TyrProAsp, and TrpProAsp, whose affinities for 3-MT are 5 to 20 times greater than that of ProAlaAsp, gave similar results, being unable to detect 3-MT at 25 μM concentration (Fig. S50†).
We earlier demonstrated that lowering the AuNP concentration (i.e., by reaching conditions of excess of analyte) can mitigate this problem since it decreases the fraction of analyte bound to the nanoparticles.24 Under these conditions, however, the available binding sites are scarce, and they are readily saturated when the analyte concentration increases. This strongly limits the interval of concentration where the sensor response is proportional to the analyte amount. Indeed, in the experiments performed with 25 μM SO3–AuNP, 3-MT could be detected with signal intensities above the LOD threshold at all the concentrations tested, but no difference in signal intensity was observed when the 3-MT concentration was doubled from 50 to 100 μM (Fig. 6b).
Nanoparticles with tuned affinity (lower, but not too low) are expected to overcome this problem. Indeed, when used at high enough concentration, they grant a larger range of analyte concentration values where the fraction of analyte bound is small enough to avoid relevant signal broadening, but its overall amount is large enough to allow detection. Fig. 6a confirms indeed that, when ProAlaAsp was used at 500 μM, clear STD signals were detected at all analyte concentrations explored (25, 50, and 100 μM). Their intensity increased linearly with the analyte concentration maintaining the S/N values always above the LOD threshold (Fig. 6b). It is noteworthy that 25 μM is the lowest concentration so far detected by nanoparticle-assisted NMR chemosensing using only 2 nm gold nanoparticles as receptors and standard NMR instrumentation. On the other hand, in the experiments performed with 25 μM ProAlaAsp, it was not possible to detect 3-MT at any concentration (Fig. 6b), since under these conditions the amount of analyte bound is too small to produce relevant signals.
Conclusions
In this work we have demonstrated the effective computational evaluation of an AuNP library targeted at the detection of 3-methoxytyramine at low concentrations. The association of a fast computational screening method with the design of an easy-to-synthesize peptide-based ligand allowed us to encompass a broad spectrum of chemical diversity and to validate experimentally the computational prediction.The resulting AuNPs featured the optimal properties to allow the identification of 3-MT in complex solutions, with a low LOD and large dynamic range. Remarkably, the nanoparticle assisted NMR-chemosensing detection of small molecule analytes at 25 μM, using only small nanoparticles and standard instrumentation, is unprecedented. Such a detection limit is already suitable for the detection of some cancer marker metabolites in urine but still too high for 3-MT, whose concentration in healthy subjects is around 1 μM.37,38 Still, detection in concentrated samples prepared, for example, by solid phase extraction is feasible.
In addition, the approach here reported can be applied for a time-effective “on-demand” design, synthesis, and testing of self-organized nanoreceptors for any application of molecular recognition beside chemosensing.
A further refinement of the computational screening would require the availability of larger sets of experimental affinity data. However, the experimental determination of the nanoparticle–guest binding constants remains a challenging task, due to both the size distribution of the nanoparticle samples and to the multivalent nature of the binding sites. Despite such limitations, this computational–experimental combined approach remains capable of swiftly directing the design of nanoreceptors with target properties.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
SFU, LR, MDV, FM conceptualized the project. SFU developed the computational model and analysed the computational data with LR and JW. GZ developed the nanoparticles synthetic protocols. FR developed the NMR experiments. GZ, AC and BM synthesized the nanoparticles. AC and BM performed the NMR experiments under the supervision of FR. MDV and FM supervised the project and were responsible for funding acquisition. The manuscript was written with contributions from SFU, LR, FR, FM, MDV. All authors approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Italian Association for Cancer Research (AIRC) under the Investigators Grants scheme (IG 25003). Also, M. D. V. thanks AIRC for financial support (IG 30631). The authors thank Andrea Melotti and Giovanni Maggioni for their assistance in the synthesis of some nanoparticles.Notes and references
- S. Sarkar, P. Ballester, M. Spektor and E. A. Kataev, Angew. Chem., Int. Ed., 2023, 62(28), e202214705 CrossRef CAS PubMed.
- J. Li, P. Nowak and S. Otto, J. Am. Chem. Soc., 2013, 135, 9222–9239 CrossRef CAS PubMed.
- J. J. BelBruno, Chem. Rev., 2019, 119, 94–119 CrossRef CAS PubMed.
- K. Eersels, P. Lieberzeit and P. Wagner, ACS Sens., 2016, 1, 1171–1187 CrossRef CAS.
- J. Manhas, H. I. Edelstein, J. N. Leonard and L. Morsut, Nat. Chem. Biol., 2022, 18, 244–255 CrossRef CAS PubMed.
- M. R. Dunn, R. M. Jimenez and J. C. Chaput, Nat. Rev. Chem., 2017, 1, 0076 CrossRef CAS.
- M.-C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- N. A. Kotov, Science, 2010, 330, 188–189 CrossRef CAS PubMed.
- F. Mancin, L. J. Prins, F. Rastrelli and P. Scrimin, in Supramolecular Chemistry in Water, Wiley, 2019, pp. 413–447 Search PubMed.
- M. F. Matus and H. Häkkinen, Nat. Rev. Mater., 2023, 8, 372–389 CrossRef CAS.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS PubMed.
- G. Zhang, Nanotechnol. Rev., 2013, 2, 269–288 CAS.
- M. M. Sherard, Q. M. Dang, S. C. Reiff, J. H. Simpson and M. C. Leopold, ACS Appl. Nano Mater., 2023, 6, 8367–8381 CrossRef CAS PubMed.
- D. J. Mikolajczak, A. A. Berger and B. Koksch, Angew. Chem., Int. Ed., 2020, 59, 8776–8785 CrossRef CAS PubMed.
- J. Lou-Franco, B. Das, C. Elliott and C. Cao, Nanomicro. Lett., 2021, 13, 10 Search PubMed.
- N. Peruffo, M. Bruschi, B. Fresch, F. Mancin and E. Collini, Langmuir, 2023, 39, 12793–12806 CrossRef CAS PubMed.
- F. De Biasi, F. Mancin and F. Rastrelli, Prog. Nucl. Magn. Reson. Spectrosc., 2020, 117, 70–88 CrossRef CAS PubMed.
- B. Perrone, S. Springhetti, F. Ramadori, F. Rastrelli and F. Mancin, J. Am. Chem. Soc., 2013, 135, 11768–11771 CrossRef CAS PubMed.
- M.-V. Salvia, G. Salassa, F. Rastrelli and F. Mancin, J. Am. Chem. Soc., 2015, 137, 11399–11406 CrossRef CAS PubMed.
- M.-V. Salvia, F. Ramadori, S. Springhetti, M. Diez-Castellnou, B. Perrone, F. Rastrelli and F. Mancin, J. Am. Chem. Soc., 2015, 137, 886–892 CrossRef CAS PubMed.
- F. De Biasi, D. Rosa-Gastaldo, F. Mancin and F. Rastrelli, Chem. Commun., 2021, 57, 3002–3005 RSC.
- A. Cesari, D. Rosa-Gastaldo, A. Pedrini, F. Rastrelli, E. Dalcanale, R. Pinalli and F. Mancin, Chem. Commun., 2022, 58, 10861–10864 RSC.
- L. Gabrielli, D. Rosa-Gastaldo, M.-V. Salvia, S. Springhetti, F. Rastrelli and F. Mancin, Chem. Sci., 2018, 9, 4777–4784 RSC.
- F. De Biasi, D. Rosa-Gastaldo, X. Sun, F. Mancin and F. Rastrelli, J. Am. Chem. Soc., 2019, 141, 4870–4877 CrossRef CAS PubMed.
- A. K. Boal and V. M. Rotello, J. Am. Chem. Soc., 2000, 122, 734–735 CrossRef CAS.
- M. Lucarini, P. Franchi, G. F. Pedulli, C. Gentilini, S. Polizzi, P. Pengo, P. Scrimin and L. Pasquato, J. Am. Chem. Soc., 2005, 127, 16384–16385 CrossRef CAS PubMed.
- C. Gabellini, M. Şologan, E. Pellizzoni, D. Marson, M. Daka, P. Franchi, L. Bignardi, S. Franchi, Z. Posel, A. Baraldi, P. Pengo, M. Lucarini, L. Pasquato and P. Posocco, ACS Nano, 2022, 16, 20902–20914 CrossRef CAS PubMed.
- E. Pellizzoni, M. Şologan, M. Daka, P. Pengo, D. Marson, Z. Posel, S. Franchi, L. Bignardi, P. Franchi, M. Lucarini, P. Posocco and L. Pasquato, J. Colloid Interface Sci., 2022, 607, 1373–1381 CrossRef CAS PubMed.
- S. Franco-Ulloa, A. Cesari, L. Riccardi, F. De Biasi, D. Rosa-Gastaldo, F. Mancin, M. De Vivo and F. Rastrelli, J. Phys. Chem. Lett., 2023, 14, 6912–6918 CrossRef CAS PubMed.
- A. Cesari, A. M. Piras, Y. Zambito, G. Uccello Barretta and F. Balzano, Int. J. Pharm., 2020, 587, 119698 CrossRef CAS PubMed.
- I. K. Mati, W. Edwards, D. Marson, E. J. Howe, S. Stinson, P. Posocco and E. R. Kay, ACS Nano, 2021, 15, 8295–8305 CrossRef CAS PubMed.
- A. K. Chew, B. C. Dallin and R. C. Van Lehn, ACS Nano, 2021, 15, 4534–4545 CrossRef CAS PubMed.
- A. K. Chew, J. A. Pedersen and R. C. Van Lehn, ACS Nano, 2022, 16, 6282–6292 CrossRef CAS PubMed.
- L. Riccardi, L. Gabrielli, X. Sun, F. De Biasi, F. Rastrelli, F. Mancin and M. De Vivo, Chem, 2017, 3, 92–109 CAS.
- X. Sun, L. Riccardi, F. De Biasi, F. Rastrelli, M. De Vivo and F. Mancin, Angew. Chem., Int. Ed., 2019, 58, 7702–7707 CrossRef CAS PubMed.
- L. Riccardi, S. Decherchi, W. Rocchia, G. Zanoni, A. Cavalli, F. Mancin and M. De Vivo, J. Phys. Chem. Lett., 2021, 12, 5616–5622 CrossRef CAS PubMed.
- I. R. N. Verly, Y. A. H. Matser, R. Leen, R. Meinsma, M. Fiocco, J. Koster, R. Volckmann, D. Savci-Heijink, G. Cangemi, S. Barco, L. J. Valentijn, G. A. M. Tytgat and A. B. P. van Kuilenburg, JCO Precis. Oncol., 2022, 6, e2000447 CrossRef PubMed.
- I. R. N. Verly, A. B. P. van Kuilenburg, N. G. G. M. Abeling, S. M. I. Goorden, M. Fiocco, F. M. Vaz, M. M. van Noesel, C. M. Zwaan, G. J. L. Kaspers, J. H. M. Merks, H. N. Caron and G. A. M. Tytgat, Eur. J. Cancer, 2018, 90, 102–110 CrossRef CAS PubMed.
- S. Franco-Ulloa, L. Riccardi, F. Rimembrana, M. Pini and M. De Vivo, J. Chem. Theory Comput., 2019, 15, 2022–2032 CrossRef CAS PubMed.
- S. Franco-Ulloa, L. Riccardi, F. Rimembrana, E. Grottin, M. Pini and M. De Vivo, J. Chem. Theory Comput., 2023, 19, 1582–1591 CrossRef CAS PubMed.
- F. Manea, C. Bindoli, S. Polizzi, L. Lay and P. Scrimin, Langmuir, 2008, 24, 4120–4124 CrossRef CAS PubMed.
- K. S. Cameron and L. Fielding, J. Org. Chem., 2001, 66, 6891–6895 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc08758e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |



